Abstract
High abundance of c-Jun is detected in invasive breast cancer cells and aggressive breast tumor malignancies. Here, we demonstrate that a major cause of high c-Jun abundance in invasive breast cancer cells is prolonged c-Jun protein stability owing to poor poly-ubiquitination of c-Jun. Among the known c-Jun-targeting E3 ligases, we identified constitutive photomorphogenesis protein 1 (COP1) as an E3 ligase responsible for c-Jun degradation in less invasive breast cancer cells because depletion of COP1 reduced c-Jun poly-ubiquitination leading to the stabilization of c-Jun protein. In a panel of breast cancer cell lines, we observed an inverse association between the levels of COP1 and c-Jun. However, overexpressing COP1 alone was unable to decrease c-Jun level in invasive breast cancer cells, indicating that efficient c-Jun protein degradation necessitates an additional event. Indeed, we found that glycogen synthase kinase 3 (GSK3) inhibitors elevated c-Jun abundance in less invasive breast cancer cells and that GSK3β nonphosphorylable c-Jun-T239A mutant displayed greater protein stability and poorer poly-ubiquitination compared to the wild-type c-Jun. The ability of simultaneously enforced expression of COP1 and constitutively active GSK3β to decrease c-Jun abundance in invasive breast cancer cells allowed us to conclude that c-Jun is negatively regulated through the coordinated action of COP1 and GSK3β. Importantly, co-expressing COP1 and active GSK3β blocked in vitro cell growth/migration and in vivo metastasis of invasive breast cancer cells. Gene expression profiling of breast tumor specimens further revealed that higher COP1 expression correlated with better recurrence-free survival. Our study supports the notion that COP1 is a suppressor of breast cancer progression.
Introduction
c-Jun is a member of the activating protein 1 family of transcription factors [1]. By forming heterodimers with other members of activating protein 1 family, c-Jun regulates the expression of a variety of genes important for diverse cellular functions including cell growth, cell migration, and invasion [2]. As one of the first identified protooncogenes, extensive studies have been exerted to characterize the role of c-Jun in cancer development including breast cancer. An early study showed that forced c-Jun expression was able to convert noninvasive/hormone-dependent breast cancer MCF7 line to an invasive and hormone-resistant line [3]. A later study using clinical breast tumor specimens revealed that c-Jun was detected in the invasive front of breast tumors and its level correlated with increased angiogenesis [4]. The role of c-Jun to promote breast tumor progression and metastasis is supported by two recent in vivo studies: 1) depletion of c-Jun reduced cell migration and invasion of ErbB2-induced mammary tumors in ErbB2 mammary tumor transgenic mouse model [5] and 2) overexpression of c-Jun was sufficient to confer nonmetastatic breast cancer cells with the capability to metastasize [6]. An active role of c-Jun in breast tumorigenesis may lie in its ability to promote cell proliferation [7] and migration/invasion [8].
High abundance of c-Jun is detected in various aggressive tumor types [9,10]. Immunohistochemistry revealed that c-Jun level was often low and present in only few cells of normal and benign breast tissues; in contrast, immunoreactivity of c-Jun was detected at high intensity and usually detected in a significant percentage of cells in breast carcinoma specimens [11]. With the limited number of human breast cancer cell lines, we previously showed that the amount of c-Jun is much higher in invasive lines than less invasive ones [8]. Although the abundance of c-Jun may be governed at transcriptional, posttranscriptional, translational, and posttranslational levels [2], the mechanism behind elevated c-Jun level in invasive breast cancer cells is not well understood.
Under the normal circumstances, c-Jun protein is known to be highly unstable [12] and its level can be regulated through a ubiquitination/proteasome-dependent mechanism [13]. Ubiquitin E3 li-gases that can add poly-ubiquitin chain on c-Jun include constitutive photomorphogenesis protein 1 (COP1) [14], cullin 4 (CUL4) [15], F-box and WD repeat domain containing 7E3 ubiquitin proteinligase (FBW7) [16], Itchy E3 ubiquitin protein ligase (ITCH) [17], mitogen-activated protein kinase kinase kinase 1 (MEKK1) [18], and sensitive to apoptosis, zinc ring finger protein (SAG) [19]. COP1 is unique from others because of its dual ability to act as an E3 ligase as well as an adaptor to recruit substrate to de-etiolated homolog 1/damage-specific DNA binding protein 1/cullin 4/ring-box 1, E3 ubiquitin protein ligase (DET1/DDB1/CUL4/RBX1) ubiquitin ligase complex [15]. The expression of E3 ligases often displays tissue specificity and varies in various types and stages of cancers; therefore, it is of great interest to investigate whether one or more of these c-Jun-targeting E3 ligases are involved in the regulation of c-Jun protein abundance in breast cancer cells.
Function of c-Jun has been shown to be regulated by phosphorylation. Casein kinase II phosphorylates c-Jun, leading to the suppression of c-Jun activity [20]. In contrast, c-Jun N-terminal kinases (JNKs) activate c-Jun by directly phosphorylating c-Jun [21]. Interestingly, JNKs also activate c-Jun-targeting E3 ligase ITCH, thereby establishing a negative feedback loop [17]. Recognition of c-Jun by E3 ligase FBW7 and subsequent degradation necessitate c-Jun first being phosphorylated by glycogen synthase kinase 3 beta (GSK3β) [22]. However, it is unknown whether c-Jun phosphorylation is a prerequisite for its degradation by COP1 and others.
The objective of this study was to answer how the protein level of c-Jun is regulated in breast cancer cells. By performing cycloheximide-chasing experiment, we found that c-Jun protein was much more stable in invasive breast cancer cells than in less invasive ones. Prolonged c-Jun protein stability was due to much less c-Jun being poly-ubiquitinated in invasive breast cancer cells. To characterize the mechanism mediating c-Jun protein degradation, we showed that either depleting COP1 or inhibiting GSK3 activity prevented polyubiquitination of c-Jun and increased c-Jun abundance in less invasive breast cancer cells. Moreover, only simultaneous enforcement of COP1 and constitutively active GSK3β (GSK3βS9A) expression was able to effectively downregulate c-Jun in invasive breast cancer cells. These results indicate that both high COP1 expression and GSK3β-mediated phosphorylation were necessary for c-Jun protein turnover. This notion was supported by the observation that non-GSK3βphosphorylable c-Jun was significantly more stable than wild-type c-Jun. Importantly, we showed that co-expressing COP1 and GSK3βS9A suppressed both in vitro cell growth/migration and in vivo metastasis of invasive breast cancer cells. Gene expression analysis of breast tumor specimens further revealed that COP1 mRNA expression is an independent positive prognostic factor of recurrence-free survival of patients with breast cancer.
Materials and Methods
Cell Culture, Inhibitors, and siRNAs
All cell lines used in the study were purchased from American Tissue Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS in a humidified atmosphere containing 5% CO2 at 37°C. Inhibitors used in the study were PYR-41 from Calbiochem (San Diego, CA); MG132 and Chloroquine from Tocris (Bristol, United Kingdom); SP600125, SB216763, and 4,5,6,7-tetrabromobenzotriazole (TBB) from Sigma (St Louis, MO). All E3 ligase-targeting siRNAs and control siRNA were purchased from Thermo Scientific (West Palm Beach, FL).
Determination of c-Jun Protein Stability
Overnight cultured MCF7, MDA-MB-231, MDA-MB-436, and T47D cells were co-transfected with plasmids encoding human influenza hemagglutinin (HA)-tagged c-Jun and green fluorescent protein (GFP) for 1 day. Cells were trypsinized and reseeded for 12 hours followed by addition of 10 µg/ml cycloheximide. At varying time points, cells were harvested and cell lysates were subjected to immunoblot analysis to detect HA-tagged c-Jun and GFP using respective antibodies. The intensities of bands were quantified by Odyssey Image System (LI-COR Biosciences, Lincoln, NE).
