Abstract
The equilibrium unfolding process of ferric horse heart cytochrome c (cyt c), induced by guanidinium hydrochloride (GdHCl), was studied using UV-vis absorption spectroscopy, resonance Raman spectroscopy and vibrational coherence spectroscopy (VCS). The unfolding process was successfully fit using a three-state model35 which included the fully folded (N) and unfolded (U) states, along with an intermediate (I) assigned to a Lys bound heme. The VCS spectra revealed for the first time several low frequency heme modes that are sensitive to cytochrome c unfolding: γa (~50 cm−1), γb (~80cm−1), γc (~100cm−1), and vs(His-Fe-His) at 205 cm−1. These out-of-plane modes have potential functional relevance and are activated by protein-induced heme distortions. The free energies for the N-I and the I-U transitions at pH 7.0 and 20°C were found to be 4.6 kcal/M and 11.6 kcal/M, respectively. Imidazole was also introduced to replace the methionine ligand so the unfolding can be modeled as a two-state system. The intensity of the mode γb~80 cm−1 remains nearly constant during the unfolding process, while the amplitudes of the other low frequency modes track with spectral changes observed at higher frequency. This confirms that the heme deformation changes are coupled to the protein tertiary structural changes that take place upon unfolding. These studies also reveal that damping of the coherent oscillations depends sensitively on the coupling between heme and the surrounding water solvent.
Keywords: Heme ruffling, vibrational coherence spectroscopy, protein folding, vibrational damping, cytochrome c
Introduction
Cytochrome c (cyt c) is a small, but very important heme protein, that shuttles electrons between cytochrome c reductase and cytochrome c oxidase in the mitochondrial membrane in order to facilitate respiration.1,2 It has also recently been discovered that cyt c plays an important role in the cellular apoptosis process. In the early stages of apoptosis, cyt c acts as a catalyst for peroxidation of cardiolipin. Cardiolipin peroxidation leads to permeabilization of the outer mitochondrial membrane and releases cyt c and other proapoptotic proteins into the cytoplasm, which initiates the formation of the apoptosome and the caspase cascade that leads to cell death.3,4
The heme group (Fe-protoporphyrin IX) is the functional center of cyt c. The heme iron is axially coordinated to His18 (proximal ligand) and Met80 (distal ligand) in its native solution state. The porphyrin ring is also covalently anchored to the protein by two thioether linkages with Cys 14 and Cys 17 forming a CXXCH motif that is a unique feature of all c-type hemes5. In cyt c, the residues between the two Cys are alanine and glutamine at positions 15 and 16, and H represents the histidine ligand at position 18.
Due to its stable protein structure, cyt c is an ideal model and is of great interest for protein folding/unfolding studies where various dynamic intermediates have been characterized.6–10 The Soret band excited resonance Raman (RR) spectra are known to be highly sensitive to the heme configuration11,12 and the high frequency “marker bands” (ν4, ν3, ν2, and ν10) have been used to characterize the heme ligation status of various unfolding intermediates.13–17 The low frequency RR spectra (200cm−1 – 800cm−1) contain many out-of-plane (OOP) modes, which would not be Raman active in the ideal D4h symmetry point group of the heme core. Many of the low-frequency modes observed in the RR spectrum of native cyt c arise from the highly ruffled and distorted structure of the heme that is imposed by the architecture of the folded protein.18 Despite numerous RR studies of cyt c unfolding, there appears to be no systematic study that has focused on how the low frequency heme vibrational modes respond to unfolding near the physiologically relevant neutral pH. In this sense, the low-frequency heme Raman modes are useful “reporters” of the protein fold and the heme distortions induced by the folding process.
In this work, we use vibrational coherence spectroscopy (VCS) and RR spectroscopy to systematically study changes in the low frequency vibrations of cyt c arising from guanidinium hydrochloride (GdHCl) induced unfolding at pH 7.0. The low frequency modes we observe below 200 cm−1 using VCS can have significant thermal populations and their interaction with the thermal bath and the surrounding amino acids can be important to the electron transport reaction mechanism. Studies of these low-frequency modes can improve our understanding of how dynamic structures relate to protein function. Generally, functionally important heme modes, such as “doming” and “ruffling”, which lie in the low frequency region below 200 cm−1, are delocalized and involve many nuclei, possibly even mixing with other modes in the surrounding protein material. Resonance Raman and infrared spectroscopies cannot reliably detect heme modes below ~150 cm−1 in the aqueous phase, due to the strong absorbance, Rayleigh and quasielastic scattering19 of water. On the other hand, the self-heterodyne nature of the VCS experiment allows it to extract the low frequency vibrational modulations of the third order polarization of the heme. We have previously investigated the low-frequency modes of a variety of heme proteins, using Soret band excitation.20–29 Unlike the higher frequency modes (> 200cm− 1), the low frequency modes (which have weaker force constants) are more easily distorted from equilibrium by the protein surroundings. These modes are activated in VCS when the protein induces symmetry breaking nonplanar heme distortions24. In addition, these modes take on a special functional significance because of their thermal accessibility. The low-frequency coherence spectra offer a unique window into how the surrounding protein environment can alter these important thermally active heme modes.
In this paper we use UV-vis spectroscopy and a previously proposed procedure30 to observe an intermediate state of unfolded cyt c in the range of 1.5 – 3 M GdHCl. Further investigations reveal that the intermediate state has little effect on the higher frequency RR spectra. However, we observe that the low frequency VCS spectra changes dramatically upon GdHCl induced equilibrium unfolding. We also investigate unfolding in the imidazole-cyt c complex, which bypasses the intermediate ligation states found in the native system. Both the axial ligands and the Soret band peak position of this complex remain fixed upon unfolding by GdHCl. In this well-controlled two-state system, the RR and VCS spectra are not affected by changing resonance conditions due to shifts in the Soret band that arise from intermediate ligation states. This allows normalization of the time domain data and helps to quantify the relative amplitudes of the coherent oscillations between the folded and unfolded states. The VCS spectra reflect a significant structural change of the heme chromophore as the protein is unfolded. This demonstrates that the protein architecture can apply forces that are large enough to modify the heme structure, at least along these lower frequency “soft” modes. Insofar as these low frequency modes are thermally accessible, and potentially involved in the activation of transition or tunneling states, it suggests how the ubiquitous heme group can have its reactivity “tuned” by different protein structures to perform such a wide ranging set of functions.5,31–34,24,35–38
Experimental Section
Sample Preparation
Horse heart cytochrome c (cyt c) and GdHCl (molecular biology grade) were purchased from Sigma-Aldrich Co. and were used without further purification. The buffer containing GdHCl was prepared using 0.05M potassium phosphate at pH 7.0±0.05. The concentration of GdHCl was determined by its refractive index.39 The imidazole-cyt c complex was prepared by adding small volume aliquots of 10 M imidazole stock solution to the total solution volume to obtain the desired concentration. Samples were freshly prepared before the spectroscopy experiments and were equilibrated for at least 30 minutes before any measurement. The absorption spectra were recorded (U-4100, Hitachi) after the preparation procedure to ensure that all chemical modifications were achieved. For VCS experiments the final concentration of protein samples was adjusted to O.D. = 1±0.05 in a 1 mm path length quartz sample cell at the selected excitation wavelength. The absorption spectra were also taken following the vibrational spectroscopy experiments to confirm the integrity of the samples during the laser exposure. All experiments were performed at room temperature.
Optical Systems
The femtosecond vibrational coherence spectroscopy system has been described in details elsewhere.40,26 Briefly, a laser pulse at 412 nm was generated and split into two arms: pump and probe. The pump arm was modulated by an acoustic-optical modulator controlled by a lock-in amplifier at 1.5 MHz. A translation stage on the probe arm controlled the delay between the pump and probe pulses. The polarization of the pump and probe beams were adjusted to be perpendicular to each other, allowing for both polarization and spatial filtering. The full width at half maximum of the pump and probe pulses was ~70 fs at the sample position. Both beams were focused into a spinning sample cell using a three inch lens in a near parallel geometry. After the sample, the beams were re-collimated and the pump light was spatially blocked and further extinguished by a polarization analyzer, so that only the probe beam, modulated by the pump, was detected.
Two detection schemes, “open band” and “detuned”, are used to obtain the VCS spectra. In the open band scheme 20,40,41,26 we employ a Si photodiode to collect the entire spectral bandwidth of the probe pulse, whereas in the detuned scheme, a monochromator is used to select a specific spectral range within the probe bandwidth (a pass band of 0.5 nm that is typically shifted 5 nm from the carrier frequency) and this is detected with a photomultiplier. The detuned scheme selectively enhances the higher frequency oscillations 42,20,43 within the full probe bandwidth and this gives improved reliability in the 200 cm−1 to 400 cm−1 region.
Resonance Raman spectra were obtained using a standard 90° light collecting geometry and a single grating monochromator (Acton SP2500i with 1800g/mm UV holographic grating, Princeton Instruments). Details of the setup have been described elsewhere.24 Samples were placed in a standard quartz cuvette (Precision Cells, Inc.) and excited with ~5mW of the 413.1 nm line form a krypton laser (Innova 300, Coherent).
Data Analysis
The experimental VCS data contain two basic components that arise from electronic and vibrational population transfer and from vibrational coherence. The population transfer component is usually dominant under resonance conditions. Important information such as ligand rebinding and vibrational cooling is carried in this component. This component must be separated to reveal the underlying vibrational coherence signal, particularly under open band conditions. The digitization of the experimental signal is done by the lock in amplifier on a 24-bit scale, which offers a sufficient dynamic range to observe the low amplitude coherence signal. The fractional change of the transmittance ΔT/T for the open band oscillatory signals was estimated to be on the order of ~10 −4 –10 −5.
To generate the power spectrum amplitudes from the time-domain oscillatory signal, we used a linear predictive singular value decomposition (LPSVD) algorithm that can fit both the monotonic background and the damped oscillations simultaneously, as described by Σiai exp(−t/τi)cos(ωit+ɸi). A few parameters can be controlled during the fitting procedure, such as the number of oscillations. The coherence coupling signal around time zero was truncated before the fitting analysis as illustrated in Fig. 6a. Data points within the truncated region (< 200 fs) were not included in the data analysis in order to eliminate the influence of the coherence coupling signal near time zero. The maximum entropy method (MEM) is not used to fit the population decay background when the coherence data is extracted. The MEM treats the vibrational coherence as noise and, as a result, the fitting procedure minimizes the sum of all residuals. But when we consider a damped oscillation, the integral of the displacement over time is not generally zero. This can lead to an unwanted distortion of the actual vibrational coherence signal amplitudes and frequencies when further frequency analysis tools such as LPSVD are applied.
