Abstract
Epithelial-mesenchymal transitions (EMT) are essential for organogenesis and triggered in carcinoma progression into an invasive state1. Transforming growth factor-β (TGF-β) cooperates with signalling pathways, such as Ras and Wnt, to induce EMT2-5, but the molecular mechanisms are not clear. Here, we report that SMAD3 and SMAD4 interact and form a complex with SNAIL1, a transcriptional repressor and promoter of EMT6, 7. The SNAIL1-SMAD3/4 complex was targeted to the gene promoters of CAR, a tight junction protein, and E-cadherin during TGF-β-driven EMT in breast epithelial cells. SNAIL1 and SMAD3/4 acted as co-repressors of CAR, occludin, claudin-3 and E-cadherin promoters in transfected cells. Conversely, co-silencing of SNAIL1 and SMAD4 by siRNA inhibited the repression of CAR and occludin during EMT. Moreover, loss of CAR and E-cadherin correlated with nuclear co-expression of SNAIL1 and SMAD3/4 in a mouse model of breast carcinoma and at the invasive fronts of human breast cancer. We propose that activation of a SNAIL1-SMAD3/4 transcriptional complex represents a novel mechanism of gene repression during EMT.
TGF-β plays dual roles in cancer by acting as a tumour suppressor in early tumour development, and paradoxically, by promoting tumour cell invasion in later stages4. Cooperation of TGF-β and other signalling pathways, such as Ras, is required for complete EMT2, 4, 5. TGF-β binding to its receptors leads to phosphorylation of SMAD2/3, which partner with SMAD4 and translocate to the nucleus where SMAD transcriptional complexes control the expression of target genes8. SMADs have low affinity for DNA and interact with DNA-binding cofactors to achieve high affinity and selectivity for specific target genes. Therefore, cells read TGF-β signals differently and the outcome of the response depends on the array of SMAD cofactors active at the time of exposure8. SMAD signalling is essential for TGF-β-induced EMT5 and SMAD4 promotes tumour cell invasion in advanced pancreatic tumours9.
The transcription factor SNAIL1 plays a key role in EMT both during development and in tumour progression10 by repressing junction components like E-cadherin11, 12, claudins, occludin13 and desmoplakin12. SNAIL1 is activated by multiple pathways6, 7 and negatively regulated by GSK-3β14, 15. SNAIL1 is specifically activated at the tumour-stroma interface16.
The documented involvement of SMADs and SNAIL1 in EMT prompted us to investigate whether these transcription factors could cooperate to regulate gene expression in TGF-β-induced EMT. In particular, we studied the regulation of the coxsackie- and adenovirus receptor (CAR), a tight junction-associated cell adhesion molecule, which is down-regulated in human cancer and in TGF-β-induced EMT17-19, and E-cadherin, an established marker of EMT. CAR is predominantly expressed in epithelial cells where it contributes to tight junction formation and barrier function17, 20. A role for CAR as a tumour suppressor has been suggested based on studies showing that loss of CAR is associated with aggressive bladder cancer21 and over-expression of CAR reduces the metastatic potential of tumour cells22. CAR is regulated by the Raf-MEK-ERK pathway during carcinoma progression23 but the transcriptional mechanisms have not been determined. Here, we used two established in vitro models of TGF-β-induced EMT to investigate the involvement of SNAIL1 and SMADs in the regulation of CAR and E-cadherin.
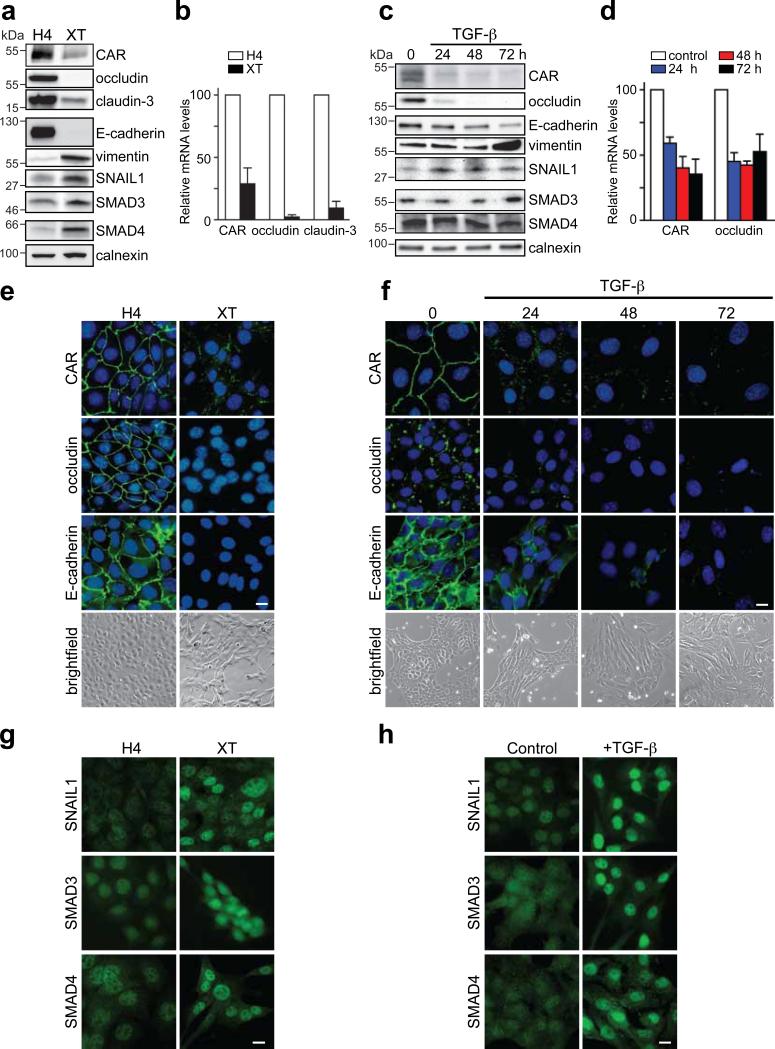
EpH4-EpRas-EpXT is a model of stable EMT in mouse breast epithelial cells induced by the cooperative actions of TGF-β and Ras24. EpXT cells had EMT characteristics with repression of E-cadherin, upregulation of vimentin (Fig. 1a, e) and N-cadherin (Fig. S1a) and a mesenchymal morphology compared to EpH4 cells (Fig. 1e). The expression of CAR, occludin and claudin-3 was also reduced in EpXT versus EpH4 cells, both at protein and mRNA levels (Fig. 1a, b and Fig. S1a), indicating that transcriptional repression was involved. The remaining CAR protein was discontinuously distributed along cell-cell contacts in EpXT cells (Fig.1e), suggesting that tight junctions were disorganized during EMT, as indicated by others13. Consistent with decreased CAR levels, EpXT cells were less sensitive to adenovirus infection compared to EpH4 cells (Fig. S1a).
