Abstract
The E3 ubiquitin ligase RING finger protein 115 (RNF115), also known as breast cancer-associated gene 2 (BCA2), has previously been reported to be overexpressed in estrogen receptor α (ERα)-positive breast tumors and to promote breast cell proliferation; however, its mechanism is unknown. In this study, we demonstrated that silencing of BCA2 by small interfering RNAs (siRNAs) in two ERα-positive breast cancer cell lines, MCF-7 and T47D, decreases cell proliferation and increases the protein levels of the cyclin-dependent kinase inhibitor p21Waf/Cip1. The protein stability of p21 was negatively regulated by BCA2. BCA2 directly interacts with p21 and promotes p21 ubiquitination and proteasomal degradation. Knockdown of p21 partially rescues the cell growth arrest induced by the BCA2 siRNA. These results suggest that BCA2 promotes ERα-positive breast cancer cell proliferation at least partially through downregulating the expression of p21.
Introduction
Protein ubiquitination is one of the crucial posttranslational modifications regulating the cell cycle progression. An ubiquitin E3 ligase, such as SCFSKP2, SCFFBW7, and APC/C, is responsible for conjugating ubiquitins to several specific proteins and usually targets them for proteasomal degradation. Among over 600 E3 ligases, a couple of them function as either oncoproteins or tumor suppressors. For example, the SKP2 oncoprotein promotes cell cycle progression by targeting the cyclin-dependent kinase (CDK) inhibitors p27Kip1 and p21Waf/Cip1 (below as p21) for ubiquitin-mediated degradation [1,2]. In contrast, the FBW7 tumor suppressor inhibits cell cycle progression through promoting the degradation of cyclin E [3,4], Myc [5,6], and Kruppel-like factor 5 (KLF5) [7]. Identification of the cancer-related E3 ligases and their substrates may provide more therapeutic targets for cancer treatment.
Previous studies showed that a novel characterized E3 ubiquitin ligase, RING finger protein 115 (RNF115), also namely breast cancer-associated gene 2 (BCA2), is overexpressed in breast cancer and its expression is significantly correlated with positive estrogen receptor α (ERα) status [8]. A recent study discovered that BCA2 is an ERα direct downstream target gene [9]. BCA2 has been demonstrated to increase proliferation of the T47D human breast cancer cell line [8]. Although several BCA2 interacting proteins, such as Rab7, tetherin, ubiquitin, UBC9, hHR23a, and 14-3-3σ, have been identified by yeast two-hybrid screening [10–12], the mechanisms by which BCA2 promotes breast cancer cell proliferation are still unclear.
The CDK inhibitor p21 controls the cell cycle progression through inhibiting the activity of several cyclin-CDK complexes [13]. Its transcription is tightly regulated by multiple transcription factors, including p53, Myc, Smad, and KLF4 [14,15]. Previous studies demonstrated that the p21 protein level is also tightly controlled by multiple E3 ligases, including SCFSkp2 [2], CRL4Cdt2 [16], APC/CCdc20 [17], MKRN1 [18], and RNF126 [19].
Since RNF126 and BCA2 have a high-sequence identity [20], we hypothesize that BCA2 promotes ERα-positive breast cancer cell proliferation through targeting p21 for ubiquitin-mediated degradation. In this study, we demonstrated that depletion of BCA2 significantly increased the p21 protein levels and decreased DNA synthesis in two ERα-positive breast cancer cell lines, BCA2 promotes p21 ubiquitination and degradation in an E3 ligase activity-dependent manner, and depletion of p21 partially rescued the BCA2 small interfering RNA (siRNA)-induced growth arrest. These findings suggest that BCA2 promotes ERα-positive breast cancer cell proliferation partially through targeting p21 for ubiquitin-mediated degradation.
Materials and Methods
Cell Lines and Cell Culture
MCF-7 and T47D cells were purchased from American Type Culture Collection (Manassas, VA) and cultured according to American Type Culture Collection recommended condition. Human embryonic kidney HEK293FT cells (R700-07; Invitrogen, Carlsbad, CA) were maintained in Dulbecco's modified Eagle's medium (Hyclone), supplemented with 10% FBS, 1% penicillin, and 1% streptomycin.
Plasmids, siRNAs, and Transfection
The Flag-tagged wild-type (WT) and RING domain mutant (C228 and C231 residues were mutated into A) constructs of BCA2 have been described in our previous study [10]. The mammalian HA-p21-expressing plasmid was kindly provided by Dr Tiebang Kang [21]. For constructing glutathione S-transferase (GST)-BCA2-expressing plasmids, the full-length and truncated sequences of BCA2 were inserted into the pEBG vector through BamHI/NotI sites. The siRNAs targeting the BCA2 gene were synthesized by Ambion (Austin, TX), and the targeting sequences were AGACAAUACCACAACAACAtt (siBCA2-A#, S26038) and GAAUAUAUAUGUCCCAGAUtt (siBCA2-B#, S26036), respectively. The siRNA targeting the firefly luciferase gene (siLuc) was used as the negative control. Lipofectamine 2000 (Invitrogen) was used to transfect plasmids and siRNAs according to the manufacturer's instructions.
