Summary
Argonautes are the central protein component in small RNA silencing pathways. Of the four human Argonautes (hAgo1–4) only hAgo2 is an active slicer. We determined the structure of hAgo1 bound to endogenous copurified RNAs to 1.75 Å resolution and hAgo1 loaded with let-7 miRNA to 2.1 Å. Both structures are strikingly similar to the structures of hAgo2. A conserved catalytic tetrad within the PIWI domain of hAgo2 is required for its slicing activity. Completion of the tetrad combined with a mutation on a loop adjacent to the active site of hAgo1 results in slicer activity that is substantially enhanced by swapping in the N domain of hAgo2. hAgo3, with an intact tetrad, becomes an active slicer by swapping the N domain of hAgo2, without additional mutations. Intriguingly, the elements that make Argonaute an active slicer involve a sophisticated interplay between the active site and more distant regions of the enzyme.
Introduction
Binding of a small RNA to an Argonaute family protein essentially defines the RNA interference (RNAi) pathway, as these elements are the essential components of the RNA-induced silencing complex, RISC. The central role of Argonaute in RNAi processes (Cenik and Zamore, 2011; Joshua-Tor and Hannon, 2011) was underscored dramatically with its identification as “Slicer” - the entity in RISC, which performs cleavage of the mRNA guided by an siRNA (Song et al., 2004). The PIWI domain of Argonaute was shown to resemble an RNase H enzyme, consistent with the biochemistry of the slicing reaction (Martinez and Tuschl, 2004; Schwarz et al., 2004). A DDH catalytic triad in the active site was found to be critical for slicing in human Argonaute-2 (hAgo2) (Liu et al., 2004; Rivas et al., 2005). Indeed, all that is needed for target cleavage is hAgo2, a guide strand and Mg2+ (MacRae et al., 2008; Rivas et al., 2005).
Structures of Thermus thermophilus Argonaute (TtAgo) bound to DNA guide strands and DNA/RNA hybrid duplexes of varying length (Wang et al., 2008a; Wang et al., 2009; Wang et al., 2008b) provided a great deal of insight into the conformational changes in both the protein and the RNA that accompany the action of the enzyme at the heart of the RISC effector complex. In the absence of the mRNA substrate, the guide strand is anchored at its 5’ end to a binding site in the Mid domain identified earlier (Ma et al., 2005; Parker et al., 2005), while the 3’ end binds to the PAZ (Wang et al., 2008b). Two structures with substrates of different lengths (Wang et al., 2008a; Wang et al., 2009) illustrate both the range of motions that the PAZ domain can undergo in the catalytic cycle as well as the dissociation of the 3’ end of the guide from the PAZ domain upon binding a perfectly paired target (Tomari and Zamore, 2005).
Recently, the structures of hAgo2 (Schirle and MacRae, 2012) and an Argonaute from the yeast Klyveromyces polysporus (KpAgo) (Nakanishi et al., 2012) were determined in complex with a mixture of heterogeneous RNA copurified from the expression host, followed by hAgo2 in complex with miR-20a (Elkayam et al., 2012) reviewed in (Kuhn and Joshua-Tor, 2013; Sasaki and Tomari, 2012). All these structures are very similar to their prokaryotic counterparts despite the fact that a biological function for the prokaryotic enzymes has not yet been established. The study of KpAgo identified a fourth catalytic residue, a glutamate that completes the active site DEDH tetrad (DEDD in KpAgo) as previously suggested (Nowotny et al., 2005).
All human Argonautes function in the microRNA (miRNA) pathway and associate with a similar repertoire of RNA and protein cofactors to carry out gene silencing although only hAgo2 is an active slicer (Liu et al., 2004; Meister et al., 2004). hAgo2 mutants that lack slicer activity are embryonic lethal in mice suggesting a specialized role for Ago2 in mammals (Liu et al., 2004). Of the remaining hAgos, hAgo1 has an arginine in place of the active-site histidine in the DEDH tetrad, while hAgo4 is missing one of the aspartates in addition to an arginine at least in part explaining their lack of slicing. Surprisingly, hAgo3 has an intact catalytic tetrad, yet is still inactive for slicer activity. Therefore, it appears that essential determinants for slicer activity lie outside of the active site.
Here we show that restoring an intact catalytic DEDH tetrad does not activate slicing in hAgo1. The lack of activity is not explained by gross conformational differences as the structure of the inactive slicer hAgo1 in complex with the let7 miRNA has a nearly identical structure to the active hAgo2 in complex with miR20a. Only by introducing a DEDH tetrad into hAgo1 and an additional point mutation within a loop adjacent to the active site does hAgo1 become a slicer enzyme. To our surprise the N domain of hAgo2 plays an important role in slicing as swapping in the N domain of hAgo2 is sufficient to activate hAgo3 and substantially enhances activated hAgo1.