Figure 1. Loss of junction proteins is associated with nuclear accumulation of SNAIL1 and SMAD3/4 during EMT in breast epithelial cells.
Analysis of the expression of CAR, occludin, claudin-3, E-cadherin, vimentin, SNAIL1 and SMAD3/4 during EMT in EpH4/EpXT (a, b, e, g) and NMuMG (c, d, f, h) cells. (a, c) Immunoblot analysis of protein expression during EMT. A time course experiment showing differences in expression in NMuMG cells treated with TGF-β (10 ng/ml) (c). Calnexin was used as a loading control. Full scans are shown in Supplementary Information, Fig. S5. (b, d) Semi-quantitative analysis of mRNA levels during EMT. A time course experiment showing differences in NMuMG cells treated with TGF-β. Data in b and d represent mean ± SEM of three independent experiments. (e, f) Immunofluorescence analysis of the expression and localization of junction proteins (green) during EMT. Nuclei were visualized with DAPI staining (blue). Morphological evidence of EMT is shown in bright-field images. Bars indicate 40 μm. (g, h) Immunofluorescence analysis of SNAIL1 and SMAD3/4 during EMT. NMuMG cells were treated with TGF-β for 12 hours. Bars indicate 40 μm.
NMuMG cells are frequently used as a model of inducible and transient TGF-β mediated EMT25. TGF-β treatment of NMuMG cells resulted in EMT with down-regulation of CAR, E-cadherin and occludin, increased expression of vimentin and cell elongation (Fig. 1c, d, f), as previously shown18. Similar to EpXT cells, CAR distribution shifted from a continuous to a discontinuous pattern at cell-cell contacts after TGF-β-treatment (Fig. 1f).
To determine the involvement of SNAIL1 and SMAD3/4 in the repression of CAR and other junction proteins in TGF-β-mediated EMT, we analyzed the expression and intracellular localization of these transcription factors. The expression of SNAIL1, SMAD3 and SMAD4 was increased in EpXT compared to EpH4 cells, at protein (Fig. 1a) and mRNA levels (Fig. S1b). TGF-β treatment of NMuMG cells resulted in increased expression of SNAIL1 but did not significantly alter the expression of SMAD3 and SMAD4 (Fig. 1c and S1c).
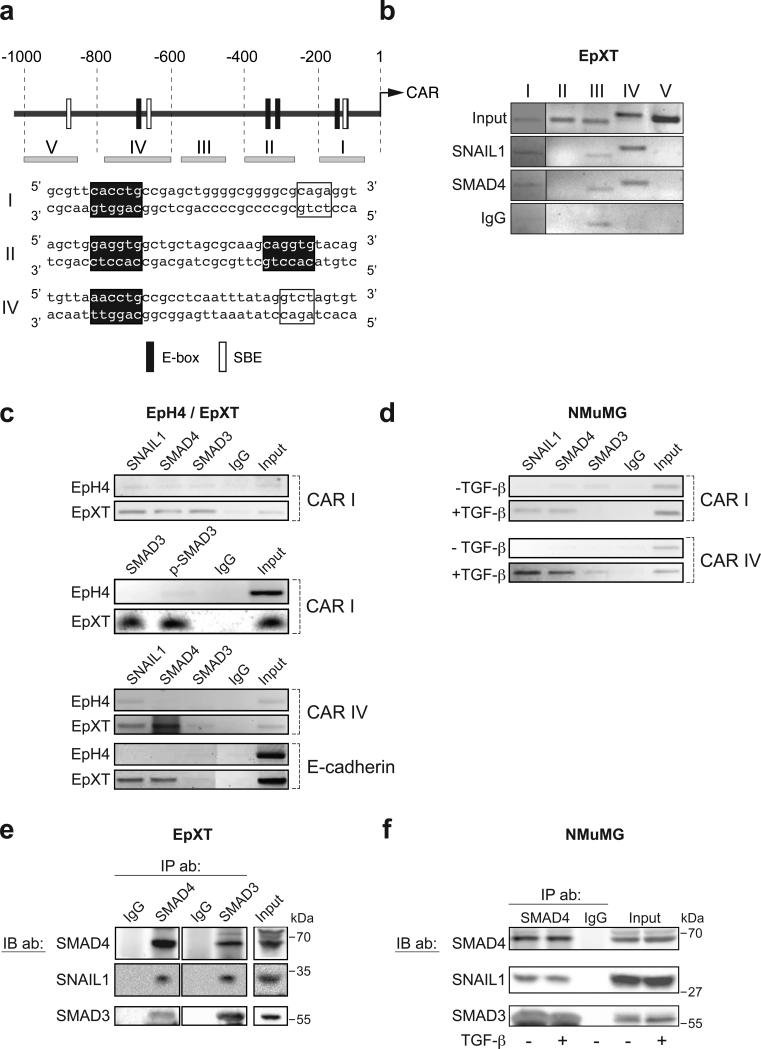
SNAIL1 and SMAD3/4 were diffusely localized in the cytosol and nucleus in EpH4 and untreated NMuMG cells but concentrated in the nuclei of EpXT and TGF-β-treated NMuMG cells (Fig. 1g, h). Nuclear translocation of SNAIL1 and SMAD3/4 occurred rapidly after TGF-β exposure and followed similar kinetics (data not shown). Computer-based sequence analysis showed that the mouse CAR promoter contained putative SNAIL1 sites (E-boxes) and SMAD binding elements (SBEs) (Fig. 2a). To test whether SNAIL1 and /or SMAD3/4 could interact with the CAR promoter we performed chromatin immunoprecipitation (ChIP) analysis using primers for five different regions of the CAR promoter (Fig. 2a, regions I-V). In EpXT cells, SNAIL1 and SMAD4 specifically interacted with regions I and IV (Fig. 2b). The E-box in region I (CACCTG) conforms to the consensus SNAIL1 binding site (CANNTG) while the E-box in region IV (CAGGTT, reverse strand) differed slightly from the consensus sequence. However, CAGGTT sequences have previously been reported to confer binding sites for SNAIL126. The E-boxes and SBEs in regions I and IV were located in close proximity to each other (16-18 base pairs apart), suggesting that SNAIL1 and SMAD3/4 could bind DNA in a cooperative fashion.