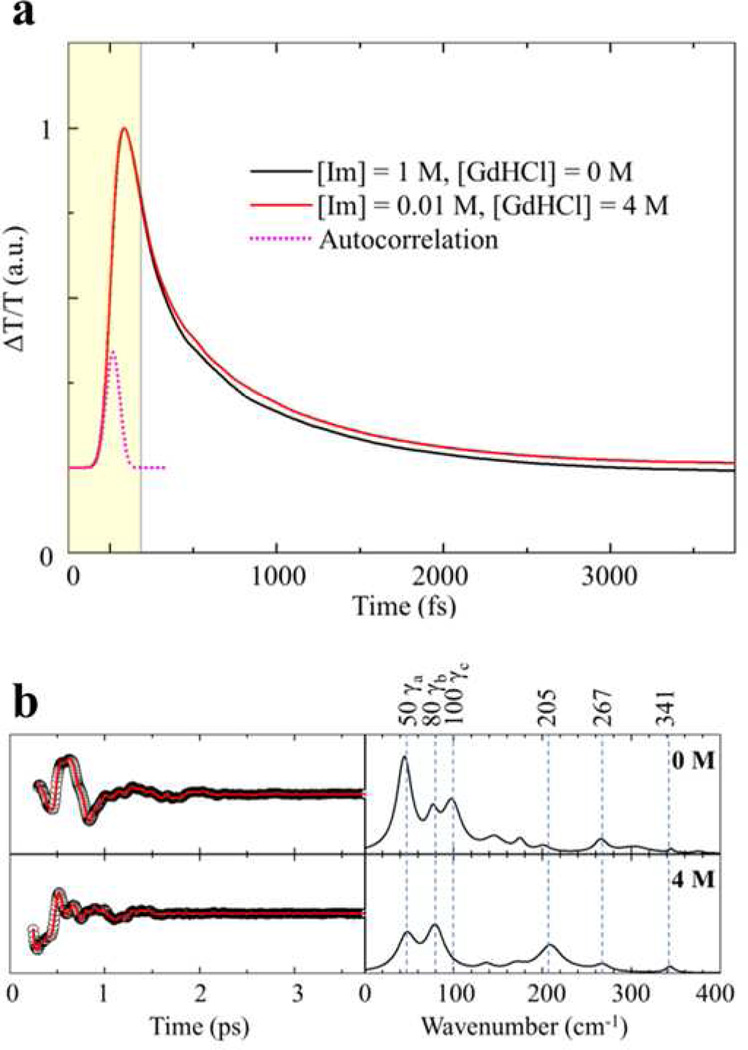
Figure 6.
(a) Open band optical responses for the folded and unfolded ferric cyt c-imidazole complex. Both traces were normalized at the peak of the coherence spike and the data within shaded window were truncated and were not included in the LPSVD fitting procedure. (b) VCS signals and power spectrum amplitudes for the same samples as shown in panel (a). The left panel displays the oscillatory components (black circles) and the LPSVD fits (red lines). The right panel shows the corresponding power spectrum amplitudes. The amplitudes of the power spectra are normalized using the kinetic traces shown in panel (a).
Results
Equilibrium unfolding of cyt c
The Soret band region of cyt c is displayed in Fig. S1 of the Supporting Information (SI) as a function of GdHCl concentration at pH 7.0 showing the absorption spectra as unfolding takes place under equilibrium conditions. The lower part of the figure shows the difference spectra of the unfolded cyt c with respect to the absorption spectrum of native cyt c. The peak of the Soret band blue-shifts from 409 nm to 406.5 nm as the GdHCl concentration is increased, reflecting the heme ligation switch from Met80 in native cyt c to either a His33 or His26 ligand as the protein is unfolded by GdHCl.44,7,10 The equilibrium unfolding of cyt c at pH 7.0 is not a simple two-state process45, as shown in the difference spectra of Fig. S1, where no clear isosbestic point is observed as the concentration of GdHCl is increased. Using 1H NMR spectra, Russell and Bren reported46 an intermediate state in which Lys (at position 79, 73 or 72) replaces native Met80 ligand under the conditions of 1.5–2.3 M GdHCl at pH 7.0 and 30 °C. A similar misligated intermediate state with Lys ligation is also observed under alkaline conditions.13
We adopt here the three-state model proposed by Latypov et al.45, which includes the equilibrium native state (N), an intermediate state (I), and an unfolded state (U). This allows all absorption spectra to be fitted consistently. The details of the fitting procedure and the results can be found in the SI. The Soret band maximum of the I-state is found at 407 nm and its population reaches a maximum (~80%) at 2.5 M GdHCl. Latypov et al.45 observed a similar unfolding intermediate state with a Soret band peak at the same position for cyt c at pH 5.0. The I-state is suggested to be similar to the “A” state47, a compact yet nonnative state, in which the iron-Met80 bond is broken and replaced by an unknown ligand with concomitant loss of the 695nm absorption band, which is attributed to a Met80 sulfur → Fe (III) charge transfer. There is also a low Trp59 florescence yield, indicating quenching as a result of a short Trp to heme distance.47,45.
The resonance Raman spectra of native cyt c has been assigned by Hu et al.18. At pH 7.0, the heme iron in native cyt c is 6-coordinate low spin (6cLS) and is characterized by its high frequency resonance Raman marker bands ν4, ν3, ν2 and ν10. The heme remains 6cLS when cyt c is fully unfolded by GdHCl at pH 7 with His33 (~80%) or His26 (~20%) replacing the distal Met80 heme ligand.44 Upon unfolding the heme marker bands downshift by ~2−4 cm−1, due to changes in the heme core size. This is the result of a decreased heme deformation associated with the Met-His ligand-switch.15,12
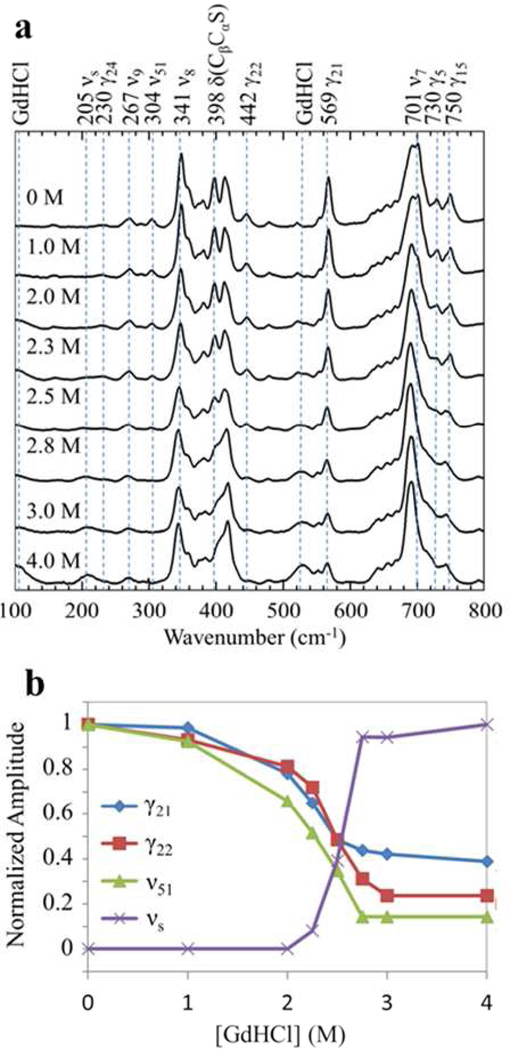
The low frequency RR spectral region (~200–800 cm−1) of native cyt c is more complicated. In Fig. 1a, we show the low frequency spectra of cyt c at pH 7.0 as unfolding takes place. There is no axial ligand mode present in the spectra.18 Most of the in-plane modes are relatively stable as the protein is unfolded, however, some in-plane modes lose their intensities such as ν51 (304 cm−1) and ν7 (701 cm−1).18 On the other hand, all of the out-of-plane (OOP) modes display a drastic loss of intensity (the 522 cm−1 mode (γ12) overlaps the broad GdHCl peak at 524 cm−1, so the change of its amplitude is not readily observable). The other OOP modes include: 750 cm−1 (γ15), 730 cm−1 (γ5), 569 cm−1 (γ21), 442 cm−1 (γ22), 398 cm−1 (δ(CβCaS)), and 230 cm−1 (γ24). Table 1 presents a summary of some of the key low frequency Raman active modes. The γ15 mode has B2u symmetry, γ5 has A2u symmetry, and the γ21, γ22, γ24 modes are of Eg symmetry in the D4h point group. The Raman activity of these OOP modes is enhanced in native cyt c, due to symmetry lowering induced by nonplanar heme distortions along the low frequency OOP mode coordinates. An interesting anomaly in the spectra is the emergence of a mode at 205 cm−1 that gains intensity upon unfolding.
Figure 1.
a) Low frequency resonance Raman spectra (100 – 800 cm‒1) of ferric hh cyt c as it undergoes equilibrium unfolding at pH 7.0 induced by GdHCl, which is varied from 0 M (folded) to 4 M (unfolded). The spectra are normalized with respect to the 1008 cm‒1 GdHCl peak (not shown). b) Relative amplitude changes of selected unfolding sensitive modes as a function of GdHCl concentration. The relative changes are normalized to unity at the maximum value for each mode.
Table 1.
Low-frequency modes sensitive to cyt c unfoldinga. Assignment of resonance Raman modes is based on Ref. 18.
| Mode Assignment | VCS | Raman | Intensity change upon unfolding |
|---|---|---|---|
| γa, significant ruffling content | 45–50 cm−1 | − | |
| γb, unassigned | 73–78 cm−1 | 0 | |
| γc, mixed with ruffling | 97–114 cm−1 | − | |
| νs, symmetric bis-His stretch | 203–205 cm−1 | 205 cm−1 | + |
| γ24, γ(CαCm) | 230 cm−1 | − | |
| ν9, δ(CδC1)sym | 264–269cm−1 | 267 cm−1 | 0 |
| ν51, δ(CβC1)asym | 304 cm−1 | − | |
| ν8, ν(FeN) | 341–351cm−1 | 341 cm−1 | 0 |
| δ(CβCαS) | 398 cm−1 | − | |
| γ22, pyrrole swivel | 442 cm−1 | − | |
| γ21, pyrrole foldsym | 569 cm−1 | − | |
| ν7, δ(pyrrole deform)sym | 701 cm−1 | − | |
| γ5, pyrrole foldsym | 730 cm−1 | − | |
| γ15, pyrrole foldsym | 750 cm−1 | − |
The intensity changes are denoted as “+” for increase and “–” for decrease, while “0” indicates little change.
