Abstract
The persistent expression of lactase into adulthood in humans is a recent genetic adaptation that allows the consumption of milk from other mammals after weaning. In Europe, a single allele (−13910∗T, rs4988235) in an upstream region that acts as an enhancer to the expression of the lactase gene LCT is responsible for lactase persistence and appears to have been under strong directional selection in the last 5,000 years, evidenced by the widespread occurrence of this allele on an extended haplotype. In Africa and the Middle East, the situation is more complicated and at least three other alleles (−13907∗G, rs41525747; −13915∗G, rs41380347; −14010∗C, rs145946881) in the same LCT enhancer region can cause continued lactase expression. Here we examine the LCT enhancer sequence in a large lactose-tolerance-tested Ethiopian cohort of more than 350 individuals. We show that a further SNP, −14009T>G (ss 820486563), is significantly associated with lactose-digester status, and in vitro functional tests confirm that the −14009∗G allele also increases expression of an LCT promoter construct. The derived alleles in the LCT enhancer region are spread through several ethnic groups, and we report a greater genetic diversity in lactose digesters than in nondigesters. By examining flanking markers to control for the effects of mutation and demography, we further describe, from empirical evidence, the signature of a soft selective sweep.
Main Text
Lactase, the enzyme that digests the milk sugar lactose, persists into adult life in approximately 35% of the world’s population.1 This genetic trait of adult lactase persistence (LP [MIM 223100]) is a recent human adaptation permitting those who carry it to use animal milk more readily as a source of nutrition. This LP phenotype is in contrast to the ancestral mammalian phenotype shared by most of the human population where lactase is downregulated before adulthood.
LP is attributable to nucleotide changes in a regulatory region that acts as an enhancer of the expression of the gene encoding lactase, LCT (MIM 603202). This LCT enhancer is located in intron 13 of the neighboring gene, MCM6 (MIM 601806), immediately upstream of LCT (for review see Ingram et al.1). LP in Europe is generally attributable to a single allele (−13910∗T, rs4988235) that seems to have been under strong directional selection in the last 5,000–10,000 years.2 The evidence for selection comes from the fact that the allele lies on an extended haplotype3,4 with low microsatellite diversity and is at a significantly higher frequency than expected for the age of the allele.1,4–6 Tests of haplotype homozygosity and population differentiation in genome-wide studies that focus on European samples show that the region of chromosome 2 containing LCT has one of the highest “signatures” of selection.7 In Africa and the Middle East, the situation is more complicated and three additional alleles (−13907∗G, rs41525747; −13915∗G, rs41380347; −14010∗C, rs145946881) in the same LCT enhancer region have been reported to cause lactase persistence.8–16 In Tanzania and Kenya, one particular allele, −14010∗C, is at high frequency and gives a significant signal of positive selection in tests of haplotype homozygosity, with expansion of the −14010∗C allele dated to approximately 3,000–7,000 years ago.16 In some cases, however, including the Jaali from Sudan and the Somali camel herders from Ethiopia, several different LP alleles are associated with lactase persistence in the same ethnic group.15 Indeed, we observed that in these groups, the sequence of the enhancer region was much more diverse in lactose digesters than in nondigesters. Analysis of mitochondrial DNA, Y chromosome, and autosomal microsatellite variations demonstrated that this difference in diversity was not attributable to hidden population stratification.15 Although it is possible that not all of the described enhancer alleles are functional, we speculated that this difference in diversity had been influenced by what has been described as a soft selective sweep—the phenomenon by which several different variants of similar function rise in frequency simultaneously.17–19 Such soft selective sweeps might not be detectable by published methods of genome-wide detection of selection.20,21
Here, we examine genetic diversity of the LCT enhancer in a larger lactose-tolerance-tested cohort (>350 individuals), which consists of volunteers from several ethnic groups in Ethiopia. We aim to determine whether any further genetic variants are likely to be functional, whether the different Ethiopian groups in our cohort carry private mutations, and whether the pattern of increased diversity of the enhancer region in lactose digesters that we detected previously is widespread in Ethiopia. We examine this diversity in relation to that of flanking markers to control for the effects of mutation and demography to describe this signal of selection.