Figure 2. SNAIL1 and SMAD3/4 interact and are recruited to CAR and E-cadherin promoters during EMT.
(a) Diagram indicating putative SNAIL1 sites (E-box) and SMAD-binding elements (SBE) in different regions (I-V) of the 1000 bp genomic DNA sequence upstream of the ATG in the CAR gene (upper panel). Sequences of E-box and SBE sites in regions I, II and IV are shown. (b) Chromatin immunoprecipitation (ChIP) analysis of regions I-V was performed with antibodies against SNAIL1 and SMAD4. (c) ChIP analysis of the interaction of SNAIL1 and SMAD3/4 with the CAR (regions I and IV) and E-cadherin promoters in EpH4 versus EpXT cells. The interaction of phosporylated SMAD3 (p-SMAD3) with region I of the CAR promoter was analyzed. (d) Interaction of SNAIL1, SMAD3/4 with the CAR promoter (regions I and IV) in untreated or TGF-β treated NMuMG cells (12 h). (e, f) Co-immunoprecipitation (Co-IP) analysis of the interaction of SNAIL1 with SMAD4 and SMAD3 in EpXT cells (e) and in untreated or TGF-β treated NMuMG cells (12 h) (f). Full scans are shown in Supplementary Information, Fig. S5.
Subsequently, we compared the interaction of SNAIL1 and SMAD3/4 with the CAR promoter under EMT versus non-EMT conditions. SNAIL1 and SMAD3/4 interacted with regions I and IV in EpXT and TGF-β-treated NMuMG cells but were less detectable in EpH4 and untreated NMuMG cells (Fig. 2c, d). SMAD3 interacted with region I of the CAR promoter in its phosphorylated form (Fig. 2c).
Putative SBEs were found adjacent to E-boxes also in E-cadherin, occludin and claudin-3 promoters (Fig. S2f), suggesting that cooperative binding of SNAIL1 and SMADs could be a general mechanism of action of these transcription factors. Additionally, a SMAD4-specific binding site was recently mapped to a location close to E-boxes in the E-cadherin promoter27. ChIP analysis showed that SNAIL1 and SMAD4, and to a lesser extent SMAD3, interacted with this particular region of the E-cadherin promoter in EpXT, but not in EpH4 cells (Fig. 2c). Thus, during TGF-β-induced EMT, SNAIL1 and SMAD3/4 were translocated to the nucleus and targeted to regions within CAR and E-cadherin promoters containing adjacent E-boxes and SBEs, suggesting that they may interact and cooperatively regulate gene expression.
To study whether SNAIL1 interacted with SMAD3/4, we performed co-immunoprecipitation (Co-IP) studies. In whole cell extracts from EpXT cells, we found SNAIL1 in complex with SMAD3 and SMAD4 (Fig. 2e), and in nuclear extracts, SMAD4 in complex with SNAIL1 (Fig. S1d). The interaction between SNAIL1 and SMAD3/4 was not restricted to EpXT cells but also found in NMuMG (Fig. 2f) and EpH4 cells (data not shown). This suggested that while the nuclear accumulation and recruitment of SNAIL1 and SMAD3/4 to CAR and E-cadherin promoters was specific to EMT, the interaction was not.
