Abstract
Double-strand breaks (DSBs) in budding yeast trigger activation of DNA damage checkpoints, allowing repair to occur. Although resection is necessary for initiating damage-induced cell cycle arrest in G2, no role has been assigned to it in the activation of G1 checkpoint. Here we demonstrate for the first time that the resection proteins Sgs1 and Exo1 are required for efficient G1 checkpoint activation. We find in G1 arrested cells that histone H2A phosphorylation in response to ionizing radiation is independent of Sgs1 and Exo1. In contrast, these proteins are required for damage-induced recruitment of Rfa1 to the DSB sites, phosphorylation of the Rad53 effector kinase, cell cycle arrest and RNR3 expression. Checkpoint activation in G1 requires the catalytic activity of Sgs1, suggesting that it is DNA resection mediated by Sgs1 that stimulates the damage response pathway rather than protein-protein interactions with other DDR proteins. Together, these results implicate DNA resection, which is thought to be minimal in G1, as necessary for activation of the G1 checkpoint.
Keywords: Yeast, DNA damage, Checkpoint activation, G1, Sgs1, Exo1, DNA resection
1. Introduction
Cellular DNA is constantly exposed to endogenous and exogenous insults, often resulting in damage. The most deleterious type of damage is the chromosomal double strand break (DSB), which can lead to translocations and loss of genetic information. To maintain genomic integrity, cells activate the DNA damage response (DDR), which is a set of coordinated pathways that promote DNA repair while protecting the cell from further damage [1–3]. Upon generation of DSBs, components of the DDR pathway localize to the damage site, forming chromatin domains called ionizing radiation induced foci (IRIF) that mediate signal transduction. IRIF can be visualized under the microscope by monitoring relocalization of fluorescently tagged factors that localize to the site such as yeast Rad9 or its metazoan ortholog 53BP1 [4, 5].
Typically, cell cycle progression is transiently arrested in response to DSBs, permitting repair to occur and thereby minimizing loss of genomic integrity due to replication or segregation of damaged DNA [6]. In budding yeast Saccharomyces cerevisiae, these cell checkpoints cause delays during G1, S, or G2/M [7, 8]. Relative to G1, the G2/M checkpoint is more sensitive to damage and generates a more robust response [9]. The S phase checkpoint is associated with slowed replication [10]. According to the prevailing model for checkpoint activation in budding yeast [11], the Mre11-Rad50-Xrs2 (MRX) complex serves as a damage sensor and is rapidly recruited to the DSB site [12, 13]. In parallel, the MRX complex collaborates with the nuclease Sae2 to process DNA ends, permitting 5' end resection by the Exo1 and Dna2/STR (Sgs1-Top3-Rmi1) nucleases to generate 3' overhangs [14–17]. The ssDNA generated from this resection becomes coated with the heterotrimeric replication protein A (RPA) complex consisting of Rfa1, 2, and 3, which then facilitates assembly of the 9-1-1 clamp to the DSB site [18, 19]. Then, the PIKK kinase Mec1 and its adaptor protein Ddc2 are recruited to the site via interactions with RPA and the 9-1-1 clamp [19–21]. In parallel, MRX recruits a second PIKK kinase, Tel1, to the break [22, 23] where it phosphorylates histone H2A at Ser129 to generate γH2A, facilitating recruitment of checkpoint mediators to the damage site [24–26]. The adaptor protein Rad9 localizes to the DSB site by interacting both with the S129-phosphorylated H2A and K79-methylated histone H3 (H3 K79me) [27–31]. There, Rad9 is phosphorylated by Mec1 and recruits the effector kinase Rad53, also phosphorylated by Mec1, leading to its activation [32, 33]. In a critical amplification step, the activated Rad53 dissociates and phosphorylates downstream effectors of the checkpoint activation cascade leading to cell cycle arrest.
While 5' DNA resection and chromatin modification appear to work cooperatively to enable a proper DNA damage response, each process seems to play a distinct role. DNA end resection is a two-step process: an initial short-range resection mediated by MRX and Sae2, and a subsequent long-range resection mediated by Exo1 and Dna2-STR acting in parallel. Cells lacking both Sgs1 and Exo1 are defective in DNA resection and activation of the G2/M checkpoint [14, 15, 17]. The checkpoint arrest in response to DNA damage in G1 is relatively insensitive to radiation and transient in comparison to G2/M [34, 35]. Formation of 3' overhangs in G1 is markedly slower than in G2/M [9], ascribed to the inactive state of cyclin dependent kinase Cdc28 [36]. While cells expressing nuclease-defective Exo1 or helicase-defective Sgs1 are G2/M checkpoint deficient [17], no similar effect has been documented for G1. Instead of DNA resection, previous studies have indicated that chromatin modifications are necessary for activation of the G1 checkpoint but not the G2/M checkpoint [26, 30, 37]. Even so, the limited resection in G1 is likely sufficient to recruit RPA to the DSB sites [9, 34, 38], suggesting a potential role in activation of the G1 checkpoint.
That Mec1 is necessary for the G1 arrest led us to reexamine a relationship between resection and the G1 checkpoint [39]. Here we show that G1-arrested sgs1Δ exo1Δ cells are sensitive to DNA damage and unable to activate RNR3 when exposed to ionizing radiation (IR). We find that the loss of Sgs1 and Exo1 significantly impairs activation of Rad53 and cell cycle arrest in irradiated cells. Interestingly, IR-induced generation of γH2A is maintained in these mutants, indicating that Sgs1 and Exo1 are not required for the chromatin modification cascade of G1 checkpoint. This is supported by the inability of the Ddc2-Rad53 fusion protein, which bypasses the requirement for Rad9, to rescue the sgs1Δ exo1Δ cells. These findings indicate that although resection is not as extensive in G1 as in G2, it could be necessary for G1 checkpoint activation.
2. Materials and methods
2.1. Strains, plasmids, growth conditions, and yeast transformations
All yeast strains (as listed in Table 1) were constructed in the W303 background unless noted. Yeast cells were grown in standard rich YPD media (1% yeast extract, 2% peptone, 2% glucose) or SC (synthetic media with 2% glucose) lacking appropriate amino acids for selection. Plasmids used are listed in Table 2 and were grown in E. coli competent DH5α and transformed into yeast as described [40–42].
Table 1.