Results
Structure of human Argonaute-1
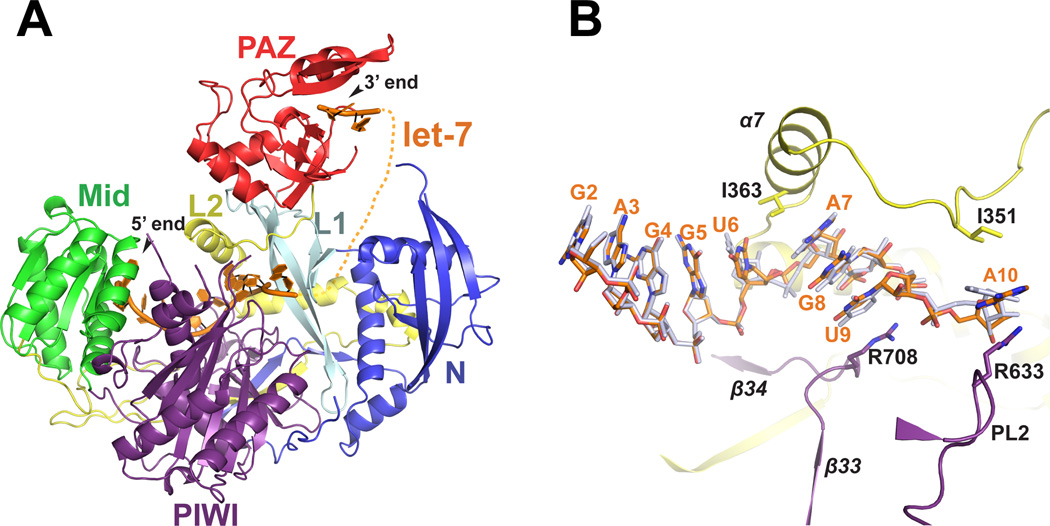
We expressed and purified human Argonaute-1 (hAgo1) from baculovirusinfected Sf9 insect cells. hAgo1 copurifies with endogenous 20 nt small RNAs although small amounts of unloaded hAgo1 can be isolated by cation-exchange chromatography (Elkayam et al., 2012). We obtained crystals of hAgo1 containing endogenous Sf9 20 nt small RNAs and determined its structure to 1.75 Å resolution (hAgo1-endoRNA). Furthermore, we loaded the fraction of unloaded hAgo1 with a 22 nt single-stranded let-7 miRNA and determined the structure of the complex to 2.1 Å (hAgo1-let-7) (Figure 1A, Figure S1, Table S1). hAgo1 displays the same domain architecture found in all Argonaute proteins, namely the four primary domains N, PAZ, Mid and PIWI with two linker regions L1 and L2 (Figure 1A). The N, Mid and PIWI domains of hAgo1 form a base upon which the PAZ domain sits, much like a duck with the PAZ domain resembling the head of the duck and the other three domains the body of the duck. As noted previously (Boland et al., 2011; Elkayam et al., 2012; Nakanishi et al., 2012; Schirle and MacRae, 2012), hAgo1 contains several insertions relative to prokaryotic Argonaute structures (Song et al., 2004; Wang et al., 2008b; Yuan et al., 2005). These insertions occur in each of Argonaute’s primary domains as well as the L2 linker and are likely to play important biological roles as they potentially interact with guide and target RNA and possibly other protein partners.
Figure 1. Structure of hAgo1 in Complex with let-7.
(A) Overall structure of hAgo1 in complex with let-7 guide RNA. The individual domains of hAgo1 are labeled and color coded. Let-7 miRNA is shown as an orange cartoon. Nucleotides 1–10 stretch from the MID domain and pass through L2, the PIWI domain and L1. A dashed line indicates the projected path of the disordered nucleotides 11–20. Nucleotides 21 and 22 are modeled in the PAZ domain. (B) Magnified view of the path of the seed region of let-7 bound to hAgo1. The let-7 miRNA bound to hAgo1 is shown as orange sticks. The superimposed miR20a from the hAgo2-miR20a complex (PDB 4F3T) is shown as grey sticks. Structural elements are colored according to the domain colors in panel A. Residues that make kinks in the let–7 miRNA are shown as sticks. See also Figure S1.
Interactions of hAgo1 with human let-7 and endogenous Sf9 RNA
The structure of the hAgo1-let-7 complex has interpretable electron density for 5’-nucleotides 1 to 10 and 3’-nucleotides 21 and 22 (Figure 1A and S1A). Weak electron density for the unmodeled nucleotides is observed after nucleotide 10 that traverses the channel between L1 and the N domain and leads up to the PAZ domain similar to the hAgo2-miR20a complex (Elkayam et al., 2012). To achieve accurate slicing, Argonaute proteins require the guide strand to be 5’ phosphorylated (Rivas et al., 2005). In the hAgo1-let-7 complex the 5’ phosphate and the U1 base are held in place by a conserved network of interactions from protein residues of the Mid and PIWI domains to exquisitely place the guide in the proper position as observed in all Argonaute-RNA complexes (Figure 1A and 1B and Figure S1) (Elkayam et al., 2012; Frank et al., 2010; Ma et al., 2005; Nakanishi et al., 2012; Parker et al., 2005; Schirle and MacRae, 2012; Wang et al., 2008b). The identity of the U1 base is “read” by the nucleotide specificity loop through a specific hydrogen bonding arrangement consisting of peptide backbone interactions from G522 and T524 in addition to a water molecule (Figure S1B) (Frank et al., 2010). The bases of nucleotides 2–6, which comprise the majority of the seed sequence, are stacked with their edges surface-exposed to initiate target binding (Figure 1B). As in hAgo2, I363 from α7 of the L2 linker creates a kink in let-7 between bases 6 and 7, interrupting the near A-form RNA conformation of nucleotides 2–6. The bases of nucleotides 7–9 of let-7 resume stacking until a second kink is introduced by R708, which stacks against U9. The A10 base is sandwiched between R633 and I351, introducing yet another kink in the RNA as it extends toward the PAZ domain (Figure 1B).
Most of the direct interactions between nucleotides 2–6 of let-7 and hAgo1 are mediated by side-chain and main-chain specific interactions from both the Mid and PIWI domains with the phosphate backbone of let-7 (Figure S1A). Following the kink between bases 6 and 7, numerous residues from the PIWI domain and L1 and L2 linkers direct the path of the phosphate backbone of nucleotides 7 through 10 (Figure 1B and Figure S1A).
hAgo1 exhibits six RNA-specific interactions with the ribose 2’-OH of nucleotides U1, G2, G4, G5, A7 and A8 of the let-7 seed sequence. These interactions are direct or water-mediated through side-chain and main-chain atoms of hAgo1. All RNA-specific interactions in the hAgo1-let-7 complex are identical to the hAgo2-miR20a complex (Figure S1A), except for one 2’-OH interaction that is lost: in the hAgo2-miR20a structure the 2’-OH of U6 forms a water-mediated interaction with the side chain of T368 and the backbone amide of A369, both from the L2 linker and T759 from PIWI. In contrast, the conformation of the U6 is slightly altered in the hAgo1-let-7 structure, likely due to the shift of α7 in the L2 linker toward the N domain (Figure 1B).
Nucleotides 21 and 22 of let-7 are bound to the PAZ domain where an identical set of interactions are as described previously (Elkayam et al., 2012; Ma et al., 2004), however, unlike in the hAgo2-miR20 structure, the terminal two bases are stacked (Figure 1A and Figure S1A).