We summarize in Fig. 1b the relative amplitude changes of γ21, γ22, ν51 and the 205 cm−1 mode, where the Raman signal is normalized using the GdHCl peak at 1008 cm−1. There are some small intensity changes (20% – 30% change) for γ21, γ22, and ν51 at [GdHCl] ≤ 2 M that indicate local environment changes associated with the formation of the I-state. However, the largest intensity change, especially the appearance of the 205 cm−1 mode, happens between 2–3 M GdHCl. This transition is consistent with a growing population of the U-state as shown in Figs. S2 and S3 of the SI. When the protein is unfolded, the altered tertiary structure relaxes its constraint on the heme and it adopts a more planar structure. As a result, the OOP Raman modes, which are enhanced in native cyt c due to the significantly ruffled geometry, are weakened24. The 398 cm−1 mode δ(CβCaS) is the bending involving the β-carbon, the sulfur atom at Cys14(17), and the carbon atom bridging them. It can be taken as a characteristic peak for the formation of the native tertiary structure of the heme pocket.17 The weakening of the γ21 peak also characterizes a less ruffled heme distortion and is also a good marker for unfolding.47,17 However, it is worth noting that the γ21 mode as a distortion marker appears to apply only to c-type hemes. In other heme systems, such as NP4-CN24, NP-H2O24, and LPO48, this mode is not observed even though strong ruffling distortions are present.
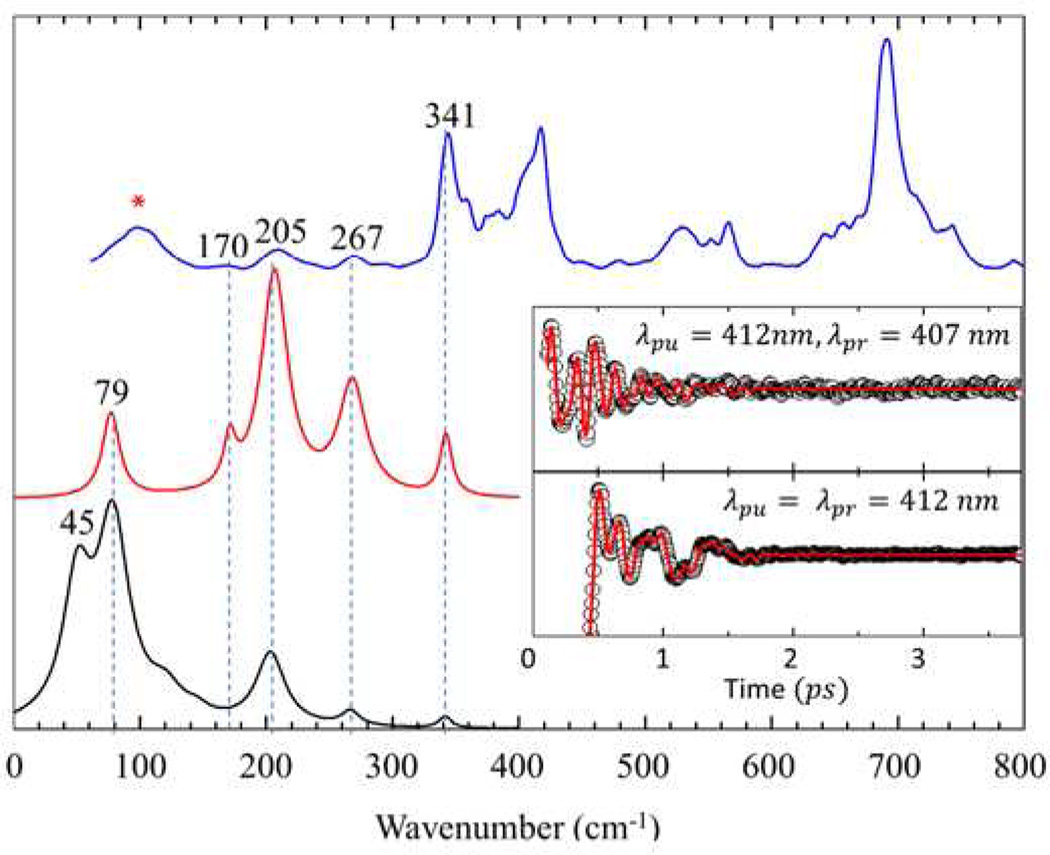
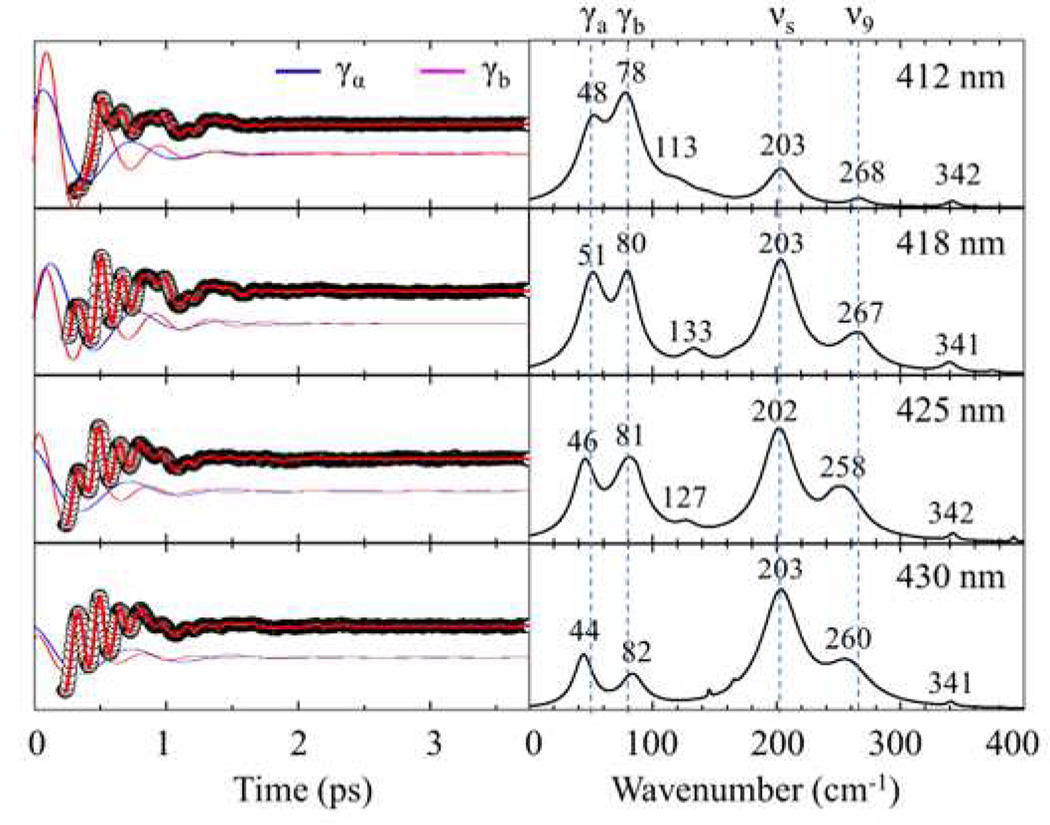
Vibrational coherence spectra of the fully unfolded cyt c (4M GdHCl at pH 7.0) are displayed in Fig. 2. The top trace shows resonance Raman spectra of unfolded cyt c, while the lower curves display the detuned (middle) and open band (lower) coherence spectra. The time domain oscillatory signals are shown in the insert. There are good correlations between the resonance Raman, detuned coherence, and open band coherence spectra. The modes at 205 cm−1, 267 cm−1 (v9), and 341 cm−1 (ν8) are present in all three spectra and display a very good correlation. The shoulder near 170 cm−1 in the detuned spectrum is not observed in the open band spectra. This might be due to its weak intensity, which allows detection only when using the enhancement obtained from the selective wavelength detection scheme. This figure demonstrates clearly that the VCS and Raman spectrum share the same mode selection rules43.
Figure 2.
Correlations between Raman and coherence spectra for unfolded ferric cyt c with 4M GdHCl at pH 7.0. The Raman spectrum (blue) was measured with excitation at 413.1 nm, whereas the open band (black) and detuned (red) coherence spectra (displaced below the Raman spectrum) were measured at a carrier wavelength of 412 nm. The detuned coherence data were collected with a 0.5 nm spectral window, detuned 5 nm to the blue of the carrier wavelength. The insets show the time domain VCS data (circles) for the detuned (upper) and open band (lower) conditions. The LPSVD fits are shown as red lines in the inserts. The Raman peak marked with “*” corresponds to a GdHCl peak.