Immunoprecipitation
To determine c-Jun poly-ubiquitination in breast cancer cells, HA-tagged c-Jun and Flag-tagged ubiquitin were co-transfected into MDA-MB-231, MDA-MB-436, MCF7, and T47D cells for 1 day followed by the treatment of MG132 for 8 hours. Cells were lysed in buffer containing 1% sodium dodecyl sulfate, 150 mM NaCl, and 10 mM Tris-HCl (pH 8.0) followed by 10-minute boiling. Samples were sheared with a sonication device and diluted in buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100. After a 30-minute centrifugation at 20,000g, the resulting supernatants were immunoprecipitated with HA monoclonal antibody (mAb) and immunoprecipitates were subjected to immunoblot analysis to detect Flag-ubiquitin using Flag mAb.
Cell Fractionation
Cell fractionation was performed as we previously described [23]. Briefly, cells were lysed in Buffer A [10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM PMSF, and 0.5 mM DTT] followed by centrifugation at 2000g for 10 minutes to obtain a crude nuclear pellet and a cytoplasmic supernatant (cytoplasmic fraction). The crude nuclear pellets were purified further by resuspension in Buffer B [20 mM Hepes (pH 7.9), 25% (vol/vol) glycerol, 0.45 M NaCl, 1.5 mM MgCl2, 1 mM EDTA, 0.5 mM PMSF, and 0.5 mM DTT]. The lysates were subjected to high-speed centrifugation for 1 hour to obtain nuclear fraction.
Lentiviral Constructs and Packaging
Coding sequences of COP1 and GSK3βS9A mutant were sub-cloned into pCDH Lentivector (System Biosciences, Mountain View, CA). Lentiviruses were packaged in 293FT cells according to the manufacturer's instruction.
Nuclear Proteasome Activity Assay
Nuclear 20S proteasome activities were determined using overnight cultured cells with the aid of 20S Proteasome Assay Kit (Cayman Chemical, Ann Arbor, MI) that measures the ability of the fractions to cleave proteasome-specific substrate SUC-LLVY-AMC. Briefly, cells were resuspended in Buffer A [10 mM Hepes, 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, and 1 mM PMSF (pH7.9)] and incubated on ice for 25 minutes followed by adding 10% NP-40 to the final concentration of 1%. Cells were vortexed for 15 seconds and then microcentrifuged at 5000g for 5 minutes. The collected pellets (cell nucleus) were analyzed for 20S proteasome activity.
Immunofluorescence Staining
Cells were culture on collagen-coated glass coverslips overnight, then fixed with 4% paraformaldehyde and permeabilize with 1% Triton X-100 for 15 minutes. To visualize c-Jun and proteasome S5A subunit, coverslips were incubated with c-Jun mAb and S5A polyclonal antibody followed by incubation with fluorescein isothiocyanate or rhodamine-labeled secondary antibodies. The fluorescence staining was visualized under a fluorescence microscope (Axiovert 200M; Zeiss, Munich, Germany). 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain nucleus of breast cancer cells.
Cell Proliferation Study
To determine the ability of enforced COP1 and GSK3βS9A expression to affect cell proliferation, MDA-MB-231 and MDAMB-436 cells were transduced with empty lentiviral vector or vector containing COP1 or GSK3βS9A for 3 days. Transduced cells were subjected to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay to evaluate cell proliferation as previously described [23]. Briefly, 5 x 104 cells were seeded into 24-well culture plates and allowed to grow for 1 to 3 days before the addition of MTT. Cell proliferation was determined by reading plates at 595 nm after dissolving MTT formazan with DMSO.
Cell Migration Assay
The ability of enforced COP1 and GSK3βS9A expression to affect cell migration of MDA-MB-231 and MDA-MB-436 cells was determined using 24-well formatted collagen-coated transwell as we previously described [8]. Briefly, 1 x 105 cells in 100 µl of serum-free medium were added into the upper chamber of each transwell and allowed to migrate for 4 hours. Nonmigrating cells in the upper chambers were removed by cotton swabs, and migrating cells on the lower surface of the chamber were stained using crystal violet. Amount of migrating cells was determined by counting the stained cells under a phase-contrast microscope.
Zebrafish Metastatic Model
The effect of combined expression of COP1 and GSK3βS9A was evaluated by a recently developed zebrafish metastatic model [24]. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Georgia Regents University. Briefly, control or COP1/GSK3βS9A-expressing MDA-MB-231 cells were first labeled with fluorescent dye CM-Dil (Life Technologies, Carlsbad, CA), and approximately 200 labeled cells were then microinjected into the perivitelline space of 48-hour post fertilization (hpf) zebrafish embryos using a pressure microinjector. The embryos were kept at 34°C and then imaged under anaesthetic by confocal microscopy at 48-hour post injection.
Statistical Analysis
Statistical analyses of laboratory studies were performed by either Student's t test or analysis of variance. To determine the potential correlation between COP1 expression and clinicopathologic parameters, we obtained data of COP1 expression in microarray analysis and clinical features of 126 patients with breast cancer who received tamoxifen monotherapy, chemotherapy, and radiation therapy from the Gene Expression Omnibus database (91 samples from GSE18229 and 35 samples from GSE40954). The cutoff value of high and low COP1 expression groups was based on the median value. Associations between COP1 expression and clinicopathologic parameters were determined using Pearson χ2 test. In univariate survival analysis, the Kaplan-Meier method and log-rank test were used to compare recurrence-free survival curves between high and low COP1 expression groups. In multivariate survival analysis, the Cox proportional hazard regression model was used to identify important factors on disease-free survival. To identify these factors, an initial model with only COP1 expression was considered and a particular clinicopathologic parameter was added to the model at a time. By repeating this procedure, the final regression model with important factors was identified by likelihood ratio test. P < .05 was considered statistically significant.
