Abstract
Recent studies in our laboratories have demonstrated that a helical polypeptide (17H6), equipped with a histidine tag and a helical alanine-rich, glutamic-acid-containing domain, exhibits pH-responsive assembly behavior useful in the production of polymorphological nanostructures. In this study, the histidine tag in these polypeptides was replaced by polyethylene glycol (PEG) with different molecular masses (5 kDa, or 10 kDa), and the self-association behavior of 17H6 and the PEGylated conjugates was characterized via dynamic light scattering (DLS), small angle neutron scattering (SANS), and cryogenic transmission electron microscopy (cryo-TEM). DLS experiments illustrated that the polypeptide and its PEG-conjugates undergo reversible assembly under acidic conditions, suggesting that the aggregation state of the polypeptide and the conjugates is controlled by the charged state of the glutamic acid residues. Nanoscale aggregates were detected at polypeptide/conjugate concentrations as low as 20 μM (∼0.3–0.5 mg ml−1) at physiological and ambient temperatures. Scattering and microscopy results showed that the size, the aggregation number, and the morphology of the aggregates can be tuned by the size and the nature of the hydrophilic tag. This tunable nature of the morphology of the aggregates, along with their low critical aggregation concentration, suggests that PEG-alanine-rich polypeptide conjugates may be useful as drug delivery vehicles in which the alanine-rich block serves as a drug attachment domain.
Introduction
The self assembly of amphiphilic block copolymers in solution has offered a versatile approach to prepare a variety of nanostructures such as micelles, nanospheres, nanocapsules and polymersomes.1 The driving force in the formation of these nanostructures is the differing solubility properties of the solvophilic and solvophobic block(s) in a selective solvent. The lack of solubility of the solvophobic block(s) drives the self-association of those block(s), while the solvophilic block(s) maintain(s) the dispersion of the self-assembled structure in the solution phase by acting as a barrier between the solvophobic block(s) and the solvent.2–4 It is well known that the morphology and size of the resulting self-assembled structures can be tuned chiefly by the nature and composition of the blocks and solvent properties.5–10 Alternatively, solution temperature7,11–15 and pH,11,14,16 additives,5,6,8,12,16,17 copolymer concentration,7,9 and processing route16–24 have also been reported to change the size and the morphology of the aggregates.
Owing to the diversity in the manipulation of these structures as well as their ability to solubilize/stabilize solvophobic molecules, self-assembled block copolymers have been proposed for and employed in many applications. For example, in the cosmetics industry, pluronic micelles serve as fragrance delivery systems.25 Block copolymer aggregates have also been employed in the synthesis and stabilization of metal oxides26 and inorganic nanoparticles27 and in fluorescence resonance energy transfer (FRET) systems.28 Many studies have focused on the application of block copolymers in drug delivery.29–32 Biocompatible block copolymers have gained interest in this area as they form nontoxic scaffolds for insoluble drugs. Additionally, the erosion properties of biodegradable polymers have also been utilized to control drug release rates.33,34 In these biocompatible systems, polyethylene glycol (PEG) has become a common choice for a hydrophilic block, as it increases plasma half-life of the resultant block copolymer due to the ‘stealthy’ nature of PEG.32,35 Polylactic acid/polylactic-co-glycolic acid,29,33,36 polycaprolactone33,37 and polyamino acids38–41 have been common choices as biodegradable hydrophobic blocks, and/or as blocks for attachment of hydrophobic drugs. Covalent attachment of hydrophobic drugs to reactive polyamino acids such as polylysine and polyaspartate renders the polypeptide domain hydrophobic and reduces burst release of the drug compared to the simple encapsulation of drug (i.e. through hydrophobic interactions). Moreover, the nature of the chemical bond between the scaffold and the drug can be used to control drug release rate. In addition, the ability to tune the size of such complexes (e.g., to sizes <100 nm) is advantageous in applications including cancer therapy.38,39,42
Despite their inherent biodegradability, facile synthesis, and modification,43 block copolymers based on genetically engineered polypeptides have not been exploited in the delivery of hydrophobic drugs as frequently as their synthetic polymer counterparts. Of the recombinantly derived block copolypeptides, elastin-like polypeptides (ELPs) are the most studied polypeptide as a drug carrier due to their lower critical solution temperature (LCST)-like behavior. For example, thermosensitive nanoparticles based on diblock ELPs have been prepared as nanocarrier systems.45 Most of the studies have focused on ELPDOX conjugates, which have been shown to be effective tumortargeting systems.46–48
We have reported recombinant alanine-rich, glutamic-acid-containing helical polypeptide scaffolds that can be chemically modified with saccharides, and have used these scaffolds to manipulate multivalent interactions.49,50 These polypeptides comprise both an alanine-rich block and a decahistidine tag; detailed characterization of a polypeptide with the highest glutamic acid density (17H6) showed that it exhibits pH-responsive self-association that can yield nanostructures with different morphologies. As determined via dynamic light scattering (DLS) investigations, the polypeptide aggregates have a hydrodynamic radius of ∼10–20 nm and reversibly dissociate into monomers upon deprotonation of the glutamic acid residues.51 In contrast, the polypeptide (in the same concentration range) in pH 7.4 phosphate-buffered saline (PBS) exhibited a lack of aggregation, indicated by very low scattering in the DLS experiment. At pH 7.4, deprotonation of the glutamic acid residues likely provides charge-charge repulsions that increase the solubility of the alanine-rich block and prevent self-association of the polypeptide. We hypothesized that 17H6 acts as an amphiphilic block copolymer comprising the positively charged histidine tag and the alanine-rich domain that becomes more hydrophobic upon protonation of glutamic acids at acidic pH.