Semiquantitative Reverse Transcription-Polymerase Chain Reaction
The mRNA levels of BCA2, p21, p53, and p27 were examined by semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) in MCF-7 and T47D. After transfected by siLuc and siBCA2, total RNAs were prepared from cultured cells using TRIzol reagents (15596-026; Invitrogen). RT of 1 µg of total RNA was performed using the iScript cDNA Synthesis Kit (170-8891; Bio-Rad, Hercules, CA). The primers for RT-PCR were given as follows: primers for BCA2, forward—5′-TGTCCCAGATGTGAATCAGG-3′ and reverse—5′-TGGAGATCTGTCAGGACGAG-3′; primers for p21, forward— 5′-GCGATGGAACTTCGACTTTGT-3′ and reverse—5′-GGGCTTCCTCTTGGAGAAGAT-3′; primers for p27, forward—5′-AGACGGGGTTAGCGGAGCAA-3′ and reverse—5′-TCTTGGGCGTCTGCTCCACA-3′; primers for p53, forward—5′-CTGGCCCCTGTCATCTTCTG-3′ and reverse—5′-CGGCTCATAGGGCACCAC-3′; primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as a loading control, forward—5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse—5′-TGGGTGGAATCATATTGGAACA-3′. PCR on cDNA products was performed using the following parameters: predenaturation at 94°C for 2 minutes, 94°C for 30 seconds, annealing at 55°C for 30 seconds, elongation at 72°C for 40 seconds, running 25 cycles, and final elongation at 72°C for 7 minutes. The PCR products were run on 2% agarose gels and examined with an ImageQuant LAS4000 (GE Healthcare, Piscataway, NJ).
Western Blot Analysis
Cells were washed once with pre-cold phosphate-buffered saline (PBS) solution and lysed in cell lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, supplemented with 1x protease inhibitor cocktail] on ice for 30 minutes. The total protein concentration was analyzed with Bio-Rad DC Protein Assay Kit (500-0116), and equal amounts of proteins were run on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels. After transferring to polyvinylidene difluoride (PVDF) membranes and blocking with 5% nonfat milk, the targeted proteins were immunoblotted with specific primary antibodies against BCA2 [8], p53 (554169; BD Biosciences, San Jose, CA), p27 (610241; BD Biosciences), p21 (2947; Cell Signaling Technology, Danvers, MA), ERα (sc-7207; Santa Cruz Biotechnology, Dallas, TX), Flag tag (F3165; Sigma, St Louis, MO), HA tag (sc-805; Santa Cruz Biotechnology), GST (G7781; Sigma), and GAPDH (25778; Santa Cruz Biotechnology), respectively. To probe different target proteins in the same membrane, the Restore Western Blot Stripping Buffer (21059; Pierce, Rockford, IL) was used to strip the primary and secondary antibodies.
Flow Cytometry
T47D cells were digested by trypsin-EDTA solution, collected by centrifugation at 500g for 5 minutes, and washed once with PBS. The cells were treated with BD Cytofix/Cytoperm buffer (51-2090KZ; BD Pharmingen) for 20 minutes at 4°C. The cells were washed twice with PBS and then incubated with PBS solution containing 0.1 mg/ml propidium iodide (PI) and 2 mg/ml RNase A for 30 minutes at 37°C. The cells were analyzed for DNA contents by flow cytometry using an Accuri C6 Flow Cytometer (BD Biosciences), and the data were analyzed using FlowJo software.
Immunoprecipitation and GST Pull-Down
For exogenous interaction between p21 and Flag-BCA2, cell lysates were directly incubated with anti-Flag M2 affinity gel (A2220; Sigma) overnight at 4°C. For endogenous protein interaction, cell lysates were first incubated with anti-p21 antibody or rabbit IgG (sc-2028; Santa Cruz Biotechnology) and then incubated with Protein A/G plus-agarose beads (sc-2003; Santa Cruz Biotechnology). For GST pull-down assay, cell lysates were directly incubated with Glutathione Sepharose 4B (52-2303-00; GE Healthcare) overnight at 4°C. The precipitates were washed four times with 1 ml of lysis buffer, boiled for 10 minutes with 1x SDS sample buffer, and subjected to Western blot (WB) analysis.