Results
Ubiquitin/Proteasome-Dependent Mechanism Controls c-Jun Abundance in Breast Cancer Cells
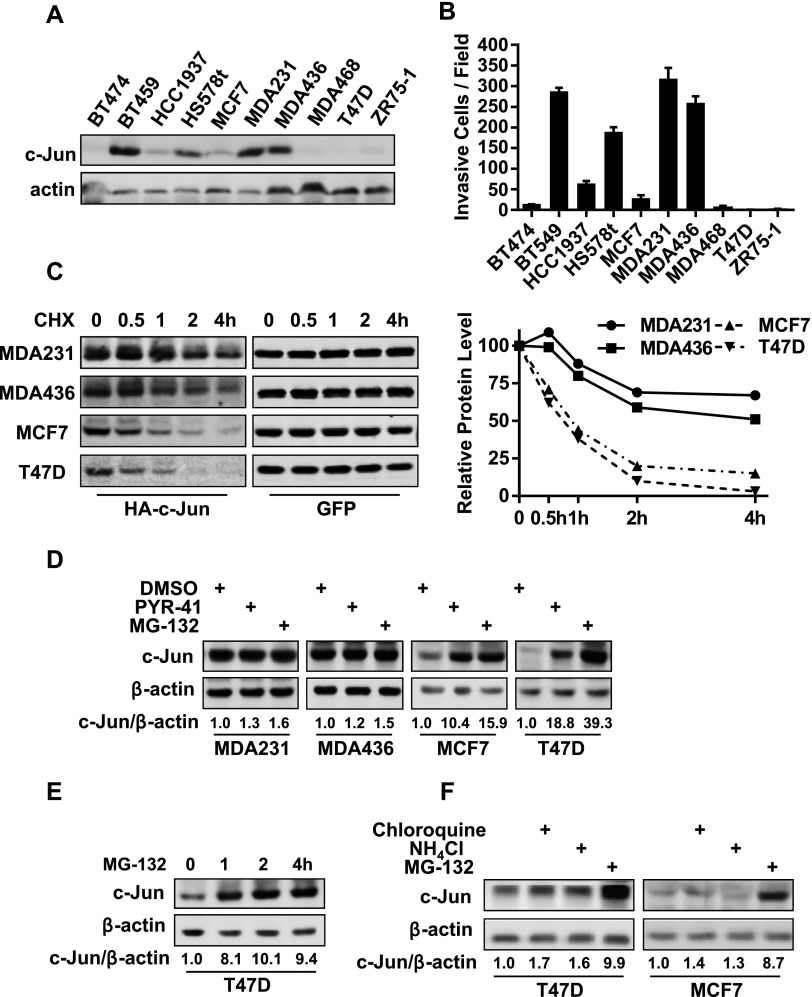
With limited human breast cancer cell lines, we previously showed that the abundance of c-Jun was much higher in invasive breast cancer cells than in less invasive ones [8]. To extend this early finding, we performed immunoblot analysis to examine c-Jun abundance in 10 established breast cancer cell lines that included 5 invasive and 5 less invasive lines (Figure 1B). High level of abundance of c-Jun was seen in all invasive lines, while it was low or barely detectable in less invasive cell lines (Figure 1, A and B). To determine whether there was any difference in c-Jun protein stability between invasive and less invasive lines, we co-transfected HA-tagged c-Jun and GFP into two invasive (MDA-MB-231 and MDA-MB-436) and two less invasive lines (MCF7 and T47D) followed by performing cycloheximide-chasing experiment to measure the stability of HA-tagged c-Jun protein in these lines. HA-tagged c-Jun was relatively stable and we detected no more than 40% reduction in the amount of HA-tagged c-Jun in MDA-MB-231 and MDA-MB-436 cells after 2 hours of cycloheximide addition (Figure 1C). In contrast, more than 80% of HA-tagged c-Jun was degraded in the same time period in T47D and MCF7 cells (Figure 1C). Because GFP exhibited similar protein stability in both invasive and less invasive lines (Figure 1C), these results indicate that greater c-Jun protein stability is a contributing factor to high c-Jun abundance in invasive breast cancer cells.
Figure 1.
Abundance of c-Jun in breast cancer cells is determined by its protein stability. (A) Immunoblot analysis of c-Jun in breast cancer cell lines. (B) In vitro invasion of breast cancer cell lines. Data are the means ± SE (n = 3). (C) Cells were co-transfected with HA-tagged c-Jun and GFP for 24 hours followed by the addition of 20 µg/ml cycloheximide for varying times. Cells were lysed, and cell lysates were subjected to immunoblot analysis to detect HA-tagged c-Jun and GFP. The relative HA-tagged c-Jun amount was standardized by GFP. (D) Cells were treated with vehicle (DMSO), 50 µM PYR-41, or 10 µM MG-132 for 8 hours and then lysed for immunoblot analysis to detect c-Jun and β-actin. (E) T47D cells were treated with 10 µM MG-132 for varying times and then lysed for immunoblot analysis to detect c-Jun and β-actin. (F) Cells were treated with 50 µM chloroquine, 10 mM NH4Cl, or 10 µM MG-132 for 8 hours and then lysed for immunoblot analysis to detect c-Jun and β-actin.
c-Jun protein can be degraded through a ubiquitin/proteasome-dependent mechanism in various cell types, including melanoma and lung cancer cells [25,26]. To determine whether ubiquitin/proteasome system plays a role in regulating c-Jun abundance in breast cancer cells, we treated MCF7, MDA-MB-231, MDA-MB-436, and T47D cells with either ubiquitin E1 ligase inhibitor PYR-41 [27] or proteasome inhibitor MG132 for 8 hours. Immunoblot analysis showed that treatment of either inhibitor markedly increased the amount of c-Jun in less invasive cells (MCF7 and T47D), while it did not significantly alter c-Jun abundance in invasive cells (MDA-MB-231 and MDA-MB-436; Figure 1D). MG132-induced accumulation of c-Jun protein was very rapid; 1 hour of MG132 treatment elevated more than eight-fold of c-Jun level in T47D cells (Figure 1E). These results suggest that ubiquitin/proteasome system is responsible for efficient c-Jun protein degradation in less invasive breast cancer cells. Because lysosome system can regulate protein stability, we treated MCF7 and T47D cells with lysosome inhibitors chloroquine and NH4Cl for 8 hours. Immunoblot analysis showed that neither significantly altered c-Jun level (Figure 1F), thus excluding the participation of lysosome in c-Jun protein degradation.
Status of c-Jun Poly-ubiquitination Determines c-Jun Abundance in Breast Cancer Cells
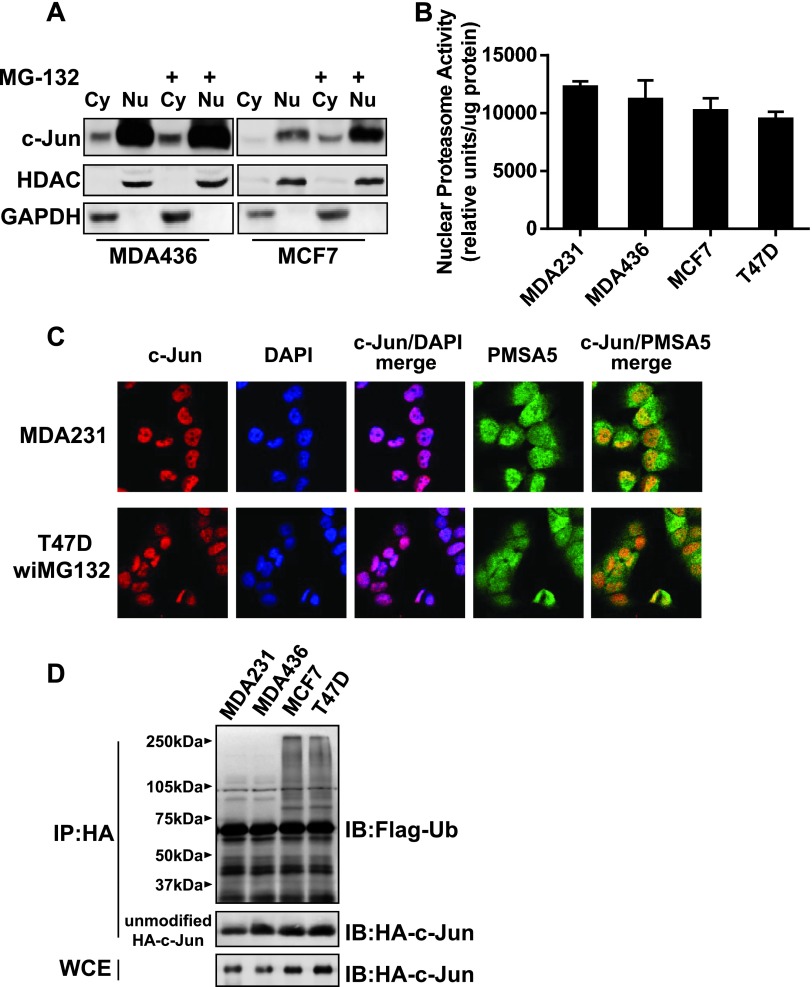
To determine whether the difference in c-Jun protein stability is linked to the variation in proteasome activity between invasive and less invasive cells, we first examined the subcellular localization of c-Jun in MCF7 and MDA-MB-436 cells. Immunoblot analysis of their cytoplasmic and nuclear fractions showed that c-Jun was similarly present in the nucleus of both lines, and the nuclear localization of c-Jun was not affected by MG132 treatment (Figure 2A). We next measured the proteasome activities in the nucleus of MCF7, MDA-MB-231, MDA-MB-436, and T47D cells. Up to 1-hour reaction period, we did not observe significant difference in nuclear proteasome activity among these lines (Figure 2B). Immunofluorescence staining further showed a similar pattern of c-Jun and proteasomal PMSA5 co-localization in MDA-MB-231 and MG132-treated T47D cells (Figure 2C). Identical pattern was also observed in MDA-MB-436 and MG132-treated MCF7 cells (Figure W1). These results indicate that the difference in c-Jun protein stability between invasive and less invasive breast cancer cells is not caused by the variation in nuclear proteasome activity or co-localization of c-Jun with the proteasome.