In the scope of the present study, we aimed to determine if it was possible to manipulate the association of the polypeptide domain via the conjugation of hydrophilic polymers of various molecular masses. Accordingly, the histidine tag of the polypeptide was cleaved and replaced by PEG blocks (with molecular masses of 5 kDa or 10 kDa) and the self-association behavior of the polypeptide and the PEG-modified polypeptides (PEG5K-c17H6 and PEG10K-c17H6) was investigated. The sequences and the molecular masses of the polypeptides and the conjugates are given in Table 1. The nanostructures of 17H6 and the conjugates under acidic conditions were characterized via dynamic light scattering, small angle neutron scattering, and cryogenic transmission electron microscopy. Our results illustrate basic principles for manipulating aggregate structures with variations in copolymer composition. The outcomes of this study have potential use in the development of polypeptide-based drug carrier systems.
Table 1. Sequences and molecular masses of the polypeptides and the conjugates.
| Notation | Sequence | M (kDa) |
|---|---|---|
| 17H6 | MGH10SSGHIHM(AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGMG | 14.8 |
| c17H6 | (AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGS/S-lactonea | 12.4 |
| PEG5K-c17H6 | (mPEG5K)-(AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGS/S-lactone | 17.4 |
| PEG10K-c17H6 | (mPEG10K)-(AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGS/S-lactone | 22.4 |
Reaction of cyanogen bromide with methionine yields a mixture of homoserine and homoserine lactone.44
Experimental
Materials
The polypeptide 17H6 with a sequence given in Table 1 was expressed using BL21(DE3)pLysS type E. coli expression host, and purified using nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography as described elsewhere.49,52 The histidine tag of 17H6 was cleaved by the reaction with cyanogen bromide (CNBr) to yield the cleaved polypeptide, c17H6 (Table 1). Propionaldehyde-functionalized PEG (5 kDa or 10 kDa) was conjugated to the N-terminus of c17H6 via Schiff base formation and subsequent reduction. A simplified route for the conjugate synthesis is given in Fig. S1 in the ESI.† The conjugates were purified using anion exchange chromatography followed by size exclusion chromatography. The synthesis of the monoPEGylated conjugates was confirmed by gel permeation chromatography (GPC) and matrix-assisted laser desorption/ionisation-time of flight (MALDI-TOF) mass spectroscopy, given with the details of synthesis and purification protocol sin Top et al. 2011.53 Phosphate bufferat pH 2.3 was prepared using 10 mM o-phosphoric acid (Fisher) with salt (140 mM sodium chloride (NaCl) and 10 mM potassium chloride (KCl) (Fisher)). Phosphate buffered saline (PBS) at pH7.4, containing the same NaCl and KCl concentrations as pH 2.3 buffer, was prepared using a dry-blend buffer pack (Thermo Scientific Inc. (Rockford, IL)).
Dynamic light scattering
Dynamic light scattering (DLS) experiments were carried out using a BI9000AT autocorrelator and a BI200SM goniometer (Brookhaven Instruments Corp., Holtsville, NY) at 532 nm. Samples were prepared by direct dissolution in the appropriate buffer followed by filtration with a 0.22 μm Millex syringe-driven filter unit (Millipore, Bedford, MA). Concentrations of the samples were confirmed by UV measurements and were based on a calibration curve constructed from samples with concentrations accurately determined via amino acid analysis. For the estimation of the apparent hydrodynamic diameter values, samples were prepared in 10 mM phosphate buffer including salt (140 mM NaCl and 10 mM KCl) at pH 2.3. Correlation functions were recorded at multiple angles and at 20 °C and 37 °C after equilibrating the sample for at least 15 min at the appropriate temperature. Average diffusion coefficients (D) and polydispersity-index values were estimated using a second-order cumulant fit of the time correlation of the scattering decay, with apparent hydrodynamic diameter values calculated from (D) using the Stokes–Einstein equation. Further details of the analysis are described elsewhere.51 In the pH responsiveness measurements, a 50 μM sample was prepared in phosphate-buffered saline (PBS) at pH 7.4, and the average scattering intensity was recorded. Then, the pH of the solution was adjusted to 2.3 using concentrated HCl, the solution was filtered and scattering intensity was measured again. Two more intensity measurements were taken after adjusting the solution pH to 7.4 with concentrated NaOH, followed by readjustment of the solution pH to 2.3. In each pH adjustment cycle a few to several ml of acid or base solution was added to ∼1 ml solution to minimize dilution effects.