Yeast strain list
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATa ade2-1 can1-100 ura3-1 leu2-3, 112 his3-11, 15 trp1-1 | [65] |
| SKY 2850 | W303-1A, rad9Δ∷KANMX6 | [34] |
| SKY 3073 | W303-1A, exo1Δ∷KANMX6 | This Study |
| SKY 3074 | W303-1A, sgs1Δ∷TRP1 | This Study |
| SKY 3075 | W303-1A, sgs1Δ∷TRP1 exo1Δ∷KANMX6 | This Study |
| SKY 2939 | W303-1A, hta1S129A∷his3MX6 hta2S129A∷TRP1 | [26] |
| SKY 2866 | W303-1A, RAD53∷13Myc-KANMX6 | [34] |
| SKY 2868 | W303-1A, RAD53∷13Myc-KANMX6 rad9Δ∷KANMX6 | [34] |
| SKY 3076 | W303-1A, RAD53∷13Myc-HIS3MX6 exo1Δ∷KANMX6 | This Study |
| SKY 3077 | W303-1A, RAD53∷13Myc-HIS3MX6 sgs1Δ∷TRP1 | This Study |
| SKY 3078 | W303-1A, RAD53∷13Myc-HIS3MX6 sgs1Δ∷TRP1 exo1Δ∷KANMX6 | This Study |
| SKY 3079 | W303-1A, RFA1-K45E | This Study |
| SKY 2998 | W303-1A, RAD53∷3xFLAG rad9Δ∷KANMX6 | [62] |
| SKY 3080 | W303-1A, RAD53∷3xFLAG-URA3 RAD9∷13Myc-KANMX6 | This Study |
| SKY 3110 | W303-1A, mec1Δ∷TRP1 sml1Δ∷HIS3 | [66] |
| SKY 3111 | W303-1A, tel1Δ∷ KANMX6 | [66] |
| SKY 3112 | W303-1A, mec1Δ∷TRP1 sml1Δ∷HIS3 tel1Δ∷ KANMX6 | [66] |
| W303-1A/X SKY 3113 | MATa/α ade2-1/ade2-1 can1-100/can1-100 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 trp1-1/trp1-1 | This Study |
| SKY 3114 | W303-1A/X, exo1Δ∷KANMX6/exo1Δ∷KANMX6 | This Study |
| SKY 3115 | W303-1A/X, sgs1Δ∷TRP1/sgs1Δ∷TRP1 | This Study |
| SKY 3116 | W303-1A/X, sgs1Δ∷TRP1/sgs1Δ∷TRP1 exo1Δ∷KANMX6/exo1Δ∷KANMX6 | This Study |
Table 2.
Plasmid list
2.2. Genetic manipulations
Complete knockouts and C-terminal epitope tagging of genomic proteins were generated by PCR-based gene replacement as described [43]. Genetic manipulations including mutations, deletions and epitope-tagging were confirmed by PCR and/or DNA sequencing. Expression of epitope-tagged proteins was confirmed by Western blotting. To reduce the influence of suppressor or enhancer mutations arising in genetically unstable backgrounds, haploids were mated to generate diploids, which were sporulated and analyzed for proper segregation of genotypes.
2.3. G1-checkpoint activation assay (αF/Noc Trap Assay)
The αF/Noc trap assay was performed as previously described [30, 34]. Briefly, overnight cultures of MATa yeast cells grown to saturation were diluted to 0.2 OD600 and then arrested in G1 using 10μM α-factor (αF) (WHWLQLKPGQPNleY) for 2.5 hr. DNA damage was induced by exposure to 300 Gy ionizing radiation in a Gammacell 220 20Co source (Nordion), after which cells were released from arrest by washing once with water. Aliquots were transferred to Trap Media (YPD supplemented with 10μM αF + 15μg ml−1 Nocodazole) at timed intervals. Time of G1 exit was scored by counting budded and non-budded cells under phase contrast to determine percentage of budded cells at each time point.
2.4. Western blot analysis to detect phosphorylated Rad53 and histone H2A
For Rad53 and histone H2A phosphorylation assays, cells were arrested in G1 with 10 μM αF for 2 h and then exposed to ionizing radiation (300 Gy). Sample aliquots are collected, treated with 0.2N NaOH for at least 5 min, resuspended in 1x SDS sample buffer (125mM Tris-HCl pH 6.8, 20% glycerol, 4% SDS, 1.45 M β-mecaptoethanol), incubated at 95°C for 5 min and centrifuged to remove cell debris. These lysates were separated on NUPAGE 3–8% TA and 4–12% Bis Tris gels (Invitrogen), transferred to nitrocellulose or PVDF membrane and probed with 9E10 mouse anti-MYC (1:500, Santa Cruz), mouse anti-FLAG M2 (1:500, Sigma), rabbit anti-yeast histone H2A phospho-Ser129 (1:1000, Millipore), or YL1/2 rat anti-tubulin (1:2000, Millipore) antibodies. Appropriate HRP conjugated secondary antibodies (1:5000, GE) were used and detected via chemiluminescence (SuperSignal West Pico, Thermo).
2.5. β-galactosidase Assays
Overnight cultures of cells transformed with an RNR3-lacZ reporter plasmid [44] were diluted and then arrested in G1 using 10 μM αF for 2 h and treated with 300 Gy ionizing radiation. After 45 min, cells were lysed via bead beating and 100 μg protein was resuspended in Z-buffer (16.1 g Na2HPO4, 5.5 g NaH2PO4, 0.75 g KCl, 0.246 g MgSO4, 2.7 ml β-mercaptoethanol, H2O to 1 liter, pH 7.0) to give 1ml assay solution. 200 μL of 4 mg ml−1 ONPG (o-nitrophenyl-β-D-galactoside) was added to initiate the reaction, which was incubated at 30°C. The time it took the samples to turn a faint yellow was noted and 500 μl of 1M Na2CO3 was added to quench the reaction prior to taking OD420 readings. β-galactosidase activity was calculated by using the equation [45]:
where OD420 is the optical density of o-nitrophenol at 420nm. 1.7 corrects for the reaction volume. 0.0045 is the optical density of 1nmole ml−1 o-nitrophenol, protein concentration is expressed as mg ml−1, extract volume is assayed in ml and time is in minutes. Thus, specific activity expressed as nmoles min−1 mg−1 of protein.
2.6. Cell Viability/DNA damage sensitivity assay
Overnight cultures of cells were diluted to 0.2 OD600 and arrested in G1 or G2/M using αF or nocodazole respectively. As diploid cells are not responsive to αF, they were arrested in G1 by starvation in media lacking a nitrogen source [46]. Cells were serially diluted ten-fold, spotted onto YPD plates and then exposed to 300 Gy IR, 50 Jm−2, or 100 Jm−2 UV. All plates were incubated at 30°C for two days prior to imaging.