Figure 2.
The extent of poly-ubiquitination in c-Jun is different between invasive and less invasive breast cancer cells. (A) Cells were treated with 10 µM MG132 or left untreated for 8 hours. Cells were lysed for the preparation of cytoplasmic (Cy) and nuclear (Nu) fractions, and fractions were analyzed by immunoblot analysis to detect c-Jun and histone deacetylase 1 (HDAC1; indicator of nuclear fraction). (B) Proteasome activities of breast cancer cell lines. Data are the means ± SE (n = 3). (C) Immunofluorescence staining of c-Jun and PMSA5 in MDA-MB-231 and 10 µMMG-132-treated T47D cells. DAPI was used for nuclear staining. (D) Cells were co-transfected with HA-tagged c-Jun and Flag-tagged ubiquitin for 1 day followed by 10 µM MG-132 treatment for 8 hours. Cells were lysed, cell lysates were immunoprecipitated with HA mAb, and immunoprecipitates were subjected to immunoblot analysis to detect Flag-tagged ubiquitin (Ub).
Subsequently, we investigated the status of c-Jun poly-ubiquitination in MG132-treated MCF7, MDA-MB-231, MDA-MB-436, and T47D cells that had been co-transfected with HA-tagged c-Jun and Flag-tagged ubiquitin. Immunoblot analysis of HA immunoprecipitates with anti-Flag mAb showed that HA-tagged c-Jun was robustly poly-ubiquitinated in MCF-7 and T47D cells, whereas the poly-ubiquitination of HA-tagged c-Jun was very weak in MDA-MB-231 and MDA-MB-436 cells (Figure 2D). These results suggest that the effectiveness of c-Jun being poly-ubiquitinated determines the abundance of c-Jun in breast cancer cells.
COP1 E3 Ligase Is Required for c-Jun Degradation in Breast Cancer Cells
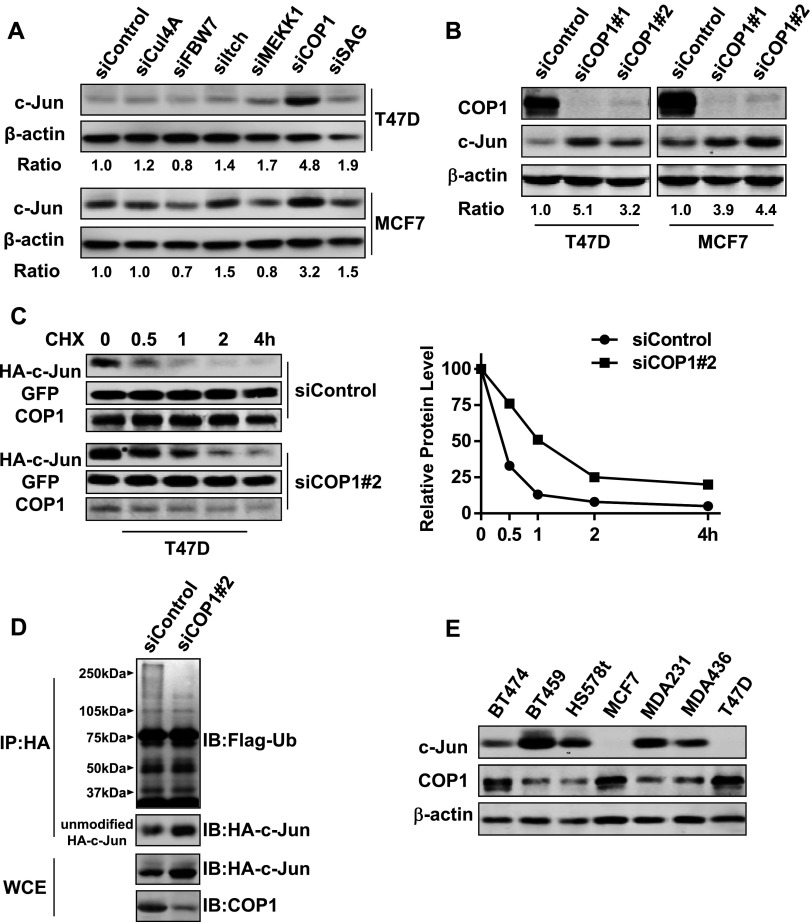
c-Jun is the substrate of CUL4, FBW7, ITCH, MEKK1, COP1, and SAG E3 ligases. To identify the E3 ligase that regulates c-Jun abundance in breast cancer cells, we treated MCF7 and T47D cells with predesigned siRNA pool for each of these E3 ligases. While all of them effectively knocked down their respective targets (Figure W2), only COP1 siRNA pool markedly elevated the amount of c-Jun in both cell lines (Figure 3A). To confirm the results generated with siRNA pool, MCF7 and T47D cells were treated with two sequence distinct COP1 siRNAs for 3 days. Immunoblot analysis showed that both of siRNAs increased c-Jun abundance (Figure 3B). We next examined the effect of COP1 knockdown on c-Jun protein stability by introducing HA-tagged c-Jun into both control and COP1-knockdown T47D cells. Cycloheximide-chasing experiment showed that HA-tagged c-Jun was more stable in COP1-knockdown T47D cells than in control T47D cells (Figure 3C). Immunoprecipitation experiment further revealed that the extent of c-Jun poly-ubiquitination was significantly reduced in COP1knockdown cells compared with the control cells (Figure 3D).
Figure 3.
COP1 is the E3 ligase mediating c-Jun turnover in breast cancer cells. (A) T47D and MCF7 cells were transfected with 50 nM siRNA pool against CUL4, FBW7, ITCH, MEKK1, COP1, or SAG for 3 days. Cells were then lysed, and cell lysates were analyzed by immunoblot analysis to detect c-Jun, COP1, and β-actin. (B) T47D and MCF7 were transfected with two sequence-distinct COP1 siRNAs (50 nM) and then lysed for immunoblot analysis to detect c-Jun, COP1, and β-actin. (C) T47D cells were transfected with 50 nM control or COP1 siRNA for 2 days and then co-transfected with HA-tagged c-Jun and GFP for 1 day. After adding 20 µg/ml cycloheximide for varying times, cells were harvested and cell lysates were analyzed by immunoblot analysis to detect HA-tagged c-Jun, GFP, and COP1. The relative HA-tagged c-Jun amount was standardized by the amount of GFP. (D) T47D cells were first treated with 50 nM control or COP1 siRNA for 2 days and then co-transfected with HA-tagged c-Jun and Flag-tagged ubiquitin for 1 day. Cells were treated with 10 µM MG-132 treatment for 8 hours and subjected to immunoprecipitation with HA mAb. Immunoprecipitates were analyzed by immunoblot analysis to detect Flag-tagged ubiquitin (Ub). (E) Immunoblot analysis of c-Jun and COP1 in breast cancer cell lines.