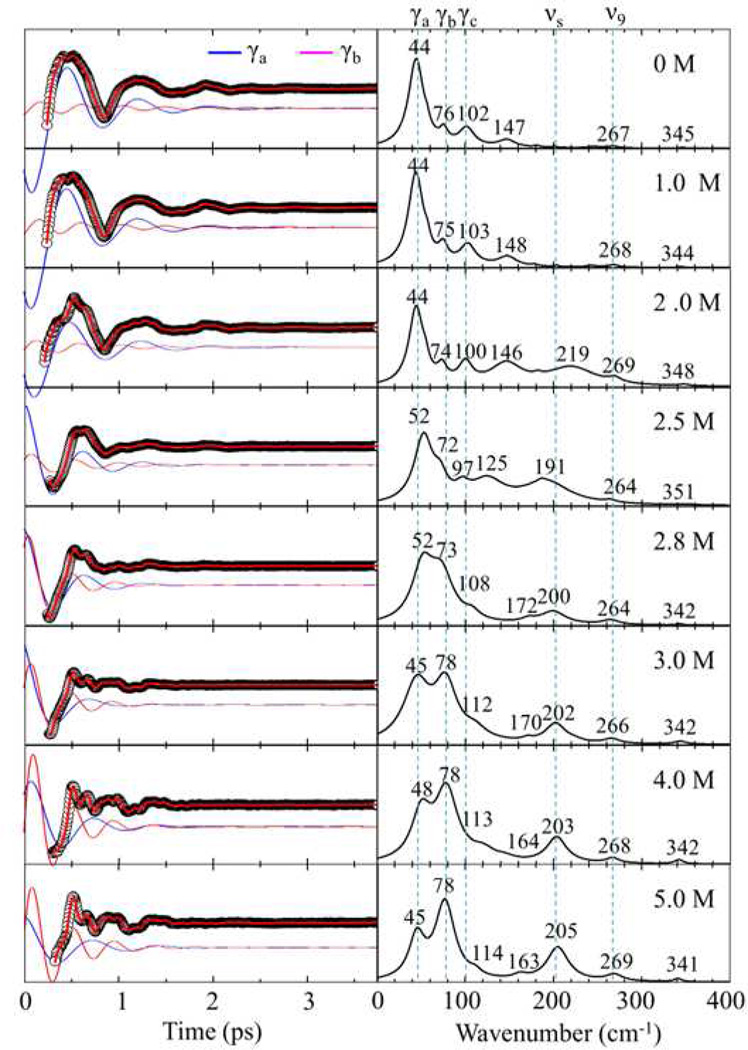
Figure 3 displays the open band VCS oscillations and the LPSVD-derived spectra as cyt c undergoes equilibrium unfolding with increasing GdHCl concentration from 0–5.0 M at pH 7.0. The amplitude of the oscillations and the frequency domain spectra have been placed on a common scale. For native cyt c, the mode at 45–50 cm−1 (γa) dominates the oscillatory signal as shown in the top panel. In addition, there are two smaller peaks present at 76–80 cm−1 (γb) and at 102 cm−1 (γc). On the other hand, when the protein is unfolded, there is a clear change in the oscillatory pattern showing that the dominating low frequency oscillation, γa, fades away and higher frequency oscillations gain relative intensity. For example, the mode at ~80 cm−1 (γb), the 205 cm−1 mode, and the 269 cm−1 mode dramatically gain intensity relative to the ~50 cm−1 (γa) and 102 cm−1 (γc) modes. Additional spectra that help to reveal changes in absolute intensity are presented below for the two-state unfolding of imidazole bound cyt c.
Figure 3.
Open band VCS spectra of ferric cyt c measured at 412 nm with a 70 fs laser pulses as GdHCl is added to induce equilibrium unfolding. The concentration of GdHCl varies between 0– 5 M. The left panel displays the oscillatory components (black circles) and the LPSVD fits (red lines). The blue and magenta oscillations represent the individual ~45–50 cm−1 (γa) and ~75–80 cm−1 (γb) oscillations, which are offset from the raw data for display. The right panel shows the amplitudes of the corresponding power spectrum. The amplitudes of all spectra and oscillations are given in arbitrary units.
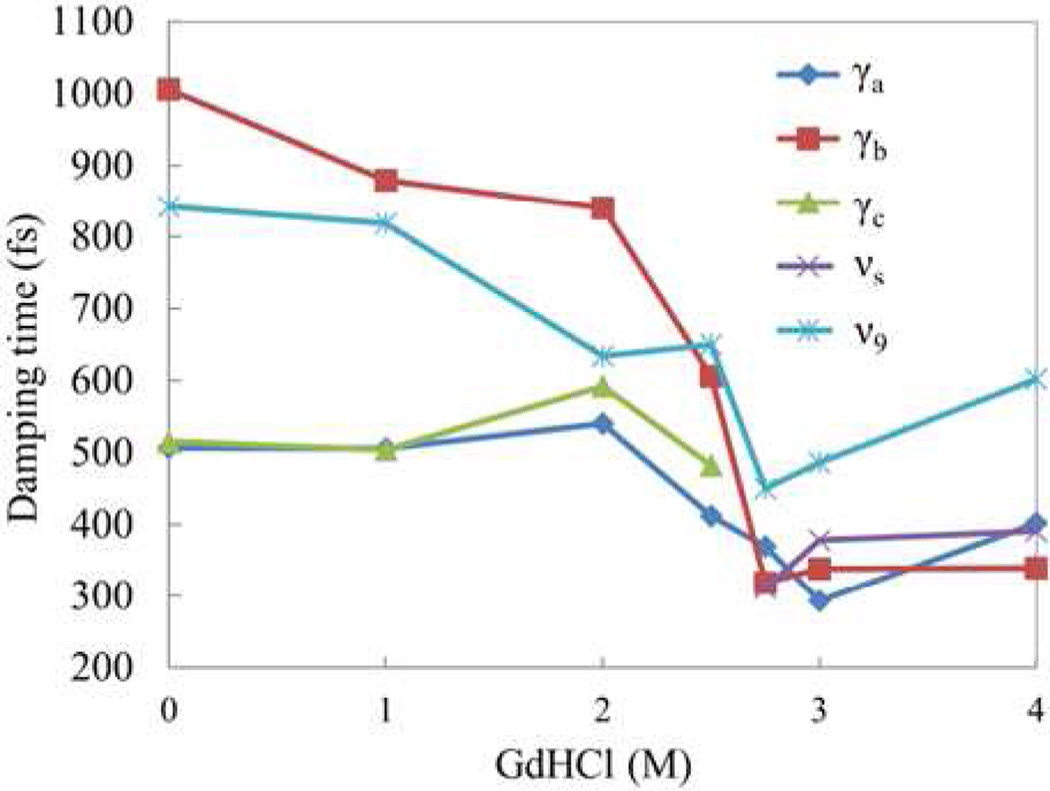
In Fig. 4 we present the damping constants of the coherent modes as a function of GdHCl concentration. For [GdHCl] ≤ 2 M, γa and γc show nearly identical damping times τ ~500 fs, while the damping time for γb is near τ~1000 fs. Interestingly, when the protein is unfolded ([GdHCl] ≥ 3 M), the damping times of all OOP modes converge to ~350 fs while the in-plane mode ν9 (267cm−1) maintains a longer damping time constant of ~600 fs. The minimum value of the damping constants appears at 2.8–3 M GdHCl, where the protein may be undergoing a fast exchange between N/I and U states. The faster damping time is what accounts for the broadened low frequency spectral lineshapes observed in Fig. 3 with 2.8 M and 3M GdHCl.
Figure 4.
A plot of the damping time constants of ferric cyt c as a function of GdHCl concentration for the 5 principal components of the VCS spectra shown in Fig. 3. The time constants are obtained by LPSVD fitting. Because γc is not present in the unfolded state, and the 205 cm−1 mode is not present in the folded state, their damping times at the corresponding concentrations are not shown.
Figure 5 shows a VCS “excitation profile” of unfolded cyt c. The excitation wavelength was tuned from 412 nm to 430 nm to systematically examine the low frequency mode dependence on the changing resonance conditions. The phase and damping time constants for each mode are summarized in Table 2. While the phases remain in the same quadrant and the damping times stay relatively fixed to within ~ 0.1 ps, the relative amplitudes of γa and γb compared to the 205 cm−1 and 269 cm−1 modes are much weaker when tuning away from the 406 nm Soret band peak of unfolded cyt c. This suggests that the lower frequency components of the signal, γa and γb, are more strongly peaked closer to the Soret maximum than are the higher frequency modes as is predicted theoretically41,49. Excitation profile studies of low frequency modes in native cyt c (work in preparation) reveal a mode at 60 cm−1 that is greatly enhanced when tuning the excitation wavelength from 412 nm to 435 nm. This demonstrates that the excitation profile of ferric cyt c with its native Met80 ligand is fundamentally different from the unfolded (bis-His ligated) protein and supports the idea that the 60 cm−1 mode is related to underlying charge transfer states that are present when Met is a ligand.50
Figure 5.
Open band VCS spectra of unfolded ferric cyt c (at 4 M GdHCl) measured with 70 fs laser pulses at 412 nm, 418 nm, 425 nm and 430 nm. The left panel displays the oscillatory components (black circles) and LPSVD fits (red lines). The blue and magenta oscillations represent the fits to the individual ~50 cm−1 (γa) and ~80 cm−1 (γb) modes and are shifted for clarity. The right panel shows the amplitudes of the corresponding power spectra. The phases and damping times of these modes are given in Table 2.
Table 2.
The phase and damping time of low frequency modes of unfolded ferric cyt c.
| λpump | γa | γb | νs | |||
|---|---|---|---|---|---|---|
| Phase (degree) |
Damping (fs) |
Phase (degree) |
Damping (fs) |
Phase (degree) |
Damping (fs) |
|
| 412nm | −38 | 395 | −66 | 341 | 54 | 387 |
| 418 nm | −76 | 403 | −69 | 488 | 29 | 333 |
| 425 nm | −10 | 492 | −53 | 337 | 45 | 315 |
| 430 nm | −4 | 574 | −99 | 456 | 5 | 284 |
Equilibrium unfolding of the cyt c-imidazole complex
In order to better understand the changes in the VCS spectra of native cyt c as it is unfolded, we performed a comparative study of the cyt c-imidazole complex. In this complex, which behaves like a two-state system, an exogenous imidazole ligand replaces Met80 leading to conformational changes of the backbone near residue Met80 as well as the repositioning of some residues in the distal cavity51. The large displacement of these residues accounts for imidazole’s low binding affinity: 29 M−1 at 18°C 52. On the other hand, when cyt c is unfolded, the heme is exposed to the solution and the barriers for exogenous ligand binding are greatly reduced. As a result, the binding affinity is much higher in the unfolded state and imidazole binds at 0.01 M concentration. As with imidazole-bound microperoxidase 8 (MP8), when the heme is exposed to solvent, even the proximal His18 can be replaced by imidazole if the concentration is high enough (4 M)53.
Fig. S4a shows the Soret absorption band of the folded and unfolded cyt c-imidazole complex at different imidazole concentrations. The peak of Soret absorption band does not change when the complex is unfolded in 4M GdHCl at 0.01 M imidazole concentration, but the peak shifts about 1.5 nm to the red at higher imidazole concentration (4M), indicating replacement of the proximal His. As a result, we compare the sample for folded cyt c in 1M imidazole (no GdHCl) with the unfolded sample (4M GdHCl) in 0.01 M imidazole. These conditions generate the same ligation status (His18 and imidazole) in the two samples and there is no obvious shift of Soret absorption maximum. Interestingly, the full-width-at-half-maximum (FWHM) of the Soret absorption band of the unfolded imidazole complex is slightly narrower (less than 5%) compared to the folded form.