In Vivo Ubiquitination Assays
The in vivo ubiquitination assay was performed as a previous protocol [22]. Briefly, after transfecting with WT or mutant Flag-BCA2, HA-p21, and 6x His-Myc-Ub expression plasmids (a gift from Prof Raymond J. Deshaies of Caltech, Pasadena, CA) and treating with 10 µM MG132 to inhibit the p21 degradation, 293FT cells were lysed in warm SDS lysis buffer [50 mM Tris-HCl (pH 7.5), 1% SDS, 1 mM EDTA]. The lysates were boiled for 10 minutes and were diluted 10-fold with dilution buffer [50 mM Tris-HCl (pH 7.5), 1% SDS, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, supplemented with 1x protease inhibitor cocktail]. The protein lysates were mixed with Ni-nitrilotriacetic acid (NTA) agarose (30210; Qiagen, Germantown, MD) for 2 hours in a shaker at 4°C. The precipitates were washed five times with washing buffer [50 mM Tris-HCl (pH 7.4), 1% SDS, 1 mM EDTA, 1 mM DTT, 0.5% NP-40] and were boiled for 10 minutes with 1x SDS sample buffer. The protein samples were subjected to WB analysis.
Cell Proliferation and Cell Viability Assays
Cell proliferation of MCF-7 and T47D cells were performed with the EdU incorporation assay using Click-iT EdU Alexa Fluor 647 Imaging Kit (10340; Invitrogen) according to the manufacturer's guidelines. The cell viability was performed with the Sulforhodamine B (SRB) assay.
Cycloheximide Chase Assays
To determine the half-life of p21 protein, cycloheximide (CHX) chase assays were performed following previous protocols [19]. After transfection for 48 hours, cells were treated for 0, 15, 35, and 60 minutes with 50 µg/ml CHX and collected for WB analysis.
Statistical Analysis
All experiments were performed at least three times, and data were performed as the means ± SD. The differences between any two groups were analyzed by t test. The P values of less than .05 were considered statistically significant.
Results
BCA2 Promotes Cell Proliferation in the MCF-7 and T47D Breast Cancer Cell Lines
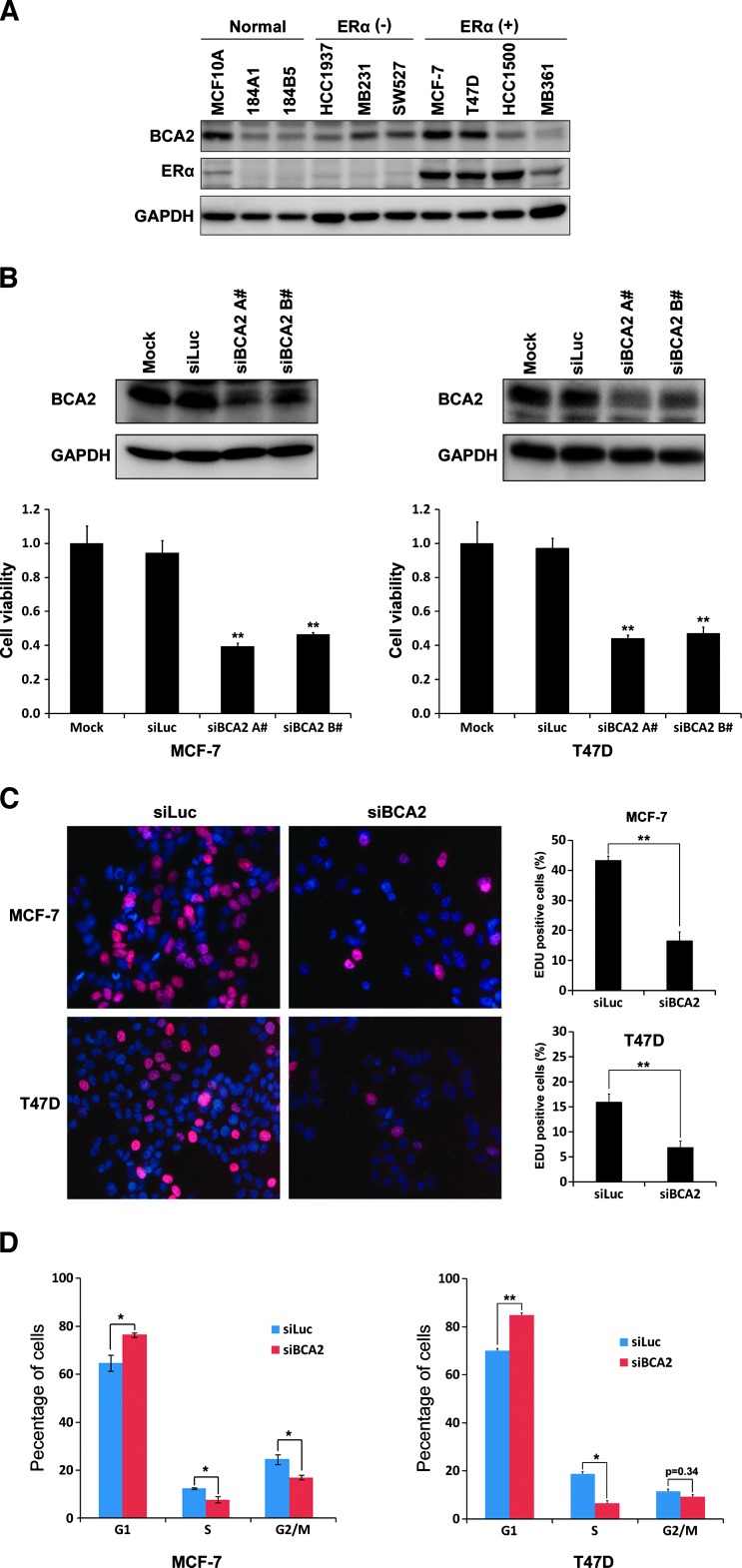
Previous studies showed that the BCA2 protein is highly expressed in ERα-positive breast cancer cell lines and tumors [8]. We first examined the BCA2 protein levels in two immortalized breast epithelial cell lines, three ERα-negative cancer cell lines, and four ERα-positive cancer cell lines by WB analysis. Consistent with a previous report [19], the BCA2 protein was highly expressed in two ERα-positive breast cancer cell lines MCF-7 and T47D (Figure 1A). Following this, we knocked down BCA2 in MCF-7 and T47D by two different siRNAs, siBCA2-A# and siBCA2-B# (Figure 1B), and measured cell growth by the SRB assay. Comparing to the mock and siLuc control, BCA2 