To substantiate our findings, we investigated a potential functional link between c-Jun and COP1 among breast cancer cell lines. Immunoblot analysis revealed that lines with relatively high c-Jun abundance (BT549, Hs578t, MDA-MB-231, and MDA-436) displayed much less COP1 than those with relatively low c-Jun abundance (BT474, MCF7 and T47D; Figure 3E). This inverse correlation between c-Jun and COP1 abundance implicates that COP1 negatively regulates the abundance of c-Jun in breast cancer cells.
c-Jun Protein Degradation Is Sensitive to the Inhibition of GSK3 Activity
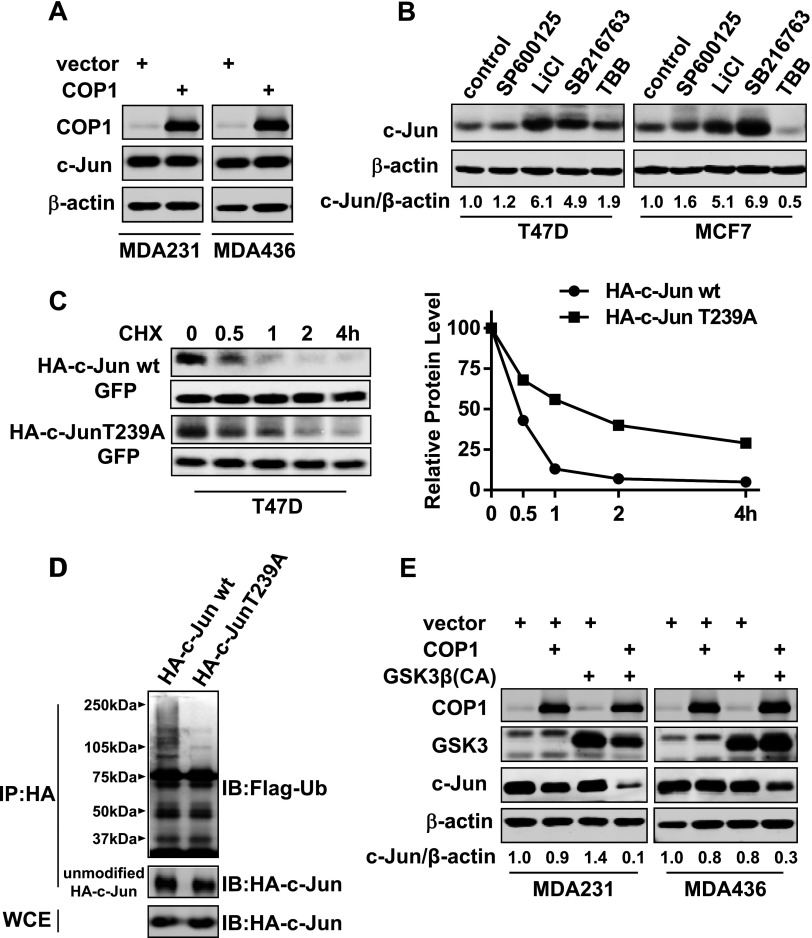
The inverse correlation between c-Jun and COP1 abundance prompted us to test whether enforcing COP1 expression in invasive breast cancer cells could downregulate the amount of c-Jun. We lentivirally introduced COP1 into MDA-MB-231 and MDA-MB-436 cells, and populations of transduced cells were analyzed for c-Jun abundance. Immunoblot analysis showed that COP1 overexpression was unable to decrease the level of c-Jun (Figure 4A), suggesting that an event additional to high COP1 expression is required for rapid c-Jun protein degradation.
Figure 4.
Activity of GSK3β is required for efficient c-Jun protein turnover in breast cancer cells. (A) COP1 was lentivirally overexpressed in MDA-MB-231 and MDA-MB-436 cells. Populations of cells were lysed for immunoblot analysis to detect COP1, c-Jun, and β-actin. (B) T47D and MCF7 cells were treated with 10 µM SP600125, 10 mM LiCl, 10 µM SB216763, or 50 µM TBB for 12 hours and then lysed for immunoblot analysis to detect c-Jun and β-actin. (C) HA-tagged c-Jun or c-Jun-T239A was co-transfected into T47D cells with GFP for 1 day followed by the addition of 20 µg/ml cycloheximide for varying times. Cells were harvested, and cell lysates were analyzed by immunoblot analysis to detect HA-tagged c-Jun and GFP. The relative HA-tagged c-Jun amount was standardized by the amount of GFP. (D) T47D cells were co-transfected with HA-tagged c-Jun and Flag-tagged ubiquitin for 1 day and then treated with 10 µM MG-132 for 8 hours. Cells were lysed, and cell lysates were subjected to immunoprecipitation with HA mAb. Immunoprecipitates were analyzed by immunoblot analysis to detect Flag-tagged ubiquitin (Ub). (E) MDA-MB-231 and MDA-MB-436 cells were lentivirally transduced with COP1 and constitutively active GSK3β either alone or together for 3 days. Cells were lysed, and cell lysates were subjected to immunoblot analysis to detect COP1, GSK3, c-Jun, and β-actin.
The activities of CK2, JNK, and GSK3 have been shown to regulate c-Jun activity or stability [17,20–22]. Especially, recognition of c-Jun by E3 ligase FBW7 necessitates GSK3-mediated phosphorylation of c-Jun [22]. To test whether phosphorylation by CK2, JNK, or GSK3 was required for c-Jun turnover, MCF7 and T47D cells were treated with specific inhibitors to JNK (SP600125), GSK3 (LiCl and SB216763), or CK2 (TBB). Immunoblot analysis showed that both GSK inhibitors but neither JNK nor CK2 inhibitor led to significant increase in the amount of c-Jun (Figure 4B), indicating the necessity of GSK3 activity for the instability of c-Jun protein. We next generated HA-tagged non-GSK3β-phosphorylable c-Jun (c-Jun-T239A) by performing Thr→Ala mutagenesis at amino acid residue 239 of c-Jun, the GSK phosphorylation site. Cycloheximide-chasing experiment showed that HA-tagged c-Jun-T239A was significantly more stable than HA-tagged c-Jun (Figure 4A), and depletion of COP1 did not affect the stability of HA-tagged c-Jun-T239A (Figure W3). Compared to HA-tagged c-Jun, HA-tagged c-Jun-T239A was poorly poly-ubiquitinated (Figure 4D). These results suggest that GSK3β-mediated phosphorylation is a prerequisite for efficient polyubiquitination of c-Jun and its subsequent degradation.
We next enforced the expression of COP1 or constitutive active GSK3β (GSK3βS9A) either alone or together in MDA-MB-231 and MDA-MB-436 cells. Although either alone did not reduce the amount of c-Jun, their combination diminished more than 90% and 70% of c-Jun in MDA-MB-231 and MDA-MB-436 cells, respectively (Figure 4E). These results suggest that COP1 and GSK3β work together to facilitate effective c-Jun protein degradation in breast cancer cells.
