SUMMARY
Protein function is regulated by diverse posttranslational modifications. The mitochondrial sirtuin SIRT5 removes malonyl and succinyl moieties from target lysines. The spectrum of protein substrates subject to these modifications is unknown. We report systematic profiling of the mammalian succinylome, identifying 2,565 succinylation sites on 779 proteins. Most of these do not overlap with acetylation sites, suggesting differential regulation of succinylation and acetylation. Our analysis reveals potential impacts of lysine succinylation on enzymes involved in mitochondrial metabolism; e.g., amino acid degradation, the tricarboxylic acid cycle (TCA) cycle, and fatty acid metabolism. Lysine succinylation is also present on cytosolic and nuclear proteins; indeed, we show that a substantial fraction of SIRT5 is extra-mitochondrial. SIRT5 represses biochemical activity of, and cellular respiration through, two protein complexes identified in our analysis, pyruvate dehydrogenase complex and succinate dehydrogenase. Our data reveal widespread roles for lysine succinylation in regulating metabolism and potentially other cellular functions.
INTRODUCTION
Protein posttranslational modifications (PTMs) represent a key means of regulating cellular processes and diversifying the proteome. In recent years, high-resolution mass spectrometry (MS) has enabled the identification of an array of novel PTMs. Lysine (Lys) residues, in particular, are targets of numerous PTMs; e.g., acetylation, methylation, biotinylation, ubiquitination and ubiquitin-like modifications, butyrylation, propionylation, crotonylation, malonylation, and succinylation (Berger, 2007; Chen et al., 2007; Peng et al., 2011; Tan et al., 2011; Xie et al., 2012; Zhang et al., 2011). Comprehensive identification of the targets of these PTMs as well as the characterization of their functional impacts remain at an early stage.
Among Lys PTMs, acetylation (Kac) is by far the most well characterized. Our work, as well as that of others, has extensively characterized the cellular acetylome (Chen et al., 2012; Choudhary et al., 2009; Hebert et al., 2013; Kendrick et al., 2011; Kim et al., 2006; Schwer et al., 2009; Zhao et al., 2010). Though this modification was originally characterized on nuclear histones, it is now clear that reversible acetylation is a major regulatory mechanism for a diverse spectrum of cellular proteins in multiple cellular compartments. A high fraction of acetylated proteins are mitochondrial metabolic enzymes. Acetylation is a major regulator of most or all metabolic pathways in a manner that is conserved from bacteria to mammals (Finley et al., 2011; Hallows et al., 2011; Hebert et al., 2013; Hirschey et al., 2011; Smith et al., 2011; Starai et al., 2002; Wang et al., 2010; Zhang et al., 2009; Zhao et al., 2010).
Sirtuin family NAD+-dependent Lys deacetylases regulate key biological processes in mammals, including many aspects of metabolism (Finkel et al., 2009). Three mammalian sirtuins (SIRT3, SIRT4, and SIRT5) localize mostly or exclusively to the mitochondrial matrix. SIRT3 represents the major mitochondrial deacetylase activity. SIRT3 deficiency in the mouse results in dramatically increased acetylation of numerous mitochondrial proteins (Lombard et al., 2007), leading, in turn, to multiple cellular and organismal defects (Lombard et al., 2012). Less is known regarding the functions of SIRT4 and SIRT5, which, unlike SIRT3, do not impact bulk mitochondrial protein acetylation (Lombard et al., 2007). SIRT5 promotes urea cycle function via the regulation of carbamoyl phosphate synthase (CPS1) (Nakagawa et al., 2009; Ogura et al., 2010) and purine metabolism via urate oxidase (Nakamura et al., 2012).
It is well known that a mass shift from MS analysis can only elucidate the elemental formula of a compound and cannot resolve its structural isomers. In addition, alternative chemical and biochemical methods should be used to fully establish the modification. In the case of a mass shift of 100.016 Da, two structural isomers, succinyllysine and methylmalonyllysine, may exist. The existence of Lys succinylation had been suggested in the literature (Acker et al., 2009; Kawai et al., 2006; Rosen et al., 2004). However, these studies were not sufficient to fully establish the chemical structure of the modification. Although an initial publication determined the correct elemental composition of Lys succinylation, the studies were not sufficient to distinguish the succinyl group from its structural isomers (Rosen et al., 2004). Moreover, there were no studies using immunochemistry, high-performance liquid chromatography (HPLC) coelution, or tandem MS (MS/MS) of synthetic peptides to elucidate whether the mass shift was caused by a succinyl group or its isomers. A second publication used an MS approach that was insufficiently accurate to definitely characterize the elemental nature of the modification and also did not distinguish the structural isomers (Kawai et al., 2006). Although an antibody against succinyllysine was described in the paper, the specificity of this antibody was not characterized. A more recent study employing an 1H nuclear magnetic resonance (NMR)-based approach did not include chemical shift and integration of hydrogen atoms for structural elucidation, nor did it fully label critical NMR peaks to distinguish succinyl from its isomer-methylmalonyl groups (Acker et al., 2009). In addition, supporting evidence (e.g., 13C NMR or HPLC coelution of synthetic peptides or immunoblot with a well-characterized antibody) to conclusively identify Lys succinylation was lacking.
Recently, we identified and fully validated Lys succinylation and malonylation as PTM pathways, implying an ancient conserved function (Peng et al., 2011; Zhang et al., 2011). These modifications are also present on histones, suggesting possible roles in regulating chromatin structure and function (Xie et al., 2012). Our work, as well as that of others, has identified SIRT5 as the only known enzyme to date catalyzing Lys desuccinylation and demalonylation (Du et al., 2011; Peng et al., 2011; Zhang et al., 2011). Although succinylation regulates activities of two metabolic enzymes, isocitrate dehydrogenase (IDH) and CPS1, a global picture of how this modification may affect cellular functions is lacking.
We report global analysis of Lys succinylation in mammals. Succinylation is widespread among diverse mitochondrial metabolic enzymes and also on extramitochondrial cytosolic and nuclear proteins. We test the impact of SIRT5 on two substrates identified in this analysis, the pyruvate dehydrogenase complex (PDC) and succinate dehydrogenase (SDH). SIRT5 suppresses the biochemical activities of, and respiration mediated by, both of these complexes. Thus, Lys succinylation is a functional modification with the potential to impact mitochondrial metabolism and diverse extramitochondrial processes.