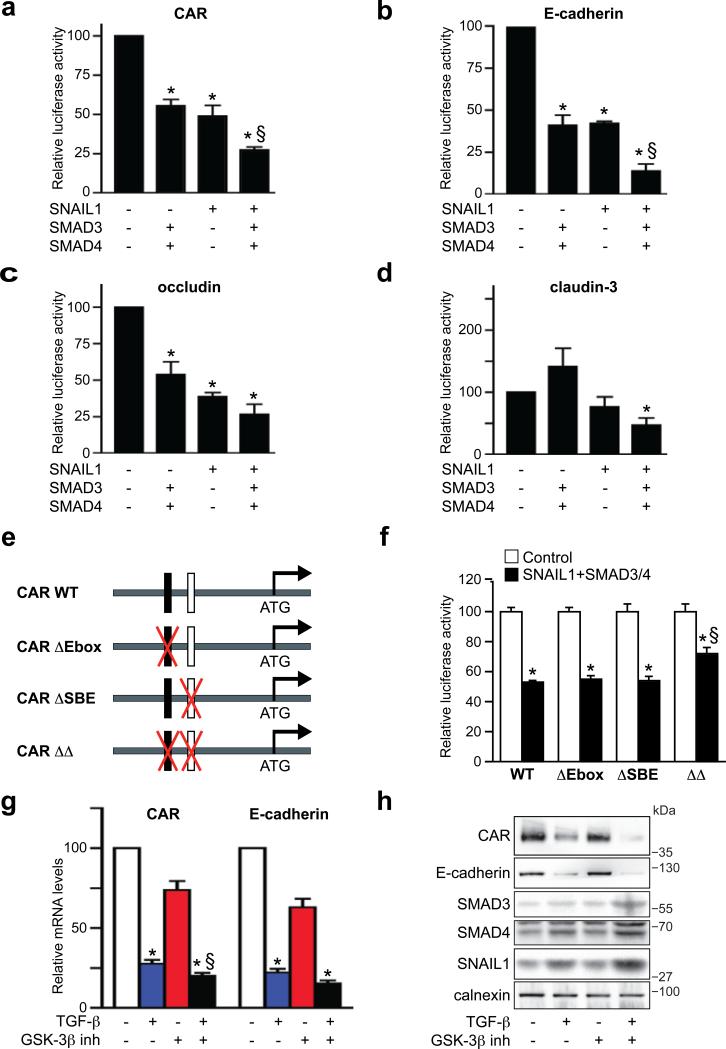
The capacity of SNAIL1 and SMAD3/4, separately or in combinations, to regulate the activity of CAR, E-cadherin, occludin and claudin-3 promoters was analyzed by reporter assays in EpH4 cells. SNAIL1 repressed CAR, E-cadherin and occludin promoter activities by 40-60% (Fig. 3a-c) while SMAD3 or SMAD4 alone did not cause significant repression (Fig. S2a-c). However, in combination, SMAD3 and SMAD4 caused, similar to SNAIL1, 50% repression of all three promoters (Fig. 3a-c). The claudin-3 promoter was repressed by SNAIL1 (20%) (Fig. 3d) but activated by SMAD3 and SMAD4 (50-100%), both in combination (Fig. 2d), and separately (Fig. S2d).
Figure 3. SNAIL1 and SMAD3/4 act as transcriptional co-repressors.
(a-d) Reporter assays showing SNAIL1 and SMAD3/4 repression of CAR, E-cadherin, occludin and claudin-3 promoters. Data represent mean ± SEM of four independent experiments. *, P < 0.05, different from negative control. §, P < 0.05, different from SNAIL1. (e, f) Mutational analysis of Ebox and SBE sites in region I of the CAR promoter. Reporter constructs containing wild-type CAR (CAR WT), E-box (CAR ΔEbox) or SBE (CAR ΔSBE) mutations, or double mutations (CAR ΔΔ) were generated and used to analyze the importance of these sites in mediating repression by SNAIL1 and SMAD3/4. Data indicate mean ± SEM and are representative of three independent experiments. *, P < 0.05, different from control. §, P < 0.05, different from WT. (g, h) Effect of GSK-3β inhibition on the repression of CAR and E-cadherin mRNA (g) and protein (h) during TGF-β induced EMT (24 h) in NMuMG cells. SNAIL1 and SMAD3/4 proteins were also analyzed (h). Data represent mean ± SEM of four independent experiments. *, P < 0.05, different from untreated cells. §, P < 0.05, different from TGF-β alone. Full scans are shown in Supplementary Information, Fig. S5.
The combination of SNAIL1 and SMAD3/4 repressed CAR, E-cadherin and occludin promoters by 70-80%, which was significantly more efficient than SNAIL1 or SMAD3/4 separately (Fig. 3a-c). The claudin-3 promoter was also significantly repressed (50%) by the combination of SNAIL1 and SMAD3/4 (Fig. 3d), indicating that SNAIL1 converted SMAD3/4 from transcriptional activators to repressors. Similar results were obtained for the human E-cadherin promoter (Fig. S2e) and for the mouse CAR promoter in NMuMG and 293 cells (data not shown), showing that the co-repressor effect of SNAIL1 and SMAD3/4 was not cell-type specific.
To study the importance of specific SNAIL1 and SMAD binding sites for the cooperative repression of SNAIL1 and SMAD3/4 we generated wild-type (WT) and mutant (single or double mutants) reporter constructs of region I of the CAR promoter (Fig. 3e). The WT and mutant promoters had comparable activities when transfected into EpH4 cells (Fig. 3f). The WT and single mutant promoters were similarly repressed by SNAIL1 and SMAD3/4. However, the double mutant promoter was significantly less repressed compared to the WT promoter, indicating the importance of the adjacent Ebox and SBE sites for the cooperative effects of SNAIL1 and SMAD3/4 (Fig. 3f).
The stability and nuclear activity of SNAIL1 is negatively regulated by phosphorylation via GSK-3β14, 15, which is inhibited by multiple pathways including Ras/PI3K and Wnt signalling. We hypothesized that stabilization of SNAIL1 via inhibition of GSK-3β would result in more potent repression of junction proteins during TGF-β-induced EMT. To test this, we treated NMuMG cells with TGF-β in the presence or absence of a GSK-3β inhibitor. The GSK-3β inhibitor alone repressed CAR and E-cadherin mRNA levels by 20-30% (Fig. 3g) but did not affect the expression or localization of CAR, E-cadherin, SNAIL1 or SMAD3/4 proteins (Fig. 3g, h and Fig. S3a, b). However, the combination of TGF-β and the GSK-3β inhibitor resulted in more potent repression of CAR and E-cadherin (Fig. 3g, h) and increased expression of SNAIL1 and SMAD3/4 protein (Fig. 3h). RNA levels were less affected (Fig. S3a), indicating that the increases in SNAIL1 and SMAD3/4 were due to enhanced protein stability and that an increased pool of SNAIL1 and SMAD3/4 transcription factors available to form a complex results in more efficient repression of junction proteins during EMT.