Small angle neutron scattering
Small angle neutron scattering (SANS) experiments were carried out using 30 m instruments, NG3 or NG7, at the NIST Center for Neutron Research (NCNR), National Institute of Standards and Technology (NIST), Gaithersburg, MD. Samples were prepared in D2O- containing buffers to ensure the contrast between the H-rich samples and D2O and to minimize incoherent scattering. 1 or 2 mm pathlength, demountable titanium cells were used, and the temperature of the cells was controlled using a 10CB 10-position sample holder with a NESLAB circulating bath. Neutrons at λ = 6 Å (with a wavelength spread, Δλ/λ, of 0.14) were employed in the scattering experiments. Data acquisition was performed using a 64 cm × 64 cm 2-D detector at three different instrument configurations: 1, 4, and 13 m detector distances (with focusing lenses at 13 m only). The data were reduced using Igor Pro software (WaveMetrics, Lake Oswego, OR) with the SANS Reduction v5 macro developed at the NCNR.54
Cryogenic transmission electron microscopy
Cryogenic transmission electron microscopy (cryo-TEM) observations of the samples were performed at 120 kV using a Tecnai 12 microscope with a Gatan cryo-holder system (FEI Inc., Hillsboro, OR). A 100 μM sample was prepared in the phosphate buffer, including salt at pH 2.3 as described above. Approximately 5 μL of sample was applied on a lacey carbon film coated on a copper grid in a Vitrobot vitrification system (FEI Inc., Hillsboro, OR). After blotting and plunging of the specimen in liquid ethane, the vitrified sample was transferred to the cryoholder in a cryo-transfer stage immersed in liquid nitrogen. The cryoholder temperature was maintained below −170 °C to prevent sublimation of vitreous water during imaging. The dimensions of the aggregates were determined using ImageJ55 (U.S. National Institutes of Health, Bethesda, MD) by taking the average of 20 measurements.
Results and discussion
pH-responsive assembly
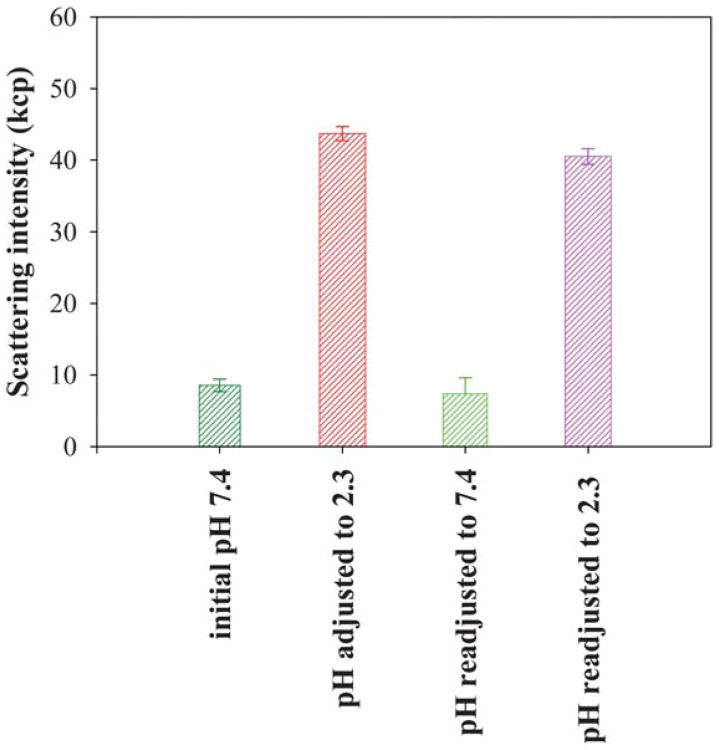
The pH responsiveness of PEG5K-c17H6 was tested by measuring the average intensity of 532 nm laser light scattering (90° scattering angle); representative results are shown in Fig. 1. Initially, in PBS buffer at pH 7.4 (above the pKa of Glu), the scattering intensity from 50 μM polypeptide solution of the conjugate is low (similar to the value of scattering intensity of the buffer) but upon a reduction in pH (below the pKa of Glu) the scattering intensity increased markedly suggesting the formation of aggregates; this aggregation is suggested to be reversible based on the observation of similar scattering results with repeated variations of solution pH (Fig. 1). These data indicate that the PEG conjugate retains the pH responsiveness observed for 17H6 alone,51 corroborating the supposition that self-assembly is controlled by the ionization state of the glutamic acid residues.
Fig. 1.
Comparison of average laser light scattering intensity of PEG5K-c17H6 as a function of pH.