The Ethiopian participants were university students who undertook lactose-tolerance testing by a breath hydrogen method. Ethical approval was obtained from the University of Addis Ababa and UCLH (ref 01/0236). Each volunteer provided fully informed consent for the project and completed a questionnaire relating to their self-declared ethnicity, ancestry, language (also language and ethnicity of parents and grandparents), and questions about health and milk-drinking habits. Buccal samples were collected from all participants, followed by DNA extraction22 and sequencing. Data were obtained from 370 individuals. Volunteers were classified as lactose digesters, nondigesters, intermediate/indeterminate digesters, or hydrogen nonproducers via a breath-hydrogen lactose-tolerance test (LTT) as described by Ingram and colleagues.14,15 Data from 14 individuals were excluded from the association study because of recent antibiotic use (which could potentially affect lactose-tolerance test results) or known family relationship between participants, resulting in a data set from 356 individuals.
A total of 139 (39%) of the volunteers from this Ethiopian cohort were classified as lactose digesters and 188 (53%) as nondigesters. A total of 22 (6%) gave intermediate measurements, and 7 (2%) produced no breath hydrogen throughout the test.
The volunteers were classified according to self-declared cultural identity (ethnicity). Volunteers with both parents and known grandparents from the same ethnic group were classified as Amhara, Oromo, Tigre, or Wolayita, and those with other or mixed ancestry were classified as “other” but clustered as far as possible by language family.
We sequenced a 500 bp DNA fragment across the LCT enhancer (for primers, cycling, and sequencing conditions, see Table S1 available online) and recorded a total of 13 variable sites. None of the loci showed significant deviation from Hardy-Weinberg equilibrium (data not shown). None of the frequent derived alleles were private to any particular ethnic group (see Table S2).
Counts of the derived enhancer alleles for all lactose digesters are shown in Table 1. The strong association of −13907∗G and −13915∗G with digester status is clearly confirmed in this cohort. Of the other previously characterized variants, −14010∗C was present in digesters only but at too low a frequency to give statistical significance in this cohort and −13010∗T was present in one digester who also carried a second enhancer allele. In this study, we show a statistically significant association of the −14009∗G variant with lactose-digester status (p = 0.0024, with a Bonferroni threshold of p < 0.004 for 12 tests). Alleles −13730∗G, −13806∗G, and −13913∗G were more frequent in nondigesters than in digesters, which indicates that these alleles do not cause LP. In this context, −13913∗G is of particular interest because it is located very close to other known functional variants. All other alleles were too rare to assess.
Table 1.
Allele Counts of Variants in Intron 13 of MCM6 and Association with Lactose-Digester Status
| Digester Status | n (2 Allele Carriers Excluded) | −14011C>T | −14010G>C | −14009T>G | −13957A>G | −13938C>T | −13915T>G | −13913T>G | −13910C>T | −13907C>G | −13806A>G | −13800G>T | −13730T>G | −13603C>T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs/ss Number | rs4988233 | rs145946881 | ss820486563 | ss820496514 | ss831883152 | rs41380347 | rs41456145 | rs4988235 | rs41525747 | ss820496565 | rs144412793 | rs4954492 | rs56348046 | |
| H2NP | 14 | 0 | 0 | 2 | 1 | 0 | 3 | 0 | 0 | 4 | 0 | 0 | 1 | 0 |
| Intermediate | 44 | 0 | 0 | 3 | 0 | 1 | 5 | 2 | 0 | 2 | 2 | 0 | 2 | 0 |
| Digester | 278 (196) | 1 (0) | 3 (2) | 22 (13) | 1 | 0 | 74 (34) | 2 (1) | 1 (0) | 44 (30) | 6 (1) | 0 | 8 (2) | 2 (0) |
| Nondigester | 376 (374) | 1 (0) | 0 | 6 | 1 | 0 | 6 | 7 | 0 | 2 (1) | 10 | 1 | 17 | 2 |
| p value | 1 | 0.076 | 0.00008∗ | 1 | NA | 9.63 × 10−24∗ | 0.313 | 0.43 | 1.91 × 10−16∗ | 0.801 | NA | 0.310 | 1 | |
| p value (2 allele carriers excluded) | NA | (0. 117) | 0.00242∗ | NA | NA | 9.50 × 10−12∗ | 0.274 | NA | 5.06 × 10−14∗ | 0.108 | NA | 0.0264 | NA |
The results reported here are from a total of 356 volunteers (712 chromosomes). To test association of the alleles independently, counts were also made after exclusion of all individuals who carry two different derived alleles (numbers in parentheses). Fisher’s exact test two-sided p values are shown for the comparison of digesters and nondigesters. Asterisks (∗) indicate statistically significant association after Bonferroni correction for 12 tests (threshold p = 0.004). Note that −14011∗T is present in one digester, but the second carrier, despite being a nondigester, also carried −13907∗G, so was presumably suffering from secondary loss of lactase. The location of −14011∗T immediately next to −14010G>C suggests the possibility of function.