2.7. Microscopy
Overnight cell cultures expressing RFA1-GFP were diluted to 0.2 OD600 and arrested in G1 using 10 μM αF for 2 h. DNA damage was induced by exposure to 300 Gy IR. Cells were then washed twice and resuspended in fresh Tris buffered saline, TBS (10 mM Tris, 150 mM NaCl). After subsequent sonication, the number of G1 cells with foci was counted using an Olympus DSU Spinning Disk Confocal microscope equipped with a back-thinned EM-CCD Hamamatsu camera at 100× and 150×.
3. Results
3.1. Sgs1 and Exo1 are required for viability of cells exposed to DNA damaging agents during their G1 and G2 phases
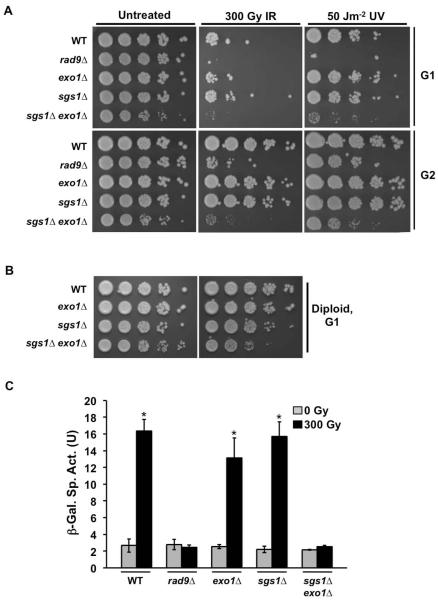
Previous work has shown that resection is required for activation of the G2/M checkpoint and initiation of homologous recombination (HR), the preferred pathway for DSB repair after completion of replication. In contrast, non-homologous end joining (NHEJ), which is independent of resection is the pathway of choice for repair in G1 [47–49]. As such, resection proteins Sgs1 and Exo1 are not expected to be important for the DDR in G1 cells. To explore this, we generated cells lacking SGS1, EXO1 or both SGS1 and EXO1 and tested their viability after exposure to DNA damage in both G1 and G2/M. Cells were arrested in G1 with αF mating pheromone or G2/M with nocodazole, then ten-fold serial dilutions plated on rich media and exposed to 300 Gy ionizing radiation (IR) or 50 Jm−2 ultraviolet radiation (UV).
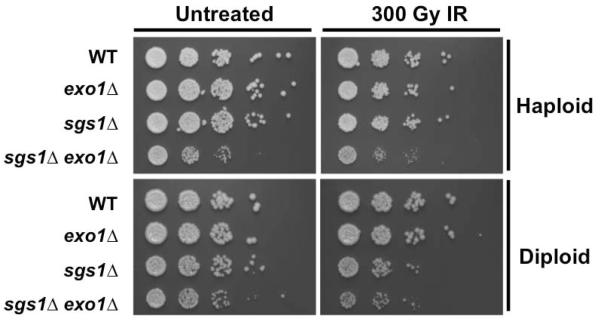
A significant difference between the G1 and G2/M phases of the cell cycle is the availability of a sister chromatid to serve as a template for recombinational repair. Consistent with expectations, we observed that wildtype (WT) G1 cells are more sensitive to IR or UV-induced damage than G2/M cells (Fig. 1A). As expected in G2/M cells, whereas the sgs1Δ and exo1Δ single mutants displayed sensitivity similar to WT, sgs1Δ exo1Δ double mutants were significantly less tolerant than WT (Fig. 1A). Surprisingly, this was also true in G1-arrested cells, indicating that both Sgs1 and Exo1 are required for fitness of cells that experienced DNA damage during G1 (Fig. 1A). To address whether the presence of a template influences this apparent significance of Sgs1 and Exo1 in G1, we generated diploid mutant cells, in which a homologous chromosome would be available for repair throughout the cell cycle. When assayed for sensitivity, WT and single mutant sgs1Δ/sgs1Δ or exo1Δ/exo1Δ diploids arrested in G1 by starvation displayed minimal sensitivity to 300 Gy IR, much like haploids arrested in G2/M. In turn, the sgs1Δ/sgs1Δ exo1Δ/exo1Δ double mutant diploids arrested in G1 displayed increased DNA damage sensitivity, but displayed greater tolerance than sgs1Δ exo1Δ haploids arrested in G2/M (Fig. 1B).
FIG. 1.
G1-arrested sgs1Δ exo1Δ mutants are sensitive to DNA damage. (A) Viability of haploid cells exposed to DNA damaging agents during G1 and G2 phases. Spot assays were performed on G1- and G2-arrested cells. Overnight cultures were diluted to 0.2 OD600 and arrested in G1 or G2 with αF (10 μM) and nocodazole (15 μg ml−1) respectively. Ten-fold serial dilutions were plated on solid media and exposed to IR (300 Gy), UV (50 Jm−2), or mock-treated. Plates were incubated at 30°C for 2 days prior to image capture. (B) Viability of diploid cells exposed to DNA damaging agents during G1 phase. Overnight cultures were diluted to 0.2 OD600 and arrested in G1 by incubation for 4 hrs in media lacking a nitrogen source. Ten-fold serial dilutions were plated on solid media and exposed to IR (300 Gy) or mock-treated. Plates were then incubated at 30°C for 2 days prior to image capture. (C) Activation of RNR3 promoter in response to DNA damage response (DDR) in G1 cells. Overnight cultures were diluted and arrested in G1 using αF. Cells were mock treated or exposed to 300 Gy IR. 45 mins after irradiation, protein samples were extracted and assayed for β-galactosidase activity by spectroscopic measurement of cleaved ONPG product. A measurement of the β-galactosidase activity is shown. Error bars represent standard deviation from three independent experiments (*, P<0.05).
This observation that Sgs1 and Exo1 are required for tolerance of G1 cells to DNA damage suggests a defect in the G1 DDR in the double mutants. To test this hypothesis, we examined DDR competency by using a RNR3 promoter lacZ reporter assay to monitor transcriptional output of the pathway [44]. Upon exposure of G1-arrested WT cells to IR, we observed a significant increase in β-galactosidase activity indicative of RNR3 promoter induction that was absent from DDR-defective rad9Δ cells. These sgs1Δ and exo1Δ single mutants displayed RNR3 activation much like WT after irradiation, while the sgs1Δ exo1Δ double mutant phenocopied rad9Δ (Fig. 1C). These results indicate that Sgs1 and Exo1 have a critical G1 function for DNA damage signal transduction.