In the hAgo1-endoRNA structure, we modeled nucleotides 1–9 as 5’-AAUAUUAAA-3’, using a 1.75 Å resolution difference map to place U in density that could only accommodate a pyrimidine and an A into density resembling a mixture of purine and pyrimidine. Clear crystallographic evidence for both a purine and pyrimidine in position 1 led us to model an A in the 5’-binding pocket, although a U would fit as well. Indeed, mammalian Argonautes have a preference for a 5’ U and to a lesser extent A (Czech and Hannon, 2011). To our surprise, the adenine base adopts a syn conformation around the glycosidic bond (Figure S1C). This is in contrast to the A1 modeled in the anti conformation in hAgo2 bound to endogenous RNAs (Schirle and MacRae, 2012) and the isolated hAgo2 MID domain in complex with AMP (Frank et al., 2010). The syn conformation of the A1 base is stabilized by stacking with Y527 and a base-specific interaction between the N1 of the adenine ring and the side-chain of Y813 from the PIWI domain.
To more closely examine the copurified Sf9 RNA we sequenced RNA extracted from purified hAgo1, hAgo2 and hAgo3. All of the hAgos were enriched for 20 nt RNAs (Figure S1D and E) as reported for hAgo2 (Elkayam et al., 2012). More than 70% of 20 nt RNAs bound to hAgo1–3 have a U or A as the 5’ nucleotide, although hAgo2 has a significant preference for U over A, in contrast to hAgo1 and 3 which have an equal distribution of U and A (Figure S1F). These sequencing data support our interpretation of the electron density in the hAgo1-endoRNA structure.
Taken together, all amino acids that contact guide RNA are absolutely conserved between hAgo1 and hAgo2 (Figure S1A). Clearly, the manner in which hAgo1 binds guide RNA fails to explain its lack of slicer activity.
Structural comparison of hAgo1 with hAgo2
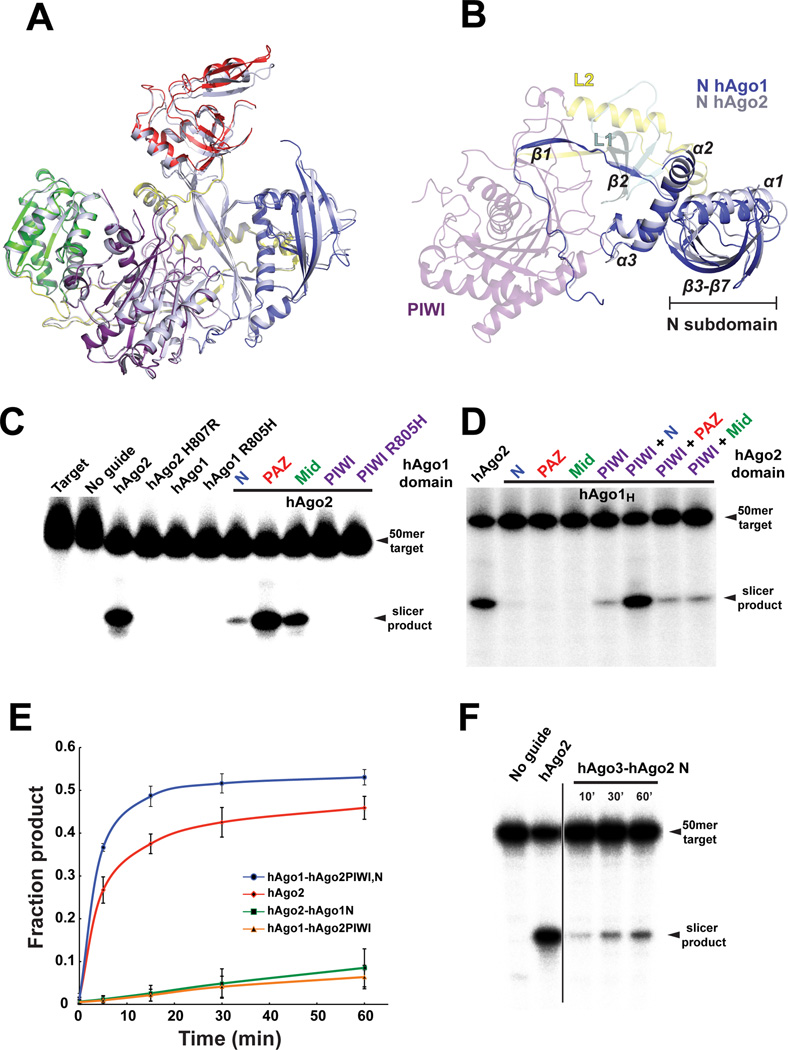
hAgo1 and hAgo2 are 84% identical in their primary sequence yet only hAgo2 is an active slicer. To better understand this difference in activity, we conducted an indepth structural analysis. First we looked for any differences in each of the domains that could explain the lack of hAgo1 slicer activity. A pairwise structural alignment of each hAgo1 domain in isolation with its counterpart in hAgo2 reveals extraordinary structural homology (Figure 2A and Figure S2C–H). Since the relative orientation between the Mid and PIWI domains are nearly identical in both structures, and in fact in all Argonaute structures determined to date, we chose to further examine relative domain movements occurring between the hAgo1-let-7 and hAgo2-miR20a complexes by alignment of the two structures based on their Mid-PIWI lobes (Figure 2A). We noticed that residues K49 to S137 (β3, α1, β4–7) of the N domain form a subdomain that is slightly twisted with a 3 Å translation toward the PIWI domain in hAgo1 compared to that observed in hAgo2 (Figure 2A and 2B). Residues 18–48 and 138–173 of the N domain align well with hAgo2 as they are mainly involved in the interface with L1, L2 and the PIWI domain. This N subdomain appears to be making contact with let-7 in our structure (Figure 1A) analogous to the hAgo2-miR20a complex (Elkayam et al., 2012) and will certainly interact with guide/target duplexes (Wang et al., 2009). The L1 linker position in hAgo1 is essentially unchanged while the PAZ domain is twisted up and away from the PIWI domain analogous to the duck looking up and behind its shoulder. The movement of the N and PAZ domains in hAgo1 relative to the Mid-PIWI lobe is assisted by a ∼1 Å translational shift of residues D356 to G396 composing α7–9 of the L2 linker towards the N domain (Figure 2A and 2B).
Figure 2. Effect of Argonaute domains on Slicer activity.