To determine whether −14009∗G has a functional effect on the LCT enhancer activity, the −14009∗G mutation was introduced by site-directed mutagenesis into the 450 bp LCT enhancer region (−14,133 bp to −13,684 bp) and inserted into the pGL3hLPH1085 promoter construct with luciferase as a reporter, as previously described.8,11 Transfections were conducted with Caco-2 cells and the results are shown in Figure 1. A significant increase in luciferase activity was observed for the −14009∗G construct in comparison with the ancestral sequence. This increase is comparable with the increase observed for the −14010∗C variant (used here as a positive control), strongly supporting function, and our studies in progress confirm alteration of transcription factor binding (A.L., J.T.T., D.M.S., and colleagues, unpublished data). The fact that as many as six individuals who carry the allele were diagnosed as nondigesters does hint, however, that this allele may behave less efficiently than the others in vivo.
Figure 1.
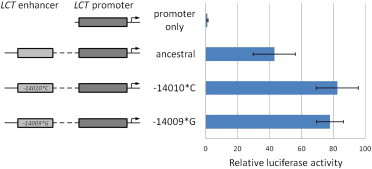
Transfection Experiment Showing the Effect of the −14009∗G Variant on the Enhancer Activity in Caco-2 Cells
Promoter/enhancer constructs containing either the ancestral sequence, the −14009∗G variant, or the −14010∗C variant LCT enhancer sequence were transfected into Caco-2 cells together with a CMV-driven β-galactosidase expression plasmid. The cells were harvested and analyzed for luciferase and β-galactosidase activities 9 days after transfection. The luciferase expressions were corrected for transfection efficiency via the β-galactosidase activities. The −14010∗C construct was used as a positive control. The bar chart shows luciferase expression relative to the “promoter only” construct (lacking the enhancer) and the error bars represent SDs (n = 8). Both the −14009∗G and −14010∗C enhancer sequences lead to an increased luciferase expression, which is significantly different from the ancestral sequence (−14009∗C/−14010∗G; p = 0.000032 and 0.000038, respectively).
Several individuals carried one of the known LP variants and yet were classified as lactose nondigesters, whereas other individuals (n = 17) were classified as digesters, yet had no variant in the LCT enhancer. At least part of this disparity is probably attributable to one of the known limitations of this phenotypic test—the lack of control for loss of enzymatic function in the gut resulting from secondary damage.23 However, the lactose digesters with no enhancer variation and confirmed hydrogen production are more difficult to explain and might be real, suggesting that a different causal mechanism could be at work. A mutation in a different cis-acting region could be responsible for LP in these individuals, or perhaps even a trans-acting factor (although trans-acting variants are less likely to be causal because such mutations are more likely than cis-acting variants to have a pleiotropic effect24). Epigenetic influences could also play a role in the persistence of lactase in some of these individuals.