3.2. Sgs1 and Exo1 are required for IR-induced phosphorylation of Rad53 in G1 cells but not for chromatin modification
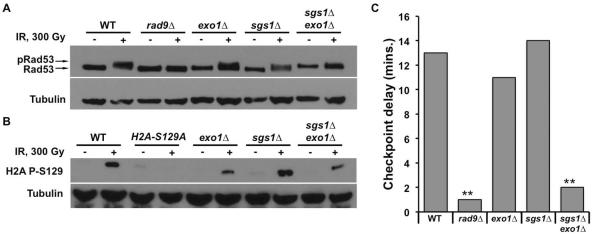
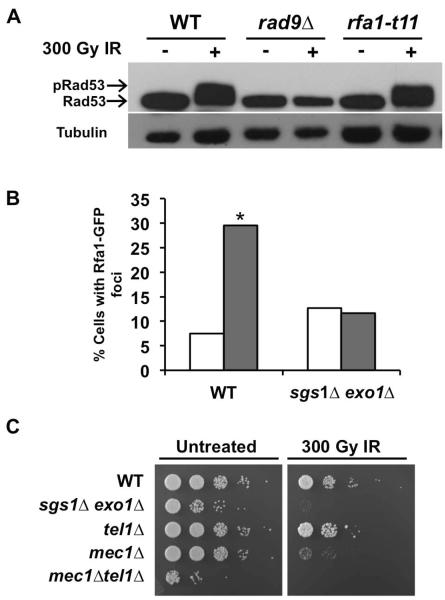
To further characterize this G1 DDR defect in sgs1Δ exo1Δ cells, we examined the activation state of Rad53 in response to DNA damage. Phosphorylation of Rad53 serves as a primary biochemical readout for checkpoint activation and occurs upstream of transcriptional induction in the DDR pathway. On SDS-polyacrylamide gels, phosphorylated Rad53 displays slower migration, producing a subtle mobility shift detectable by Western blotting. Upon irradiation of αF-arrested G1 cells, a Rad53 phosphorylation mobility shift was observed in WT, exo1Δ, and sgs1Δ cells (Fig. 2A). Consistent with the lack of RNR3 promoter activation, neither the rad9Δ mutant nor the sgs1Δ exo1Δ double mutant demonstrated an appreciable Rad53 mobility shift upon irradiation (Fig. 2A).
FIG. 2.
Sgs1 and Exo1 are required for activation of the G1 checkpoint in response to ionizing radiation. (A) IR-induced phosphorylation of Rad53. Cells expressing myc-tagged Rad53 were arrested in αF for 2 h. Aliquots were taken prior to and 15 min after mock or IR-treatment, and then prepared for Western blot via NaOH treatment. Rad53 was detected with anti-myc antibody while anti-tubulin antibody was used as a loading control. (B) Phosphorylation of H2A in response to irradiation of G1 cells. Overnight cultures were arrested in αF and the cells mock or IR-treated. Samples were prepared for Western blot using NaOH and H2A pS129 was detected using the yeast specific phosphoH2A antibody. Specificity for the antibody was assessed using a nonphosphorylable H2A-S129A mutant. Anti-tubulin antibody was used as a loading control (C) αF/Noc trap assay for G1 cell cycle arrest. Cells were arrested for 2 h in G1 using αF. They were mock or IR-treated and then released from αF arrest by washing in rich media. G1 exit was assayed by monitoring the percentage of budded cells under microscope. Time of G1 exit was taken as the time it took 10% of the unbudded cells to bud. Data is represented as the delay in time to bud upon irradiation. (**, P<0.05, n=3)
Phosphorylation of H2A at S129 (γH2A) adjacent to the DSB is an early event in the checkpoint activation cascade. To determine a role for Sgs1 and/or Exo1 in DSB-induced chromatin modification, we assayed for IR-induced formation of γH2A. G1-arrested cells were irradiated and tested for phosphorylation of H2A at the S129 residue via Western blot. Despite the failure of sgs1Δ exo1Δ cells to promote Rad53 phosphorylation in G1, H2A S129 phosphorylation was readily detected upon irradiation (Fig. 2B), suggesting that Sgs1 and Exo1 are not required for γH2A formation in G1. Nonetheless, H2A phosphorylation appeared attenuated in both the single exo1Δ and double sgs1Δ exo1Δ mutants.
3.3. Sgs1 and Exo1 are required for activation of the G1 cell cycle checkpoint in response to IR
To examine the functional roles of Sgs1 and Exo1 in activation of G1 DNA damage checkpoint response, we tested whether Sgs1 and Exo1 are necessary for damage-induced cell cycle arrest in G1. G1-arrested cells were irradiated to induce DNA damage, and the cells were monitored for IR-induced delay in G1 exit. This was accomplished by removing aliquots of cells at intervals after irradiation and incubating them with αF mating pheromone and nocodazole, thereby arresting cells that remained in G1 as unbudded cells bearing mating projections, while any cells that had progressed to S phase accumulated as large budded cells [7, 34]. Thus, the time-course of G1 exit could be determined based on the percentage of budded cells at each time point. A persistent population of αF sensitive cells after irradiation indicated the slower G1 exit due to a DNA damage checkpoint delay. To enable proper interpretation of the results, we ensured that at time 0, unirradiated and irradiated samples had similar percentages of G1-arrested cells (Fig. S1). We observed that upon 300 Gy irradiation of G1 cells, WT, exo1Δ and sgs1Δ single mutants exited G1 only after a delay of 10 min or more compared to unirradiated cells (Fig. 2C). This response was absent in checkpoint-defective rad9Δ cells as expected, but sgs1Δ exo1Δ double mutants also exhibited little or no delay in G1 exit after irradiation (Fig. 2C).