RESULTS
Lysine Succinylation Proteome in Mouse Cells
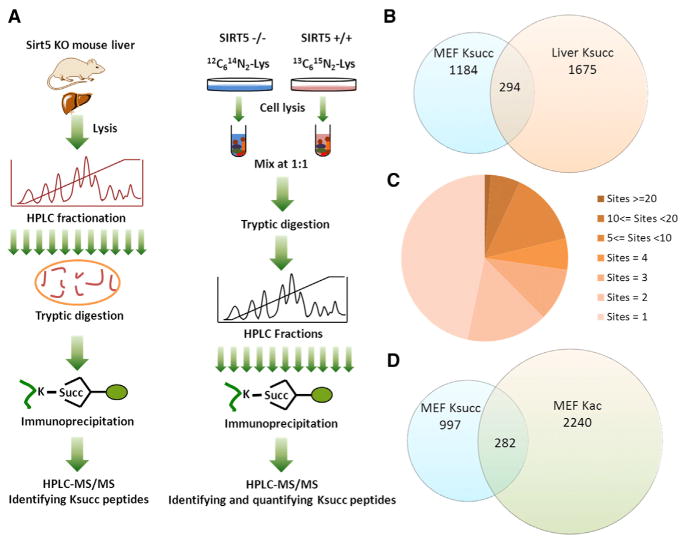
To identify Lys-succinylated (Ksucc) proteins and sites regulated by SIRT5, we performed two sets of large-scale proteomic analyses. First, we carried out proteomics screening of Lys succinylated peptides in Sirt5 knockout (KO) mouse liver using protein fractionation, affinity enrichment, and HPLC-MS/MS analysis (Figure 1A, left panel, and Figure S1A available online). This study provided a comprehensive list of Ksucc substrates in an organ important for energy metabolism. To study dynamics of Ksucc in response to SIRT5, the only enzyme currently known to regulate Ksucc status, we quantified the Ksucc proteomes between wild-type (WT) and Sirt5 KO mouse embryonic fibroblasts (MEFs) using stable isotope labeling of amino acids in cell culture (SILAC) and MS (Figures 1A, right panel, and S1B). We identified 1,675 sites from 436 proteins in liver tissue (Table S1B) and 1,184 Lys succinylation sites from 505 proteins in MEFs (Table S1A) with a false discovery rate (FDR) of less than 1% at protein, peptide, and site levels. Furthermore, we removed those peptides with additional stringent cutoff criteria (see Supplemental Experimental Procedures). After removing overlapping sites, we identified a total of 2,565 Lys succinylation sites from 779 proteins in mouse cells (Figures 1B and 1C). We found that some proteins were highly succinylated, including CPS1 with 47 sites and hydroxyacyl-coenzyme A dehydrogenase (HADHA) with 32 sites (Figure 1C and Tables S1A–S1B). Among the identified Ksucc sites, only 294 sites overlapped between MEFs and mouse liver cells (Figure 1B).
Figure 1. Profiling Lys Succinylation Proteome in Mouse Cells.
(A) Schematic representation of experimental workflow for the identification of Lys succinylation substrates in Sirt5 KO mouse liver (left) and SILAC quantification of Lys succinylation in WT and Sirt5 KO MEFs (right) (see also Figure S1).
(B) Venn diagram showing the total numbers of Lys succinylation sites identified in MEFs and in mouse liver tissue and their overlap (see also Figure S2).
(C) Pie chart showing the distribution of the number of Ksucc site identifications per protein.
(D) Venn diagram showing overlap between quantifiable Lys succinylation sites and Lys acetylation sites in MEFs.
To investigate the extent to which Lys succinylation and Lys acetylation sites overlapped, we quantified Lys acetylation substrates in the Sirt5 KO and WT SILAC MEFs using HPLC fractionation, immunoaffinity enrichment, and HPLC-MS/MS analysis. We identified 2,329 nonredundant Lys acetylation sites with an FDR less than 1% and with additional stringent cutoff criteria (see Supplemental Experimental Procedures) (Table S1C). To compare Ksucc sites and Kac sites in MEFs, we extracted all quantified Ksucc and Kac sites from the two proteomics studies. We found that only 282 sites overlapped between Ksucc and Kac sites identified in MEFs (Figure 1D).
To gain functional insight into how Ksucc may regulate cellular function, we assessed Ksucc levels in whole-cell extracts and mitochondria in the liver and kidney from male and female mice under fed and fasting conditions (Figures S2A–S2H). We found that Ksucc levels rose in response to fasting in both tissues, and this change was more marked in the liver than the kidney. Similar trends were apparent for acetylation. Thus, Lys succinylation might contribute to the fasting response.
Quantitative Analysis of Lys Succinylation and Acetylation Sites in Response to Sirt5 KO
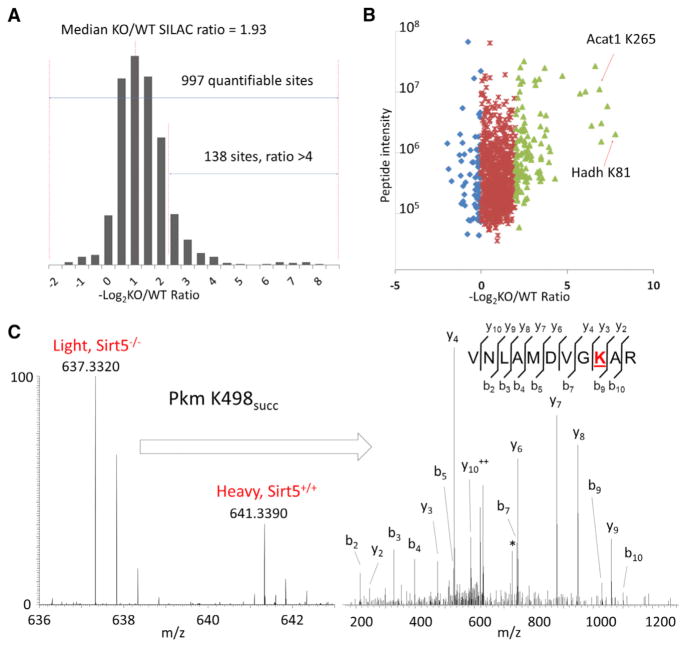
Using SILAC, we quantified the dynamics of Lys succinylation sites in WT and Sirt5 KO MEFs. Out of 1,184 Ksucc sites, 997 sites were quantifiable (Figure 2A), and more than 90% of these showed increasing abundance in Sirt5 KO cells with an average KO/WT SILAC ratio of 3.61 and a median KO/WT SILAC ratio of 1.93. Significantly, over 12% of the sites showed over 4-fold increase in abundance in SIRT5-deficient cells (Figure 2A). These data showed that SIRT5 deficiency significantly impacted Ksucc levels, suggesting that SIRT5 is a central regulator of Lys succinylation in mammalian cells, analogous to the role of SIRT3 in regulating overall Lys acetylation in mitochondria (Hebert et al., 2013; Lombard et al., 2007; Smith et al., 2011). To investigate whether SIRT5 significantly impacted Lys acetylation in vivo, we compared the quantitative changes of Lys acetylation between WT and Sirt5 KO MEFs. Consistent with previous reports (Du et al., 2011; Lombard et al., 2007; Peng et al., 2011), our data showed that SIRT5 deficiency does not significantly impact global Lys acetylation and has an average KO/WT SILAC ratio of 1.04 (Figure S3A).
Figure 2. Quantitative Analysis of Lys Succinylation Proteome.