In an initial analysis of the diversity of the LCT enhancer region, we found greater sequence diversity in the digesters than in the nondigesters (Nei’s H for digesters 0.73 versus 0.023 for nondigesters; π 0.002 for digesters versus 0.0005 for nondigesters), which is consistent with our previous findings in Ethiopian Somali camel herders.15 To further explore the causes of this greater diversity in digesters, we sought to control for the possible effects of demography and mutation rate. To do this, we sequenced two control regions either side of the LCT enhancer that were unlikely in most cases to have been separated from the enhancer by recombination since the onset of the spread of these alleles (for primers, cycling, and sequencing conditions see Table S1).
We selected a 500 bp fragment 16 kb upstream of the LCT enhancer in intron 4 of MCM6 and a 361 bp fragment 13 kb downstream of the enhancer, approximately 1 kb upstream of LCT exon 1. Assuming a recombination rate of 0.5 cM/Mb observed in families for that region (UCSC Genome Browser), 5% or fewer of the chromosomes are likely to have recombined during the last 300 generations (5,000 years). Intron 4 of MCM6 is likely to have a similar chromatin state to that of intron 13 and, therefore, a similar vulnerability to germline mutation, thus doubling as a control region with similar mutation rate.
The subsequent analyses were conducted on samples with complete sequence data for all three regions, irrespective of phenotype classification. From the previously collected cohort of Ethiopian Somali camel herders,15 81 individuals were included for comparison. The frequencies of sequence variants in the two control regions for digesters and nondigesters are shown in Table S3.
Haplotypes were inferred with the computer program PHASE,25 and the results were then checked by visual inspection of the data. The haplotypes that were present in this cohort, excluding those haplotypes with fewer than three occurrences, are shown in Figure 2 in relation to our previous designations.27 Figure S1 details the full set of haplotypes and indicates nonrecombinant and recombinant chromosomes. Only 7% of chromosomes show evidence of historic recombination between the flanking regions and the frequency of detectable recombinants is lower for the chromosomes carrying the derived enhancer alleles (3.9%). As previously reported, −13915∗G is found on a C, or a closely related haplotype (of which 85 out of 88 are identical across all three regions), and −13907∗G is on an extended A, or closely related haplotype, in 62 out of 63 cases (Figures 2 and S1). The −14009∗G allele occurred in 34 out of 36 cases on a haplotype that has a deletion at −942, which, according to our previous haplotype designations, could be an O, S, T, U, or X type haplotype. Typing position 5579 of the LCT cDNA sequence (rs2278544) of a number of the chromosomes with the −14009∗G showed that they carry a C nucleotide, suggesting that this is most probably an X, rather than the more common U, haplotype.27
Figure 2.
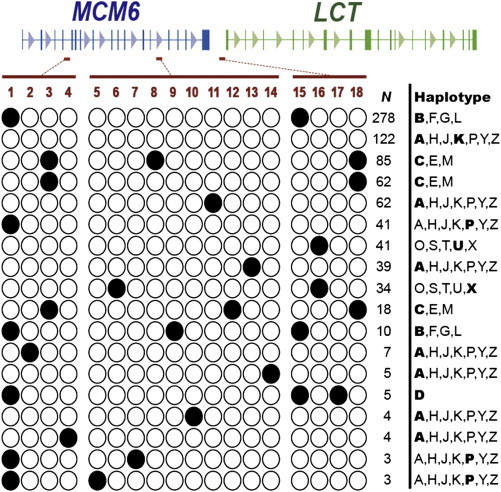
Phased Haplotypes for the Lactase Enhancer and Two Flanking Regions, in Intron 4 of MCM6 and 1 kb Upstream of LCT in a Cohort of 422 Ethiopian Individuals with No Known Shared Ancestry to the Grandparental Level, for whom Full Sequencing Data Were Available
Haplotypes that occurred <3 times (n = 21) are omitted. The variable sites are numbered and refer to the following chromosome positions relative LCT; 1, −30210∗C; 2, −30203ins; 3, −30182∗G; 4, −29949∗C; 5, −14010∗C; 6, −14009∗G; 7, −13957∗G; 8, −13915∗G; 9, −13913∗C; 10, −13910∗T; 11, −13907∗G; 12, −13806∗G; 13, −13730∗G; 14, −13603∗T; 15, −958∗T; 16, −942del; 17, −875∗A; 18, −678∗G. n indicates the number of chromosomes for which that haplotype was inferred. The lettered haplotypes that are shown refer to the haplotypes previously reported by Hollox et al.27 Those that are shown in bold are the most likely haplotype according to previously reported distributions and examination of additional alleles (unpublished data).