3.4. The helicase function of Sgs1 is required for G1 checkpoint activation
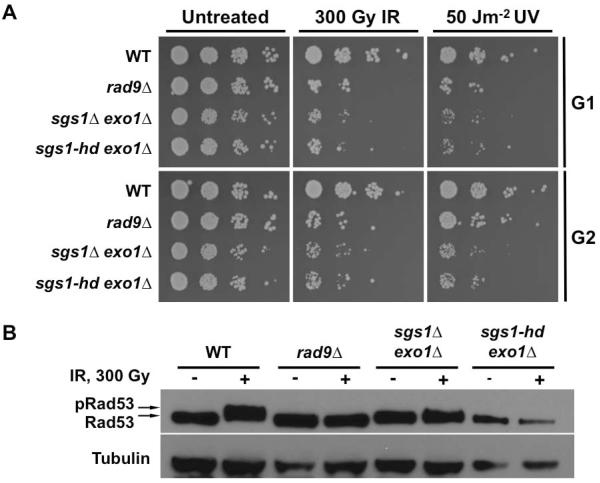
Sgs1 has both scaffolding and enzymatic functions that are important for checkpoint activation in G2/M. Sgs1 has been shown to interact with RPA to promote checkpoint in the S phase while its helicase activity is required for ssDNA generation at the damage site in G2 [50, 51]. To determine whether the helicase activity of Sgs1 is important for the response of G1 cells to DNA damage, a helicase defective Sgs1 mutant, sgs1-hd, was introduced into the sgs1Δ exo1Δ double mutant to generate sgs1-hd exo1Δ cells. First we assessed the viability of these cells after exposure to DNA damaging agents. We found that the sgs1-hd exo1Δ cells phenocopied the sgs1Δ exo1Δ mutant, as reflected in their increased sensitivity to IR and UV (Fig. 3A). We also assayed the sgs1-hd exo1Δ cells for IR-induced Rad53 phosphorylation by monitoring its migration patterns on Western blots. No appreciable Rad53 mobility shift was observed in the sgs1-hd exo1Δ mutants, much like the sgs1Δ exo1Δ cells (Fig. 3B). These results indicate that the helicase activity, rather than scaffolding function of Sgs1, mediates the response of G1 cells to DNA damage.
FIG. 3.
Catalytic function of Sgs1 is required for its role in G1 checkpoint. (A) Viability assay for DNA damaged G1 and G2 cells. Yeast cell cultures arrested with αF or nocodazole were treated with IR or UV. Ten-fold serial dilutions were spotted on solid media and then exposed to irradiation. All plates were incubated at 30°C for 2 days prior to image capture. (B) DNA damage-induced Rad53 phosphorylation. Cells expressing myc-tagged Rad53 and helicase-defective Sgs1 (sgs1-hd) in exo1Δ sgs1Δ background were arrested with αF and exposed to IR. Aliquots taken prior to and 15 min after irradiation were prepared for Western blot using NaOH. Rad53-myc was detected with anti-myc antibody while anti-tubulin antibody was used as a loading control. A mobility shift in the Rad53-myc band indicates phosphorylation.
3.5. Rad9-independent localization of Rad53 to the DNA damage site is insufficient to restore G1 checkpoint in cells lacking Sgs1 and Exo1
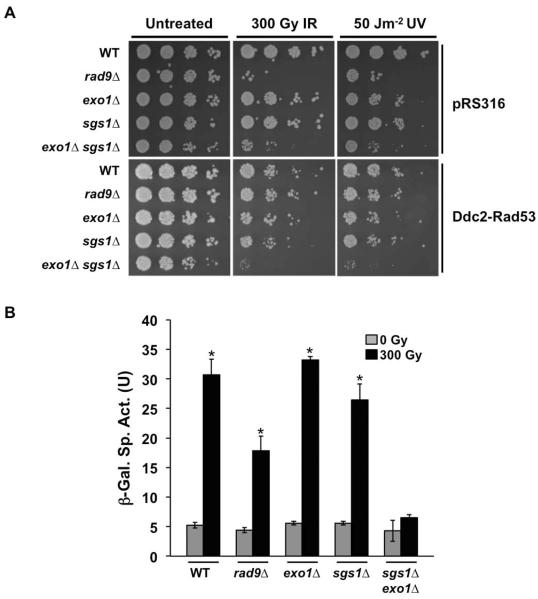
By recruiting Rad53 to the damage site, Rad9 plays a central role of linking damage sensors and signal transducers with the effectors of the activation cascade. This key function of Rad9 is not limited to G2/M but has been shown to be essential for activation of the DDR in G1 [7, 30]. Rad9 is recruited to the DSB site, via its interactions with γH2A and other chromatin modifications, where it is phosphorylated by Mec1 that has been recruited via its Ddc2 partner to ssDNA coated with RPA. Subsequently the phosphorylated Rad9 recruits Rad53 for activation by Mec1 [30, 52, 53]. Significantly, a Ddc2-Rad53 fusion protein has been shown to bypass the requirement for Rad9 in the G2/M checkpoint by making Rad53 recruitment and activation solely dependent on 5' end resection to form ssDNA coated by RPA [54]. Significantly, expression of the Ddc2-Rad53 fusion also restored DNA damage tolerance to rad9Δ cells exposed to IR or UV in G1 (Fig. 4A), indicating the accumulation of functionally significant levels of ssDNA. In turn, Ddc2-Rad53 failed to suppress the DNA damage sensitivity of sgs1Δ exo1Δ mutants exposed to IR or UV in G1. This result is consistent with a requirement for Sgs1 and Exo1 in 5' end resection in G1.
FIG. 4.
Ddc2-Rad53 fusion protein does not rescue sgs1Δ exo1Δ double mutants from DDR defect. (A) Viability assay for G1 cells expressing Ddc2-Rad53. Cells transformed with Ddc2-Rad53 or empty vector (pRS316) were arrested in αF then mock-treated or exposed to DNA damaging agents (IR or UV). Ten-fold serial dilutions were spotted on solid media and then exposed to irradiation. All plates were incubated at 30°C for 2 days. (B) IR-induced activation of RNR3 promoter transcription in G1 cells expressing Ddc2-Rad53. Overnight cultures were diluted and then arrested in G1 using αF. Cells were Mock treated or exposed to 300 Gy IR. 45 mins after irradiation, protein samples were extracted and β-galactosidase activity measured by spectroscopy. A measurement of the β-galactosidase activity is shown. Error bars represent standard deviation from three independent experiments (*, P<0.05).
To confirm that Sgs1 and Exo1 function upstream of Rad53 in the DDR cascade, we assayed for damage-induced RNR3 transcription in G1 cells expressing the Ddc2-Rad53 construct. Upon exposure to IR, β-galactosidase activity was partly restored in DDR-defective rad9Δ cells, although to a level lower than observed in WT, exo1Δ and sgs1Δ cells. In contrast to rad9Δ cells, expression of Ddc2-Rad53 did not rescue RNR3 promoter activation in sgs1Δ exo1Δ cells, consistent with a lack of ssDNA sufficient to activate the checkpoint cascade (Fig. 4B). Altogether, these results indicate that Sgs1 and Exo1 function in parallel to the chromatin-Rad9 branch of the cascade but upstream of the resection-dependent Ddc2-Mec1 pathway.