Using the normal coordinate structural decomposition (NSD) method,33,34,54 we can quantitatively characterize the nonplanar heme distortions. NSD describes out-of-plane distortions by using the lowest frequency OOP mode of each symmetry type. In our analysis, we use the optimized planar ferrous porphine (including the iron atom) with D4h symmetry as the reference structure. The displacement along each normal coordinate was calculated in the mass-weighted coordinate space using the scalar product (X - X0) · Qα, where X and X0 are the mass-weighted atomic coordinates of the input and reference structures, and the unit vectors, Qα, are taken from the mass-weighted normal modes, α, of the reference structure. Fig. S4b in the SI displays the NSD analysis of the out-of-plane heme core distortions for native cyt c (PDB# 1HRC) and its imidazole complex based on the NMR structure (PDB #1FI7).51 Here it must be pointed out that the NMR structure lacks resolution and there are ambiguities in the heme conformation due to lack of protons in the carbon core. However, the NOE data51 does include the protons on the meso carbons, which can be used to estimate the distortion along the ruffling coordinate. The imidazole and His protons, along with knowledge of the mean porphyrin plane position, should also allow a reasonable estimate of the doming distortions. With these caveats, we note that the NSD of the imidazole complex also suggests a strong ruffling distortion of ~ 2.5 amu1/2Å, which is somewhat less than the ~ 3.5 amu1/2Å found in native cyt c. It also displays some saddling (~ 1.5 amu1/2Å) and doming (~ 1 amu1/2Å) distortions. The negative sign for doming suggests that the iron is displaced toward the distal (imidazole bound) side.
Given the potential ambiguity of the NMR structure for imidazole bound cyt c, we compare the RR spectra of the folded and unfolded imidazole complex in Fig. S4c. The RR spectrum of the folded complex is very similar to native cyt c. Especially noteworthy are the strong unfolding sensitive modes at ~400 cm−1 (δ(CβCαS)) and 569 cm−1 (γ21). When we compare to the RR spectrum of native cyt c in the top panel of Fig. 1a, we see that the native cyt c, which has larger ruffling distortion, shows stronger 569 cm−1 mode. This supports the argument that γ21 is enhanced by the ruffling distortion of c-type heme.47 When the complex is unfolded, these two modes lose their intensities, and the 205 cm−1 band emerges, indicating that the more planar heme structure associated with the unfolding helps to enhance the 205 cm−1 mode. Such an effect could arise if the 205 cm−1 mode undergoes significant mode mixing and becomes “diluted” when the heme is in a more highly distorted state inside the folded protein. Generally, the changes associated with unfolding the imidazole-cyt c complex are very consistent with the observations for equilibrium unfolding of native cyt c.
However, in order to gain a better perspective on how the low frequency modes of cyt c and its imidazole complex change during unfolding, we would like to be able to measure the oscillation intensities in an absolute sense. Unfortunately, under different experimental conditions, the absolute strength of the signal is not easily comparable and an internal standard would be useful to better normalize the amplitudes of the oscillations. The VCS experiment uses a near parallel, but still a crossed beam geometry, i.e. there is a small angle between the direction of the pump and probe beams focused on the sample.26 This means that the detection of the self-heterodyned signal is very sensitive to absorbance of the sample and alignment of the optics (beam overlap within the sample). The intensity at time zero provides an inherently strong signal that is detected when the pump and probe pulses overlap in time. This signal has been studied in detail in prior work55,56 so that the detected signal57 at t > 0 can, in principle, be normalized by the signal intensity at time zero, which can act as an internal standard. The signal at time zero will be approximately invariant so long as key experimental variables, including the sample OD, are kept constant. Under these conditions, the amplitude of the coherent oscillations can be normalized to the height of the signal at time zero so that the VCS spectra of different samples can be compared.
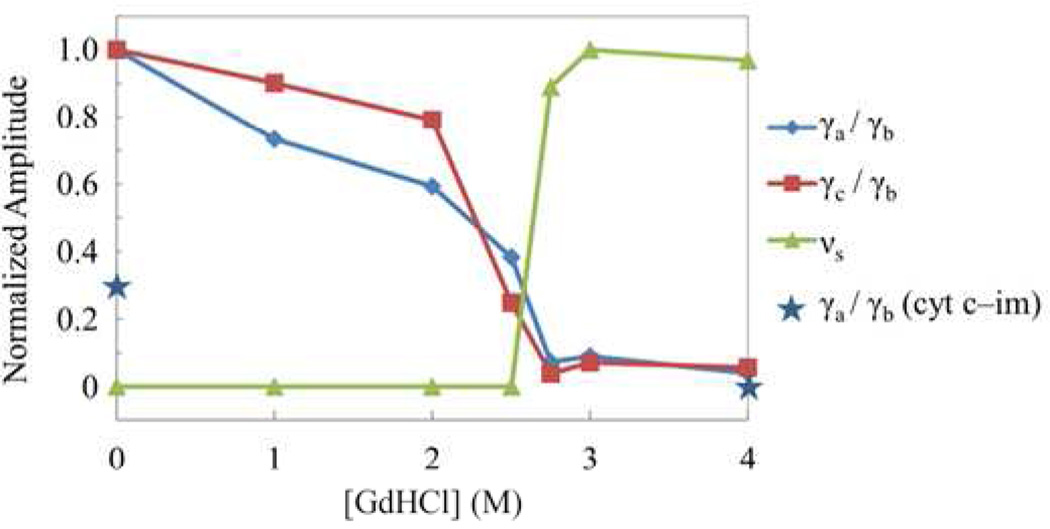
Fig. 6a shows the optical response of the folded and unfolded cyt c-imidazole complex. For this pair of samples, the ligand coordination remains unaltered, the peak of the Soret band is fixed and the change in its width is less than 5%. Assuming the intensities of the signals at time zero are the same in the two cases, we normalize the traces to unity at t=0. In Fig. 6b, the amplitude of the oscillations and the VCS spectra are scaled with the same normalization. We see that when the complex is unfolded, the amplitude of the 80 cm−1 mode (γb) is roughly unchanged, while the 50 cm−1 mode (γa) loses more than half of its intensity. The mode at 100 cm−1 (γc) totally disappears upon unfolding, while the 205 cm−1 mode emerges, and the modes at 267 cm−1 (v9) and 341 cm−1 (v8) stay relatively constant in agreement with the RR spectra. Note that the LPSVD spectra extracted for the folded and unfolded cyt c have mode frequencies for γa, γb, γc, 205 cm−1, ν8 and ν9 that agree to within 2 cm−1.
Application of this normalization method to the equilibrium unfolding of native cyt c (without added imidazole) is difficult, because the unfolding process involves denaturant concentration dependent populations of three states: N, I and U. Both the I- and U-states have Soret absorption peaks near 406 nm, however, the N-state has the Soret peak at 409 nm. However, because the γb mode amplitude stays approximately the same as the imidazole complex unfolds, we have elected to use it as a reference to characterize the amplitude changes of the other modes in native cyt c. In Fig. 7 we plot the ratio of γa/γb, γc/γb, and 205 cm−1/γb as a function of GdHCl concentration. The amplitude ratios are normalized to unity at the maximum value of each ratio. Figure 7 shows that the low frequency mode intensities reflect major changes in heme structure taking place upon unfolding of native cyt c. It can also be seen that the VCS data (Fig. 7) are very consistent with the RR results (Fig. 1b).
Figure 7.
Plot of the relative amplitude changes for the unfolding-sensitive low-frequency modes of native cyt c as a function of GdHCl concentration. The amplitude ratios with respect to γb, which is relatively insensitive to unfolding, are plotted. The relative change is normalized to unity at the maximum value for each mode.
Discussion
Mode Assignments
The combination of the low frequency RR spectra and VCS allows the full low frequency Raman spectral density of cyt c to be observed following GdHCl induced unfolding. The main unfolding sensitive modes below 800 cm−1 include: γa, γc, γ24, ν51, δ(CβCαS), γ22, γ21, ν7, γ5, γ15 along with the νs(His-Fe-His) mode at 205 cm−1 (see Table 1). Because heme structural information for the unfolded protein is lacking (no crystal structure), specific mode assignments in the unfolded state are difficult. However, the simultaneous change of both the Raman and VCS modes implies systematic changes in the heme deformation take place. As discussed by Kubo et al., the amplitude of the oscillation of a given low-frequency OOP normal mode of the heme is proportional to the square of its distortion from the planar state, which helps us to understand the Raman activity of “soft” low frequency modes.24
In unfolded cyt c, the heme adopts a more planar structure as indicated by the reduced intensity of the OOP modes (Fig. 1b, Fig. 7); i.e., the strong ruffling distortion of the heme in native cyt c is significantly relaxed. From the NSD analysis of native cyt c shown in Fig. S4b, the heme deformation is mainly along the ruffling coordinate with only very small contributions from other normal coordinates, such as saddling. Thus, the enhanced amplitude of γa in native cyt c should be due to the lowering of porphyrin symmetry mainly along the ruffling normal coordinate. Considering γa is the dominant mode in the VCS spectra, we assign it to have a large contribution from ruffling motion. The assignment of the γa mode at ~45–50 cm−1 to ruffling suggests that the c-type heme ruffles at a slightly lower frequency than found for the b-type heme in myoglobin and nitrophorin (NP4) where it was assigned to a mode near 60 cm−1. However, in the NP4 study24, it was noted that the ruffling mode “softens” from 72 to 53 cm−1 as the ruffling distortion was increased systematically using several different heme complexes. Because the heme in cyt c involves a ruffling distortion that is slightly larger than found for NP4-CN, where the ruffling frequency was assigned at 53 cm−1, the value for the frequency near 45–50 cm−1 in cyt c appears to be very consistent with what was found in the nitrophorin system24.
Based on the NSD analysis in the SI, we assign γc to be a mode with a lesser amount of ruffling admixture, possibly with some saddling or waving content. It is not fully understood why the amplitude of γb remains roughly constant during the unfolding process. The data suggest that γb involves a mode with an inherent deformation that is relatively unaffected by the protein folding process. It is noteworthy that Shelnutt et al.58 used the four-coordinate Ni-substituted heme in microperoxidase11 and cyt c to show that a significant level of porphyrin OOP distortion, due to forces associated with the CXXCH pentapeptide, is retained even when cyt c is unfolded. The intensity invariance of γb is likely related to such a distortion, which is relatively isolated from the protein folding process. Because the intensity of the coherent signal depends quadratically on the magnitude of the distortion24, the observed reduction in the γa and γc intensities by ~90% is consistent with residual distortions in the unfolded state that are ~32% of the native amount. This amount of residual distortion is less than predicted using an optimized microperoxidase vacuum structure58. On the other hand, it seems possible that the interaction between the guanidinium and the carbonyl groups may weaken the H-bonds that constrain the CXXCH pentapeptide58, resulting in additional relaxation of the heme distortions in the GdHCl unfolded structure.