siRNAs significantly decreased the cell growth in both cell lines (Figure 1B). Because BCA2 knockdown did not induce cell death in these cells (data not shown), we examined the DNA synthesis by the EdU incorporation experiments. As expected, the DNA synthesis of both cell lines was dramatically inhibited by BCA2 siRNAs compared to siLuc (Figure 1C). Cell cycle analysis indicated that MCF-7 and T47D cells were mainly arrested in the G1 phase (Figure 1D). These results indicate that endogenous BCA2 promotes cell proliferation in ERα-positive breast cancer cells.
Figure 1.
Knockdown of BCA2 inhibits breast cancer cell proliferation, DNA synthesis, and cell cycle. (A) WB analysis of BCA2 protein levels in a panel of immortalized breast epithelial cell lines and ERα-negative and ERα-positive breast cancer cell lines. GAPDH was used as a loading control. (B) MCF-7 and T47D cells were transfected for 72 hours with either Lipofectamine 2000 reagent as mock, control siLuc RNA, siBCA2-A#, or siBCA2-B#. The knockdown effect was assessed by WB analysis, and cell viability was examined using the SRB assay (**P < .01 compared to siLuc). (C) After silencing of BCA2 by siBCA2-A#, MCF-7 and T47D cells were pulse labeled with EdU Alexa Fluor 647 for 4 hours and were stained the nuclei with 10 µg/ml PI. The percentage of positive cells incorporated with EdU were shown in the histograms (right side, **P < .01). All quantitative values were from triplicate experiments. (D) After silencing of BCA2 by siBCA2-A#, MCF-7 and T47D cells were harvested, fixed, stained with PI solution, and analyzed by flow cytometry. The percentage of arrest G1 phase cells was shown in the histograms (*P < .05 and **P < .01).
BCA2 Suppresses the p21 Expression at the Protein Level but Not the mRNA Level
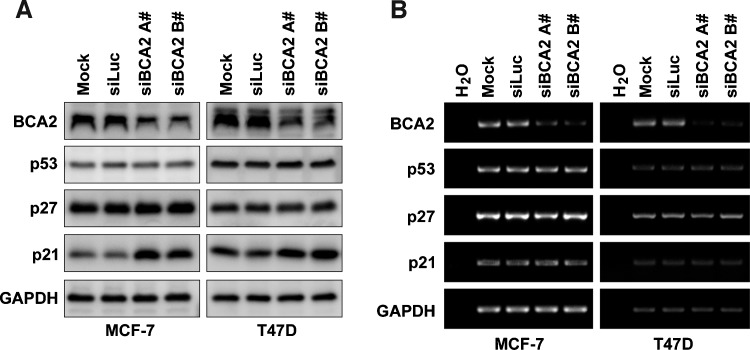
To study the mechanism by which BCA2 promotes breast cancer cell proliferation, we first investigated the protein levels of cell cycle-regulatory proteins p21 and p27, because of the homology of BCA2 and RNF126, which has recently been shown by us to target p21 for degradation [19]. Indeed, silencing of BCA2 in MCF-7 and T47D specifically increased the p21 protein levels but not the p27 and p53 protein levels (Figure 2A). To determine whether BCA2 regulates p21 at the mRNA level, we first evaluated the mRNA levels of p21 by using semiquantitative RT-PCR and found that there were no significant changes for p21 after BCA2 was knocked down in both MCF-7 and T47D (Figure 2B). The result was further confirmed by real-time PCR (Figure W1). Thus, BCA2 is likely to regulate the p21 expression at the protein posttranslational level.