As an additional check that the enhancer region in intron 13 is not intrinsically more mutable than the other regions that we have examined, we compared the divergence of the three regions from primate sequences by using the sequences available from the UCSC Human Genome Browser. We observed that the divergence of the intron 13 region is in fact less than that of the other two regions. For example, in comparison with the rhesus macaque, the values obtained were 5.9%, 5.2%, and 6.3% for control region 1, the enhancer sequence, and control region 2, respectively.
We then compared haplotype diversity (H) and nucleotide diversity (π) of all three sequence regions (Table 2) in the digesters and nondigesters from the Amhara, Tigre, Oromo, and Wolayita and the previously collected Somali. Individuals of mixed ancestry or those belonging to groups with fewer than 20 individuals in our data set, for the purposes of this analysis were grouped together, irrespective of language family, as “all other Ethiopians.”
Table 2.
Comparative Diversity Data for the Three Regions Sequenced
|
Region |
Control Region 1-Intron 4 MCM6 |
Enhancer - Intron 13 MCM6 |
Control Region 2-Upstream of LCT |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnic Group | Digester Status | n | Seg Sites | N Haplo | H | π | Seg Sites | N Haplo | H | π | Seg Sites | N Haplo | H | π |
| Amhara | nondigester | 62 | 3 | 4 | 0.672 | 0.0019 | 4 | 5 | 0.185 | 0.00038 | 3 | 5 | 0.694 | 0.0026 |
| digester | 32 | 2 | 3 | 0.669 | 0.0017 | 5 | 6 | 0.595 | 0.00141 | 3 | 4 | 0.736 | 0.0028 | |
| Oromo | nondigester | 78 | 3 | 4 | 0.556 | 0.0013 | 5 | 6 | 0.261 | 0.00055 | 4 | 6 | 0.671 | 0.0027 |
| digester | 54 | 3 | 5 | 0.706 | 0.0021 | 7 | 8 | 0.705 | 0.00185 | 4 | 6 | 0.716 | 0.0029 | |
| Tigre | nondigester | 30 | 3 | 4 | 0.522 | 0.0014 | 1 | 2 | 0.067 | 0.00013 | 3 | 4 | 0.646 | 0.0023 |
| digester | 48 | 2 | 3 | 0.635 | 0.0015 | 4 | 5 | 0.704 | 0.00182 | 3 | 4 | 0.668 | 0.0023 | |
| Wolayita | nondigester | 24 | 2 | 3 | 0.565 | 0.0013 | 1 | 2 | 0.083 | 0.00017 | 3 | 4 | 0.663 | 0.0022 |
| digester | 16 | 2 | 3 | 0.667 | 0.0016 | 4 | 5 | 0.717 | 0.00180 | 3 | 4 | 0.742 | 0.0027 | |
| Somali | nondigester | 98 | 3 | 4 | 0.559 | 0.0013 | 3 | 4 | 0.223 | 0.00046 | 3 | 4 | 0.588 | 0.0021 |
| digester | 32 | 3 | 4 | 0.688 | 0.0017 | 5 | 6 | 0.714 | 0.00182 | 3 | 4 | 0.718 | 0.0026 | |
| Other | nondigester | 162 | 4 | 5 | 0.621 | 0.0015 | 9 | 10 | 0.312 | 0.00067 | 4 | 5 | 0.686 | 0.0025 |
| digester | 132 | 2 | 4 | 0.652 | 0.0016 | 10 | 11 | 0.748 | 0.00203 | 4 | 5 | 0.732 | 0.0029 | |
Using the fully phased data set, measures of genetic diversity were calculated with DnaSP software, using those samples with a clear diagnosis of either lactose-digester or -nondigester status. Note that for the enhancer region, the numbers of segregating sites, numbers of haplotypes, and calculated values for H (haplotype heterozygosity) and Π (nucleotide diversity) are in each case larger in the digesters than nondigesters. These analyses were conducted on samples for which uniform ancestry was recorded and a single additional group was composed of all others. Two simple indel polymorphisms (one in intron 4 and one in the upstream LCT region) are considered as SNPs in the calculations. N = number of chromosomes. Detailed statistics are shown in Table S4.