The 205 cm‒1 mode is somewhat anomalous because it gains intensity upon unfolding. Its appearance as well as its increasing amplitude with increasing [GdHCl] is very consistent in both the VCS and RR spectra. Based on the work of Mitchell et al.59, this mode can be assigned to the symmetric stretch, νs( His-Fe-His), of the bis-histidine complex that is formed in the unfolded state. This is also consistent with conclusions based on RR studies of MP8 with added imidazole.53 Additional measurements (data not shown) indicate that the depolarization ratio for this mode is ~1/3, which could arise from coupling to only a single (x or y) component of the transition dipole. This might be associated with asymmetry induced by the relative histidine-heme orientations.
Damping of Coherent Motion
The damping times of the coherent oscillations are summarized in Fig. 4, which provides a direct probe of the effects of protein folding and the heme exposure to solvent fluctuations. The damping of the coherence signal has three main sources: spectral inhomogeneity, vibrational lifetime, and “pure” dephasing; the latter being due to a disparity in the coupling of the surrounding fluctuations to the individual vibrational eigenstates involved in the coherent superposition. Previous studies have demonstrated that the low frequency coherences of the heme within a protein matrix have a much longer damping time than when exposed to solvent22. Further temperature dependent studies of the damping time of low frequency modes in horseradish peroxidase (HRP) suggested that pure dephasing is an important source of the coherence decay within the protein environment.22 Figure 4 demonstrates that the low frequency heme coherences can have significantly different damping times, presumably related to the specifics of how they are coupled to the environment. The nearly identical damping for γa and γc between 0 − 2M GdHCl suggests that they may couple to the protein with a very similar mechanism in the folded state. Low frequency oscillations involve the cooperative motion of many atoms in the heme and even extend into the protein. It is likely that the low frequency oscillations (γa and γc) in cyt c involve ruffling and/or saddling motions that involve the linkage between the pyrrole rings of the heme and the two thioether bridges linked to the protein. Thus, there exists strong coupling between these modes and the protein matrix leading to shorter decay times. In addition, it has been shown that a major heme energy relaxation “doorway” involves the heme proprionate groups, which facilitate heme-solvent interaction in the folded state of myoglobin.60,61 Similar interactions could be involved in the dephasing of low-frequency heme modes.
In contrast to γa and γc, the mode assigned as γb has an intensity that is roughly independent of the cyt c folding process and it shows a ~ 1 ps damping time (Fig. 4), which is typical of most low frequency oscillations in a ferric heme.20–22,40 Unfolding has a dramatic impact on the damping time of this mode, reducing it to ~300fs and suggesting a more solvent-exposed heme environment. Generally, the low frequency OOP modes have a consistent damping constant of ~350 fs when unfolded. This can be contrasted with the ~600 fs damping time of the ν9 in-plane mode at 267 cm−1. The damping times in the fully unfolded state are comparable with previous measurements on solvent-exposed ferric heme models, where the measured damping times were about ~ 500 fs.40
Folding
The protein folding process is often interpreted by energy landscape theory where the protein first collapses and then, through the search of native contacts gradually, forms a native folded structure within a funnel-like energy landscape.62–64 The presence of misligated heme traps in cyt c slows the folding kinetics; for example, when His 33 (or His 26) binds to the heme iron44. This bis-histidine trap has been proposed by Bren et al., to be on the folding pathway, helping to guide the folding process toward the native structure.46 The folding process of cyt c involves the sequential folding of five pseudocooperative units (foldons) in order of descending free energy: the N- and C- terminal helices, the 60s helix, the β sheets (residues 37–39 and 58–61), the 71–85 loop (Met80 loop) and the 40–57 loop, along with the omega loop containing His 26 and 33, which does not display foldon properties65–68,63,64.
The Met80 loop unfolding represents an early step on the major cyt c unfolding pathway with a free energy of ~6 kcal mol−1.69,63 The presence of the I-state (heme coordination with Lys 73 or Lys 79 in the Met80 loop) under various very mild denaturation conditions is associated with the destabilization of Met80 loop. From the global fitting of the equilibrium unfolding absorption spectra at pH 7 and 20°C (see SI), we determine the free energy (ΔGNI) for the transition from N to I to be 4.6 ± 0.3 kcal/mol and the ΔGIU from I to U to be 11.6 ± 0.4 kcal/mol, with a total change of free energy, ΔGtotal=16.2 ± 0.5 kcal/mol. The value of ΔGtotal at pH 7.0 and 20°C is higher than ΔGtotal under other experimental conditions. Bai et al.70,65 reported ΔGtotal=12.8 kcal/mol at pH 7.0 and 30°C. However, since 1–3 kcal/mol decrease of ΔGtotal is expected for a temperature rise from 20°C to 30°C at pH 7.0, this is consistent with our observations.70 Latypov et al.45 reported ΔGtotal=9.8 kcal/mol at pH 5.0, but the ~6 kcal/mol drop in ΔGtotal can be accounted for by the expected reduction of unfolding free energy in going from pH 7 to pH 5.71 The value for ΔGtotal found here is also larger than the 11.2 kcal/mol found for urea-induced unfolding at pH 7.0 and 30°C72. In addition to the lowering of the unfolding free energy due to the temperature increase, a difference between urea and GdHCl is also expected due to the fact that GdHCl is an ionic denaturant.45,73 On the other hand, the value of ΔGNI = 4.62 kcal/mol obtained here is close to the 4.0 kcal/mol found using urea to induce the partial unfolding to form the intermediate state72. It is also near to the ~6 kcal/mol unfolding free energy of the Met80 loop deduced from previous hydrogen exchange experiments 70,65,71 using a limited concentration range for GdHCl (0–2.0 M). These results strongly suggest that the N to I transition is correlated with the Met80 loop unfolding.
Despite the change of axial ligand in going from the N to I state as the Met80 loop unfolds, there are only very small amplitude changes in the low frequency RR and VCS modes for [GdHCl] ≤ 2M. This suggests that the destabilized Met80 loop has only limited “responsibility” for the highly ruffled heme structure. Comparing Fig. 1b and Fig.7 with Fig.S3, we see that the changes of the low frequency Raman and VCS modes follow the population profile of the U-state, displaying a midpoint of 2.7 M (Fig. 1b and 7) and saturating at higher GdHCl concentrations. The midpoint concentration is nearly the same as found using the Soret band titration method (CNU =2.65 kcal/mol, see SI). This indicates that heme structural changes in cyt c are strongly correlated with the global unfolding that forms the U-state. Among the 5 foldon units in cyt c, the terminal helices have the highest unfolding free energy and this corresponds to the last step in unfolding (and the first step in refolding), i.e. its unfolding free energy and midpoint concentration define the free energy and midpoint concentration of global unfolding.71,63 The CXXCH motif, which is covalently connected to the porphyrin, is found on the N-terminal. The unfolding of this unit leads to a structural change of the CXXCH motif and this affects the deformation of porphyrin ring. Thus, our results suggest that cyt c controls the distortion of the heme through its CXXCH motif, which is consistent with prior studies74,58.
Lipid Binding
The structure changes associated with cyt c binding to model membrane systems (polyanions75, phospholipid vesicles76 and electrodes77) as well as intact mitochondria78 has also been investigated. The studies of model membrane systems demonstrated that cyt c undergoes a tertiary structural change upon binding. Such a change, either due to electrostatic binding or hydrophobic binding, again results in the loss of the Met80 ligand76, which is replaced by either a His residue or water. The γ21 peak intensity near 569 cm−1 is generally reduced, which indicates partial relaxation of the ruffled heme structure79. Studies using intact mitochondria78 also reported that γ21 has completely disappeared when cyt c is bound in the living mitochondrial membrane. This suggests that, when cyt c binds to the membrane, it might both unfold and disrupt the H-bonds constraining CXXCH pentapeptide58, thus inducing more heme relaxation than is achieved using chemical denaturants alone. It appears that the relaxed heme structure may, in fact, be very important for the in vivo function of cyt c. This conclusion is also supported by recent studies of cardiolipin bound cyt c80.
Summary
In summary, we find that low-frequency modes at ~50 cm−1, ~80 cm−1, and ~100 cm−1, as well as certain higher frequency Raman modes, especially the νs( His-Fe-His) mode at 205 cm−1, and the γ21 mode (569 cm−1) directly track the unfolding of cyt c. We infer that the primary heme structural change associated with the fully unfolded protein involves the ruffling distortion and we assign the mode near 45–50 cm−1 (γa) as having significant ruffling content. We also observe that the unfolded state generally leads to more rapid decoherence and shorter damping times, presumably due to increased coupling between the heme and the solvent. Finally, we use a simple 3-state thermodynamic model to generate the free energy for unfolding in the steps N-I and I-U. The relative intensity changes of the low-frequency heme modes indicate that the protein-induced heme distortions, specifically involving the ruffling coordinate, are significantly (but not completely) relaxed during the final I to U unfolding transition. More importantly, the binding of cyt c to natural lipid membranes appears to relax the heme distortions even further.
Supplementary Material
Acknowledgment
This work is supported by grants from the NSF (MCB-744738) and the NIH (DK35090).