3.6. Cells lacking Sgs1 and Exo1 show sensitivity to IR similar to mec1Δ mutants and do not localize Rfa1-GFP to DNA damage induced foci
The single stranded DNA binding complex RPA binds to 3' ssDNA overhangs generated by DNA resection at the DSB site and Mec1 kinase is recruited to the damage site by Ddc2 [21], mediated by its interaction with the RPA subunit Rfa1 [55]. It is conceivable that Sgs1 and Exo1 regulate the interaction between Ddc2 and RPA and hence the Ddc2-Rad53 fusion protein could not restore DDR defects in sgs1Δ exo1Δ cells. First we tested whether Rad53 activation in G1 is dependent on Rfa1-Ddc2 interaction by using rfa1-t11, a mutant allele described to have impaired binding to Ddc2 [55]. In G1-arrested rfa1-t11 cells, Rad53 could still be phosphorylated upon exposure of the cells to IR (Fig. 5A), similar to what had been observed in G2/M-arrested cells [56–58]. Even if the Rfa1-Ddc2 interaction may not be compromised in the sgs1Δ exo1Δ double mutant, we looked further upstream to test whether Sgs1 and Exo1 regulate RPA recruitment to the site. WT and sgs1Δ exo1Δ cells expressing Rfa1-GFP were arrested in G1 and then exposed to IR. As previously described [38], WT cells displayed a significant increase in foci formation when irradiated. However, sgs1Δ exo1Δ cells showed no significant increase in foci formation upon irradiation (Fig. 5B). These results suggest that the G1 DDR defects in sgs1Δ exo1Δ are due to a lack of RPA recruitment, which is mediated by the presence of ssDNA.
FIG. 5.
Sgs1 and Exo1 appear to interact with factors along the resection-dependent Mec1 activation pathway. (A) DNA damage-induced Rad53 phosphorylation. Cells expressing FLAG-tagged Rad53 and rfa1-t11 were used. Diluted overnight cultures were arrested with αF and exposed to IR. Aliquots taken prior to and 15 min after irradiation were prepared for Western blot using NaOH. Rad53-myc was detected with anti-FLAG antibody while anti-tubulin antibody was used as a loading control. A mobility shift in the Rad53-FLAG band indicates phosphorylation. (B) Rfa1-GFP foci formation in response to IR. WT and sgs1Δ exo1Δ cells were transformed with Rfa1-GFP plasmids. αF-arrested cultures were mock or IR-treated then resuspended in TBS. Number of G1 cells with foci was scored using confocal fluorescence microscopy. 100 cells were counted for each cell type and treatment (*, P< 0.05). White bars-0 Gy; Grey bars-300 Gy. (C) IR sensitivity of G1-arrested Mec1/Tel1 mutants relative to G1-arrested sgs1Δ exo1 cells. Overnight cultures of the indicated strains were diluted to 0.2 OD600 and arrested in G1 using αF (10 μM). Ten-fold serial dilutions were plated on solid media (YPD) and then exposed to IR (300 Gy) or mock-treated. Plates were incubated at 30°C for 2 days prior to image capture.
DNA resection generates ssDNA, which serves as a landing dock for Mec1 at the DSB foci [21]. With Sgs1 and Exo1 being required for generation of ssDNA, it would be expected that sgs1Δ exo1Δ mutants may behave similarly to mec1Δ in response to DNA damage. We tested this by comparing the sensitivity of these mutants to irradiation in G1. As anticipated, sgs1Δ exo1Δ double mutants displayed similar sensitivity to the mec1Δ mutants (Fig. 5C).
3.7. Prolonged G1 delay restores DNA damage tolerance to sgs1Δ exo1Δ mutants
The increase in sensitivity to IR-induced DNA damage observed in cells lacking Sgs1 and Exo1 may derive from lack of proper DDR in G1, leading to defects in G1 checkpoint and repair. Alternatively, this sensitivity might arise from DNA damage checkpoint or repair defects in the subsequent S phase or mitosis. To differentiate among potential mechanisms, WT, sgs1Δ, exo1Δ, and sgs1Δ exo1Δ cells were arrested in G1 with αF, irradiated and then maintained in G1 with αF for four hours. The prolonged G1 delay largely restored DNA damage tolerance to the sgs1Δ exo1Δ mutants (Fig. 6). The same experiment was performed in diploid cells using nitrogen starvation to arrest the cells in G1 and then to block cell cycle progression after irradiation. With the exception of sgs1Δ/sgs1Δ cells, which appeared to show a slight increase in sensitivity, WT, exo1Δ/exo1Δ, and sgs1Δ/sgs1Δ exo1Δ/exo1Δ cells displayed similar sensitivity (Fig. 6).
FIG. 6.
Prolonged G1 arrest restores DNA damage tolerance to cells lacking Exo1 and Sgs1. Overnight cultures of haploid cells were diluted to 0.2 OD600 and arrested in G1 with αF (10 μM). Cells were exposed to 300 Gy IR or mock-treated and then held in αF (10 μM) for 4 additional hours. Ten-fold serial dilutions were plated on solid media and were incubated at 30°C for 2 days prior to image capture. For diploid cells, overnight cultures were diluted to 0.2 OD600 and arrested in G1 by incubation for 4 hours in media lacking a nitrogen source. After exposure to either 300 Gy IR or mock-treatment, cells were held in G1 for a further 4hrs. At this point, cells were plated in ten-fold serial dilutions on solid media. Plates were incubated at 30°C for 2 days prior to image capture.
4. Discussion
Our current view of activation of DNA damage responses in budding yeast involves a critical role throughout the cell cycle for Mec1 phosphorylation of the effector kinase Rad53, which activates downstream pathways including DNA damage checkpoints [8, 59]. Previously examined during G2/M checkpoint response, Mec1 recruitment to DNA double strand breaks requires generation of ssDNA by regulated 5' end resection. Cells lacking both Sgs1 helicase and Exo1 nuclease display a defect in end resection, conferring a G2/M checkpoint deficiency [14, 15, 17]. A second factor positively regulating resection is the cyclin dependent kinase Cdc28. Cdc28 activity is high in G2/M and its inhibition both prevents 5' end resection and impairs DNA damage signaling [36, 60]. Notably, Cdc28 activity is absent from cells arrested in G1, providing a simple mechanism for the observed low levels of resection [36]. Indeed, 5' end resection is commonly considered dispensable for DNA damage signaling in G1. Nonetheless, that the response to DNA damage in G1 is dependent on Mec1 and characterized by Rad53 phosphorylation and a transient cell cycle delay creates a paradox. To explore this further, we examined the checkpoint defect in sgs1Δ exo1Δ cells in G1 and G2/M. Surprisingly, G1-arrested sgs1Δ exo1Δ cells were sensitive to DNA damage and defective in checkpoint activation. Holding sgs1Δ exo1Δ mutants in G1 after irradiation partly restored DNA damage tolerance, thereby placing the activity of Sgs1 and Exo1 in G1 and in DNA damage checkpoint signaling rather than DNA repair. We also found that the helicase activity of Sgs1, which is required for its role in resection, was vital to the function of Sgs1 in mediating G1 Rad53 activation. These findings indicate that 5' end resection is required for checkpoint activation in G1 cells, despite the absence of Cdc28 activation. Interestingly, contrary to our findings, prior work by Giannattasio et al. described a G1 DNA damage checkpoint deficiency in exo1Δ mutants treated with UV [61]. An explanation may lie in a requirement for DNA damage processing by excision repair to initiate DNA damage signaling after UV irradiation in G1, as previously shown [34].