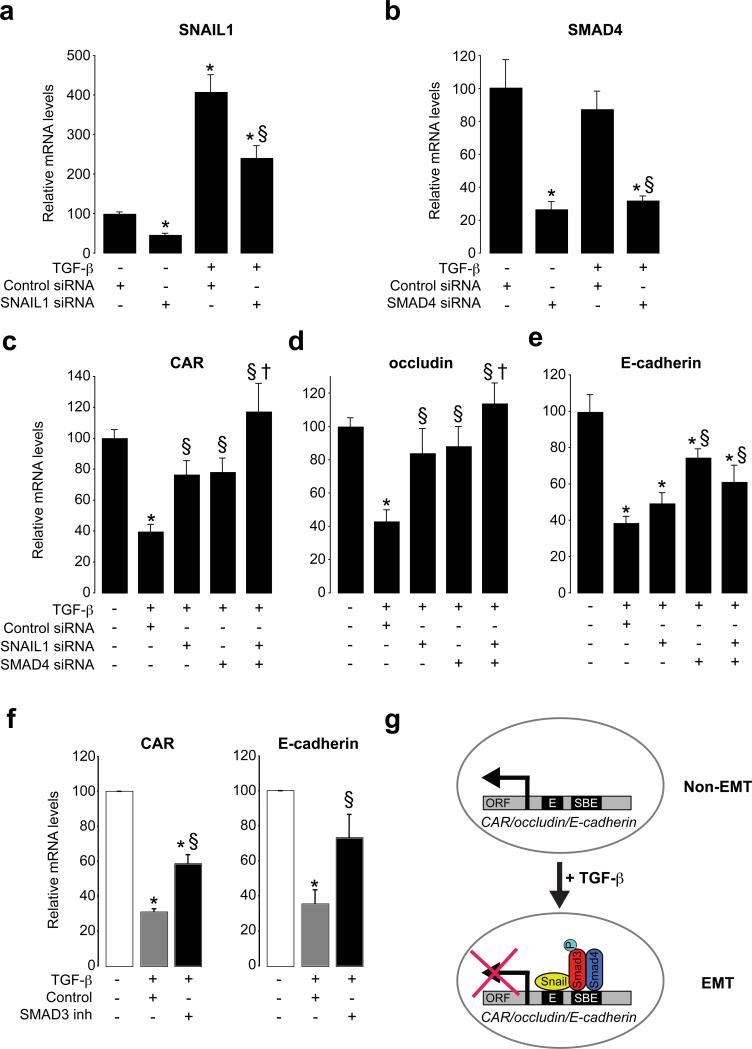
To further determine the involvement of SNAIL1 and SMAD3/4 in the repression of CAR and other junction proteins during TGF-β-induced EMT, we used RNA interference to silence these transcription factors in NMuMG cells. Using SNAIL1 and SMAD4 siRNAs, SNAIL1 was repressed by 50% and SMAD4 by 70%, respectively, both under baseline conditions and after TGF-β (Fig. 4a, b). Silencing of SNAIL1 significantly rescued the repression of CAR and occludin (Fig. 4c, d) after TGF-β. E-cadherin was marginally rescued by SNAIL1 siRNA (Fig. 4e), suggesting the repression of CAR and occludin was more dependent on SNAIL1. SMAD4 silencing resulted in significant rescue of CAR, occludin and E-cadherin (Fig. 4c, d, e) repression after TGF-β. Since our previous results showed that SNAIL1 and SMAD3/4 functioned as co-repressors, we studied if the combination of SNAIL1 and SMAD4 siRNAs was more potent in rescuing TGF-β-induced repression. Indeed, co-silencing of SNAIL1 and SMAD4 resulted in complete rescue of the repression of CAR and occludin after TGF-β (Fig. 4c, d). For E-cadherin, no additive rescue effect of using both SNAIL1 and SMAD4 siRNAs was observed (Fig. 4e). The involvement of SMAD3 was determined by using a specific inhibitor of SMAD3 (SIS3), which caused significant rescue of both CAR and E-cadherin repression after TGF-β (Fig. 4f).
Figure 4. SNAIL1 and SMAD3/4 play essential and cooperative roles as transcriptional repressors of junction components during EMT.
(a, b) RNA analysis of the effect of siRNA mediated silencing of SNAIL1 and SMAD4 in NMuMG cells in untreated and TGF-β treated NMuMG cells (24 h). (c-e) Effect of SNAIL1 and SMAD4 siRNAs on CAR, occludin and E-cadherin mRNA levels during TGF-β induced EMT. Data indicate mean ± SEM and are representative of three independent experiments. *, P < 0.05, different from untreated cells. §, P < 0.05, different from TGF-β + control siRNA. †, P < 0.05, different from TGF-β + SNAIL1 siRNA. (f) Effect of SMAD3 inhibition on CAR and E-cadherin mRNA levels during TGF-β induced EMT. *, P < 0.05, different from untreated cells. §, P < 0.05, different from TGF-β treatment. Data represent mean ± SEM of three independent experiments. (g) Cooperation of SNAIL1 and SMAD3/4 transcription factors during TGF-β-induced EMT. SNAIL1 and SMAD3/4 interact and form a transcriptional repressor complex, which is targeted to adjacent Eboxes (E) and SBE sites in genes encoding junction proteins like CAR, occludin and E-cadherin. As a result, these genes are repressed.
Together, these results demonstrate that SNAIL1 is a cofactor for SMAD3/4 and that SNAIL1-SMAD3/4 complexes during TGF-β-induced EMT are recruited to CAR, occludin and E-cadherin promoters, where Eboxes and SBEs are located in close proximity to each other (Fig. 4g). As a result, the transcription of these genes is repressed. The repression of E-cadherin in NMuMG cells was less dependent on SNAIL1 compared to CAR and occludin, which is in agreement with published data indicating that other transcriptional repressors, such as SIP1 and δEF1, repress E-cadherin in NMuMG cells during TGF-β-induced EMT28. However, since SNAIL1 was incompletely blocked, one cannot exclude the possibility that SNAIL1 contributes to the repression of E-cadherin in NMuMG cells.