(A) Histogram showing the ratio distribution of quantifiable Lys succinylation sites between Sirt5 KO and WT MEFs (see also Figure S3).
(B) Scatter plot showing the quantification of Lys succinylation sites in relation to peptide intensities.
(C) Full MS and MS/MS spectra for the identification and quantification of K498 succinylation on Pkm. b and y ions indicate peptide backbone fragment ions containing the N and C terminal, respectively. ++ indicates doubly charged ions, and * indicates loss-of-amine ions. Succinylated Lys is underlined and colored in red.
To investigate whether SIRT5 deficiency significantly impacted global protein expression, we quantified protein levels in WT and Sirt5 KO MEFs using SILAC and HPLC-MS/MS. Our analysis quantified 3,944 nonredundant protein groups with 5,109 proteins (Table S1D). Our data showed that about 85% of the proteins (3,348 out of 3,944) showed less than 1-fold change in abundance in Sirt5 KO MEFs (SILAC Sirt5 KO/WT ratio between 0.5 and 2) (Figure S3B), and normalization by protein quantification ratios did not significantly alter the quantification ratio distribution of Lys succinylation sites (Figure S3A).
In SIRT5-deficient MEFs, 28 Ksucc sites showed a more than 10-fold increase in abundance (Figure 2B). Hydroxyl-coenzyme A dehydrogenase (mitochondrial HADH) is an essential enzyme in lipid metabolism with major activity in the oxidation of 3-hydroxybutyryl-CoA. Our data indicate that the HADH amino acid residue K81, which is adjacent to a CoA binding site at K80, is succinylated, and its modification increased by more than 200-fold in Sirt5 KO fibroblasts (about 74-fold after normalization with protein abundance changes). Mitochondrial acetyl-CoA acetyltransferase is a critical enzyme in ketone body metabolism that conjugates acetyl-CoA into acetoacetyl CoA. Succinylation at K265, located near a CoA-binding site (K260) increased in abundance by more than 120-fold in Sirt5 KO cells (about 104-fold after normalization with protein abundance changes).
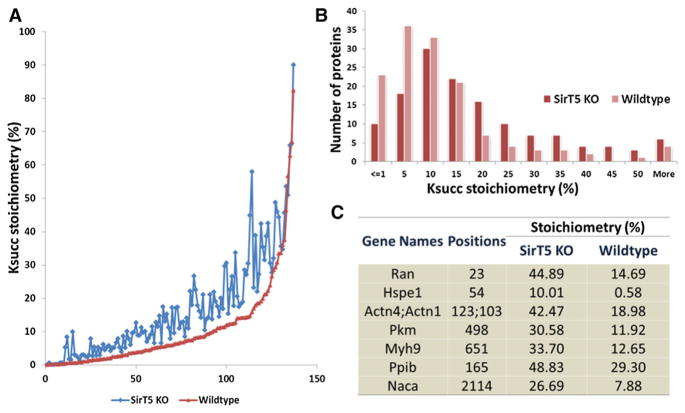
To calculate the absolute stoichiometry of Lys succinylation, we modified a previously reported algorithm (Olsen et al., 2010) that used SILAC quantification ratios of modified and un-modified peptides as well as corresponding proteins. In this method, the stoichiometries of those Ksucc sites can be calculated if they meet several criteria (see Supplemental Experimental Procedures) (Olsen et al., 2010). With this consideration, we were able to calculate absolute stoichiometries of 137 Ksucc sites in both WT and Sirt5 KO MEFs with a dynamic range from 0.02% to 90%, out of which 134 sites showed increased Ksucc stoichiometries in SIRT5 deficiency (Figure 3A and Table S1E). Our analysis showed that 32% of the Ksucc sites had stoichiometries greater than 10% in Sirt5 WT cells, whereas this figure increased to 56% in Sirt5 KO cells (Figures 3B–3C). For example, GTP-binding nuclear protein (RAN) is a GTPase involved in protein and RNA transport between the nucleus and cytoplasm. Our data showed that the K23 site on RAN, located within one of the GTP-binding regions, is Lys succinylated, and its stoichiometry increased from 15% to 45% in Sirt5 KO cells. Pyruvate kinase isozyme M1/M2 (PKM) is a critical glycolytic enzyme with an established role in cancer cell Warburg metabolism (Vander Heiden et al., 2009). Recent studies identified additional functional roles in the nucleus as a protein kinase regulating histone H3 and STAT3 phosphorylation (Gao et al., 2012; Yang et al., 2012). Our data revealed that a total of seven sites on PKM were Lys succinylated, among which the stoichiometry of K498 site increased from 12% to 31% in Sirt5 KO MEFs (Figures 2C and 3C).
Figure 3. Absolute Stoichiometries of Lys Succinylation Sites in Sirt5 KO and WT MEF Cells.
(A) Comparison of stoichiometries of Ksucc sites in WT (red dots) and Sirt5 KO MEF cells (blue dots).
(B) Comparison of distributions of Ksucc stoichiometries between WT (pink bars) and Sirt5 KO ells (red bars).
(C) List of representative Ksucc sites with significant changes in absolute stoichiometries between WT and Sirt5 KO cells.
Flanking Sequence and PTM Correlation Analysis
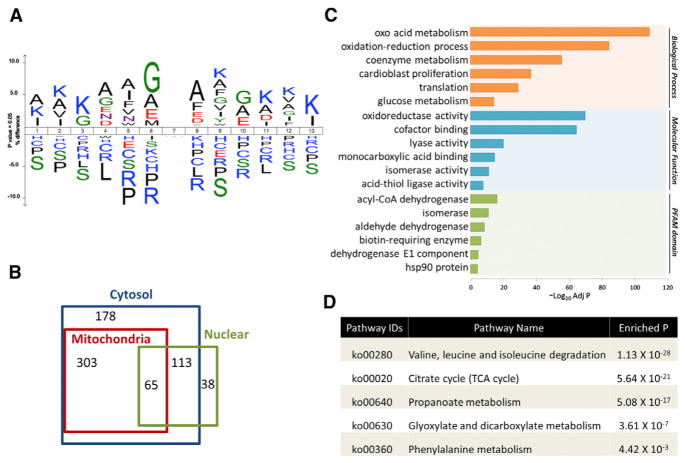
To assess whether SIRT5 shows structural preferences for its enzymatic activity, we evaluated the flanking sequences of Ksucc sites identified from both MEFs and liver tissues. The data showed a preference for Gly or Ala at the −1 position and Ala at the +1 position (Figure 4A). Arg is largely depleted at both the −1 and +1 position. Accordingly, motif analysis identified AK, GK, KF, and KA motifs as significantly overrepresented for Lys succinylation sites (Bonferroni corrected p < 0.05) (Figure S4). This is in contrast to recent studies demonstrating that Kac sites targeted by SIRT3 tend to be near positively charged amino acids (Hebert et al., 2013; Smith et al., 2011).