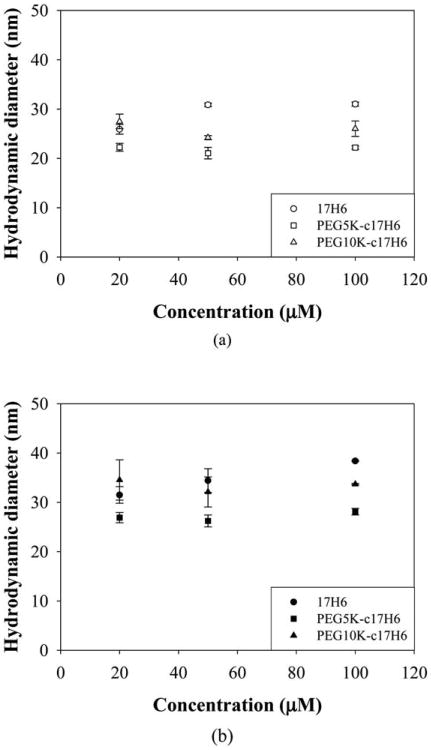
Average diffusion coefficients and polydispersity index (PDI) values of the aggregates were determined using a second order cumulant fit of the DLS data. Representative fits for PEG5K-c17H6 and PEG10K-c17H6 (100 μM on a monomer basis) at 20 °C, in the pH 2.3 buffer containing isotonic salt, are given Fig. S2 and S4 (ESI†), respectively, and indicated good agreement between the experimental data and the cumulant fits; the residuals of the fits were small and randomly scattered (data not shown) confirming the validity of the fit. PDI values ranged between 0.1 and 0.3; no correlation was observed between PDI and temperature or concentration or nature of the block copolymer. Diffusion coefficients were estimated from the slope of the plots of decay rate versus the square of the scattering vector (Γ vs. q2; Fig S3 and S5, ESI†). Γ vs. q2 was linear with an intercept of zero (within statistical uncertainty) for all sample conditions, consistent with an isotropic aggregate morphology and lack of contributions to Γ from rotational diffusion.56 Hydrodynamic diameter (DH) values of the polypeptide and the conjugates were obtained from the fitted diffusion coefficients at sample concentrations of 20, 50 and 100 μM, at 20 °C and 37 °C; results are shown in Fig. 2a and Fig. 2b, respectively. Similar hydrodynamic diameter values (∼20–30 nm, Fig. 2a) were obtained for 17H6 and the conjugates with little or negligible dependence on polypeptide concentration as observed in other block copolymer systems57,58 over select concentration ranges. DH values at 37 °C, ranging between ∼25 and 40 nm (Fig. 2b), were observed to be slightly larger than those at 20 °C.
Fig. 2.
Concentration dependence of hydrodynamic diameters of 17H6 (circles), PEG5K-c17H6 (squares), and PEG10K-c17H6 (triangles) in pH 2.3 buffer containing isotonic salt at (a) 20 °C (open symbols), and (b) 37 °C (closed symbols).
Various trends have been observed for other block copolymer systems, depending on the chemical nature of the copolymers. For a polystyrene-block-poly(N-vinylpyrrolidone) (PS-PNVP) block copolymer, for example, no change in DH was observed between 10 and 40 °C.57 A decrease in DH with T was obtained for thermosensitive poly(2-cinnamoylethyl methacrylate)-block-poly(N-isopropylacrylamide) (PCEMA-PNIPAAm) due to the collapse of PNIPAAm.7 Similarly, a significant decrease in DH was observed upon increasing temperature for polystyrene-block-poly(ethylene oxide) (PS-PEO) copolymers due to the morphological change from vesicles to worm like micelles.13 However, c17H6 is likely more flexible compared to those core-forming polymers, which may allow chain exchange so that aggregate size can vary with temperature. Morphological changes or expansion of corona chains may be another explanation of the temperature dependence of DH for the alanine-rich copolymers. Indeed, it has been shown that the apparent volume of PEG chains increases with increasing temperature, due to disordering of tightly bound water.58 Regardless of the detailed reasons for these modest differences as a consequence of changes in temperature, our results show that the protonated alanine-rich blocks support the formation of nano-size aggregates as long as a hydrophilic block is attached. The pH dependence of the self-association of the polypeptide and the conjugates indicates that they can form stable aggregates when the repulsions between the glutamic acids are eliminated. Hence, attachment of hydrophobic drugs to the glutamic acid residues should drive aggregation of the polypeptide/conjugates independent of pH. These aggregates form at relatively low concentrations (as low as 20 μM (∼0.3–0.5 mg ml−1)) and maintain their integrity at physiological temperatures, suggesting that these association processes may be relevant to the assembly of drug delivery vehicles.