The importance of the SNAIL1-SMAD3/4 complex for EMT in cultured breast epithelial cells encouraged us to identify in vivo evidence of cooperation of SNAIL1 and SMAD3/4 in invasive breast cancer. First, we used a mouse model of subcutaneously grown D2F2 breast carcinoma cells overexpressing human HER-2 (D2F2/E2), which results in the establishment of tumours with invasive properties and capacity to metastasize to the lungs29. Established tumours were composed of morphologically distinct compartments containing cells with either an epithelial or mesenchymal morphology (Fig. 5a). Cells within both compartments were positive for HER-2, indicating that the mesenchymal-like cells had originated from the initial pool of injected tumour cells that had undergone EMT-like changes during tumour formation in vivo. In agreement with this, mesenchymal cells were positive for vimentin while the epithelial cells expressed CAR and E-cadherin (Fig. 5a). SNAIL1 and SMAD4 were not detected in cells within the epithelial compartment but were co-expressed in the nuclei of cells within the mesenchymal compartment (Fig. 5b). This supported a role for the SNAIL1-SMAD3/4 complex in the induction of EMT in this subcutaneous mouse model of breast carcinoma.
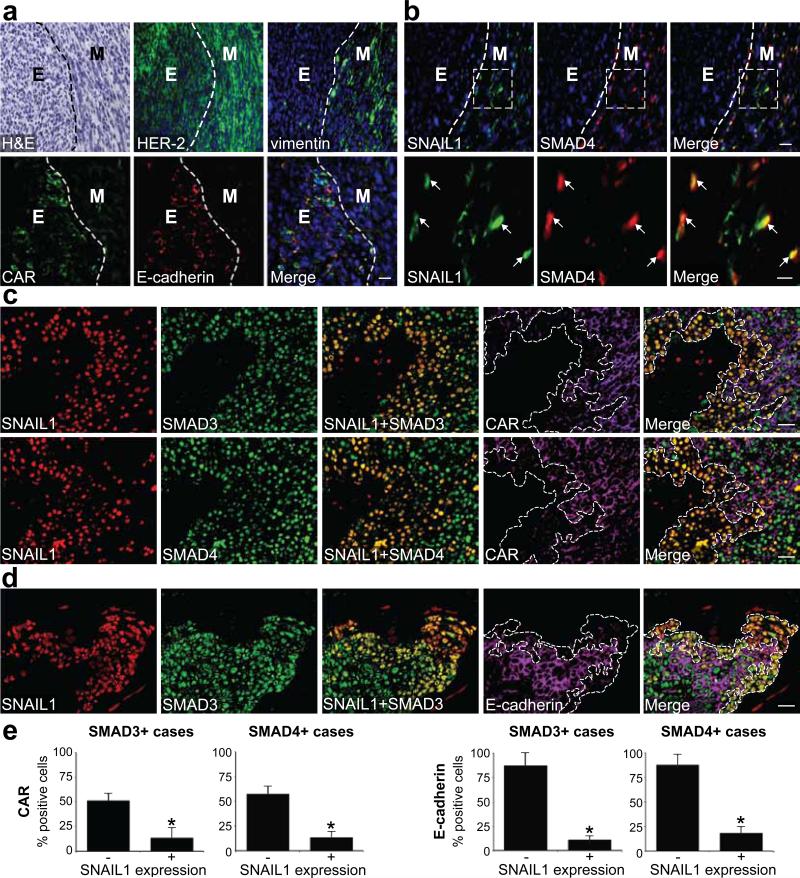
Figure 5. Loss of junction proteins correlates with nuclear co-expression of SNAIL1 and SMAD3/4 in mouse breast carcinomas and at the invasive front in human breast cancer.
(a, b). Immunofluorescence analysis of the expression of HER-2, vimentin, CAR, E-cadherin, SNAIL1 and SMAD4 in subcutaneously grown mouse D2F2/E2 breast carcinomas. Tumour compartments with epithelial (E) or mesenchymal (M) morphology (separated by dashed lines) were identified by hematoxylin and eosin (H&E) staining. Boxes in b indicate areas magnified in lower panels. Arrows indicate nuclear co-expression of SNAIL1 and SMAD4. Scale bars = 50 μm or 20 μm (b, lower panels). (c, d) Immunofluorescence analysis of the expression of SNAIL1, SMAD3/4, CAR and E-cadherin in sections of invasive human ductal breast carcinomas. (c) Representative images showing correlation of nuclear co-expression of SNAIL1 and SMAD3 (upper panels) or SMAD4 (lower panels) with loss of CAR expression (outlined regions). Scale bars = 30 μm. (d) Representative images showing correlation between nuclear co-expression of SNAIL1 and SMAD3 and loss of E-cadherin expression (outlined regions). Scale bars = 30 μm. (e) Quantification of the percentage of SMAD3+ cases and SMAD4+ cases of tumour cells expressing CAR, E-cadherin and SNAIL1. *, P = 0.026 (CAR), P = 0.012 (E-cadherin), different from SNAIL1 negative cells in each graph. 100 cells per field in 10 different fields of each tumour (n=8) were quantified.