Figure 4. Characterization of Lys Succinylation Proteome.
(A) Icelogo representation shows flanking sequence preferences for all Lys succinylation sites (see also Figure S4).
(B) Venn diagram showing cellular compartment distribution of Ksucc proteins.
(C) Bar graphs showing representative ontology annotations enriched with Ksucc proteome (see also Figure S5).
(D) Table of all KEGG pathways involving succinyl-CoA in mice showing the enrichment of each pathway in Ksucc proteins (see also Table S3).
We compared our data sets to the public databases of known PTMs in UniProt (http://www.uniprot.org). In addition to Lys acetylation, we identified 18 Ksucc sites overlapped with other modifications (Table S2), including ubiquitination and previously reported Lys malonylation sites. Furthermore, we identified 92 Ksucc sites located within five residues in flanking distance to various types of PTMs (Table S2), including phosphorylation, ubiquitination, and lipid modification sites. Among all Ksucc sites colocalized with phosphorylation sites, a surprisingly high percentage (18%) colocalized near known or predicted phosphotyrosine residues.
Cellular Localization of the Lys Succinylation Proteome
SIRT5 is thought to localize primarily to mitochondria (Lombard et al., 2007; Michishita et al., 2005; Nakagawa et al., 2009; Schlicker et al., 2008). An intriguing question is whether SIRT5 regulates Lys succinylation on extramitochondrial proteins. We performed cellular compartment analysis of all Lys-succinylated proteins identified from Sirt5 KO liver tissue and MEFs. As expected, a significant portion of cytosolic Ksucc proteins localized exclusively or partially to mitochondria (p = 2.3 × 10−112, Fisher’s exact test) (Figure 4B). Furthermore, we compared our data set with known mitochondria genes annotated in the Mito-Carta database (Pagliarini et al., 2008). Sixteen percent of mitochondrial proteins were Lys succinylated in MEFs, whereas 23% of mitochondrial proteins were Lys succinylated in liver tissue. Altogether, 29% of mitochondrial proteins were Lys succinylated, suggesting potentially important roles of Lys succinylation in regulating mitochondria functions. Also, we identified similar numbers of proteins that were not annotated as mitochondrial (Figure 4B), including ribosomes and histone proteins H2A, H2B, H3, and H1, many of which have significantly upregulated Lys succinylation upon Sirt5 KO (Table S3). Lys acetylation is known to be abundant on nuclear proteins. In contrast, only 38 Lys-succinylated proteins identified in this study were previously annotated as exclusively nuclear.
Functional Annotation of the Lys Succinylome
To understand the biological functions of Lys succinylation substrates, we performed enrichment analysis with the Gene Ontology annotation database. Consistent with previous reports (Du et al., 2011; Peng et al., 2011), our data showed that Lys-succinylated proteins are significantly enriched in cellular metabolic process with specific enrichment in oxoacid metabolism (adjusted p = 1.6 × 10−110), oxidation reduction process (adjusted p = 8.2 × 10−86), and coenzyme metabolism (adjusted p = 5.4 × 10−57) (Figure 4C, upper panel). Accordingly, the molecular functions carried out by Lys succinylated proteins included cofactor binding (adjusted p = 1.2 × 10−65) and oxidoreductase activity (adjusted p = 4.2 × 10−71) (Figure 4C, middle panel). In addition, we also found that Lys-succinylated proteins were enriched for translational processes (adjusted p = 3.4 × 10−30) because of the identification of succinylated ribosome proteins. Analysis of the PFAM domain database showed that Lys-succinylated proteins were enriched for acyl-CoA dehydrogenase (adjusted p = 5.3 × 10−17), isomerase (adjusted p = 9.2 × 10−12), and aldehyde dehydrogenase (adjusted p = 2.9 × 10−9) (Figure 4C, bottom panel).
To understand the cellular pathways involving SIRT5-regulated Lys succinylation, we performed enrichment analysis of KEGG pathways. Our data showed that, in Sirt5 KOs, 37 of 51 proteins in the Val-Leu-Ile degradation pathway, as well as over 80% of all proteins in the tricarboxylic acid (TCA) cycle, and 60% of all proteins in fatty acid metabolism were Lys succinylated (Figures S5A–S5C). Interestingly, we found that nearly all annotated pathways involving succinyl-CoA in mice were enriched with Lys-succinylated proteins, including the Val-Leu-Ile degradation pathway (adjusted p = 1.1 × 10−28), the TCA cycle (adjusted p = 5.6 ×10−21), and propionate metabolism (adjusted p = 5.1 × 10−17) (Figure 4D).
SIRT5 Is Present Extramitochondrially
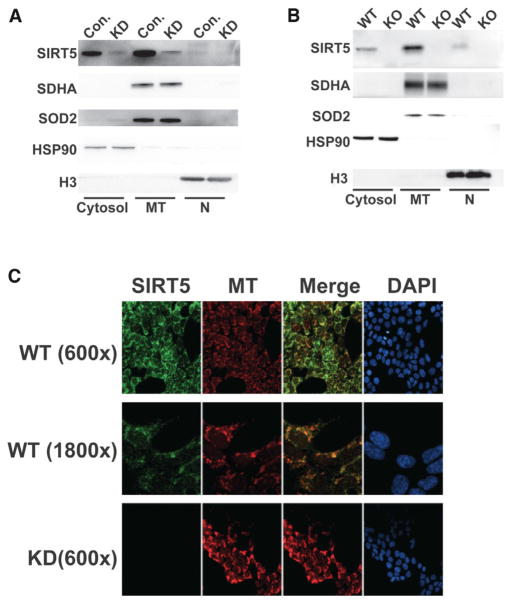
These data suggested that a portion of SIRT5 might exist extramitochondrially, regulating Lys succinylation outside this organelle. Subcellular fractionation revealed that a significant fraction of endogenous SIRT5 was present cytosolically both in human cells (Figure 5A) and mouse liver (Figure 5B). A very small amount of SIRT5 was also detected in the nuclear fraction, which was particularly evident in the liver samples (Figure 5B). To confirm these results, we performed immunofluorescence (IF) and confocal microscopy to localize endogenous SIRT5 (Figure 5C). Consistent with our fractionation studies, SIRT5 (green) partially colocalized with MitoTracker Red to produce a yellow signal. However, a detectable fraction of SIRT5 was present cytosolically, as evidenced by a green signal that was not apparently localized to mitochondria. No SIRT5 signal was evident in the Sirt5 knockdown (KD) control cells. Thus, SIRT5 is present both mitochondrially and extramitochondrially, a finding which is consistent with the proteomic results.
Figure 5. Subcellular Localization of Human and Mouse SIRT5.
(A and B) Subcellular fractionation of control and Sirt5 KD HEK 293T cells (A) and WT and Sirt5 KO mouse livers
(B). Fractionation controls were SDHA and SOD2 (mitochondria), HSP90 (cytosol), and histone H3 (nucleus). MT, mitochondria; N, nucleus.