(A) Structural superposition of hAgo1 and hAgo2 based on the MID–PIWI lobe. hAgo1 is shown as a cartoon with the same color scheme as Figure 1A. hAgo2 is shown as a grey cartoon. (B) The same alignment in panel A but with a 90° rotation around the horizontal axis. All of the domains are colored as in panel A, but with the MID and PAZ domains deleted for clarity. Secondary structure elements in the N domain that interact with the L1, L2 and PIWI domain are indicated as are the elements composing the subdomain of the N that move in hAgo1 with respect to hAgo2. (C) Slicer assays for WT hAgo1, hAgo2, the mutants of the catalytic tetrad and chimeric hAgo2 constructs swapped with inactive hAgo1 domains. (D) Slicer assay of hAgo1 chimeras comprised of single and double domain swaps from hAgo2. (E) Plot of fraction product cleaved over a 60–minute time course showing enhanced slicing by the hAgo2 N domain. The standard error deviation from the mean of three individual replicates is plotted with data points connected by a smooth curve in Excel. (F) Slicer assay for hAgo3-hAgo2 N domain chimera. Time points were taken every 10, 30 and 60 minutes. A line designates an irrelevant lane that was cropped. Slicer assays shown in panels C, D and F are representatives of at least three individual replicates. See also Figure S2.
hAgo2 PIWI domain activates hAgo1
As there were no obvious structural differences that could account for differential activity between hAgo1 and hAgo2, we initiated biochemical studies to identify the specific hAgo1 defect that prevents slicing. We purified hAgo mutants using a two-step protocol, coupling Strep-affinity with cation-exchange chromatography to obtain homogenous samples for our slicer assays (Figure S3). To confirm that our recombinant hAgos behave similarly to immunopurified Argonautes from human cells, we assayed wild type hAgo2 and hAgo1 for slicer activity by loading hAgo with a miR20a guide strand (Figure S3) and measured slicing of a radiolabeled complimentary target. As reported previously, hAgo2 cleaved the target while hAgo1 did not (Figure 2C) (Liu et al., 2004; Meister et al., 2004). Even though hAgo1 possesses an incomplete catalytic tetrad (DEDR vs. DEDH), completing this tetrad by a R805H mutation does not rescue hAgo1 slicer activity (Figure 2C). Therefore, an incomplete catalytic tetrad in the active site of hAgo1 does not fully explain its lack of slicing. Interestingly, the substitution of R805H in hAgo1 is necessary, but not sufficient for slicing activity, as a H807R mutation in hAgo2 abrogates its activity (Figure 2C) (Rivas et al., 2005). Since a complete catalytic tetrad is essential for slicing in hAgos we mutated R805H in hAgo1 (referred to herein as hAgo1H).
Next, we conducted domain-swapping experiments by substituting each functional domain of hAgo2 with the equivalent one in hAgo1. The N, PAZ and Mid domains of hAgo1 can individually substitute for their hAgo2 counterparts and support hAgo2 slicing (Figure 2C). Both hAgo1 linkers L1 and L2 can support slicing in hAgo2 as well (data not shown). In contrast, exchange of the PIWI domain of hAgo1 or hAgo1H into hAgo2 abrogates slicing, providing strong evidence that other factors in addition to the incomplete DEDH tetrad are responsible for the slicer defect of hAgo1 (Figure 2C). Taken, together, the domain swap experiments demonstrated that each of the hAgo1 domains is supportive of slicing, with the exception of the PIWI domain, consistent with their structural similarities.
Given that the PIWI domain of hAgo1H deactivates hAgo2 slicing we conducted the reciprocal experiment and created an active hAgo1 chimera by substituting the active PIWI domain of hAgo2 into the non-slicer hAgo1. Indeed, the hAgo2 PIWI domain swap alone is sufficient to activate slicing in hAgo1 (Figure 2D). Neither the N, PAZ or Mid domains of hAgo2 support slicing when swapped individually into hAgo1H. However, the active hAgo1 chimera only restored ∼10% of wild type (WT) hAgo2 slicer activity (Figure 2E).
The N domain of hAgo2 is optimized for slicing
The N, PAZ and Mid domains of hAgo1 can support slicing when swapped into hAgo2. However, we noticed a strong suppression of hAgo2 activity in the chimera containing the N domain of hAgo1 (Figure 2C). In fact, replacing the hAgo2 N domain with the one from hAgo1 leads to a ∼90% reduction of WT hAgo2 activity (Figure 2C and 2E). The activity of this chimera is similar to the hAgo1 chimera containing the PIWI domain of hAgo2 (Figure 2C and 2D). To test whether the N domain of hAgo2 is playing a role in slicing we made double domain swaps in hAgo1, by exchanging the N, PAZ and Mid domain in combination with the hAgo2 PIWI domain. By swapping the N and PIWI domains of hAgo2 into hAgo1 we were able to convert hAgo1 into an active slicer with activity similar to WT hAgo2 (Figure 2D and 2E). Neither the hAgo2 PAZ-PIWI nor Mid-PIWI domain swap into hAgo1 showed enhancement of slicing (Figure 2D). To further exemplify this point we swapped the hAgo2 N domain into hAgo3, which is not an active slicer despite having a complete catalytic tetrad. The N domain chimera of hAgo3 is activated by the hAgo2 N domain (Figure 2F), although the activity is weak compared to hAgo2 and the hAgo1 chimera with the N and PIWI of hAgo2 (Figure 2D). It is important to note that the hAgo2 N domain alone is not capable of activating hAgo1 (Figure 2D).