SANS analysis of aggregates
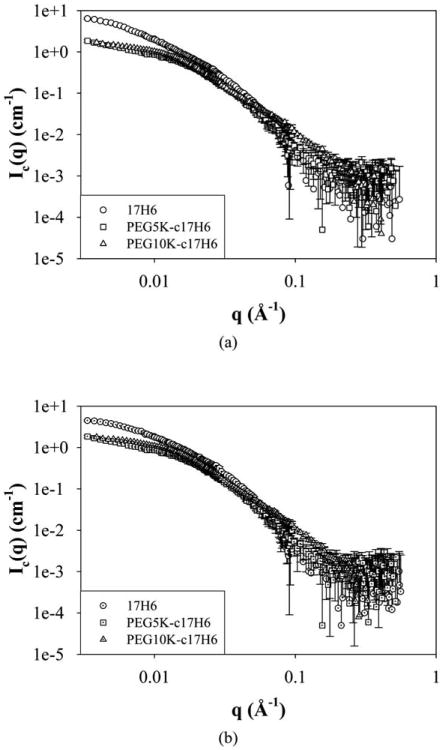
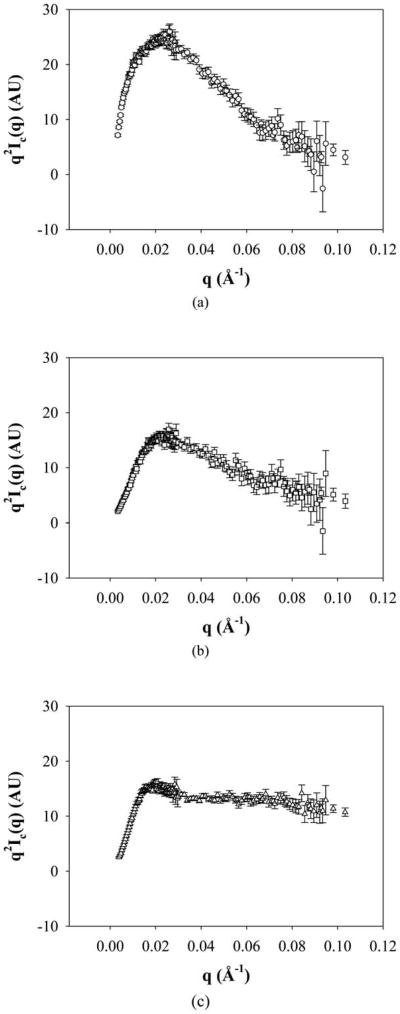
Preliminary investigations with SANS were used to evaluate structural parameters such as the radius of gyration, morphology and aggregation number of the aggregates. SANS data were collected at 100 μM sample concentrations so as to maximize scattering intensities (see also below), and the samples were prepared in phosphate buffer containing 150 μM salt at acidic pD (below the pKa of Glu) in D2O. Since the actual scattering intensities of the samples are low, incoherent scattering becomes considerable. For this reason, the intensity of the background scattering (B) was subtracted from the actual scattering intensities (I) to obtain coherent scattering intensities (Ic) and in all data analyses (Debye and Kratky plots and power law analyses) coherent scattering intensity values were used.59 SANS curves for 17H6 and the conjugates (with background scattering correction) at 20 °C, and 37 °C are given in Fig. 3. Radius of gyration values and scattering intensity values at q = 0 were determined using a Debye plot, which has been used in the analysis of surfactant and copolymer aggregates.60,61 For q < 0.01, the Debye equation can be given as:
Fig. 3.
SANS curves of 17H6 (circles), PEG5K-c17H6 (squares), and PEG10K-c17H6 (triangles) in pH 2.3 buffer at (a) 20 °C (open symbols), and (b) 37 °C (dotted symbols).
| (1) |
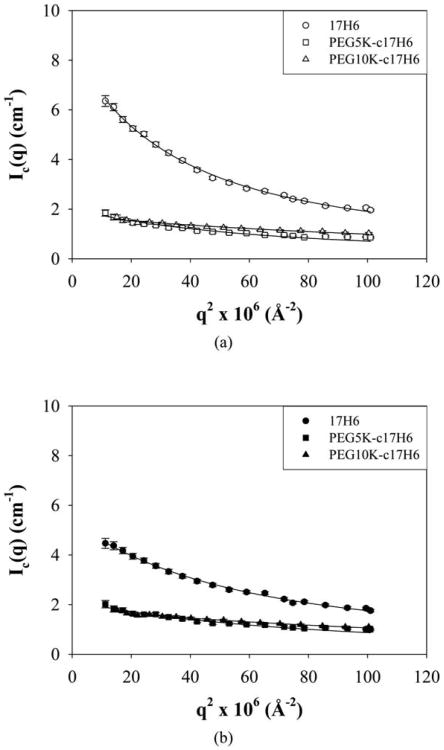
where x = (qRg)2, I0 is the scattering intensity at q = 0, and Rg is the radius of gyration.60 Debye plots of the samples are given in Fig. 4 and estimated I0 and Rg values are given in Table 2. Rg values were estimated to be ∼14–28 nm ranging as a function of the nature of the hydrophilic block and temperature. Rg/RH values (characteristic ratios) were used to estimate the architecture of the aggregates and are given in Table 2. It is commonly accepted that characteristic ratio values of 0.778, 1.505, and >2 indicate hard spheres, structures with a random-coil organization like that of Gaussian chains, and rigid rods respectively.62,63 Comparisons of these values with the characteristic values determined for the polypeptide and conjugates at room temperature (Table 2) yield insights into the structures formed by the aggregation of these macromolecules. 17H6 and PEG5K-c17H6, with characteristic ratios of approximately 1.8 and 1.9, respectively, are suggested to adopt semi-flexible elongated structures. In contrast, PEG10K-c17H6, with a characteristic ratio near 1, is suggested to form spherical structures; these results are consistent with what is anticipated based on the increasing length of the hydrophilic block presumably biasing towards greater surface curvature for the aggregates. The Rg/RH values for all polypeptides decrease near physiological temperature, suggesting that both the polypeptide and the conjugates form more spherical structures. Temperature-induced structural changes, from elongated structures to spheres, have also been observed for other block copolymer systems such as PS-PEO,13 and polystyrene-block-polyisoprene (PS-PI)64 micelles. The morphological change in the PS-PI systems was attributed to the increase in excluded volume parameter of PI and the decrease in the surface free energy for PS in heptane as temperature increases.64 Thus, it can be speculated that any temperature-induced structural changes of the conjugates may be due to the increase in the volume of PEG chains upon increasing temperature58.