(C) Confocal microscopy analysis of SIRT5 localization in control and Sirt5 KD cells along with MitoTracker Red (MT).
Protein Interaction Networks of Lys Succinylation Proteome
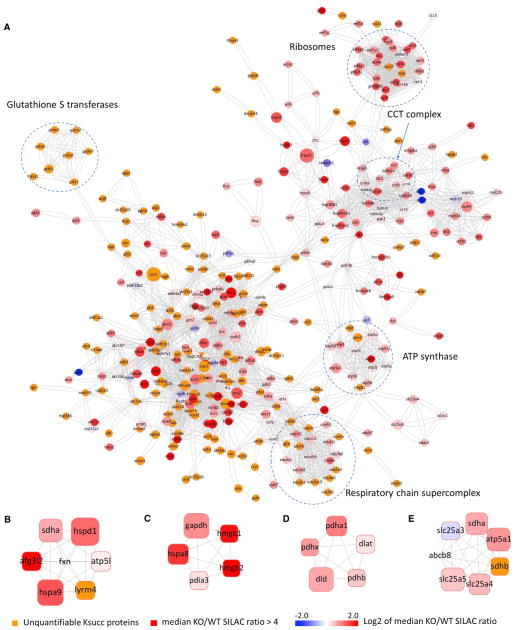
We visualized the protein-protein interaction networks of Lys-succinylated proteins on the basis of the STRING database (Jensen et al., 2009). Our data set showed a vast, highly connected network with clusters of nodes extending out to form protein complex patterns (Figure 6A). Using the MCODE tool (Bader and Hogue, 2003), we identified a number of highly connected subnetworks among Lys-succinylated proteins, including the F1F0 ATP synthetase, respiratory chain supercomplexes (complexes I, III, and IV), glutathione S transferases, ribosomes, and the chaperonin-containing TCP1 complex (Figure 6A). The first two protein networks are localized to mitochondria, whereas the last three are mainly cytosolic. Quantitative analysis showed an increase in Lys succinylation among most protein networks in SIRT5 deficiency, and some Ksucc proteins were only identified in Sirt5 KO MEFs or mouse liver tissue (Figure 6A).
Figure 6. Interaction Network and Protein Complex Analysis of Lys Succinylation Proteome.
(A–E) Protein-protein interaction network on the basis of the STRING database (v8.3) (A) and visualized in Cytoscape and representative protein complexes enriched in Lys succinylation proteome: Frataxin complex (B), HMGB1-HMGB2-HSC70-ERP60-GAPDH complex (C), PDC (D), and SDH-mABC1-PIC-ANT-ATPase complex (E). Each node is color coded to represent the Log2 of the median SILAC KO/WT ratio for each protein. The size of the node corresponds to the relative number of Ksucc sites identified on each protein.
Protein Complex Analysis
To identify complexes regulated by SIRT5 and Lys succinylation, we performed protein complex enrichment analysis with the manually curated CORUM core complex database (Ruepp et al., 2008) for mammalian proteins. Because only a limited number of mouse complexes were annotated and experimentally confirmed, all of the mammalian protein complexes in the database were taken into consideration on the basis of the assumption of evolutionary conservation of core protein complexes among mammalian species. Our analysis showed over 20 enriched protein complexes from Lys-succinylated proteins (adjusted p < 0.01). In addition to the protein complexes identified by interaction network analysis, we identified significant enrichment of Lys succinylation in the frataxin complex (Figure 6B, adjusted p = 1.8 × 10−11), the HMGB1-HMGB2-HSC70-ERP60-GAPDH complex (Figure 6C, adjusted p = 3.8 × 10−10), the PDC (Figure 6D, adjusted p = 3.6 × 10−10), and the SDH-mABC1-PIC-ANT-ATPase complex (Figure 6E, adjusted p = 7.7 × 10−7), suggesting potential impacts of Lys succinylation and SIRT5 enzymatic activity in regulating physiological functions of these protein complexes (see below). Lys acetylation is known to be involved in the regulation of macromolecular complexes, many of which are located in the nucleus and involved in transcription-related processes (Choudhary et al., 2009). In contrast, we were not able to identify any nuclear protein complexes that were enriched among Ksucc proteins.
Kinetics of Lys Succinylation
To assess succinylation turnover, we pulse labeled HeLa cells with D4-isotopic succinate and collected cells to compare the Ksucc heavy-to-light abundances at 0, 8, and 24 hr. Cells were lysed and subjected to a single immune-affinity enrichment with an antibody against Ksucc. Enriched peptides were analyzed by HPLC-MS/MS, and raw data from each time point were analyzed with MaxQuant (v.1.3.0.5) with the specification of Lys succinylation and Lys D4 succinylation as variable modifications. In-house-developed scripts were used to extract maximum peak intensities of each Lys-succinylated (IL) or D4-succinylated (IH) peptide. The labeling ratios of Ksucc peptides were calculated as IH/IL, and the percentage of unlabeled Lys-succinylated peptide was calculated as IL*100%/(IL +IH). From these data, we were able to identify 15 Ksucc peptides in both light and heavy form from all three time points (Table S4). We performed fuzzy c-means clustering analysis of the labeling ratios from all 15 peptides and identified four core clusters (Figure S6A).
Each cluster clearly showed different dynamic profiles in Ksucc turnover upon pulse labeling. For simple calculation of the turnover rate, we selected Cluster 2, in which the main Ksucc sites showed nearly linear increase in labeling ratios with increasing labeling time. We performed a linear regression analysis on the percentage of the Lys D4 succinylation (heavy form) and the percentage of the Lys H4 succinylation (light form) at the three time points (0, 8, and 24 hr) for all the major Ksucc sites in cluster 2. Our data showed that the average rate of Lys D4 succinylation incorporation (or the average rate of the Lys H4 succinylation loss) is about 0.92% per hr for the Ksucc sites in Cluster 2 (Figure S6B). Thus, Lys succinylation may regulate functions of its targets on a relatively slow time frame (hr to days). However, the modification of specific sites may occur much more rapidly.
SIRT5 Suppresses Pyruvate Dehydrogenase Complex Activity
As proof of principle that the SIRT5 targets identified in our studies can be functionally impacted by succinylation, we tested the role of SIRT5 in regulating activities of two metabolic protein complexes, PDC and SDH. PDC catalyzes oxidization of pyruvate to acetyl-CoA for use in the TCA cycle. PDC consists of E1α and E1β heterotetramers surrounding a core consisting of E2, E3, and E3bp subunits (Patel and Korotchkina, 2006). PDC activity is tightly regulated. E1α phosphorylation by pyruvate dehydrogenase kinases represses PDC activity and represents a major known mechanism of PDC regulation. Pyruvate dehydrogenase phosphatases dephosphorylate E1α and activate PDC.