Recent studies of G1 DNA damage checkpoint response have focused on Rad9 recruitment and activation, giving formation of γH2A and regulation of other chromatin modifications a critical role [26, 27, 30, 62]. With the unexpected finding that sgs1Δ exo1Δ cells are G1 checkpoint deficient, it was also striking that the formation of γH2A after DNA damage is maintained. Also suggesting that chromatin modification may be necessary but not sufficient, we observed that a Ddc2-Rad53 fusion protein can bypass the G1 DNA damage checkpoint defect in rad9Δ mutants but cannot rescue sgs1Δ exo1Δ cells. These results argue that Sgs1 and Exo1 function in parallel to mediate the formation of ssDNA in G1 and that their activity is critical for G1 checkpoint response, likely via recruitment of Mec1 to phosphorylate Rad53, much like in G2/M. This is corroborated in part by our finding that sgs1Δ exo1Δ and mec1Δ mutants share similar sensitivity to irradiation in G1.
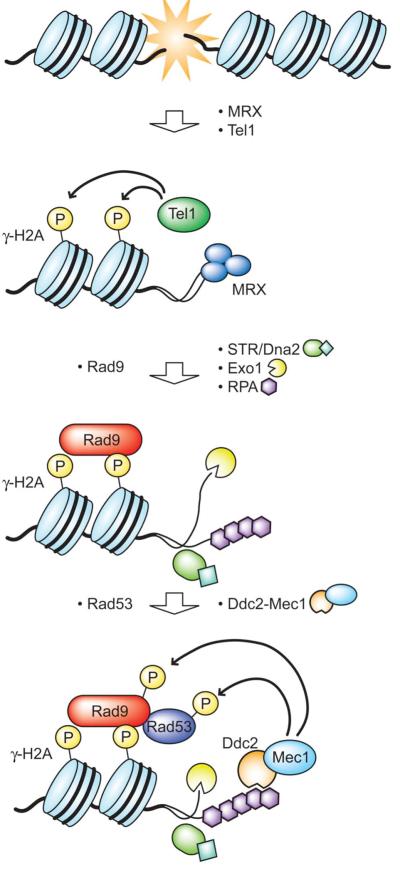
Results from this study, together with our previous findings that chromatin modification is necessary for G1 checkpoint [27, 30, 34, 62], have led us to a revised model for G1 checkpoint activation that likens it to the G2/M checkpoint cascade (Fig. 7). Tel1 is recruited by MRX to the DSB site, where it phosphorylates H2A to generate γH2A. This mediates localization of Rad9, which subsequently recruits Rad53 to the site. Activation of Rad53 is dependent on Mec1, which is recruited in parallel. At the DNA damage site, MRX equally promotes resection mediated by Exo1 and the Dna2/STR (Sgs1-Top3-Rmi1) complex. The short stretch of ssDNA generated is coated with RPA, which interacts with Ddc2-Mec1 to mediate its localization to the site. Once localized, Mec1 phosphorylates Rad53 leading to activation of the G1 checkpoint [19–21].
FIG. 7.
A model suggesting that the G1 checkpoint is activated in a similar manner to G2. In response to DSB, Sgs1 of the STR (Sgs1-Top3-Rmi1) complex and Exo1 are required for localization of RPA to the DSB site. In turn, RPA mediates Ddc2-Mec1 recruitment leading to phosphorylation of Rad53 by Mec1.
In response to DSB in G1, cells initially attempt to repair the damage by NHEJ, which although less accurate than HR, occurs quickly enough to negate the requirement for a cell cycle delay [48]. However, in the event of significant unresolved damage, the G1 checkpoint is activated via phosphorylation of Rad53. Our model suggests that activation of the G1 checkpoint is surprisingly similar to G2/M activation, and requires the resection proteins Sgs1 and Exo1. This raises the question of why cells might require resection for response to DNA damage in G1. Beyond a role in signaling, we infer that DNA resection is unlikely to enhance damage repair in haploid cells in G1. Perhaps, a short resected overhang serves to tag damaged DNA sites to be recognized and repaired in the subsequent S phase, where formation of sister chromatids and activation of Cdc28 enable repair by HR.
It is noteworthy that a high dose of ionizing radiation is needed to activate even a short G1 checkpoint delay. Even under conditions where >90% of cells are lethally irradiated based on loss of colony formation, nearly all the cells still successfully progress into S phase [63]. We envision progression into the S phase with damaged DNA as a means for cells to access improved mechanisms for DNA damage repair. This puts the G1 checkpoint in sharp contrast to G2/M, where progressing through a subsequent anaphase with persistent DNA damage might be lethal. Toward avoiding the dire consequence of aneuploidy, the G2/M checkpoint robustly delays progression to allow repair. This contrast is reflected by the relative extent of resection in G1 versus G2/M [9, 36], where the former may be sufficient for checkpoint signaling and the latter required for repair.
When one or a few DNA double strand breaks occur in G1, DNA damage detection by MRX may be sufficient to recruit Tel1 to induce γH2A but the lesions may rapidly resolve via NHEJ [24, 49, 64]. With our new understanding, we consider the activation of Rad53 in G1 as a reporter for DNA resection as might occur upon incomplete DNA damage repair, initiating a signal that persistent damage may be passed along into S phase. In turn, successful NHEJ repair effectively terminates propagation of the damage response prior to onset of 5' end resection, preventing Mec1 recruitment and Rad53 phosphorylation. Indeed, we observed accumulation of γH2A without Rad53 activation even after lethal G1 DNA damage in sgs1Δ exo1Δ mutants, nicely separating the chromatin modification and DNA resection arms of the pathway. Our data suggest a temporal and functional distinction between these two mechanisms, potentially indicating a similar separation of roles throughout the cell cycle where chromatin and ssDNA signaling may collaborate to promote an effective and appropriate DNA damage response.
Supplementary Material
Highlights
Yeast 5' end resection factors Sgs1 and Exo1 mediate G1 DNA damage tolerance.
G1 activation of Rad53 after irradiation depends on 5' end resection.
G1 DNA damage checkpoint requires both resection and chromatin modification.