Simultaneously Enhancing COP1 Expression and Elevating GSK3β Activity Suppress Breast Tumorigenesis
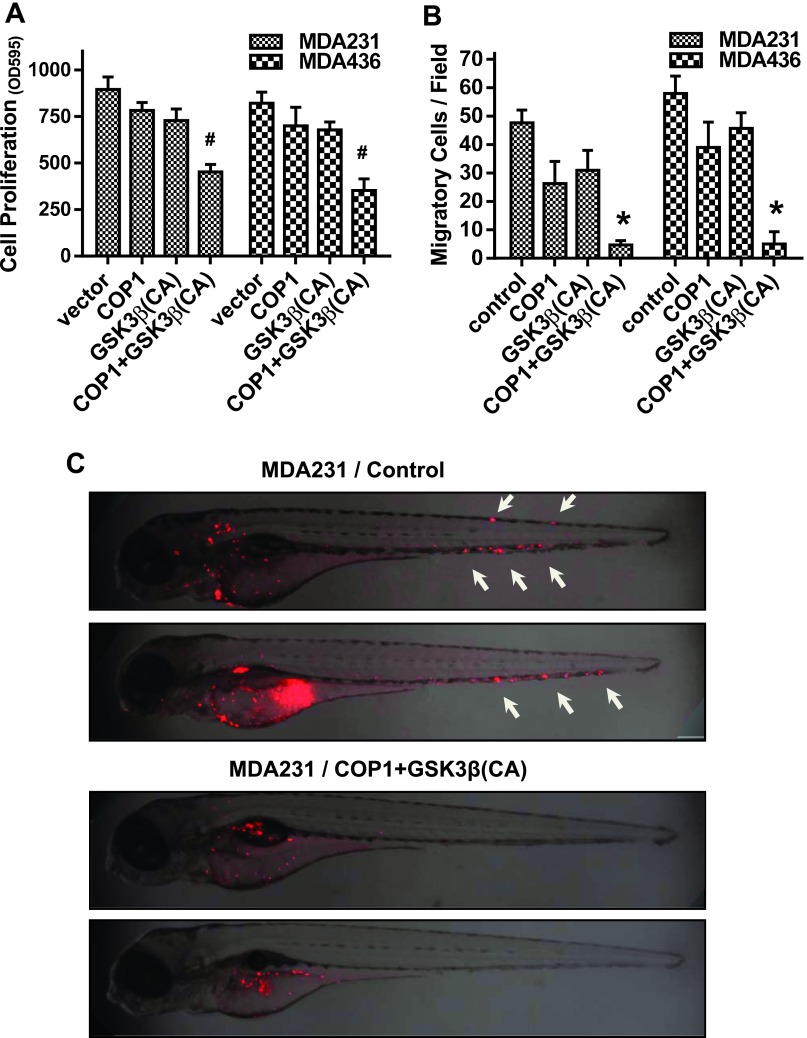
We previously showed that the presence of c-Jun is essential for cell migration and metastasis of invasive breast cancer cells [8]. Others have also reported that c-Jun promotes breast cancer cell proliferation by controlling the expression of cyclins and E2F [7]. We thus hypothesized that the reduced c-Jun abundance by combined expression of COP1 and constitutively active GSK3β would lead to the suppression in breast tumorigenesis. To test this hypothesis, we lentivirally introduced COP1 and GSK3βS9A either alone or in combination into MDA-MB-231 and MDA-MB-436 cells. MTT assay showed that raising COP1 level or enforcing GSK3βS9A expression alone moderately reduced cell proliferation, whereas their co-expression led to much greater degree of inhibition in cell proliferation (Figure 5A). Transwell migration assay showed that forced co-expression of COP1 and GSK3βS9A blocked about 90% of cell migration, whereas expressing either one alone inhibited only 20% to 40% of cell migration when compared to the control (Figures 5B and W4). We further analyzed the effect of combined COP1 and GSK3βS9A expression in the metastasis of MDA-MB-231 and MDA-MB-436 cells using a well-established zebrafish metastasis model [24,28,29]. Control cells and cells with the co-expression of COP1 and GSK3βS9A were microinjected into the perivitelline space of 48-hpf zebrafish embryos, and embryos were imaged after 2 days. Metastasis was detected in 44 of 48 and 46 of 56 embryos injected with control MDA-MB-231 and MDA-MB-436 cells, respectively (Figures 5C and W5). In contrast, metastasis was only seen in 10 of 54 and 4 of 42 embryos injected with MDA-MB-231 and MDA-MB-436 cells co-expressing COP1 and GSK3βS9A, respectively, while remaining embryos displayed cells at the site of injection (Figures 5C and W5). These results demonstrate that simultaneously enhancing COP1 expression and elevating GSK3β activity can effectively suppress breast cancer cell tumorigenesis.
Figure 5.
Simultaneously expressing COP1 and constitutively active GSK3β suppress breast tumorigenesis. (A) MDA-MB-231 and MDA-MB-436 cells were lentivirally transduced with COP1 and constitutively active GSK3β either alone or together for 3 days. Populations of transduced cells were then subjected to MTT assay to measure cell proliferation. Data are the means ± SE (n = 3). #P < .01 versus empty vector (control). (B) Populations of transduced cells were subjected to transwell assay to measure cell migration. Data are the means ± SE (n = 3). *P < .001 versus empty vector (control). (C) Control or COP1/GSK3βS9A-transduced MDA-MB-231 cells were labeled with fluorescent dye CM-Dil and then microinjected into the perivitelline space of 48-hpf zebrafish embryos. Embryos were imaged under a confocal microscope.
COP1 Expression Correlates with Recurrence-Free Survival of Patients with Breast Cancer
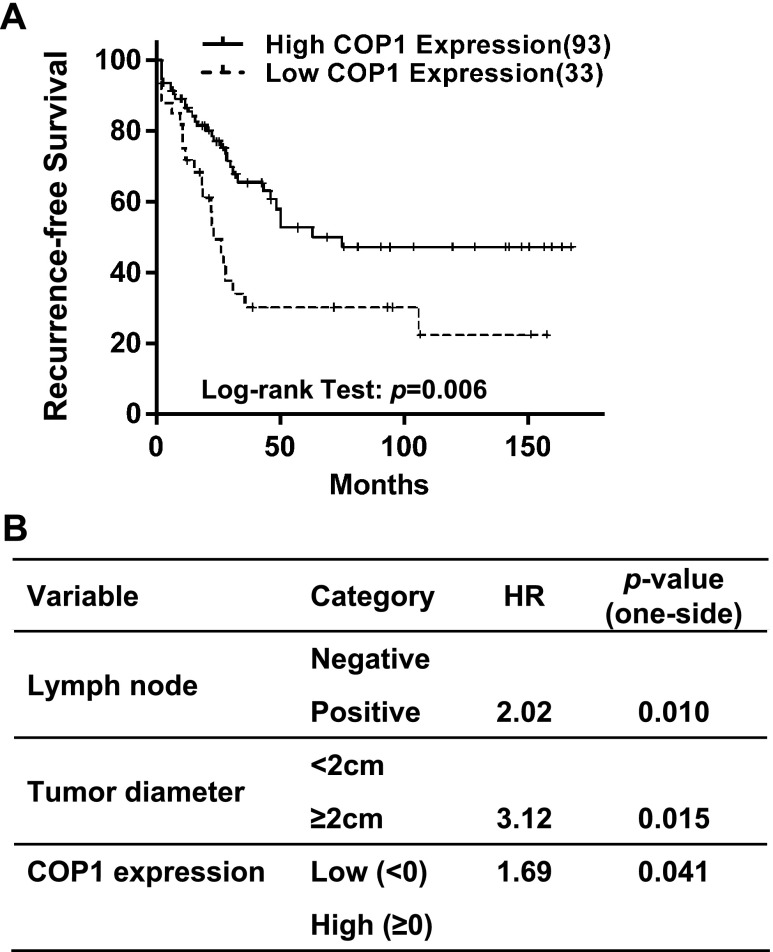
As laboratory study may not always recapitulate clinical breast malignancy, we explored the potential clinical relevance of COP1 expression in human breast cancer. To do so, we first assessed the relationship between COP1 mRNA expression and clinical features of breast tumors using microarray data of 126 patients with breast cancer obtained from the Gene Expression Omnibus. There were no significant associations between COP1 expression (divided into low and high expression groups by median) and pathologic parameters, including histologic grade, tumor diameter, estrogen and progesterone receptor expression, and lymph node status (Table W1). However, univariate survival analysis (Kaplan-Meier method, log-rank test) showed that higher COP1 expression was significantly associated with high recurrence-free survival of patients (P = .006; Figure 6A). By multivariate analysis (proportional hazard method) and encompassing basic prognostic factors such as tumor diameter, histologic grade, and lymph node status in addition to COP1 expression (initial regression model), COP1 expression status remained as an independent positive prognostic factor in the final multivariate model (P = .041, likelihood ratio test) along with tumor diameter and lymph node status, with a hazard ratio of 1.69 between patients with high and low COP1 expression (i.e., patients with low COP1 expression have 1.69 times higher hazard for the recurrence of breast cancer than those with high COP1 expression; Figure 6B). These results also support the notion that COP1 is a negative regulator of breast tumor progression.