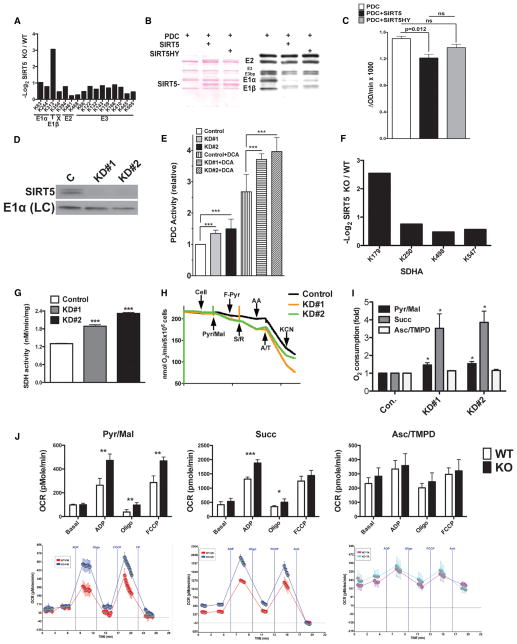
Using SILAC analysis, we identified multiple subunits of PDC as hypersuccinylated in Sirt5 KO MEFs (Figure 7A, Tables S1A and S5). Additional succinylation sites on PDC were identified in MS/MS analysis of Sirt5 KO livers (Tables S1B and S5). We immunoblotted commercially available PDC and confirmed that we could detect succinylation on multiple PDC subunits (Figure 7B). Consistent with our SILAC data, WT SIRT5 robustly desuccinylated PDC in vitro (Figure 7B); a SIRT5H158Y mutant showed hypomorphic desuccinylase activity (Nakagawa et al., 2009). Treatment of purified PDC with SIRT5 decreased holoenzyme activity in vitro (Figure 7C). These data suggested that SIRT5 negatively regulates PDC activity. To test this in vivo, we generated two independent human embryonic kidney (HEK) 293T cell lines in which SIRT5 was depleted via lentiviral small hairpin RNA (Figure 7D). Sirt5 KD resulted in elevated PDC activity (Figure 7E). This increase was synergistic with treatment with dichloroacetate (DCA), which stimulates PDC activity by reducing the phosphorylation and turnover of E1α (Morten et al., 1999; Stacpoole et al., 2008). Thus, SIRT5 and E1α phosphorylation attenuate PDC activity through distinct mechanisms.
Figure 7. SIRT5 Suppresses the Activities of Pyruvate Dehydrogenase Complex and Succinate Dehydrogenase.
(A) Quantitative Ksucc changes on PDC subunits between Sirt5 KO and control MEFs (see also Tables S1A and S5).
(B) SIRT5 desuccinylates PDC subunits in vitro. Left, ponceau S; Right, IB: Ksucc.
(C) SIRT5 suppresses PDC activity in vitro. PDC activity in reactions in (B). Data are mean ± SEM of triplicate measurements; p value designates comparison via a paired two-tailed t test.
(D) SIRT5 immunoblot in Sirt5 KD cell lines and control. LC, loading control.
(E) PDC activity in Sirt5 KD cells and controls ± 5 mM DCA. Data summarize four independent experiments, each done in quadruplicate. Error bars designate SEM; ***p < 0.001 by two-way ANOVA.
(F) Quantitative Ksucc changes on SDHA between SIRT5 KO and control MEFs (see also Tables S1A and S6 and Figure S7).
(G) Increased SDH activity in Sirt5 KD cells. Data are mean ± SEM of triplicate measurements; ***p < 0.001 by an unpaired two-tailed t test.
(H) Representative oxygen consumption in Sirt5 KD and control cell lines. Additions are as follow: cells; Pyr/Mal, pyruvate + malate; F-Pyr, fluoropyruvate; S/R, succinate + rotenone; AA, antimycin A; A/T, ascorbate + TMPD; KCN.
(I) Quantification of (H). Succ, succinate + rotenone; Asc/TMPD, ascorbate + TMPD. Data are mean ± SEM of triplicate measurements; *p < 0.05 by an unpaired two-tailed t test.
(J) Increased respiration in the presence of pyruvate or succinate but not ascorbate + TMPD in SIRT5-deficient mitochondria. Top, plotted data; bottom, raw data (blue or turquoise, Sirt5 KO; red or pink, WT control). OCR, oxygen consumption rate. Data are representative of five independent experiments performed in quadruplicate on a littermate WT/KO pair. Statistical comparisons were performed with an unpaired two-tailed t test; error bars designate SEM.
SIRT5 Suppresses SDH Activity and Cellular Respiration
The SDH complex (complex II) includes four subunits, SDHA–SDHD. SDH participates in both the TCA cycle and the electron transport chain, catalyzing the oxidation of succinate to fumarate with concomitant conversion of ubiquinone to ubiquinol. Germ-line mutations in all Sdh subunit genes can cause mitochondrial disease and tumor susceptibility syndromes (Gill, 2012). SILAC studies identified four Lys succinylation sites on the SDHA subunit of SDH (Figure 7F). Analysis of Sirt5 KO liver tissue identified several additional sites in SDHA as well as a single Lys succinylation site in SDHB (Tables S1B and S6).
First, to test the potential functional impact of SIRT5 on SDH, we assessed SDH biochemical function in Sirt5 KD cells and controls. Sirt5 KD led to a substantial increase in SDH activity (Figure 7G). This was associated with a large (roughly 3.5-fold) elevation in cellular respiration in the presence of the SDH substrate succinate (cells were permeabilized to allow compounds access to mitochondria) (Figures 7H and 7I). Increased respiration was also observed in Sirt5 KD cells in the presence of the PDC substrate pyruvate (Figures 7H and 7I). Sirt5 KD did not impact respiration in the presence of the complex IV substrate ascorbate.
To confirm the cell results, we isolated mitochondria from Sirt5 KO livers and controls and assessed for respiratory activity using various substrates (Figure 7J). SIRT5-deficient mitochondria showed significantly increased respiration in the presence of succinate and pyruvate. In contrast, no significant differences in respiration between WT and Sirt5 KO were observed with ascorbate. We conclude that SIRT5 targets PDC and SDH to suppress their biochemical activities and mitochondrial respiration driven by these complexes.
DISCUSSION
We report global analysis of the Lys succinylome in mammals. This modification is widespread in fibroblasts and hepatocytes, occurring on both mitochondrial metabolic enzymes as well as extramitochondrial proteins such as histones and ribosomes. Consistent with a role for SIRT5 in regulating Ksucc levels extra-mitochondrially, we find that a substantial fraction of SIRT5 protein is cytosolic. Our data are consistent with a major role for SIRT5 in regulating Ksucc levels, which is analogous to the principal role of SIRT3 in regulating mitochondrial Lys acetylation (Hebert et al., 2013; Lombard et al., 2007; Smith et al., 2011). Supporting functional roles for SIRT5 and succinylation, we have identified roles for SIRT5 in suppressing respiration via PDC and SDH, succinylated enzyme complexes that were identified in our studies. The succinylation of many of SIRT5 targets identified in this analysis will most likely prove functionally relevant and impact metabolism or other cellular processes. Other activities of SIRT5 (e.g., demalonylation) most likely contribute to its biological effects as well.