Figure 2.
BCA2 depletion upregulates the p21 expression at the protein level but not the mRNA level. (A) MCF-7 and T47D cells were transfected with either siLuc or siBCA2 for 72 hours. Protein levels of BCA2, p21, p27, and p53 were analyzed by WB analysis. (B) The mRNA levels of BCA2, p21, p27, and p53 were analyzed using semiquantitative RT-PCR. GAPDH was used as a loading control.
BCA2 Promotes p21 Protein Degradation through Proteasome
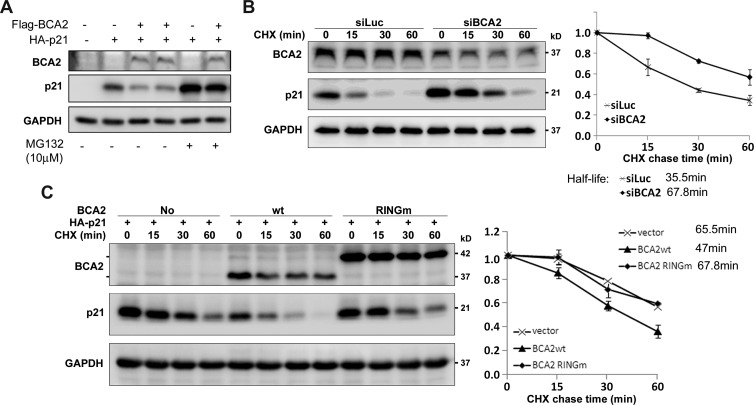
As an E3 ligase, BCA2 may promote the proteolysis of p21. To test this, we cotransfected BCA2 and p21 into HEK293FT cells and found that the p21 protein levels were dramatically downregulated by BCA2 (Figure 3A). The BCA2-mediated p21 degradation was largely blocked by the proteasome inhibitor MG132 (Figure 3A). We further measured the protein half-lives of p21 by performing the CHX chase assay. In MCF-7 cells, when BCA2 was knocked down by siRNA for 48 hours, the half-life of p21 protein was extended from 35.5 to 67.8 minutes (Figure 3B). In addition, we ectopically overexpressed Flag-BCA2 and its RING finger mutant, Flag-BCA2-RINGm [10], which does not have the E3 ubiquitin ligase activity, in HEK293FT cells. As shown in Figure 3C, WT BCA2 shortened the half-life of p21 protein from 65.5 to 47 minutes, while the RINGm BCA2 did not significantly change the half-life of p21 protein. Thus, BCA2 promotes p21 protein degradation in an E3 ubiquitin ligase activity-dependent manner.
Figure 3.
(A) BCA2 promotes p21 protein degradation through proteasome. Flag-BCA2 and HA-p21 were cotransfected into HEK293FT cells as indicated. The proteasome inhibitor MG132 (10 µM) was added to treat the cells for 8 hours if necessary. (B) After depletion of BCA2 by siBCA2-A# in MCF-7 cells, the cells were exposed to CHX for a time as indicated, and WB analysis was performed. The quantitative half-lives of p21 protein from two independent experiments are shown on the right side. (C) HEK293FT cells were cotransfected with HA-p21 and WT BCA2 or RING finger mutant BCA2 expression plasmids. The BCA2-RINGm protein migrates much slower than WT BCA2 possibly because of the protein structure difference under the electrophoresis condition. The cells were exposed to CHX for a time as indicated. The level of p21 protein was detected by WB analysis. The quantitative data from two independent experiments are shown on the right side.