Figure 3 shows the distribution of nucleotide diversity values for each region. For the enhancer region, in all cases, π was greater in the digesters than the nondigesters. We assessed the significance of the difference in π for each ethnic group by permutation analysis in which the haplotypes were drawn and assigned to digesters and nondigesters in 50,000 random permutations. A conservative two-tailed p value was computed as the proportion of permutations with an absolute π difference equal to or larger than the observed π difference between digesters and nondigesters. For the enhancer, there was a significant difference in π between digesters and nondigesters for all ethnic groups (p < 0.005) (Table S4). Similar comparison of the control sequences showed just one ethnic group with a significant π difference (the Oromo for the control region in intron 4, p = 0.0002).
Figure 3.
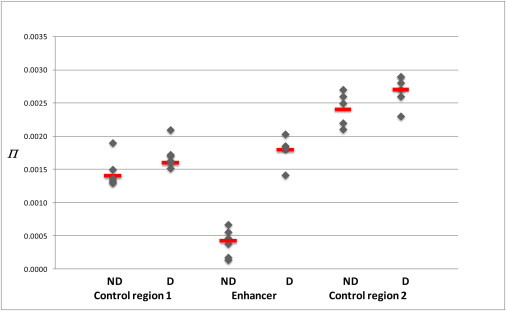
Comparison, in Nondigesters and Digesters, of Nucleotide Diversity π Measured across the Three Sequence Regions
Data points show the five ethnic groups tested: Amhara, Tigre, Oromo, Wolayita, Somali, and “other Ethiopians” as a single group. Red horizontal bars show median values. See Table 2 for n values. Abbreviations are as follows: D, digester; ND, nondigester.
Haplotype diversity of the enhancer region was also significantly greater in digesters than nondigesters for all ethnic groups (as determined by the test Hdiff;26 Table S4). This distribution is depicted in Figure S2. As for the π analysis, the only significant difference in the control sequences was the intron 4 region in Oromo (p = 0.0028). However, even if not statistically significant, it is noteworthy that in most cases the diversity of both the flanking regions is slightly lower in the nondigesters than digesters of the same group. This effect, which is particularly prominent in the Somali group, is probably a consequence of the very high prevalence throughout Ethiopia of a long haplotype that does not carry any derived LP-associated alleles (the first on Figure 1, and see the blue haplotype in Figure S3).
Thus we confirm our previous observation on the diversity of the LCT enhancer sequence region and show with a cohort of independent samples that there is significantly greater nucleotide and haplotype diversity in the enhancer region in digesters than in nondigesters.
By sequencing two flanking regions, which have followed the same demographic history as the enhancer over the last 300 generations, both control sequences have rather high diversity in nondigesters as well as in digesters. Indeed, it is the lack of diversity in the enhancer sequence of nondigesters that is noteworthy and this seems to reflect the general conservation of this sequence across primates. Between positions −14028 and −13800, the sequences are approximately 93.5% identical across humans, chimpanzee, gorilla, orangutan, gibbon, baboon, and rhesus macaque, and percentage identity declines on either side of this (see Figure S4). Thus, this sequence region may have been under evolutionary constraint because of its function as a regulatory element for lactase expression in infant mammals. This constraint appears to have been overcome in some human populations, presumably because of the benefit of allowing adult lactase expression.
This study brings the total of confirmed LP causal variants to five (−14010∗C, −14009∗G, −13915∗G, −13910∗T, and −13907∗G). Their close location, all within the most conserved part of the 450 bp segment known to have enhancer function, suggests that the functional region is smaller than the sequence that we have tested and this will be explored further in future studies.