Two point mutations in the PIWI domain can restore slicing in hAgo1
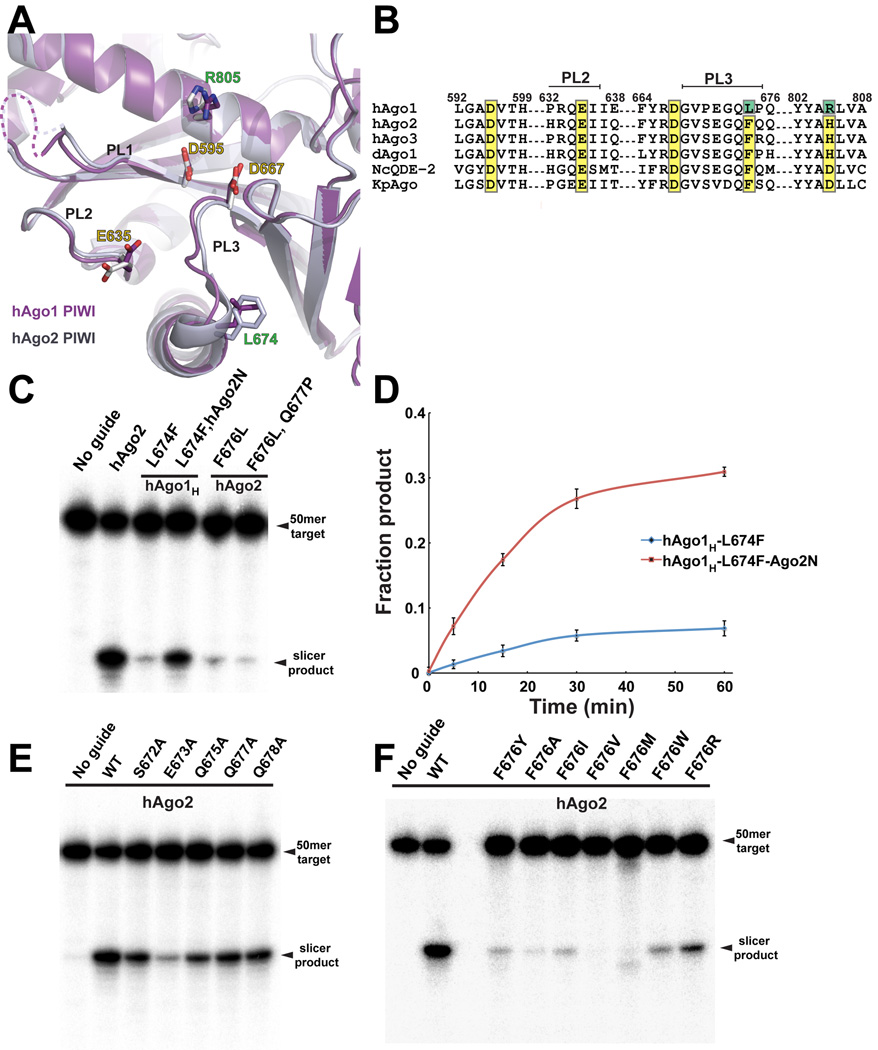
In our quest to find the minimal defect in hAgo1 slicing we mutated every nonconserved amino acid in the hAgo1H PIWI domain into the corresponding amino acid found in hAgo2 (Figure 3A and 3B, Figure S3). To further validate the results obtained with hAgo1H, we mutated the same set of amino acids in the hAgo2 PIWI domain to the corresponding residues found in hAgo1. We discovered a single mutation in the PIWI domain, L674F that rescued hAgo1H slicer activity (hAgo1HF) (Figure 3C). Correspondingly, the equivalent F676L mutation in hAgo2 drastically reduced its slicer activity (Figure 3C). The mutation of L674F in hAgo1H is sufficient to produce an active hAgo1 slicer with levels comparable to that of the hAgo2 PIWI domain swap. The hAgo2 F676L mutation reduced slicing to ∼5% of wild-type hAgo2 with a near complete lack of slicing observed from the double mutation of adjacent residues F676L and Q677P (Figure 3C). The N domain of hAgo2 complements hAgo1HF as we observe a robust enhancement in slicing suggesting that the N domain is intimately involved in some aspect of the slicing catalytic cycle (Figure 3C and 3D). Although the hAgo2 N domain enhances slicing of hAgo1HF to levels approaching WT hAgo2, the single point mutation of F676L is devastating to hAgo2 activity (Figure 3C).
Figure 3. Two Mutations in the PIWI Domain Activate hAgo1.
(A) View of the superimposed active sites in the PIWI domain of hAgo1 and hAgo2. The PIWI domain of hAgo1 is colored purple and hAgo2 is grey. Conserved active site residues are labeled with yellow text. Mutations in hAgo1 that activate slicing are labeled in green. The PIWI domain loops (PL1, PL2 and PL3) that cluster near the active site are indicated. (B) Sequence alignment focused on the catalytic tetrad residues for hAgo1, hAgo2, hAgo3, Drosophila Ago1 and Ago2 (dAgo1 and dAgo2), and two other eukaryotic Argonautes for which structures are known, NcQDE-2 and KpAgo. Conserved catalytic residues are highlighted in yellow and mutations that activate hAgo1 are highlighted in green. The PIWI domain loops PL2 and PL3 are indicated. (C) Slicer assay showing activated hAgo1H by mutation of L674F. The N domain of hAgo2 enhances the slicing by the activated hAgo1. F676L mutants of hAgo2 have a severe defect in slicing. (D) Plot of fraction product cleaved over a 60–minute time course showing the activated hAgo1H L674F and the enhancement from the hAgo2 N domain. The standard error deviation from the mean of three individual replicates is plotted with data points connected by a smooth curve in Excel. (E) Mutants of the PL3 loop residues in hAgo2 show that E673 is important for slicing. (F) Mutants of F676 in hAgo2 greatly impact slicing activity. Slicer assays shown in panels C, E and F are representatives of at least three individual replicates. See also Figure S3.
A PIWI domain loop plays a role in slicing
L674 in hAgo1 and F676 in hAgo2 are located on the C-terminus of a loop connecting β32 with α16 in the core of the PIWI domain (Figure 3A and Figure S4). We will refer to the loop harboring the L674F mutation as PIWI loop 3 (PL3) to avoid confusion with the linker L1 and L2 (Figure 3A and Figure S4). Interestingly hAgo3 has a phenylalanine at this position within the PL3 loop and yet still remains inactive. Two other loops near the active site (formerly labeled L1 and L2) with previously described roles in slicing (Nakanishi et al., 2012; Wang et al., 2009), which we now refer to as PL1 and PL2 are also present in hAgos. To probe the importance of the PL3 loop for slicing in hAgo2, we selected five amino acids within PL3 that might play a role in slicing and mutated them to alanine (Figure 3B and 3E). Only a single mutation showed decreased slicing activity, E673A (Figure 3E), although the slicing defect was modest compared to the mutation of F676L (Figure 3C and 3F). This is not surprising given that this residue is not entirely conserved in slicer active Argonautes (Figure 3B). The residues in PL3 are in position to interact with target RNA as noted earlier (Boland et al., 2011), however, mutants of E673 and Q675 in Drosophila Ago1 (Drosophila Ago1 E798 and Q800) are fully active in tethering assays in S2 cells, are loaded with guide RNA and bind GW182 (Boland et al., 2011). It seems that PL3 may not play a role in binding RNA but rather may have a more intimate role in aligning paired targets for slicing.