Our analysis of Lys succinylation and acetylation in Sirt5 WT and KO cells suggests that there is a limited overlap between the two modifications, implying different regulatory mechanisms. It is unknown how proteins are succinylated in the cell—e.g., whether this process is catalyzed by putative succinyl transferases or, instead, occurs passively by reaction of succinyl-CoA with surface-exposed Lys (Du et al., 2011; Peng et al., 2011; Zhang et al., 2011). We attempted to evaluate the latter possibility by inhibiting SDH function in order to increase intra-cellular succinate levels either by treating cells with the SDH inhibitors 3-nitropropionic acid (3-NPA; Figure S7A) or malonate (Figure S7B) over a range of concentrations or by knocking down SDHC (Figure S7C). 3-NPA treatment, but not malonate treatment or SDHC KD, resulted in a slight increase in Ksucc levels. Thus, mechanisms of Lys succinylation remain unclear. It is possible that the occurrence of acetylation and succinylation on distinct subsets of lysines occurs as a consequence of divergent mechanisms of modification. Alternatively, SIRT3 and SIRT5 may be targeted to distinct Lys sites for the removal of these modifications.
Given the reported mitochondrial localization of SIRT5, it is somewhat surprising that we identified 35 nonredundant histone Lys succinylation sites, six of those sites overlapping with our first report of histone succinylation sites in mouse cells (Xie et al., 2012). Lys succinylation on histones increases significantly in SIRT5 deficiency with an average SILAC KO/WT ratio of 5.09. Similar to previous findings, Lys succinylation sites on histones primarily localize to the C-terminal globular domains, the exception being the histone H2B K5. This is in contrast to Lys acetylation, which primarily occupies the N-terminal tails of histones, potentially suggesting a distinct epigenetic role of histone Lys succinylation in comparison to Lys acetylation. These data imply that SIRT5 functions outside mitochondria, potentially impacting chromatin functions.
We tested whether Lys succinylation sites identified in this study overlapped with known enzymatic activity sites on the basis of UniProt annotation, an important sign that Lys succinylation may regulate the function of a substrate. Sixteen Ksucc sites overlapped with known cofactor binding or catalytic sites, and 74 Ksucc sites were located within five residues in flanking distance to enzymatic activity sites (Table S7). IDH is a key metabolic enzyme catalyzing isocitrate conversion to 2-oxoglutarate. Our previous studies identified Ksucc sites on E. coli IDH (Zhang et al., 2011). Mutations in the IDH2 gene, a mammalian IDH homolog, causes D-2 hydroxyglutaric aciduria type 2 (D2HGA2) (Kranendijk et al., 2010), whereas its downregulation leads to epigenetic dysfunction via the loss of 5-hydroxymethyl-cytosine in melanoma (Lian et al., 2012). IDH1 or IDH2 mutations lead to production of the oncometabolite D2-hydroxyglutarate (D-2HG) (Yen et al., 2010). We identified a total of 15 Ksucc sites on IDH2, 2 Ksucc sites (K180 and K256) being located within five residues in flanking sequence to sites on IDH2 that are critical for catalysis (Y179 and K251, respectively). These data suggest a potential impact of Lys succinylation on the activity of IDH2, which may impact epigenetic regulation and neoplasia.
SIRT5 suppresses activities of PDC and SDH. Germline SDH mutations have been identified in the SDH subunits SDHA–SDHD and also in SDH assembly factor 2 in tumor predisposition syndromes (Hao et al., 2009; Korpershoek et al., 2011; Neumann et al., 2004; Niemann and Müller, 2000; Oudijk et al., 2013; Pollard et al., 2005; Ricketts et al., 2008; Ricketts et al., 2010). Dysregulated SDH activity in the setting of these mutations promotes the accumulation of succinate and mitochondrial reactive oxygen species, activating HIF1α and inducing angiogenesis (Selak et al., 2005). Elevated succinate levels can lead to epigenetic dysregulation (Cervera et al., 2009; Smith et al., 2007). Thus, SIRT5 could play a role in neoplasia by regulating PDC and SDH.
About 50% of overall daily caloric intake passes through PDC, highlighting the central importance of this holoenzyme in glucose metabolism (Patel and Korotchkina, 2006). The regulation of PDC activity is the major mechanism regulating glucose oxidation in mammals in vivo (Wieland et al., 1972). PDC dysfunction is linked to type 2 diabetes (T2D) and a diverse spectrum of other human pathologies, such as neoplasia and cardiac ischemia (Patel and Korotchkina, 2006). PDC activity is tightly regulated by metabolite levels as well as E1α phosphorylation. Our finding that SIRT5 desuccinylates PDC subunits to inhibit PDC activity reveals a previously unappreciated aspect of PDC regulation and raises the eventual possibility of inhibiting SIRT5 activity as an adjunct treatment in T2D. However, it is likely that the impact of SIRT5 on mitochondrial respiration and overall metabolism is much broader than repressing PDC and SDH function.
EXPERIMENTAL PROCEDURES
Animal experiments were performed in accordance with institutional guidelines at the University of Michigan. See the Supplemental Information for complete experimental details.
MEF Studies
MEFs were generated by standard methods from Sirt5 heterozygous breeding pairs in the 129-strain background. Cells were subsequently immortalized by large T antigen transduction. For SILAC, MEFs were cultured for more than six generations in media containing either “heavy” or “light” Lys.
PTM-Specific Immunoaffinity Enrichment
Enrichment of Ksucc and Kac peptides was performed with antibody-immobilized beads as previously described (Kim et al., 2006).
Enzyme Activity Assays
PDC activity was assayed with previously described methods (Jeoung et al., 2006; Schwab et al., 2005). SDH activity was assayed by previously published methods (Nadanaciva et al., 2007).
Supplementary Material
Acknowledgments
Our work was supported by National Institutes of Health (NIH) award R01GM101171 (D.B.L.), the National Technology Center for Networks and Pathways (U54GM103520; Y.Z.), the Ellison Medical Foundation (AG-NS-0583-09; D.B.L.), and the National Science and Technology Major Project of the Ministry of Science and Technology of China (2012ZX09301001-007; M.T.). J.P. and D.X.T. were supported in part by a National Institute on Aging training grant (T32AG000114). Work in the Lombard laboratory was also supported by DP3DK094292 and P30AG013283 and by pilot awards from the Michigan Diabetes Research and Training Center (P60DK020572), the Michigan Metabolomics and Obesity Center (P30DK089503), and the Claude D. Pepper Older American’s Independence Center (P30AG024824). The authors acknowledge S. Pletcher for help with statistical analysis; C. Burant and R. Harris, and members of the Lombard and Zhao labs for helpful discussions; and C. Fry and Y. Li (Cell Signaling Technology) for SIRT5 antibodies. Y.Z. is a shareholder and a member of the scientific advisory board of PTM BioLabs. J.P. and D.X.T. carried out respiration and enzymatic studies. Y.C., M.T., L.D., and Y.Z. performed the HPLC fractionation, affinity enrichment, HPLC-MS/MS, and bioinformatic data analysis. C.P., Z.X., J.P., D.T., and B.M.M.Z. generated MEF cell lines and performed immunoblot analysis. J.P., B.M.M.Z., and M.E.S. performed mouse husbandry. D.B.L. and Y.Z. conceived the project and supervised the experiments. D.B.L., Y.C., J.P., and Y.Z. wrote the manuscript.