Fig. 4.
Debye plots of 17H6 (circles), PEG5K-c17H6 (squares), and PEG10K-c17H6 (triangles) in pH 2.3 buffer at (a) 20 °C (open symbols), and (b) 37 °C (closed symbols). Debye fits are given as lines.
Table 2. Summary of DLS and SANS data.
| Sample | T (°C) | RH (nm) | Rg (nm) | Rg/RH | I0 | <y> |
|---|---|---|---|---|---|---|
| 17H6 | 20 | 15.5 ± 0.2 | 27.6 ± 0.3 | 1.78 ± 0.03 | 8.34 ± 0.09 | 266 ± 3 |
| 17H6 | 38 ± 1a | 19.2 ± 0.1 | 22.1 ± 0.3 | 1.15 ± 0.02 | 5.36 ± 0.06 | 171 ± 2 |
| PEG5K-c17H6 | 20 | 11.1 ± 0.2 | 20.9 ± 0.7 | 1.88 ± 0.08 | 2.01 ± 0.05 | 31 ± 1 |
| PEG5K-c17H6 | 38 ± 1a | 14.1 ± 0.3 | 19.1 ± 0.6 | 1.35 ± 0.06 | 2.16 ± 0.05 | 33 ± 1 |
| PEG10K-c17H6 | 20 | 13.0 ± 0.8 | 14.1 ± 0.4 | 1.08 ± 0.06 | 1.74 ± 0.03 | 14 ± 1 |
| PEG10K-c17H6 | 37 | 16.9 ± 0.1 | 14.2 ± 0.3 | 0.84 ± 0.02 | 1.88 ± 0.03 | 15 ± 1 |
Average of SANS experiment temperature (39 °C) and DLS experiments' temperature (37 °C).
I0 values obtained from the Debye plots were used to estimate aggregation number, <y>, using the following equation:
| (2) |
where Magg and Mmon are the molecular weight of aggregate and unimer respectively, NA is Avogadro's number, c is the concentration of solute, ρ and ρs are the scattering length density of solute and solvent respectively, and ν is the partial molar volume of solute.59 In the estimation of <y> values, ρ and ν of the conjugates were calculated based on the weight fraction of each block (νc17H6 = ν17H6 = 0.71 cm3 g−1, νPEG = 0.89 cm3 g−1, ρc17H6 = 1.99 × 1010 cm−2, ρ17H6 = 2.16 × 1010 cm−2, ρPEG = 0.64 × 1010 cm−2). Calculated <y> values are given in Table 2. Aggregation numbers were estimated to be in the order of ∼102 and ∼101 for the polypeptide and the conjugates, respectively, consistent with the relative RH and Rg values in Table 2. An approximate 35% drop in aggregation number for the 17H6, upon increasing temperature, was indicated from the SANS data despite similarities in the overall shape of the scattering curve, suggesting some limitations in determining the aggregation number for 17H6 from these data. The estimated high aggregation number (∼102) of the polypeptide thus may result in part from overestimated I0 values that result from the lack of a plateau in the low q regime. No such limitations were suggested in the data for the conjugates. Despite the ambiguities in quantitatively estimating aggregate numbers from the 17H6 data, it is clear from the data that the polypeptide exhibits a higher I0 value, which unambiguously indicates a higher average aggregation number. The decrease in the aggregation number with the increase in the degree of the polymerization of the solvophilic chain (NB) was expected, given the increase in steric bulk of the larger PEG chains, which favors an increase in surface curvature. The grafting distance, b, (i.e., the square root of the area per chain (b2), which is comparable to the area per head group in the case of surfactant micelles) increases with increasing degree of polymerization of the solvophilic chain, with resulting morphological changes as observed for PS-PEO65 and poly(butylene oxide-block-poly(ethylene oxide) (PBO-PEO)66 copolymers.
Kratky plots were also used to assess the structure of the aggregates and are given in Fig. 5. For 17H6 and PEG5K-c17H6, a definite peak was obtained. In contrast, PEG10K-c17H6 exhibited a slight peak and reached a horizontal asymptote at increasing q values. The peak in a Kratky plot suggests a compact structure due to strong core contrast67 as observed in folded proteins,68,69 star polymers,70,71 and micellar systems.61,67,72 Horizontal asymptotes (e.g., Ic ∼ q−2 scaling behavior at high q) have also been observed for polymer coils and micelles,61 star polymers70 and polymer brushes surrounding lamellar sheets73 and in these previous cases is due to the scattering from corona chains.74 Our results thus indicate scaling for 17H6 and PEG5K-c17H6 that is consistent with that of compact structures, while that for PEG10K-c17H6 reveals a more dominant effect of swollen PEG chains, as expected. Kratky plots of the samples at a near physiological temperature are given in Fig. S6 (ESI†) and are similar to those in Fig. 5, showing no significant temperature effect.