Second, we analyzed a series of 25 human infiltrating ductal breast carcinomas for co-expression of SNAIL1 and SMAD3/4 and loss of CAR or E-cadherin expression. Significant expression of SNAIL1 was found in 8 cases where it was confined to the nuclei of tumour cells at the invasive front. The expression of SMAD3 and SMAD4 was more ubiquitous (Fig. 5c, d). CAR and E-cadherin were detected at junctions between tumour cells except in areas of invasion, where the expression was reduced or lost. Loss of CAR and E-cadherin correlated with nuclear co-expression of SNAIL1 and SMAD3/4 (Fig. 5c, d and Fig. S4a-c). SMAD3/4 expressing tumour cells positive for SNAIL1 expressed significantly less CAR and E-cadherin compared to cells negative for SNAIL1 (Fig. 5e). These results supported cooperative roles of SNAIL1-SMAD3/4 in the induction of EMT during tumour cell invasion.
In summary, we found that SNAIL1 is a cofactor for SMAD3/4 and that these transcription factors form a transcriptional repressor complex, which, during TGF-β-induced EMT, represses CAR, occludin and E-cadherin transcription. Thus, cooperation of SNAIL1 and SMAD3/4 transcription factors affects the outcome of the TGF-β response in cells. A strong correlation was found between loss of CAR and E-cadherin and nuclear co-expression of SNAIL1 and SMAD3/4 at the invasive front of breast carcinomas in support of a role for SNAIL1-SMAD3/4 complexes in human cancer.
Methods
Cells, antibodies, plasmids, primers and reagents. See supplementary Information, Methods.
Protein analysis including immunofluorescence microscopy, immunoprecipitation and Western blotting. See supplementary Information, Methods.
Semi-quantitative real-time PCR, luciferase reporter and chromatin immunoprecipitation (ChIP) assays. See supplementary Information, Methods.
RNA Interference
Short interfering RNAs (siRNAs) were transfected into NMuMG cells grown in 12-well plates according to the protocol recommended for Dharmafect I (Dharmacon/Thermo Scientific/Nordic Biolabs, Taby, Sweden). Final concentrations of each siRNA were 200 nM. At 20 h after transfection, cells were split and after an additional 6 h, 5 ng/ml TGF-β was added to the media. Cells were harvested and analyzed at 12h after the administration of TGF-β. The following siRNA sequences were used: mouse SNAIL1 (GGUACAACAGACUAUGCAA){Shirakihara, 2007 #47}, mouse SMAD4 (ON-TARGET plus SMARTpool; UCAGGUGGCUGGUCGGAAA, GAGUGCAGUUGGAAUGUAA, GCAAUUGAGAGUUUGGUAA, UCAGUAUGCGUUUGACUUA) and control (ONTARGETplus Non-Targeting Pool).
Subcutaneous mouse breast carcinoma model
Syngeneic breast carcinomas were established by injection of 2×106 D2F2 cells stably transfected with HER-230 subcutaneously into the flank of 10 week old female Balb/c mice. Upon reaching a volume of 500 mm3, tumours were excised and frozen in OCT following perfusion through the heart with 10 ml PBS and 10 ml 2% paraformaldehyde. Frozen sections were air dried and fixed in ice-cold acetone for 10 minutes. Following stabilization in PBS for 2 × 5 min, sections were blocked using 0.5% blocking reagent in PBS (NEN Life Science Products) or using mouse-on-mouse blocking reagent (Vector Laboratories) for 1-2 hours at room temperature. After aspiration, antibodies diluted in PBS with 1% BSA (washing solution) were applied and incubated at 4°C overnight. Antibodies and dilutions used were: HER-2 (1:40; CB11-L-CE, Novocastra), vimentin (1:1; 3B4, DAKO), SMAD4 (rabbit, polyclonal Santa Cruz, 1:100), SNAIL1 (monoclonal, 1:100). Slides were washed 3 × 10 min in washing solution and subsequently incubated 2 h at room temperature with appropriate conjugated secondary antibodies (fluorescently labeled, 1:1000, Invitrogen). Following 3 × 10 min wash in washing solution, sections were mounted for microscopic evaluation in medium containing DAPI (Vectashield, Vector Laboratories).
Immunofluorescence staining of human breast carcinomas
Immunofluorescence analysis of SNAIL1, SMAD3, SMAD4, CAR and E-cadherin was performed on two sequential formalin-fixed paraffin-embedded 3μm tissue sections from 25 human breast carcinoma specimens placed on positively-charged glass slides (2 per specimen). A pathologist (FR) diagnosed all cases as infiltrating ductal carcinomas. After deparaffinization, antigen retrieval was performed by incubation in Tris-EDTA buffer, pH 9.0 (Dako, Carpinteria, CA), in a heated (97°C) water bath for 40 minutes. The tumour sections were immersed in TBS/5% BSA for 10 minutes to block non-specific binding. The first set of sections from each tumour was incubated with the following antibodies: rabbit polyclonal anti-SMAD3 (Abcam, Cambridge, UK, diluted 1:100), mouse monoclonal anti-SNAIL1 (diluted 1:5) and mouse monoclonal anti-E-cadherin (Dako, diluted 1:50) or rabbit polyclonal anti-CAR antibodies (Atlas Antibodies, Stockholm, Sweden, diluted 1:50). The second set of sections was incubated with rabbit polyclonal anti-SMAD4 (SantaCruz Biotech, Santa Cruz, CA, diluted 1:125) and the combination of SNAIL1 and E-cadherin/CAR antibodies, as described. SNAIL, CAR and E-cadherin were detected using Alexa Fluor 655 conjugated goat anti-mouse or Alexa Fluor 568 mouse anti-rabbit IgG (Molecular Probes, Eugene, OR) secondary antibodies (diluted 1:700) and SMAD3/4 by using an Alexa Fluor 488 mouse anti-rabbit IgG secondary antibody. 8 out of 25 cases were clearly positive for SNAIL1. Sections were counterstained with 4',6-diamidino-2-phenylindole dihydrochloride (DAPI, Vysis, Downers Grove, IL) to visualize cell nuclei. All incubations were performed at room temperature. Fluorescence assays were performed using a Dako Autostainer. Staining was evaluated by a pathologist (FR) using a Leica fluorescence DM2000 microscope. Pictures were obtained by Leica Application Suite v2.8.1 and color deconvolution and image composition was performed using ImageJ software (http://rsb.info.nih.gov/ij). Further details of the procedure used for immunofluorescence staining are available upon request. Fluorescence in tumour cells was scored as percentage of the expression of SMAD3, SMAD4, SNAIL, CAR and E-cadherin.