Since the F676L mutation in the PL3 loop in hAgo2 has the most severe defect in slicing we mutated F676 to investigate its role in PL3 function. Mutation of F676 in hAgo2 to small aliphatic amino acids caused a severe defect in slicing (Figure 3F) consistent with the natural change to leucine in hAgo1. Only two mutations supported modest levels of slicing when substituted for F676, a tryptophan and arginine. Mutation of F676W maintains the aromatic and hydrophobic properties of phenylalanine. Despite the presence of a positive charge, F676R is the strongest slicer of the F676 mutants we have examined, and like phenylalanine, may participate in hydrophobic interactions through its aliphatic chain and may even interact with RNA bases during target binding. Surprisingly, a F676Y mutation fails to maintain robust slicing despite the presence of an aromatic ring; apparently the hydroxyl group of tyrosine in this position is not tolerated (Figure 3F).
Discussion
Beyond the catalytic tetrad
The conservation of the catalytic tetrad in both the active slicer hAgo2 and the inactive hAgo3 already pointed toward determinants for slicer activity that are distinct from the active site residues. The conservation in structure between hAgo2 and hAgo1 described here underscores this notion (Figure 1B and 2A). Moreover, restoration of an intact catalytic tetrad in hAgo1 is insufficient to impart slicer activity (Figure 2B). Here we show that the PL3 loop together with the DEDH tetrad, are the missing determinants for hAgo1 slicer activity (Figure 3C). The loop is part of a conserved sequence insertion in eukaryotic Argonautes first described in the N. crassa QDE-2 Mid-PIWI structure (referred to as L2) (Boland et al., 2011). In the full-length Argonaute structures it was referred to as cS7 (Nakanishi et al., 2012; Schirle and MacRae, 2012). However, its role in slicing has escaped notice until now.
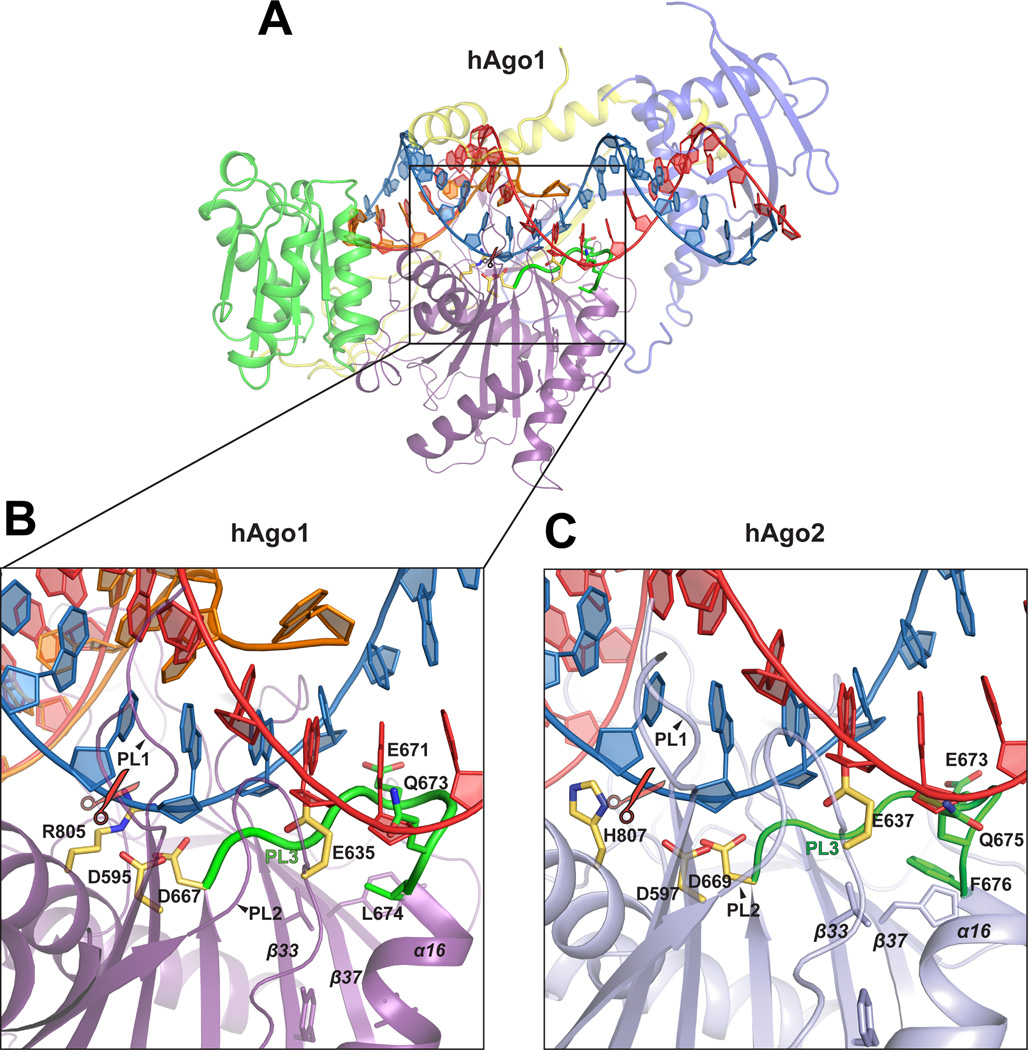
To understand the role of PL3 in slicing, we modeled an A-form RNA duplex (PDB code 3CIY) (Figure 4A and 4B) into the let-7 guide complex of hAgo1 and the miR20a guide complex of hAgo2 (Elkayam et al., 2012). The guide strand of the modeled A-form RNA duplex aligns well with bases 2–6 of the let-7 guide strand of hAgo1 (Figure 4A). As noted for hAgo2, α7 from L2, the PL1, PL2 and PL3 loops of the PIWI domain, and the N domain of hAgo1 clash with the modeled RNA duplex suggesting that major conformational changes must occur during target binding (Figure 4A) (Schirle and MacRae, 2012). Despite these clashes, the position of the scissile phosphate of the target strand aligns well with the active site in hAgo1 and hAgo2 (Figure 4B and 4C). The PL3 loop immediately follows the catalytic D667 in primary sequence (Figure 3A, 3B and 4). In the model, PL3 is positioned in the minor groove of the A-form RNA making potential contacts with nucleotides from both guide and target strands from nucleotides 11 through 14 (Figure 4A). Slicing is restored to hAgo1H by mutation of L674F, which is located on the C-terminal end of the PL3 loop near the top of α16. L674 in hAgo1 and F676 in hAgo2 stack atop the hydrophobic core within the PIWI domain formed between α16 and β33 and β37 (Figure 4A, 4B and 4C).