Figure 6.
COP1 expression is a positive prognostic factor for recurrence-free survival of patients with breast cancer. (A) Univariate survival method (Kaplan-Meier method) of patients with breast cancer indicates a strong positive prognostic value of COP1 expression (P = .006, log-rank test). (B) Multivariate survival analysis (proportional hazard method) shows a positive, independent prognostic importance of COP1 expression (P < .05, likelihood ratio test), in addition to the independent prognostic impact of tumor diameter and lymph node status. HR, hazard ratio.
Discussion
High c-Jun abundance is detected in both invasive breast cancer cell lines and specimens of aggressive breast tumors [8,11]. However, how c-Jun abundance is regulated in breast cancer cells is not well understood. In this study, we showed that c-Jun protein is much more stable in invasive breast cancer cells than less invasive ones (Figure 1), indicating that prolonged protein stability is a major cause of high c-Jun abundance in invasive breast cancer cells or aggressive breast tumor specimens [8,11]. The stability of c-Jun protein in breast cancer cells appeared to depend on whether c-Jun was efficiently poly-ubiquitinated rather than the nuclear proteasome activity or the ability of c-Jun to localize in the proteasomes (Figure 2). These findings led us to hypothesize that there is lack of c-Jun-targeting E3 ubiquitin ligase in invasive breast cancer cells. With the aid of target-specific siRNAs, we identified COP1 as an E3 ligase responsible for c-Jun turnover in breast cancer cells as COP1 depletion enhanced c-Jun protein stability and diminished c-Jun poly-ubiquitination in less invasive breast cancer cells (Figure 3). In human primary T lymphocytes, proto-oncogene c-Abl phosphorylates c-Jun, which in turn prevents c-Jun and ITCH interaction and thus stabilizes c-Jun [30]. Receptor for activated C-kinase 1 stabilizes c-Jun by releasing it from FBW7 complex [31]. In this study, we showed that the levels of c-Jun and COP1 are inversely correlated in breast cancer cells (Figure 3), implicating the low abundance of COP1 as a principal cause of prolonged c-Jun protein stability in invasive breast cancer cells.
Phosphorylation at a specific site is the prerequisite of ubiquitination for many E3 ubiquitin ligases. For example, Skp2 ubiquitinates and promotes the degradation of forkhead box protein O1 (FOXO1). However, this Skp2-mediated event requires Akt-specific phosphorylation of FOXO1 at Ser-256 [32]. c-Jun is ubiquitinated by FBW7 in various cell types, and GSK3β-mediated phosphorylation at Thr-239 is necessary for c-Jun ubiquitination to take place [22]. In this study, we showed that blocking GSK3 activity increased c-Jun abundance in less invasive breast cancer cells (Figure 4). Specifically, mutating Thr-239 to Ala stabilized c-Jun protein and prevented c-Jun polyubiquitination (Figure 4). Our data thus suggest that GSK3β-mediated phosphorylation at Thr-239 is an event required for both COP1- and FBW7-mediated c-Jun protein degradation. This possibility is further supported by the observation that simultaneously overexpressing COP1 and elevating GSK3β activity can effectively diminish c-Jun abundance in invasive breast cancer cells (Figure 4).
In vitro and in vivo studies have demonstrated a convincing role of c-Jun in breast tumorigenesis. For example, enforced expression of c-Jun confers noninvasive and hormone-dependent MCF7 cells with an invasive and hormone-resistant phenotype [3]. In MMTV-ErbB2 transgenic breast cancer model, c-Jun deficiency led to reduced cell migration, invasion, and mammosphere formation of erbB2-induced mammary tumors [5]. In this study, we showed that simultaneously raising COP1 level and elevating GSK3β activity suppressed in vitro cell growth/migration and in vivo metastasis of invasive breast cancer cells (Figure 5). Taken together, our study presents strong evidence that both high COP1 expression and GSK3β activity negatively impact breast tumor progression and metastasis.
There are evidences for both up-regulation and loss of COP1 in cancers [14,33,34]. It remains to be determined if overexpression of COP1 promotes tumorigenesis or is the consequence of tumor formation; however, mice with partial loss of COP1 function or tissue-specific COP1 ablation strongly suggest that COP1 is a tumor suppressor [14,35]. Using publicly available breast tumor gene expression microarray database, we found that reduced COP1 expression correlated with poorer disease-free survival and COP1 expression is an independent positive prognostic factor for recurrence-free survival (Figure 6). Our study adds strong evidences that COP1 is a suppressor of breast tumor progression.
Although our mechanistic study was performed with the established breast cancer cell lines, our observation that high COP1 expression correlated with better disease-free survival of patients with breast cancer supports the results generated from our experimental studies. To conclude, our study suggests that COP1 and GSK3β cooperate to promote c-Jun degradation and, hence, subsequent inhibition of breast tumor progression and metastasis.
Supplemental Information
Antibody Information
c-Jun rabbit mAb: CS-9165, Cell Signaling Technology (Danvers, MA); titer 1:1000 (immunoblot analysis)
COP1 rabbit polyclonal antibody: A300-894A, Bethyl Laboratories (Montgomery, TX); titer 1:1000 (immunoblot analysis)
GSK3 rabbit mAb: SC-5676, Cell Signaling Technology; titer 1:1000 (immunoblot analysis)
HA rabbit mAb: CS-5017, Cell Signaling Technology; titer 1:1000 (immunoblot analysis)
β-Actin mouse mAb: SC-47778, Santa Cruz Biotechnology (Santa Cruz, CA); titer 1:500 (immunoblot analysis)
Flag mouse mAb: F 3165, Sigma; titer 1:1000 (immunoblot analysis)
c-Jun mouse mAb: BD-610327, BD Biosciences (San Jose, CA); titer 1:100 (immunofluorescence staining)
PSMA5 (proteasome S5A subunit) rabbit polyclonal antibody: CS-2457, Cell Signaling Technology; titer 1:50 (immunofluorescence staining)
Information for Primers Used in Quantitative Reverse Transcription-Polymerase Chain Reaction
| COP1 F | CCACAGGTGGCAGATATTTCAA |
| COP1 R | CATTGGCCAAATCAAGGTTATCTT |
| FBW7 F | AATGGCCAAGGGCAACAA |
| FBW7 R | CATTCCTGGAGGCCTGTAGGT |
| ITCH F | GATGGCTCCAGATCCAAGGA |
| ITCH R | TGCATCTTCAGGGTCATCTGAT |
| CUL4A F | GAATCAGAGTGACTCAGGCCCTATA |
| CUL4A R | GGCCAGTAGCCCATTGTGA |
| SAG F | AACAATCGCTGCCCTCTCT |
| SAG R | CTCATTTGCCGATTCTTTGGA |
| MEKK1 F | CAAACCGCCGTGTTAACAAA |
| MEKK1 R | GGCCCTATCTGCTGCAGTAAGT |
| β-Actin F | CCAGCTCACCATGGATGATG |
| β-Actin R | ATGCCGGAGCCGTTGTC |
Sequences of COP1 siRNAs
siCOP1 #1, AAAAGAAAUGACCUGCAAUUCGATTGGATCCAATCGAATTGCAGGTCATTTC
siCOP1 #2, AAAAAAACAGCCUUGGUAUAAUATTGGATCCAATATTATACCAAGGCTGTTT
Footnotes
This work was supported by funding from National Institutes of Health HL083335, Shanghai Eastern Scholar Fund, and E-Institutes of Shanghai Municipal Education Commission Project E03008.