Footnotes
Supporting Information Available:
Supporting figures and a description of the absorption spectra fitting procedure using a 3-state unfolding model. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nall BT. Cytochrome C: A Multidisciplinary Approach. Sausalito, CA: University Science Books; 1996. [Google Scholar]
- 2.Pettigrew GW, Moore GR, editors. Cytochromes C - Biological Aspects. Berlin, Heidelberg: Springer-Verlag; 1987. [Google Scholar]
- 3.Basova LV, Kurnikov IV, Wang L, Ritov VB, Belikova NA, Vlasova II, Pacheco AA, Winnica DE, Peterson J, Bayir H, et al. Cardiolipin Switch in Mitochondria: Shutting Off the Reduction of Cytochrome C and Turning on the Peroxidase Activity. Biochemistry. 2007;46:3423–3434. doi: 10.1021/bi061854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ow YLP, Green DR, Hao Z, Mak TW. Cytochrome C: Functions Beyond Respiration. Nat. Rev. MolCell Bio. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 5.Bowman SEJ, Bren KL. The Chemistry and Biochemistry of Heme C: Functional Bases for Covalent Attachment. Nat. Prod. Rep. 2008;25 doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan CK, Hofrichter J, Eaton WA. Optical Triggers of Protein Folding. Science. 1996;274:628–629. doi: 10.1126/science.274.5287.628. [DOI] [PubMed] [Google Scholar]
- 7.Elove GA, Bhuyan AK, Roder H. Kinetic Mechanism of Cytochrome C Folding: Involvement of the Heme and Its Ligands. Biochemistry. 1994;33:6925–6935. doi: 10.1021/bi00188a023. [DOI] [PubMed] [Google Scholar]
- 8.Jones CM, Henry ER, Hu Y, Chan CK, Luck SD, Bhuyan A, Roder H, Hofrichter J, Eaton WA. Fast Events in Protein Folding Initiated by Nanosecond Laser Photolysis. Proc. Natl. Acad. Sci. USA. 1993;90:11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascher T, Chesick JP, Winkler JR, Gray HB. Protein Folding Triggered by Electron Transfer. Science. 1996;271:1558–1560. doi: 10.1126/science.271.5255.1558. [DOI] [PubMed] [Google Scholar]
- 10.Sosnick TR, Mayne L, Hiller R, Englander SW. The Barriers in Protein-Folding. Nat. Struct. Mol Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 11.Morikis D, Champion PM, Springer BA, Egeberg KD, Sligar SG. Resonance Raman Studies of Iron Spin and Axial Coordination in Distal Pocket Mutants of Ferric Myoglobin. J. Biol Chem. 1990;265:12143–12145. [PubMed] [Google Scholar]
- 12.Parthasarathi N, Hansen C, Yamaguchi S, Spiro TG. Metalloporphyrin Core Size Resonance Raman Marker Bands Revisited: Implications for the Interpretation of Hemoglobin Photoproduct Raman Frequencies. J. Am. Chem Soc. 1987;109:3865–3871. [Google Scholar]
- 13.Dopner S, Hildebrandt P, Rosell FI, Mauk AG. Alkaline Conformational Transitions of Ferricytochrome C Studied by Resonance Raman Spectroscopy. J. Am. Chem Soc. 1998;120:11246–11255. [Google Scholar]
- 14.Droghetti E, Oellerich S, Hildebrandt P, Smulevich G. Heme Coordination States of Unfolded Ferrous Cytochrome C. Biophys J. 2006;91:3022–3031. doi: 10.1529/biophysj.105.079749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oellerich S, Wackerbarth H, Hildebrandt P. Spectroscopic Characterization of Nonnative Conformational States of Cytochrome C. J. Phys. Chem B. 2002;106:6566–6580. [Google Scholar]
- 16.Takahashi S, Yeh S-R, Das TK, Chan C-K, Gottfried DS, Rousseau DL. Folding of Cytochrome C Initiated by Submillisecond Mixing. Nat. Struct. Mol Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 17.Yeh SR, Takahashi S, Fan BC, Rousseau DL. Ligand Exchange During Cytochrome C Folding. Nat. Struct. Mol Biol. 1997;4:51–56. doi: 10.1038/nsb0197-51. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, Morris IK, Singh JP, Smith KM, Spiro TG. Complete Assignment of Cytochrome C Resonance Raman Spectra Via Enzymic Reconstitution with Isotopically Labeled Hemes. J. Am. Chem Soc. 1993;115:12446–12458. [Google Scholar]
- 19.Reinisch L, Schomacker KT, Champion PM. Anomalous Resonance Enhanced Quasielastic Light Scattering of Cytochrome-C. J. Chem Phys. 1987;87:150–158. [Google Scholar]
- 20.Gruia F, Ionascu D, Kubo M, Ye X, Dawson J, Osborne RL, Sligar SG, Denisov I, Das A, Poulos TL, et al. Low-Frequency Dynamics of Caldariomyces Fumago Chloroperoxidase Probed by Femtosecond Coherence Spectroscopy. Biochemistry. 2008;47:5156–5167. doi: 10.1021/bi7025485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruia F, Kubo M, Ye X, Champion PM. Investigations of Vibrational Coherence in the Low-Frequency Region of Ferric Heme Proteins. Biophys J. 2008;94:2252–2268. doi: 10.1529/biophysj.107.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruia F, Kubo M, Ye X, Ionascu D, Lu C, Poole RK, Yeh SR, Champion PM. Coherence Spectroscopy Investigations of the Low-Frequency Vibrations of Heme: Effects of Protein-Specific Perturbations. J. Am. Chem Soc. 2008;130:5231–5244. doi: 10.1021/ja7104027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunakaran V, Benabbas A, Sun Y, Zhang Z, Singh S, Banerjee R, Champion PM. Investigations of Low-Frequency Vibrational Dynamics and Ligand Binding Kinetics of Cystathionine Beta-Synthase. J. Phys. Chem. B. 2010;114:3294–3306. doi: 10.1021/jp909700r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo M, Gruia F, Benabbas A, Barabanschikov A, Montfort WR, Maes EM, Champion PM. Low-Frequency Mode Activity of Heme: Femtosecond Coherence Spectroscopy of Iron Porphine Halides and Nitrophorin. J. Am. Chem. Soc. 2008;130:9800–9811. doi: 10.1021/ja800916d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosca F, Kumar ATN, Ionascu D, Ye X, Demidov AA, Sjodin T, Wharton D, Barrick D, Sligar SG, Yonetani T, et al. Investigations of Anharmonic Low-Frequency Oscillations in Heme Proteins. J. Phys. Chem. A. 2002;106:3540–3552. [Google Scholar]
- 26.Rosca F, Kumar ATN, Ye X, Sjodin T, Demidov AA, Champion PM. Investigations of Coherent Vibrational Oscillations in Myoglobin. J. Phys. Chem. A. 2000;104:4280–4290. [Google Scholar]
- 27.Wang W, Ye X, Demidov AA, Rosca F, Sjodin T, Cao WX, Sheeran M, Champion PM. Femtosecond Multicolor Pump-Probe Spectroscopy of Ferrous Cytochrome C. J. Phys. Chem. B. 2000;104:10789–10801. [Google Scholar]
- 28.Zhu L, Li P, Huang M, Sage JT, Champion PM. Real-Time Observation of Low-Frequency Heme Protein Vibrations Using Femtosecond Coherence Spectroscopy 3 372. Phys. Rev. Lett. 1994;72:301–304. doi: 10.1103/PhysRevLett.72.301. [DOI] [PubMed] [Google Scholar]
- 29.Zhu LY, Sage JT, Champion PM. Observation of Coherent Reaction Dynamics in Heme-Proteins. Science. 1994;266:629–632. doi: 10.1126/science.7939716. [DOI] [PubMed] [Google Scholar]
- 30.Latypov RF, Maki K, Cheng H, Luck SD, Roder H. Folding Mechanism of Reduced Cytochrome C: Equilibrium and Kinetic Properties in the Presence of Carbon Monoxide. J. Mol. Biol. 2008;383:437–453. doi: 10.1016/j.jmb.2008.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galinato MG, Kleingardner JG, Bowman SE, Alp EE, Zhao J, Bren KL, Lehnert N. Heme-Protein Vibrational Couplings in Cytochrome C Provide a Dynamic Link That Connects the Heme-Iron and the Protein Surface. Proc. Natl. Acad. Sci. USA. 2012;109:8896–8900. doi: 10.1073/pnas.1200345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobbs JD, Shelnutt JA. Conserved Nonplanar Heme Distortions in Cytochromes-C. J. Prot. Chem. 1995;14:19–25. doi: 10.1007/BF01902840. [DOI] [PubMed] [Google Scholar]
- 33.Jentzen W, Ma JG, Shelnutt JA. Conservation of the Conformation of the Porphyrin Macrocycle in Hemoproteins. Biophys. J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jentzen W, Song XZ, Shelnutt JA. Structural Characterization of Synthetic and Protein-Bound Porphyrins in Terms of the Lowest-Frequency Normal Coordinates of the Macrocycle. J. Phys. Chem. B. 1997;101:1684–1699. [Google Scholar]
- 35.Maes EM, Walker FA, Montfort WR, Czernuszewicz RS. Resonance Raman Spectroscopic Study of Nitrophorin 1, a Nitric Oxide-Binding Heme Protein from Rhodnius Prolixus, and Its Nitrosyl and Cyano Adducts. J Am Chem Soc. 2001;123:11664–11672. doi: 10.1021/ja0031927. [DOI] [PubMed] [Google Scholar]
- 36.Roberts SA, Weichsel A, Qiu Y, Shelnutt JA, Walker FA, Montfort WR. Ligand-Induced Heme Ruffling and Bent No Geometry in Ultra-High-Resolution Structures of Nitrophorin 4. Biochemistry. 2001;40:11327–11337. doi: 10.1021/bi0109257. [DOI] [PubMed] [Google Scholar]
- 37.Walker FA. Magnetic Spectroscopic (Epr, Eseem, Mossbauer, Mcd and Nmr) Studies of Low-Spin Ferriheme Centers and Their Corresponding Heme Proteins. Coord. Chem. Rev. 1999;186:471–534. [Google Scholar]
- 38.Walker FA. Nitric Oxide Interaction with Insect Nitrophorins and Thoughts on the Electron Configuration of the {Feno}(6) Complex. J. Inorg. Biochem. 2005;99:216–236. doi: 10.1016/j.jinorgbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Nozaki Y. The Preparation of Guanidine Hydrochloride. Methods Enzymol. 1972;26:43–50. doi: 10.1016/s0076-6879(72)26005-0. [DOI] [PubMed] [Google Scholar]
- 40.Gruia F, Ye X, Ionascu D, Kubo M, Champion PM. Low Frequency Spectral Density of Ferrous Heme: Perturbations Induced by Axial Ligation and Protein Insertion. Biophys. J. 2007;93:4404–4413. doi: 10.1529/biophysj.107.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar ATN, Rosca F, Widom A, Champion PM. Investigations of Ultrafast Nuclear Response Induced by Resonant and Nonresonant Laser Pulses. J. Chem. Phys. 2001;114:6795–6815. [Google Scholar]