Figure 4. PL3 Loop Plays a Role in Slicing.
(A) Structure of hAgo1 with a modeled A–form duplex RNA. Domains of hAgo1 are colored as in Figure 1A. The PAZ domain and L1 linker are omitted for clarity. Let-7 miRNA is orange, the modeled guide RNA is red and target strand blue. The location of the scissile phosphate is indicated with scissors. The active site tetrad is shown as yellow sticks. The PL3 loop is highlighted green with important residues identified in this study shown as green sticks. (B) A close-up view of panel (A) but only the PIWI domain is shown. The PL2 and PL3 loops are indicated. (C) A–form RNA modeled in hAgo2 with the same layout as panel B. See also Figure S4.
While the precise function of the PL3 loop and the role of F676 have to await structures with target mRNA, the lack of slicing by hAgo1H might be due to a catalytic defect. For mouse Ago2, mismatches at positions 12–13 between guide and target RNA have little effect on binding (Wee et al., 2012). But for fly Ago2 these mismatches show a strong catalytic defect that is especially apparent under single turnover conditions, which are similar to the in vitro slicing system described here, and the mismatches are in the vicinity of the PL3 loop in the extended RNA duplex model (Figure 4B and 4C). It is conceivable that hAgo1H binds fully paired target RNA but fails to orient the substrate properly during catalysis.
The PL3 loop is also in close proximity to the PL2 loop (Figure 4). This loop adopts a so-called “plugged in” conformation (Nakanishi et al., 2012) in all the eukaryotic Argonaute structures (Kuhn and Joshua-Tor, 2013), thereby placing the catalytic glutamate residue in a position to complete the DEDH (DEDD in KpAgo) catalytic tetrad. Slicing is lost when we mutated this glutamate (E637) to alanine in hAgo2 (data not shown). Although there are no direct contacts between residues in the PL2 and PL3 loops, it is possible that the presence of target mRNA can coordinate movements between these loops and thus regulate slicing.
The N domain is important for Argonaute slicing
The N domain was previously implicated in duplex unwinding during RISC assembly (Kwak and Tomari, 2012). It was proposed that the N domain acted as an active wedge to unwind duplexes during RISC loading and during target binding and subsequent cleavage. Together with the previous study our results highlight a clear role for the N domain in target slicing. The target strand of the modeled duplex in hAgo1 can proceed unimpeded while the guide strand clashes with nucleotides 16–20, just downstream of the base pairs that interact with the PL3 loop (Figure 4A). Despite a more open channel for target strand binding in eukaryotic Argonautes (Nakanishi et al., 2012) further movement of the N domain is likely to influence guide/target pairing and slicing. The N domain of hAgo2 activates slicing in hAgo3 and enhances slicing in hAgo1HF approaching WT hAgo2 levels (Figures 2F, 3C and 3F). We suspect that the hAgo2 N domain is optimized for dealing with fully paired RNA substrates in comparison to hAgo1 and hAgo3. Indeed, hAgo2 is the only human Argonaute that can unwind paired duplexes (Yoda et al., 2010) and may therefore be the only Argonaute that retains the ability to efficiently bind and slice complimentary targets utilizing its N domain as well. While this work was in under review, another study reported that immunopurified hAgo1 and hAgo3 chimeras containing elements from hAgo2 are active slicers (Hauptmann et al., 2013), thus corroborating the results presented here. Taken together our data show that slicing has many more nuances than previously appreciated and that events distant from the active site play equally important roles.
Experimental Procedures
Expression and purification of hAgo1 and hAgo2
hAgo2 was expressed and purified as previously described (Elkayam et al., 2012). The gene coding for full-length WT hAgo1 was codon optimized for expression in Sf9 cells (Life Technologies, Grand Island, NY) and cloned into the pFL vector of the MultiBac baculovirus expression system (Bieniossek et al., 2008) with a Two-Strep-SumoStar (TSS) tag based on the One-Strep-SumoStar (OSS) tag (Schalch et al., 2011) followed by a TEV cleavage site. Sf9 insect cells, grown in Hyclone CCM3 media (Thermo Scientific) were infected with a baculovirus expressing hAgo1 for 48–72 hours. Initial purification was done using Strep-Tactin superflow resin (IBA bioTAGnology) followed by tag removal with TEV protease. Protein was then loaded onto a Mono S 5/50 column (GE Healthcare) equilibrated with 50mM Tris, pH 8.0, 50 mM KCl and 5 mM DTT. Elution with a gradient from 50–1500 mM KCl resulted in two protein fractions, one that was bound to endogenous Sf9 RNA and a second that was RNA-free (100 µg from 10 liters of Sf9) as determined by the UV absorption ratio at 260/280nm and confirmed by RNA extraction and analysis by denaturing urea PAGE.
hAgo1-let-7 complex preparation
A 22mer derived from human let-7 with the sequence, 5’ p-UGAGGUAGUAGGUUGUAUAGUU-3’ (Dharmacon RNAi technologies) was added to the RNA-free protein fraction at a 1:1.5 (protein:miRNA) molar ratio. Excess let-7 was separated from the hAgo1-let-7 complex by size exclusion chromatography using a Superdex 200 10/300 column equilibrated with 20 mM Tris, pH 8.0, 100 mM NaCl and 5 mM DTT.