Footnotes
Supplemental Information contains Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2013.06.001.
References
- Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J Am Chem Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Cervera AM, Bayley JP, Devilee P, McCreath KJ. Inhibition of succinate dehydrogenase dysregulates histone modification in mammalian cells. Mol Cancer. 2009;8:89. doi: 10.1186/1476-4598-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics. 2012;11:1048–1062. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1a destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill AJ. Succinate dehydrogenase (SDH) and mitochondrial driven neoplasia. Pathology. 2012;44:285–292. doi: 10.1097/PAT.0b013e3283539932. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:267–277. doi: 10.1101/sqb.2011.76.010850. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung NH, Sanghani PC, Zhai L, Harris RA. Assay of the pyruvate dehydrogenase complex by coupling with recombinant chicken liver arylamine N-acetyltransferase. Anal Biochem. 2006;356:44–50. doi: 10.1016/j.ab.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Fujii H, Okada M, Tsuchie Y, Uchida K, Osawa T. Formation of Nepsilon-(succinyl)lysine in vivo: a novel marker for docosahexaenoic acid-derived protein modification. J Lipid Res. 2006;47:1386–1398. doi: 10.1194/jlr.M600091-JLR200. [DOI] [PubMed] [Google Scholar]
- Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Korpershoek E, Favier J, Gaal J, Burnichon N, van Gessel B, Oudijk L, Badoual C, Gadessaud N, Venisse A, Bayley JP, et al. SDHA immunohistochemistry detects germline SDHA gene mutations in apparently sporadic paragangliomas and pheochromocytomas. J Clin Endocrinol Metab. 2011;96:E1472–E1476. doi: 10.1210/jc.2011-1043. [DOI] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, Amiel J, Buist NR, Das AM, de Klerk JB, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science. 2010;330:336. doi: 10.1126/science.1192632. [DOI] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Tishkoff DX, Zwaans BM. Mitochondrial regulation by protein acetylation. In: Cadenas E, Orrenius S, Packer L, editors. Mitochondrial Signaling in Health and Disease. London: Taylor and Francis; 2012. pp. 269–298. [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morten KJ, Beattie P, Brown GK, Matthews PM. Dichloroacetate stabilizes the mutant E1alpha subunit in pyruvate dehydrogenase deficiency. Neurology. 1999;53:612–616. doi: 10.1212/wnl.53.3.612. [DOI] [PubMed] [Google Scholar]
- Nadanaciva S, Bernal A, Aggeler R, Capaldi R, Will Y. Target identification of drug induced mitochondrial toxicity using immunocapture based OXPHOS activity assays. Toxicol In Vitro. 2007;21:902–911. doi: 10.1016/j.tiv.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Ogura M, Ogura K, Tanaka D, Inagaki N. SIRT5 deacetylates and activates urate oxidase in liver mitochondria of mice. FEBS Lett. 2012;586:4076–4081. doi: 10.1016/j.febslet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Neumann HP, Pawlu C, Peczkowska M, Bausch B, McWhinney SR, Muresan M, Buchta M, Franke G, Klisch J, Bley TA, et al. Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations. JAMA. 2004;292:943–951. doi: 10.1001/jama.292.8.943. [DOI] [PubMed] [Google Scholar]
- Niemann S, Müller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- Ogura M, Nakamura Y, Tanaka D, Zhuang X, Fujita Y, Obara A, Hamasaki A, Hosokawa M, Inagaki N. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem Biophys Res Commun. 2010;393:73–78. doi: 10.1016/j.bbrc.2010.01.081. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Oudijk L, Gaal J, Korpershoek E, van Nederveen FH, Kelly L, Schiavon G, Verweij J, Mathijssen RH, den Bakker MA, Oldenburg RA, et al. SDHA mutations in adult and pediatric wild-type gastrointestinal stromal tumors. Mod Pathol. 2013;26:456–463. doi: 10.1038/modpathol.2012.186. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MS, Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006;34:217–222. doi: 10.1042/BST20060217. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111 012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P, Wortham N, Barclay E, Alam A, Elia G, Manek S, Poulsom R, Tomlinson I. Evidence of increased microvessel density and activation of the hypoxia pathway in tumours from the hereditary leiomyomatosis and renal cell cancer syndrome. J Pathol. 2005;205:41–49. doi: 10.1002/path.1686. [DOI] [PubMed] [Google Scholar]
- Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, Maher ER. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst. 2008;100:1260–1262. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- Ricketts CJ, Forman JR, Rattenberry E, Bradshaw N, Lalloo F, Izatt L, Cole TR, Armstrong R, Kumar VK, Morrison PJ, et al. Tumor risks and genotype-phenotype-proteotype analysis in 358 patients with germline mutations in SDHB and SDHD. Hum Mutat. 2010;31:41–51. doi: 10.1002/humu.21136. [DOI] [PubMed] [Google Scholar]
- Rosen R, Becher D, Büttner K, Biran D, Hecker M, Ron EZ. Probing the active site of homoserine trans-succinylase. FEBS Lett. 2004;577:386–392. doi: 10.1016/j.febslet.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stümpflen V, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36(Database issue):D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schwab MA, Kölker S, van den Heuvel LP, Sauer S, Wolf NI, Rating D, Hoffmann GF, Smeitink JA, Okun JG. Optimized spectro-photometric assay for the completely activated pyruvate dehydrogenase complex in fibroblasts. Clin Chem. 2005;51:151–160. doi: 10.1373/clinchem.2004.033852. [DOI] [PubMed] [Google Scholar]
- Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Smith EH, Janknecht R, Maher LJ., 3rd Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum Mol Genet. 2007;16:3136–3148. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem Biol. 2011;6:146–157. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacpoole PW, Kurtz TL, Han Z, Langaee T. Role of dichloroacetate in the treatment of genetic mitochondrial diseases. Adv Drug Deliv Rev. 2008;60:1478–1487. doi: 10.1016/j.addr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland OH, Patzelt C, Löffler G. Active and inactive forms of pyruvate dehydrogenase in rat liver. Effect of starvation and refeeding and of insulin treatment on pyruvate-dehydrogenase interconversion. Eur J Biochem. 1972;26:426–433. doi: 10.1111/j.1432-1033.1972.tb01783.x. [DOI] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.