The coexistence of all five alleles in Ethiopia goes some way to explaining the greater nucleotide diversity within the LCT enhancer in intron 13 of MCM6 in digesters. Ethiopia has been a crossroads of human migrations in the last 5,000 years since the LP alleles are likely to have come under selection, and studies on other African and Middle eastern populations (B.L.J., D.M.S., and colleagues, unpublished data) show quite different geographic distributions, with overlap in Ethiopia, suggesting that their origins are all different, but determining where these were and how they spread is likely to be difficult. The combination of mutation, large effective population size, migration, and selection has been shown to be important in generating this kind of pattern of diversity, namely parallel selection of multiple alleles of similar function, a so-called soft selective sweep.17 Here we confirm this unusual pattern of diversity in the LCT enhancer region in Ethiopia, and by testing flanking sequences we control, at a chromosomal level, for possible differences in migrational history, effective population size, and mutation rate between the digesters and nondigesters. Because increased genetic diversity in digesters is localized to the LCT enhancer, we can infer that recent selection is acting on this small relatively conserved functional sequence region. Selection has the effect of increasing the frequency of the background haplotypes on which the derived alleles occur, though the impact on diversity of the flanking sequences is not large. This pattern contrasts with that observed from the hard selective sweep seen for the “European” LCT enhancer allele, where a single extended haplotype is present in at least one copy in digesters, whereas haplotype heterozygosity is significantly higher for the nondigesters3,4,28 (D.M.S. and colleagues, unpublished data). This study, therefore, provides important pointers toward describing the sequence features of such motifs in other areas of the genome.
Acknowledgments
This work was funded by the MRC UK (MRC DTA studentship for B.L.J.), the European Union (Marie Curie ITN FP7 Framework Programme grant, LeCHE, Grant ref 215362-2 to A.L.), the Annals of Human Genetics (B.L.J. and A.L.), and Melford Charitable Trust (studentship for T.O.R.). N.B. is the settlor and senior trustee of Melford Charitable Trust. Neither N.B. nor the charitable trust has any intellectual property or other rights with respect to the results of the study. We thank Mari Wyn Burley and Ranji Araseretnam for technical help, Mirna Kovacevic and Ripudaman Bains for their help with data handling, and Ed Hollox for helpful discussion.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
DnaSP, http://www.ub.edu/dnasp/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
References
- 1.Ingram C.J., Mulcare C.A., Itan Y., Thomas M.G., Swallow D.M. Lactose digestion and the evolutionary genetics of lactase persistence. Hum. Genet. 2009;124:579–591. doi: 10.1007/s00439-008-0593-6. [DOI] [PubMed] [Google Scholar]
- 2.Itan Y., Powell A., Beaumont M.A., Burger J., Thomas M.G. The origins of lactase persistence in Europe. PLoS Comput. Biol. 2009;5:e1000491. doi: 10.1371/journal.pcbi.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulter M., Hollox E., Harvey C.B., Mulcare C., Peuhkuri K., Kajander K., Sarner M., Korpela R., Swallow D.M. The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann. Hum. Genet. 2003;67:298–311. doi: 10.1046/j.1469-1809.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 4.Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., Hirschhorn J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anagnostou P., Battaggia C., Coia V., Capelli C., Fabbri C., Pettener D., Destro-Bisol G., Luiselli D. Tracing the distribution and evolution of lactase persistence in Southern Europe through the study of the T(-13910) variant. Am. J. Hum. Biol. 2009;21:217–219. doi: 10.1002/ajhb.20851. [DOI] [PubMed] [Google Scholar]
- 6.Coelho M., Luiselli D., Bertorelle G., Lopes A.I., Seixas S., Destro-Bisol G., Rocha J. Microsatellite variation and evolution of human lactase persistence. Hum. Genet. 2005;117:329–339. doi: 10.1007/s00439-005-1322-z. [DOI] [PubMed] [Google Scholar]