BCA2 Interacts with p21 and Promotes Its Ubiquitination
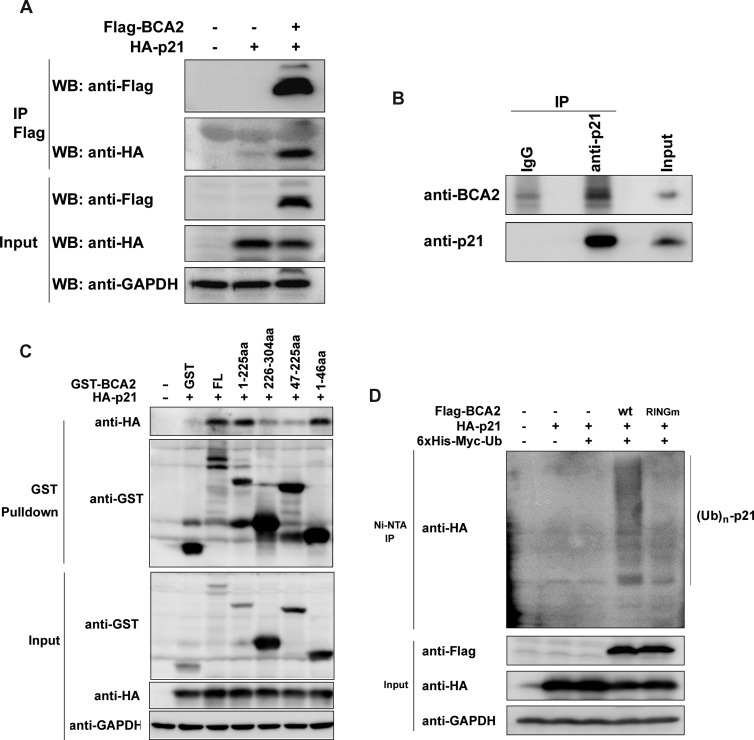
To test whether BCA2 interacts with p21, we cotransfected Flag-BCA2 and HA-p21 constructs into HEK293FT cells treated with 10 µM MG132 for 4 hours to prevent p21 degradation. When Flag-BCA2 was immunoprecipitated by the anti-Flag M2 affinity gel, HA-p21 was detected in the complex (Figure 4A). Without Flag-BCA2, the HA-p21 protein could not be immunoprecipitated (Figure 4A), indicating the specificity of interaction. To further test if endogenous proteins also interact with each other, the endogenous p21 protein was immunoprecipitated by the anti-p21 antibody in MCF-7 cells, and the endogenous BCA2 was specifically co-immunoprecipitated (Figure 4B). To investigate the detailed interaction, we constructed GST-fused WT and four truncated BCA2 expression plasmids and performed the GST pull-down assays. WT BCA2, BCA2 (1-225), and BCA2 (1-46) pulled down p21 (Figure 4C). Consistently, BCA2 (47–225) failed to interact with p21. These results suggest that BCA2 interacts with p21 through the first 46 amino acid residues (Figure 4C).
Figure 4.
BCA2 interacts with p21 and promotes its ubiquitination. (A) The HA-p21 expression construct was transiently cotransfected with an empty control or Flag-BCA2 expression plasmid into HEK293FT cells, respectively. After culturing for 48 hours, the cells were exposed to 10 µM MG132 for 4 hours and collected for immunoprecipitation experiments using anti-Flag M2 affinity gel. The samples were subjected to WB analysis using anti-Flag and anti-HA antibodies. Total cell lysates were used as the input control. (B) Cell lysates from exponentially growing MCF-7 cells were immunoprecipitated with the anti-p21 antibody or the rabbit IgG control. Precipitates were subjected to WB analysis with anti-p21 and anti-BCA2 antibodies. (C) The HA-p21 expression construct was transiently cotransfected with a pEBG vector as a control, PEBG-BCA2 as a positive control, or truncated GST-BCA2 expression plasmids in HEK293FT cells, respectively. Total cell lysates were collected 48 hours later and precipitated with Glutathione Sepharose 4B. The precipitated proteins were subjected to WB analysis using anti-HA and anti-GST antibodies. Total cell lysates were used as the input control. (D) HEK293FT cells were transiently cotransfected with a panel of expression plasmids as indicated. Forty-eight hours later, the cells were treated for 4 hours with 10 µM MG132 and lysed. The samples were precipitated with Ni-NTA agarose beads under a denaturing condition. The proteins were subjected to WB analysis with anti-HA and anti-Flag antibodies.
To determine whether BCA2 promotes the p21 protein degradation through ubiquitination, we cotransfected a panel of expression plasmids, including Flag-BCA2/Flag-BCA2 RINGm, HA-p21, and 6x His-Myc-Ub, into HEK293FT cells and examined the p21 ubiquitination. WT BCA2 obviously increased the ubiquitination level of p21 protein compared to the empty vector control and BCA2 RINGm (Figure 4D).