Yeast G1 and G2/M DNA damage checkpoints are surprisingly similar.
ACKNOWLEDGMENTS
The authors thank Drs. K. Kristjansdottir and S. Takahashi for insightful comments in preparing the manuscript, with particular appreciation to Dr. S. Takahashi for artistic contributions. We are grateful to Drs. K. Belanger, S. Brill, S. Elledge, J. Igual and D. Stern for generously providing materials used in this study.
This work was supported by NIH grant GM60443 and The Ludwig Center for Metastasis Research (SJK), The Foglia Foundation and Mr. and Mrs. Vincent Foglia (AWT, SJK) and University of Chicago MSTP/BSD (FOB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT The authors declare none.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 3.Bennett CB, et al. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(12):5613–7. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274(5293):1664–72. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 6.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–35. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 7.Siede W, Friedberg AS, Friedberg EC. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(17):7985–9. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241(4863):317–22. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 9.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27(13):1875–85. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82(5):841–7. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 11.Finn K, Lowndes NF, Grenon M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cellular and molecular life sciences : CMLS. 2012;69(9):1447–73. doi: 10.1007/s00018-011-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupnik A, Lowndes NF, Grenon M. MRN and the race to the break. Chromosoma. 119(2):115–35. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 13.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4(9):844–9. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–4. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Z, et al. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 17(12):1478–85. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravel S, et al. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–72. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majka J, et al. Replication protein A directs loading of the DNA damage checkpoint clamp to 5'-DNA junctions. J Biol Chem. 2006;281(38):27855–61. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 19.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100(24):13827–32. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24(6):891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubrana K, et al. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007;120(Pt 23):4209–20. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- 22.Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17(16):1957–62. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukunaga K, et al. Activation of protein kinase Tel1 through recognition of protein-bound DNA ends. Mol Cell Biol. 31(10):1959–71. doi: 10.1128/MCB.05157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408(6815):1001–4. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 25.Foster ER, Downs JA. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 2005;272(13):3231–40. doi: 10.1111/j.1742-4658.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 26.Javaheri A, et al. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci U S A. 2006;103(37):13771–6. doi: 10.1073/pnas.0511192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downs JA, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16(6):979–90. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14(19):1703–11. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humpal SE, Robinson DA, Krebs JE. Marks to stop the clock: histone modifications and checkpoint regulation in the DNA damage response. Biochem Cell Biol. 2009;87(1):243–53. doi: 10.1139/O08-109. [DOI] [PubMed] [Google Scholar]
- 30.Wysocki R, et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Molecular and cellular biology. 2005;25(19):8430–43. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Molecular cell. 1998;2(2):183–9. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz MF, et al. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell. 2002;9(5):1055–65. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- 33.Sweeney FD, et al. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol. 2005;15(15):1364–75. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 34.Gerald JN, Benjamin JM, Kron SJ. Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. Journal of cell science. 2002;115(Pt 8):1749–57. doi: 10.1242/jcs.115.8.1749. [DOI] [PubMed] [Google Scholar]
- 35.Siede W, et al. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics. 1994;138(2):271–81. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–7. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clerici M, et al. A Tel1/MRX-dependent checkpoint inhibits the metaphase-toanaphase transition after UV irradiation in the absence of Mec1. Mol Cell Biol. 2004;24(23):10126–44. doi: 10.1128/MCB.24.23.10126-10144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30(1):73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siede W, et al. The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene product, is required for G1 arrest following radiation treatment. Journal of bacteriology. 1996;178(19):5841–3. doi: 10.1128/jb.178.19.5841-5843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gietz D, et al. Improved method for high efficiency transformation of intact yeast cells. Nucleic acids research. 1992;20(6):1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gietz RD, et al. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11(4):355–60. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 42.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Current genetics. 1989;16(5–6):339–46. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 43.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Z, Elledge SJ. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics. 1992;131(4):851–66. doi: 10.1093/genetics/131.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amberg DC, et al. Methods in yeast genetics : a Cold Spring Harbor Laboratory course manual. ed2005 xvii. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2005. p. 230. [Google Scholar]
- 46.Gallego C, et al. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. The EMBO journal. 1997;16(23):7196–206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Frontiers in bioscience : a journal and virtual library. 2006;11:1958–76. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 48.Daley JM, et al. Nonhomologous end joining in yeast. Annual review of genetics. 2005;39:431–51. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 49.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Molecular and cellular biology. 1996;16(5):2164–73. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J, et al. Human homologues of yeast helicase. Nature. 1996;383(6602):678–9. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 51.Bjergbaek L, et al. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. The EMBO journal. 2005;24(2):405–17. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121(7):973–6. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura TM, et al. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Molecular and cellular biology. 2004;24(14):6215–30. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee SJ, Duong JK, Stern DF. A Ddc2-Rad53 fusion protein can bypass the requirements for RAD9 and MRC1 in Rad53 activation. Molecular biology of the cell. 2004;15(12):5443–55. doi: 10.1091/mbc.E04-07-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 56.Lee SE, et al. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Current biology : CB. 2001;11(13):1053–7. doi: 10.1016/s0960-9822(01)00296-2. [DOI] [PubMed] [Google Scholar]
- 57.Nnakwe CC. Chromatin dynamics in the DNA damage response. The University of Chicago; United States -- Illinois: 2009. p. 190. [Google Scholar]
- 58.Pellicioli A, et al. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Molecular cell. 2001;7(2):293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 59.Sanchez Y, et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271(5247):357–60. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 60.Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62(4):1191–243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannattasio M, et al. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Molecular cell. 2010;40(1):50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Nnakwe CC, et al. Dissection of Rad9 BRCT domain function in the mitotic checkpoint response to telomere uncapping. DNA repair. 2009;8(12):1452–61. doi: 10.1016/j.dnarep.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wysocki R, et al. CDK Pho85 targets CDK inhibitor Sic1 to relieve yeast G1 checkpoint arrest after DNA damage. Nature structural & molecular biology. 2006;13(10):908–14. doi: 10.1038/nsmb1139. [DOI] [PubMed] [Google Scholar]
- 64.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Molecular cell. 2002;10(5):1189–99. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 65.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56(4):619–30. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 66.Soriano-Carot M, Bano MC, Igual JC. The yeast mitogen-activated protein kinase Slt2 is involved in the cellular response to genotoxic stress. Cell division. 2012;7:1. doi: 10.1186/1747-1028-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belanger KD, et al. The karyopherin Kap95 and the C-termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae. DNA and cell biology. 2011;30(9):641–51. doi: 10.1089/dna.2010.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.