This article refers to supplementary materials, which are designated by Figures W1 to W5 and Table W1 and are available online at www.neoplasia.com.
References
- 1.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 2.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 3.Smith LM, Wise SC, Hendricks DT, Sabichi AL, Bos T, Reddy P, Brown PH, Birrer MJ. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene. 1999;18:6063–6070. doi: 10.1038/sj.onc.1202989. [DOI] [PubMed] [Google Scholar]
- 4.Vleugel MM, Greijer AE, Bos R, van der Wall E, van Diest PJ. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum Pathol. 2006;37:668–674. doi: 10.1016/j.humpath.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Jiao X, Katiyar S, Willmarth NE, Liu M, Ma X, Flomenberg N, Lisanti MP, Pestell RG. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. J Biol Chem. 2010;285:8218–8226. doi: 10.1074/jbc.M110.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Pu X, Shi M, Chen L, Song Y, Qian L, Yuan G, Zhang H, Yu M, Hu M, et al. Critical role of c-Jun overexpression in liver metastasis of human breast cancer xenograft model. BMC Cancer. 2007;7:145. doi: 10.1186/1471-2407-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen Q, Uray IP, Li Y, Krisko TI, Strecker TE, Kim HT, Brown PH. The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene. 2008;27:366–377. doi: 10.1038/sj.onc.1210643. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Zhu G, Li Y, Padia RN, Dong Z, Pan ZK, Liu K, Huang S. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009;69:9228–9235. doi: 10.1158/0008-5472.CAN-09-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okutomi Y, Shino Y, Komoda F, Hirano T, Ishihara T, Yamaguchi T, Saisho H, Shirasawa H. Survival regulation in pancreatic cancer cells by c-Jun. Int J Oncol. 2003;23:1127–1134. [PubMed] [Google Scholar]
- 10.Neyns B, Katesuwanasing, Vermeij J, Bourgain C, Vandamme B, Amfo K, Lissens W, DeSutter P, Hooghe-Peters E, DeGrève J. Expression of the jun family of genes in human ovarian cancer and normal ovarian surface epithelium. Oncogene. 1996;12:1247–1257. [PubMed] [Google Scholar]
- 11.Tiniakos DG, Scott LE, Corbett IP, Piggott NH, Horne CH. Studies of c-jun oncogene expression in human breast using a new monoclonal antibody, NCL-DK4. J Pathol. 1994;172:19–26. doi: 10.1002/path.1711720106. [DOI] [PubMed] [Google Scholar]
- 12.Jariel-Encontre I, Salvat C, Steff AM, Pariat M, Acquaviva C, Furstoss O, Piechaczyk M. Complex mechanisms for c-fos and c-jun degradation. Mol Biol Rep. 1997;24:51–56. doi: 10.1023/a:1006804723722. [DOI] [PubMed] [Google Scholar]
- 13.Westermarck J. Regulation of transcription factor function by targeted protein degradation: an overview focusing on p53, c-Myc, and c-Jun. Methods Mol Biol. 2010;647:31–36. doi: 10.1007/978-1-60761-738-9_2. [DOI] [PubMed] [Google Scholar]
- 14.Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, Zwolinska A, Depaepe V, Hochepied T, Skarnes WC, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest. 2011;121:1329–1343. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 16.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 17.Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 18.Xia Y, Wang J, Xu S, Johnson GL, Hunter T, Lu Z. MEKK1 mediates the ubiquitination and degradation of c-Jun in response to osmotic stress. Mol Cell Biol. 2007;27:510–517. doi: 10.1128/MCB.01355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Q, Bowden GT, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage-dependent targeting of c-Jun/AP1 and IκB-α/NF-κB. J Cell Biol. 2007;178:1009–1023. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A, Frost J, Deng T, Smeal T, al-Alawi N, Kikkawa U, Hunter T, Brenner D, Karin M. Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell. 1992;70:777–789. doi: 10.1016/0092-8674(92)90311-y. [DOI] [PubMed] [Google Scholar]
- 21.Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 22.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, Noh H, Chen H, Padia R, Pan ZK, Su SB, Jing Q, Ding HF, Huang S. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary microRNAs. Sci Signal. 2013;6:ra16. doi: 10.1126/scisignal.2003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouhi P, Jensen LD, Cao Z, Hosaka K, Lanne T, Wahlberg E, Steffensen JF, Cao Y. Hypoxia-induced metastasis model in embryonic zebrafish. Nat Protoc. 2010;5:1911–1918. doi: 10.1038/nprot.2010.150. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Bergami P, Huang C, Goydos JS, Yip D, Bar-Eli M, Herlyn M, Smalley KS, Mahale A, Eroshkin A, Aaronson S, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He H, Gu Q, Zheng M, Normolle D, Sun Y. SAG/ROC2/RBX2 E3 ligase promotes UVB-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/AP-1 and p27. Carcinogenesis. 2008;29:858–865. doi: 10.1093/carcin/bgn021. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol. 2012;14:717–726. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 29.Thomas C, Rajapaksa G, Nikolos F, Hao R, Katchy A, McCollum CW, Bondesson M, Quinlan P, Thompson A, Krishnamurthy S, et al. ERβ1 represses basal-like breast cancer epithelial to mesenchymal transition by destabilizing EGFR. Breast Cancer Res. 2012;14:R148. doi: 10.1186/bcr3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao B, Lee SM, Fang D. The tyrosine kinase c-Abl protects c-Jun from ubiquitination-mediated degradation in T cells. J Biol Chem. 2006;281:29711–29718. doi: 10.1074/jbc.M604596200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Zhu F, Li X, Dong Z, Xu Y, Peng C, Li S, Cho YY, Yao K, Zykova TA, et al. Rack1 protects N-terminal phosphorylated c-Jun from Fbw7-mediated degradation. Oncogene. 2012;31:1835–1844. doi: 10.1038/onc.2011.369. [DOI] [PubMed] [Google Scholar]
- 32.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci USA. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dornan D, Bheddah S, Newton K, Ince W, Frantz GD, Dowd P, Koeppen H, Dixit VM, French DM. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64:7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 34.Lee YH, Andersen JB, Song HT, Judge AD, Seo D, Ishikawa T, Marquardt JU, Kitade M, Durkin ME, Raggi C, et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res. 2010;70:8264–8269. doi: 10.1158/0008-5472.CAN-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitari AC, Leong KG, Newton K, Yee C, O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.