BCA2 Promotes Breast Cancer Cell Proliferation Partially through Downregulating the Expression of p21
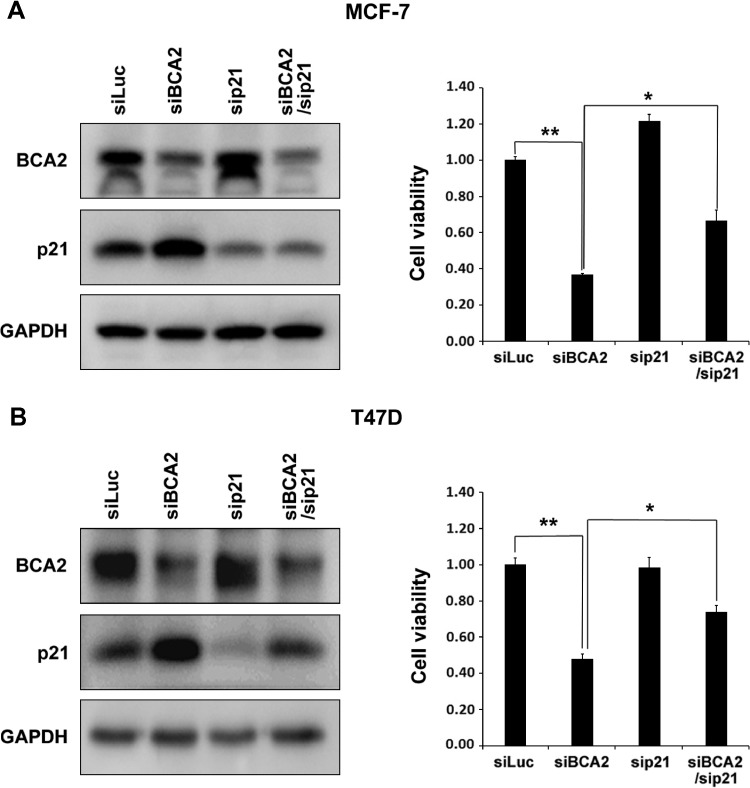
BCA2 may ubiquitinate multiple substrates. We sought to determine whether BCA2 functions through p21. Rescue experiments were performed in MCF-7 and T47D cells. Since BCA2 knockdown increased the p21 protein levels in both cell lines, we silenced the expression of both p21 and BCA2 and found that p21 knockdown partially but significantly restored the BCA2 siRNA-induced growth arrest in both cell lines (Figure 5).
Figure 5.
BCA2 promotes breast cancer cell proliferation partially through downregulating the expression of p21. MCF-7 and T47D cells were transiently transfected with siLuc, sip21, and siBCA2 siRNA, as indicated. Cell lysates were collected 72 hours later and subjected to WB analysis. Protein levels of p21 and BCA2 in MCF-7 (A) or T47D (B) were shown. The cell viability was measured by the SRB assay and shown in the graphs, respectively, (A) MCF-7 or (B) T47D (**P < .01).
Because BCA2 has been reported to be a direct target gene of ER [9], we further tested whether BCA2-mediated p21 degradation is regulated by estrogen. As shown in Figure W2, estrogen treatment for 24 hours induced degradation of ERα but only slightly increased the BCA2 and p21 protein levels in MCF-7. Knockdown of BCA2 dramatically increased the p21 protein levels independent of estrogen treatment.
Discussion
In eukaryotic cells, activities of each member of the CDK family are specifically controlled by the availability of its cyclin partners and the CDK inhibitors, such as p21, p27, and p57 [23]. Deregulation of cyclins or CDK inhibitors results in aberrant cell cycle progression and cell proliferation, eventually causing cancer initiation and progression. Ubiquitin-mediated proteasomal degradation plays crucial roles on controlling the abundance of these cell cycle regulators [24]. In this study, we demonstrated that BCA2 is a novel E3 ubiquitin ligase for p21 because BCA2 interacts with p21 and increased the p21 protein ubiquitination and degradation in an E3 ligase activity-dependent manner. Consistently, depletion of BCA2 promotes breast cancer cell proliferation.
A recent study showed that the transcription of BCA2 was induced by estrogen through ERα binding to the estrogen-responsive elements at the BCA2 gene promoter in MCF-7 cells [9]. This is consistent with our previous study in which the protein expression of BCA2 was correlated with positive ERα status. We verified that BCA2 was highly expressed in two ERα-positive breast cancer cell lines MCF-7 and T47D and further demonstrated that BCA2 promoted cell proliferation in these cells. However, we also found that the induction of BCA2 by estrogen in MCF-7 is modest (Figure W2) and depletion of BCA2 also decreased cell viability in the ERα-negative MDA-MB-231 breast cancer cell line and PC-3 prostate cancer cell line in an E3 ligase siRNA screening [19]. Thus, BCA2 may promote cell proliferation regardless of the ERα status.