Crystallization and structure determination
Crystals were grown by hanging-drop vapor diffusion after mixing 1.5 µL of protein-RNA complex at 3–5 mg/ml with 1 µL of 100 mM Tris, pH 9.0, 12.8% PEG-3350 (w/v), 9.4% 2–propanol (v/v) for hAgo1 with endogenous RNA from Sf9 cells and with 100 mM Tris, pH 9.0, 10.4% PEG-3350 (w/v) and 9.4% 2-propanol (v/v) for hAgo1-let-7 complex. For data collection, crystals were cryoprotected by transferring the crystals briefly into a reservoir solution supplemented with 25% (v/v) ethylene glycol and flash frozen in liquid nitrogen. X-ray diffraction data for hAgo1 with endogenous Sf9 RNA were collected to 1.75 Å at beamline X29 at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL). Data for the hAgo1-let-7 complex were collected to 2.1 Å resolution at beamline X25. Diffraction data were indexed, integrated and scaled using XDS (Kabsch, 2010). The structure of hAgo1 bound to endogenous Sf9 RNA was solved by molecular replacement with PHASER (McCoy et al., 2007) using the protein chain of the hAgo2 structure as a search model (PDB ID: 4F3T). The molecular replacement solution was rigid body refined in PHENIX (Adams et al., 2010) followed by simulated annealing refinement prior to manual correction in COOT (Emsley et al., 2010). Final TLS refinement of the model was done using PHENIX with manually selected TLS groups (Version 1.8.1_1168). The structure of the hAgo1-let-7 complex was refined using a similar strategy. The final structures were refined to an Rwork/Rfree of 17.2%/20.2% for hAgo1 with endoRNA and Rwork/Rfree of 17.4%/21.7% for hAgo1-let-7 complex. Figures were generated using PyMol (The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC).
Slicer assays
All Argonaute mutants were concentrated to 10 µM and analyzed by SDS-PAGE to ensure homogeneity (Figure S4). Slicer assays were performed in 10 mM Tris-HCl, pH 8.0, 100 mM KCl, 10 mM DTT, 2 mM MgCl2 and 10% glycerol. Argonaute protein (10 nM concentration) was loaded with single stranded miR20a guide strand (5-pUAAAGUGCUUAUAGUGCAGGUA-3’ (Dharmacon RNAi technologies) at a 1:1 ratio for 20 min at 27°C. 10 µL slicing reactions were initiated by mixing 5’-32P radiolabeled target RNA (5’-CGAGCAGUAAUUCUAGAACUAUACAACC-UACUACCUCACUCGAGCGGCCG-3, 1 nM final concentration) in the reaction buffer with 4 nM loaded hAgo:miR20a complex. After 60 min the reaction was Trizol extracted, precipitated in isopropanol and resuspended in formamide loading buffer followed by incubation at 95°C for 2 min. Samples were then resolved on 15% denaturing urea gels. The radiolabeled slicing products were visualized by phosphorimaging (Fujifilm FLA-5100) and quantified using ImageJ software (Schneider et al., 2012). For time course analysis the results of three experiments were quantified using ImageJ and displayed as a mean of with error bars representing ± SD. When comparing activity levels between different hAgo constructs we analyzed the mean fraction of product cleaved at the 1- hour time point. To confirm equal levels of guide RNA loading into each Argonaute mutant, 5’-32P radio–labeled guide RNA was loaded and subjected to slot-blot nitrocellulose membranes, ensuring equal levels of RISC formation (Figure S4.
Small RNA library preparation and bioinformatics analysis
Small RNAs were cloned as previously described (Brennecke et al., 2007) and sequenced using Illumina HiSeq technology. Sequencing results reported in this study can be found at GEO using accession number GSE46870 (which includes libraries GSM1139470-GSM1139477).
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with PDB codes 4KRE and 4KRF for hAgo1 with endogenous RNA and hAgo1 let–7 complex, respectively.
Supplementary Material
Highlights.
First structure of human Argonaute-1 (hAgo1) bound to let-7 miRNA.
hAgo1 becomes an active slicer utilizing elements from the N and PIWI domains of hAgo2.
hAgo3 is an active slicer upon swapping of the hAgo2 N domain .
New mechanistic insight into the essential elements of human Argonaute slicing.
Acknowledgements
We thank Jonathan Ipsaro and Claus Kuhn for critical comments on the manuscript and the Joshua–Tor laboratory for helpful comments and suggestions. We also thank Annie Héroux and Howard Robinson for help at the National Synchrotron Light Source, which is supported by the Department of Energy, Office of Basic Energy Sciences. This work was supported by the Louis Morin Charitable Trust and the Robertson Research Fund of Cold Spring Harbor Laboratory (to L.J.). G.J.H and L. J. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse–Kunstleve RW, et al. PHENIX: a comprehensive Python–based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Richmond TJ, Berger I. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci. 2008:20. doi: 10.1002/0471140864.ps0520s51. Chapter 5, Unit 5. [DOI] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Cenik ES, Zamore PD. Argonaute proteins. Current biology : CB. 2011;21:R446–R449. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5’-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nature structural & molecular biology. 2013 doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb Perspect Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, Joshua-Tor L. Eukaryotic Argonautes come into focus. Trends in biochemical sciences. 2013 doi: 10.1016/j.tibs.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nature structural & molecular biology. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Science. Vol. 305. New York, NY: 2004. Argonaute2 is the catalytic engine of mammalian RNAi; pp. 1437–1441. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5’ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Molecular cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature structural & molecular biology. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Sasaki HM, Tomari Y. The true core of RNA silencing revealed. Nature structural & molecular biology. 2012;19:657–660. doi: 10.1038/nsmb.2302. [DOI] [PubMed] [Google Scholar]
- Schalch T, Job G, Shanker S, Partridge JF, Joshua-Tor L. The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nature structural & molecular biology. 2011;18:1351–1357. doi: 10.1038/nsmb.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. Science. Vol. 336. New York, NY: 2012. The crystal structure of human Argonaute2; pp. 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Science. Vol. 305. New York, NY: 2004. Crystal structure of Argonaute and its implications for RISC slicer activity; pp. 1434–1437. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008a;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008b;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nature structural & molecular biology. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Molecular cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.