Fig. 5.
Kratky plots of (a) 17H6, (b) PEG5K-c17H6, and (c) PEG10K-c17H6 at 20 °C.
Aggregate compactness was also roughly indicated from the slopes of log Ic vs. log q curves in the intermediate and high q regions (Fig. S7, ESI†). Of note in Fig. S7 is the existence of a slope of approximately −4 for 17H6 (Fig. S7a), describing the Porod region (scattering from a sharp interface) and corroborating the compact structure for 17H6 aggregates indicated by the Kratky plots. Scattering from PEG5K-c17H6 and PEG10K-c17H6 structures (Fig. S7b and S7c), in contrast, lacked a Porod region (Fig. S7b) likely due to the lack of a sharp interface caused by the dispersion of the PEG chains in the water phase.74 Thus, Kratky plots and power law plots indicated that the aggregates compactness is regulated by the nature and the length of the hydrophilic block.
Morphology
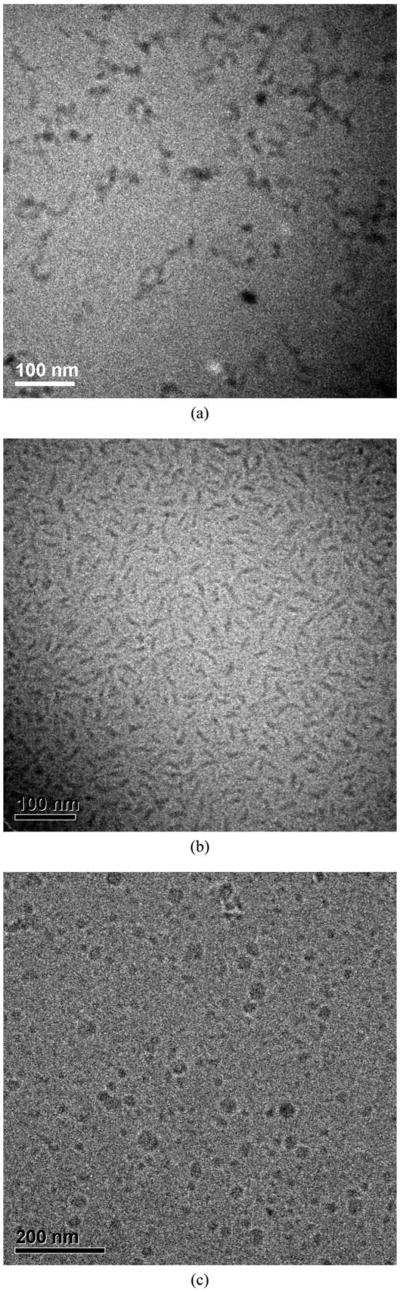
The morphologies of the aggregates were more directly visualized via cryo-TEM. Images of the aggregates are shown in Fig. 6, and the dimensions of the aggregates, estimated from the TEM data, are presented in Table 3. As it is clearly illustrated in Fig. 6a, the polypeptide 17H6 adopts irregular and elongated aggregates, as might be expected for this moderately hydrophobic polypeptide. 17H6 tends to form clusters under these conditions, perhaps due to the increase in local concentration during TEM sample preparation.75 In contrast to the elongated structures observed for 17H6 in cryo-TEM, globular structures were obtained when employing a negative-staining technique,51 likely as a result of changes in morphology during drying of these aggregation-prone polypeptides.
Fig. 6.
Cryo-TEM images of (a) 17H6, (b) PEG5K-c17H6, and (c) PEG10K-c17H6. Scale bars: (a), (b) 100 nm, and (c) 200 nm.
Table 3. Dimensions of the aggregates from cryo-TEM data.