- 42.Constantine S, Zhou Y, Morais J, Ziegler LD. Dispersed Optical Heterodyne Detected Birefringence and Dichroism of Transparent Liquids. J. Phys. Chem. A. 1997;101:5456–5462. [Google Scholar]
- 43.Kubo M, Gruia F, Benabbas A, Barabanschikov A, Montfort WR, Maes EM, Champion PM. Low-Frequency Mode Activity of Heme: Femtosecond Coherence Spectroscopy of Iron Porphine Halides and Nitrophorin 2. J. Am. Chem. Soc. 2008;130:9800–9811. doi: 10.1021/ja800916d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colon W, Wakem LP, Sherman F, Roder H. Identification of the Predominant Non-Native Histidine Ligand in Unfolded Cytochrome C. Biochemistry. 1997;36:12535–12541. doi: 10.1021/bi971697c. [DOI] [PubMed] [Google Scholar]
- 45.Latypov RF, Cheng H, Roder NA, Zhang J, Roder H. Structural Characterization of an Equilibrium Unfolding Intermediate in Cytochrome C. J. Mol. Biol. 2006;357:1009–1025. doi: 10.1016/j.jmb.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 46.Russell BS, Bren KL. Denaturant Dependence of Equilibrium Unfolding Intermediates and Denatured State Structure of Horse Ferricytochrome C. J. Biol. Inorg. Chem. 2002;7:909–916. doi: 10.1007/s00775-002-0381-z. [DOI] [PubMed] [Google Scholar]
- 47.Jordan T, Eads JC, Spiro TG. Secondary and Tertiary Structure of the a-State of Cytochrome C from Resonance Raman Spectroscopy. Prot. Sci. 1995;4:716–728. doi: 10.1002/pro.5560040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manthey JA, Boldt NJ, Bocian DF, Chan SI. Resonance Raman Studies of Lactoperoxidase. J. Biol. Chem. 1986;261:6734–6741. [PubMed] [Google Scholar]
- 49.Rosca F, Ionascu D, Kumar ATN, Demidov AA, Champion PM. Femtosecond Coherence Spectroscopy Using Spectrally Selective Differential Photodetection. Chem. Phys. Lett. 2001;337:107–116. [Google Scholar]
- 50.Negrerie M, Cianetti S, Vos MH, Martin JL, Kruglik SG. Ultrafast Heme Dynamics in Ferrous Versus Ferric Cytochrome C Studied by Time-Resolved Resonance Raman and Transient Absorption Spectroscopy. J. Phys. Chem. B. 2006;110:12766–12781. doi: 10.1021/jp0559377. [DOI] [PubMed] [Google Scholar]
- 51.Banci L, Bertini I, Liu G, Lu J, Reddig T, Tang W, Wu Y, Yao Y, Zhu D. Effects of Extrinsic Imidazole Ligation on the Molecular and Electronic Structure of Cytochrome C. J. Biol. Inorg. Chem. 2001;6:628–637. doi: 10.1007/s007750100240. [DOI] [PubMed] [Google Scholar]
- 52.Schejter A, Aviram I. The Reaction of Cytochrome C with Imidazole. Biochemistry. 1969;8:149–153. doi: 10.1021/bi00829a021. [DOI] [PubMed] [Google Scholar]
- 53.Othman S, Lelirzin A, Desbois A. Resonance Raman Investigation of Imidazole and Imidazolate Complexes of Microperoxidase - Characterization of the Bis(Histidine) Axial Ligation in C-Type Cytochromes. Biochemistry. 1994;33:15437–15448. doi: 10.1021/bi00255a026. [DOI] [PubMed] [Google Scholar]
- 54.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Nonplanar Porphyrins and Their Significance in Proteins. Chem.Soc.Rev. 1998;27:31–41. [Google Scholar]
- 55.Chachisvilis M, Fidder H, Sundström V. Electronic Coherence in Pseudo Two-Colour Pump-Probe Spectroscopy. Chem. Phys. Lett. 1995;234:141–150. [Google Scholar]
- 56.Chachisvilis M, Sundstrom V. The Tunneling Contributions to Optical Coherence in Femtosecond Pump--Probe Spectroscopy of a Three Level System. J. Chem. Phys. 1996;104:5734–5744. [Google Scholar]
- 57.Kumar ATN, Rosca F, Widom A, Champion PM. Investigations of Amplitude and Phase Excitation Profiles in Femtosecond Coherence Spectroscopy. J. Chem. Phys. 2001;114:701–724. [Google Scholar]
- 58.Ma JG, Vanderkooi JM, Zhang J, Jia SL, Shelnutt JA. Resonance Raman Investigation of Nickel Microperoxidase-11. Biochemistry. 1999;38:2787–2795. doi: 10.1021/bi982332a. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell ML, Li XY, Kincaid JR, Spiro TG. Axial Ligand and out-of-Plane Vibrations for Bis(Imidazolyl)Heme: Raman and Infrared Iron-54, Nitrogen-15, and Deuterium Isotope Shifts and Normal Coordinate Calculations. J. Phys. Chem. 1987;91:4690–4696. [Google Scholar]
- 60.Bu LT, Straub JE. Vibrational Energy Relaxation of “Tailored” Hemes in Myoglobin Following Ligand Photolysis Supports Energy Funneling Mechanism of Heme “Cooling”. J. Phys. Chem. B. 2003;107:10634–10639. [Google Scholar]
- 61.Sagnella DE, Straub JE. Directed Energy “Funneling” Mechanism for Heme Cooling Following Ligand Photolysis or Direct Excitation in Solvated Carbonmonoxy Myoglobin. J. Phys. Chem. B. 2001;105:7057–7063. [Google Scholar]
- 62.Lyubovitsky JG, Gray HB, Winkler JR. Mapping the Cytochrome C Folding Landscape. J. Am. Chem. Soc. 2002;124:5481–5485. doi: 10.1021/ja017399r. [DOI] [PubMed] [Google Scholar]
- 63.Weinkam P, Zimmermann Jr, Romesberg FE, Wolynes PG. The Folding Energy Landscape and Free Energy Excitations of Cytochrome C. Acc. Chem. Res. 2010;43:652–660. doi: 10.1021/ar9002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinkam P, Zong C, Wolynes PG. A Funneled Energy Landscape for Cytochrome C Directly Predicts the Sequential Folding Route Inferred from Hydrogen Exchange Experiments. Proc. Natl. Acad. Sci. USA. 2005;102:12401–12406. doi: 10.1073/pnas.0505274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai Y, Sosnick T, Mayne L, Englander S. Protein Folding Intermediates: Native-State Hydrogen Exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoang L, Bédard S, Krishna MMG, Lin Y, Englander SW. Cytochrome C Folding Pathway: Kinetic Native-State Hydrogen Exchange. Proc. Natl. Acad. Sci. USA. 2002;99:12173–12178. doi: 10.1073/pnas.152439199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jay R W. Cytochrome C Folding Dynamics. Curr. Opin. Chem. Biol. 2004;8:169–174. doi: 10.1016/j.cbpa.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 68.Maity H, Maity M, Englander SW. How Cytochrome C Folds, and Why: Submolecular Foldon Units and Their Stepwise Sequential Stabilization. J. Mol. Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Krishna MM, Maity H, Rumbley JN, Englander SW. Branching in the Sequential Folding Pathway of Cytochrome C. Prot. Sci. 2007;16:1946–1956. doi: 10.1110/ps.072922307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bai Y, Milne JS, Mayne L, Englander SW. Protein Stability Parameters Measured by Hydrogen Exchange. Proteins. 1994;20:4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- 71.Krishna MM, Maity H, Rumbley JN, Lin Y, Englander SW. Order of Steps in the Cytochrome C Folding Pathway: Evidence for a Sequential Stabilization Mechanism. J. Mol. Biol. 2006;359:1410–1419. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 72.Russell BS, Melenkivitz R, Bren KL. Nmr Investigation of Ferricytochrome C Unfolding: Detection of an Equilibrium Unfolding Intermediate and Residual Structure in the Denatured State. Proc. Natl. Acad. Sci. USA. 2000;97:8312–8317. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monera OD, Kay CM, Hodges RS. Protein Denaturation with Guanidine Hydrochloride or Urea Provides a Different Estimate of Stability Depending on the Contributions of Electrostatic Interactions. Prot. Sci. 1994;3:1984–1991. doi: 10.1002/pro.5560031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fee JA, Chen Y, Todaro TR, Bren KL, Patel KM, Hill MG, Gomez-Moran E, Loehr TM, Ai J, Thöny-meyer L, et al. Integrity of Thermus Thermophilus Cytochrome C552 Synthesized by Escherichia Coli Cells Expressing the Host-Specific Cytochrome C Maturation Genes, Ccmabcdefgh: Biochemical, Spectral, and Structural Characterization of the Recombinant Protein. Prot. Sci. 2000;9:2074–2084. doi: 10.1110/ps.9.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leu BM, Alatas A, Sinn H, Alp EE, Said AH, Yavas H, Zhao JY, Sage JT, Sturhahn W. Protein Elasticity Probed with Two Synchrotron-Based Techniques. J. Chem. Phys. 2010;132 doi: 10.1063/1.3332585. [DOI] [PubMed] [Google Scholar]
- 76.Oellerich S, Lecomte S, Paternostre M, Heimburg T, Hildebrandt P. Peripheral and Integral Binding of Cytochrome C to Phospholipids Vesicles. J. Phys. Chem. B. 2004;108:3871–3878. [Google Scholar]
- 77.Yue HJ, Khoshtariya D, Waldeck DH, Grochol J, Hildebrandt P, Murgida DH. On the Electron Transfer Mechanism between Cytochrome C and Metal Electrodes. Evidence for Dynamic Control at Short Distances. J. Phys. Chem. B. 2006;110:19906–19913. doi: 10.1021/jp0620670. [DOI] [PubMed] [Google Scholar]
- 78.Berezhna S, Wohlrab H, Champion PM. Resonance Raman Investigations of Cytochrome C Conformational Change Upon Interaction with the Membranes of Intact and Ca2+-Exposed Mitochondria. Biochemistry. 2003;42:6149–6158. doi: 10.1021/bi027387y. [DOI] [PubMed] [Google Scholar]
- 79.Hildebrandt P, Heimburg T, Marsh D. Quantitative Conformational Analysis of Cytochrome C Bound to Phospholipid Vesicles Studied by Resonance Raman Spectroscopy. Eur Biophys J. 1990;18:193–201. doi: 10.1007/BF02427378. [DOI] [PubMed] [Google Scholar]
- 80.Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, Pletneva EV. Conformational Properties of Cardiolipin-Bound Cytochrome C. Proc. Natl. Acad. Sci. USA. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.