- 7.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 8.Jensen T.G., Liebert A., Lewinsky R., Swallow D.M., Olsen J., Troelsen J.T. The -14010∗C variant associated with lactase persistence is located between an Oct-1 and HNF1α binding site and increases lactase promoter activity. Hum. Genet. 2011;130:483–493. doi: 10.1007/s00439-011-0966-0. [DOI] [PubMed] [Google Scholar]
- 9.Olds L.C., Ahn J.K., Sibley E. 13915∗G DNA polymorphism associated with lactase persistence in Africa interacts with Oct-1. Hum. Genet. 2011;129:111–113. doi: 10.1007/s00439-010-0898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sibley E., Ahn J.K. Theodore E. Woodward Award: lactase persistence SNPs in African populations regulate promoter activity in intestinal cell culture. Trans. Am. Clin. Climatol. Assoc. 2011;122:155–165. [PMC free article] [PubMed] [Google Scholar]
- 11.Troelsen J.T., Olsen J., Møller J., Sjöström H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003;125:1686–1694. doi: 10.1053/j.gastro.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Maravelias C., Sibley E. Lactase gene promoter fragments mediate differential spatial and temporal expression patterns in transgenic mice. DNA Cell Biol. 2006;25:215–222. doi: 10.1089/dna.2006.25.215. [DOI] [PubMed] [Google Scholar]
- 13.Enattah N.S., Jensen T.G., Nielsen M., Lewinski R., Kuokkanen M., Rasinpera H., El-Shanti H., Seo J.K., Alifrangis M., Khalil I.F. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingram C.J., Elamin M.F., Mulcare C.A., Weale M.E., Tarekegn A., Raga T.O., Bekele E., Elamin F.M., Thomas M.G., Bradman N., Swallow D.M. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum. Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 15.Ingram C.J., Raga T.O., Tarekegn A., Browning S.L., Elamin M.F., Bekele E., Thomas M.G., Weale M.E., Bradman N., Swallow D.M. Multiple rare variants as a cause of a common phenotype: several different lactase persistence associated alleles in a single ethnic group. J. Mol. Evol. 2009;69:579–588. doi: 10.1007/s00239-009-9301-y. [DOI] [PubMed] [Google Scholar]
- 16.Tishkoff S.A., Reed F.A., Ranciaro A., Voight B.F., Babbitt C.C., Silverman J.S., Powell K., Mortensen H.M., Hirbo J.B., Osman M. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermisson J., Pennings P.S. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pennings P.S., Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2006;2:e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pennings P.S., Hermisson J. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 2006;23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- 20.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 21.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman B., Smith N., Curtis C., Huckett L., Mill J., Craig I.W. DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav. Genet. 2003;33:67–72. doi: 10.1023/a:1021055617738. [DOI] [PubMed] [Google Scholar]
- 23.Mulcare C.A., Weale M.E., Jones A.L., Connell B., Zeitlyn D., Tarekegn A., Swallow D.M., Bradman N., Thomas M.G. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am. J. Hum. Genet. 2004;74:1102–1110. doi: 10.1086/421050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones B.L., Swallow D.M. The impact of cis-acting polymorphisms on the human phenotype. Hugo J. 2011;5:13–23. doi: 10.1007/s11568-011-9155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens M., Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas M.G., Weale M.E., Jones A.L., Richards M., Smith A., Redhead N., Torroni A., Scozzari R., Gratrix F., Tarekegn A. Founding mothers of Jewish communities: geographically separated Jewish groups were independently founded by very few female ancestors. Am. J. Hum. Genet. 2002;70:1411–1420. doi: 10.1086/340609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollox E.J., Poulter M., Zvarik M., Ferak V., Krause A., Jenkins T., Saha N., Kozlov A.I., Swallow D.M. Lactase haplotype diversity in the Old World. Am. J. Hum. Genet. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey C.B., Hollox E.J., Poulter M., Wang Y., Rossi M., Auricchio S., Iqbal T.H., Cooper B.T., Barton R., Sarner M. Lactase haplotype frequencies in Caucasians: association with the lactase persistence/non-persistence polymorphism. Ann. Hum. Genet. 1998;62:215–223. doi: 10.1046/j.1469-1809.1998.6230215.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