The BCA2 protein shares 46% sequence homology with RNF126, a newly characterized E3 ligase targeting p21 [19]. Both BCA2 and RNF126 share a similar protein structure based on sequence identity [20]. Both proteins interact with p21 through their N-terminal fragments and promote p21 protein ubiquitination and degradation through their RING finger domains. However, RNF126 is highly expressed in ERα-negative breast cancer cell lines but lowly expressed in ERα-positive breast cancer cell lines including MCF-7 and T47D [19]. In contrast, BCA2 is predominately expressed in ERα-positive breast cancer cell lines and tumors. Thus, they seem to play a similar role in different types of breast cancers. How BCA2, RNF126, and other E3 ligases coordinately regulate the protein level of p21 in cell type-, cell cycle-, and other context-dependent manners needs further investigation.
Besides p21, several other BCA2 interacting proteins, including Rab7, tetherin, ubiquitin, UBC9, hHR23a, and 14-3-3σ have been identified [20]. However, BCA2 appears not to target these proteins for ubiquitin-mediated degradation. It is significant to identify the physiological substrates for BCA2 to fully elucidate the function and mechanism of BCA2 in breast cancer and other diseases.
In conclusion, we demonstrated that BCA2, a potential oncogenic E3 ubiquitin ligase, promotes breast cancer cell proliferation partially through targeting p21 for ubiquitination and proteasomal degradation. These findings provide a mechanistic insight for BCA2 function in breast cancer.
Supplementary Material
Acknowledgments
We thank Tiebang Kang of Sun Yat-Sen University Cancer Center for providing the HA-p21 expression construct.
Footnotes
This study was supported by National Key Basic Research Program of China (2013CB910900), National Natural Science Foundation of China (81072162, 81120108019, 81272930, and U1132605), Top Talents Program of Yunnan Province, China (2010CI114), Science and Technological Key Project of Yunnan Province (2012FB185), and “Western Light” Talents Training Program of Chinese Academy of Sciences (to Q.K.). The authors have no conflicts of interest.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 2.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 4.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 5.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 8.Burger AM, Gao Y, Amemiya Y, Kahn HJ, Kitching R, Yang Y, Sun P, Narod SA, Hanna WM, Seth AK. A novel RING-type ubiquitin ligase breast cancer-associated gene 2 correlates with outcome in invasive breast cancer. Cancer Res. 2005;65:10401–10412. doi: 10.1158/0008-5472.CAN-05-2103. [DOI] [PubMed] [Google Scholar]
- 9.Kona FR, Stark K, Bisoski L, Buac D, Cui Q, Dou QP. Transcriptional activation of breast cancer-associated gene 2 by estrogen receptor. Breast Cancer Res Treat. 2012;135:495–503. doi: 10.1007/s10549-012-2107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res. 2008;6:1385–1396. doi: 10.1158/1541-7786.MCR-08-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyakawa K, Ryo A, Murakami T, Ohba K, Yamaoka S, Fukuda M, Guatelli J, Yamamoto N. BCA2/Rabring7 promotes tetherin-dependent HIV-1 restriction. PLoS Pathog. 2009;5:e1000700. doi: 10.1371/journal.ppat.1000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bacopulos S, Amemiya Y, Yang W, Zubovits J, Burger A, Yaffe M, Seth AK. Effects of partner proteins on BCA2 RING ligase activity. BMC Cancer. 2012;12:63. doi: 10.1186/1471-2407-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denicourt C, Dowdy SF. Cip/Kip proteins: more than just CDKs inhibitors. Genes Dev. 2004;18:851–855. doi: 10.1101/gad.1205304. [DOI] [PubMed] [Google Scholar]
- 14.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 15.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 16.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/CCdc20 controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, Song J. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhi X, Zhao D, Wang Z, Zhou Z, Wang C, Chen W, Liu R, Chen C. E3 ubiquitin ligase RNF126 promotes cancer cell proliferation by targeting the tumor suppressor p21 for ubiquitin-mediated degradation. Cancer Res. 2013;73:385–394. doi: 10.1158/0008-5472.CAN-12-0562. [DOI] [PubMed] [Google Scholar]
- 20.Burger A, Amemiya Y, Kitching R, Seth AK. Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia. 2006;8:689–695. doi: 10.1593/neo.06469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu S, Feng Z, Zhang M, Wu Y, Sang Y, Xu H, Lv X, Hu K, Cao J, Zhang R, et al. hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene. 2011;30:2219–2229. doi: 10.1038/onc.2010.596. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Zhou Z, Guo P, Dong JT. Proteasomal degradation of the KLF5 transcription factor through a ubiquitin-independent pathway. FEBS Lett. 2007;581:1124–1130. doi: 10.1016/j.febslet.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Lu Z, Hunter T. Ubiquitylation and proteasomal degradation of the p21Cip1, p27Kip1 and p57Kip2 CDK inhibitors. Cell Cycle. 2010;9:2342–2352. doi: 10.4161/cc.9.12.11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.