Statistical analysis
Values are presented as means +/− SEM with 3-6 samples (n) per group. The significance between means was assessed by analysis of variance (ANOVA), followed by the Fisher's test or the Wilcoxon Signed Ranks test (for the tumour samples) for multiple comparisons. P < 0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Dr. Carl-Henrik Heldin (Ludwig Institute for Cancer Research, Uppsala Branch) and Dr. Ola Hermansson (Department of Neuroscience, Karolinska Institute, Sweden) for fruitful discussions, Dr. Clara Francí (IMIM-Hospital del Mar, Barcelona) for the initial immunohistochemical analysis of human tumors, Anita Bergström, Silvia Menéndez and Marta Garrido for excellent technical assistance and Dr. Olov Andersson (Department of Cell and Molecular Biology, Karolinska Institute) for antibodies. Jonas Fuxe was supported by grants from the Swedish Research Council, the Swedish Wenner-Gren Foundation, the Swedish Childhood Cancer Foundation and an International Union Against Cancer (UICC), American Cancer Society International Fellowship for Beginning Investigators. Theresa Vincent was supported by the Swedish Research Council. Philip Leopold and Ronald Crystal were supported by the National Institutes of Health (NIH) by PO1 HL59312 and Antonio García de Herreros, Joan Albanell and Federico Rojo by RD06/0020/109, RD06/0020/040, FIS PI061513, SAF2006-00339 and Fundació Privada Cellex.
References
- 1.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 2.Janda E, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massague J. TGF[beta] in Cancer. Cell. 2008;134:215–230. [Google Scholar]
- 4.Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tissues Organs. 2005;179:11–23. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- 5.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 6.De Craene B, van Roy F, Berx G. Unraveling signalling cascades for the Snail family of transcription factors. Cell Signal. 2005;17:535–547. doi: 10.1016/j.cellsig.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Yook JI, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 8.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 9.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 11.Batlle E, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 12.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 13.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 14.Bachelder RE, Yoon SO, Franci C, de Herreros AG, Mercurio AM. Glycogen synthase kinase-3 is an endogenous inhibitor of Snail transcription: implications for the epithelial-mesenchymal transition. J Cell Biol. 2005;168:29–33. doi: 10.1083/jcb.200409067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 16.Franci C, et al. Expression of Snail protein in tumor-stroma interface. Oncogene. 2006;25:5134–5144. doi: 10.1038/sj.onc.1209519. [DOI] [PubMed] [Google Scholar]
- 17.Coyne CB, Bergelson JM. CAR: a virus receptor within the tight junction. Adv Drug Deliv Rev. 2005;57:869–882. doi: 10.1016/j.addr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Lacher MD, et al. Transforming growth factor-beta receptor inhibition enhances adenoviral infectability of carcinoma cells via up-regulation of Coxsackie and Adenovirus Receptor in conjunction with reversal of epithelial-mesenchymal transition. Cancer Res. 2006;66:1648–1657. doi: 10.1158/0008-5472.CAN-05-2328. [DOI] [PubMed] [Google Scholar]
- 19.Philipson L, Pettersson RF. The coxsackie-adenovirus receptor--a new receptor in the immunoglobulin family involved in cell adhesion. Curr Top Microbiol Immunol. 2004;273:87–111. doi: 10.1007/978-3-662-05599-1_3. [DOI] [PubMed] [Google Scholar]
- 20.Raschperger E, et al. The coxsackie- and adenovirus receptor (CAR) is an in vivo marker for epithelial tight junctions, with a potential role in regulating permeability and tissue homeostasis. Exp Cell Res. 2006;312:1566–1580. doi: 10.1016/j.yexcr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto K, Shariat SF, Ayala GE, Rauen KA, Lerner SP. Loss of coxsackie and adenovirus receptor expression is associated with features of aggressive bladder cancer. Urology. 2005;66:441–446. doi: 10.1016/j.urology.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita M, Ino A, Kawabata K, Sakurai F, Mizuguchi H. Expression of coxsackie and adenovirus receptor reduces the lung metastatic potential of murine tumor cells. Int J Cancer. 2007;121:1690–1696. doi: 10.1002/ijc.22852. [DOI] [PubMed] [Google Scholar]
- 23.Anders M, Christian C, McMahon M, McCormick F, Korn WM. Inhibition of the Raf/MEK/ERK pathway up-regulates expression of the coxsackievirus and adenovirus receptor in cancer cells. Cancer Res. 2003;63:2088–2095. [PubMed] [Google Scholar]
- 24.Oft M, et al. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 25.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauhin V, Lutz Y, Dennefeld C, Alberga A. Definition of the DNA-binding site repertoire for the Drosophila transcription factor SNAIL. Nucleic acids research. 1993;21:3951–3957. doi: 10.1093/nar/21.17.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nawshad A, Medici D, Liu CC, Hay ED. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Molecular biology of the cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahoney KH, Miller BE, Heppner GH. FACS quantitation of leucine aminopeptidase and acid phosphatase on tumor-associated macrophages from metastatic and nonmetastatic mouse mammary tumors. J Leukoc Biol. 1985;38:573–585. doi: 10.1002/jlb.38.5.573. [DOI] [PubMed] [Google Scholar]
- 30.Wei WZ, et al. Protection against mammary tumor growth by vaccination with full- length, modified human ErbB-2 DNA. Int J Cancer. 1999;81:748–754. doi: 10.1002/(sici)1097-0215(19990531)81:5<748::aid-ijc14>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.