| Sample | Core dimension | Average size (nm) |
|---|---|---|
| 17H6 | width | 10 ± 2 |
| 17H6 | length | 52 ± 10 |
| PEG5K-c17H6 | width | 7 ± 2 |
| PEG5K-c17H6 | length | 35 ± 4 |
| PEG10K-c17H6 | diameter | 19 ± 6 |
Striking differences in morphology, however, are observed upon the addition of PEG chains to the cleaved polypeptide. As clearly captured in Fig. 6b, PEG5K-c17H6 adopts regular and elongated structures, corroborating the estimations based on Rg/RH values. A fraction of PEG5K-c17H6 structures in the images appears spherical; these structures are likely the crosssection of the elongated structures when they lie perpendicular to the surface. The observed core widths of these elongated structures of 17H6 and PEG5K-c17H6 are not different from each other, but longer structures were observed for the polypeptide as expected from the relative Rg and RH values of the polypeptide and PEG5K-c17H6. These observations clearly show the stabilization of the smaller structures imparted by the PEG corona, and suggest opportunities to vary the morphologies of these aggregates via variations in the PEG chain length. Indeed, as illustrated in Fig. 6c, PEG10K-c17H6 exhibits a different morphology; spherical structures with an average core diameter of 19 nm, and a polydispersity greater than that of PEG5K-c17H6, are observed. Considering the Rg of PEG10K (∼3 nm), this value is consistent with the DH estimates obtained from DLS (∼26 nm) within experimental uncertainty, as solvated PEG chains are not likely to appear in cryo-TEM observations.76
The cylinder-to-sphere transformation as hydrophilic block length increases is consistent with experimental observations of other polymeric systems,77,78 and predictions based on molecular dynamics simulations.79 An unexpected result is the ∼10 nm core width of PEG5K-c17H6 versus the ∼20 nm core diameter of PEG10K-c17H6, given experimental observations and scaling relationships that predict that core diameter is inversely proportional to solvophilic chain length.5,78,80 It seems likely that PEG10K-c17H6 has a more swollen core, owing to a lower efficiency of packing of the polypeptide chains in the spherical aggregates; the lower aggregation number obtained for these structures is consistent with these speculations. Additionally, in contrast to many examples of block copolymers composed of one type of monomer in each block, the hydrophobic c17H6 block in our polymers contains different monomers—hydrophobic alanine as well as hydrophilic glutamine and glutamic acids—and it is possible that these hydrophilic components within the core blocks may be differently hydrated in aggregate structures of different morphologies.
Conclusions
The self-association behavior of a polypeptide, 17H6, containing a histidine-rich fusion tag and an alanine-rich block, was compared to the self-association of PEGylated conjugates of the alanine-rich polypeptide. The alanine-rich block, equipped with ionizable amino acids, imparts pH responsiveness to 17H6 and the conjugates. It was also shown that the histidine tag plays a solubilizing role when it is charged, in addition to its conventional function in protein purification and its observed role in the immobilization of negatively charged nanoparticles.81 At acidic pH (upon protonation of glutamic acid residues), the polypeptide and the conjugates formed aggregates with elongated or spherical morphologies depending on the nature and length of the hydrophilic block. Although only a few examples were shown in this study, our results clearly illustrate that a combination of recombinant synthesis and chemical conjugation methods may offer a wide spectrum of nanostructures with controlled size and morphology by the precise control of the block composition.
The pH-responsiveness observed in the c17H6 based polymer systems suggests opportunities for these systems in drug delivery applications. Specifically, functionalization of the reactive glutamic acids with hydrophobic molecules is likely to trigger the aggregation of the alanine-rich block, but independent of pH. Owing to the low concentrations at which the polypeptide and conjugates aggregate (aggregation concentrations lower than 0.5 mg ml−1 at both ambient and physiological temperatures), coupled with the tuning of their morphologies, PEG-c17H6 conjugates may be useful as drug carrier systems where c17H6 serves as a hydrophobic drug attachment site (similar to PEG-polyaspartate-drug conjugate systems38,39,42), or where the polypeptide domain serves as a therapeutic molecule based on its multivalent display of ligands or drugs.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (NIH) and the National Center for Research Resources (NCRR), a component of the NIH (1-P20-RR017716, 1-RO1-EB006006, and P30-RR031160 (instrument facilities)). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Preliminary studies were made possible by funding by the National Aeronautics and Space Administration (NA68-01923). This work was also partially supported by the Center for Neutron Science at University of Delaware under award #70NANB7H6178 (U.S. Department of Commerce). The authors thank Dr. Aaron P. Eberle and Dr. Paul D. Butler for acquisition of SANS data and suggestions in the SANS data analysis, respectively. The NCNR and the Center for Neutron Science (CNS) at University of Delaware are acknowledged for making SANS experiments possible. The W. M. Keck College of Engineering Electron Microscopy Laboratory at the University of Delaware is also acknowledged for support in conducting TEM experiments.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/c1sm05686g/
Contributor Information
Christopher J. Roberts, Email: cjr@udel.edu.
Darrin J. Pochan, Email: pochan@udel.edu.
Kristi L. Kiick, Email: kiick@udel.edu.
References
- 1.Letchford K, Burt H. Eur J Pharm Biopharm. 2007;65:259–269. doi: 10.1016/j.ejpb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Panagiotopoulos AZ, Floriano MA, Kumar SK. Langmuir. 2002;18:2940–2948. [Google Scholar]
- 3.Gohy JF, Lohmeijer BGG, Alexeev A, Wang XS, Manners I, Winnik MA, Schubert US. Chem –Eur J. 2004;10:4315–4323. doi: 10.1002/chem.200400222. [DOI] [PubMed] [Google Scholar]
- 4.Hillmyer MA. Science. 2007;317:604–605. doi: 10.1126/science.1144899. [DOI] [PubMed] [Google Scholar]
- 5.Zhang LF, Eisenberg A. J Am Chem Soc. 1996;118:3168–3181. [Google Scholar]
- 6.Yu YS, Zhang LF, Eisenberg A. Macromolecules. 1998;31:1144–1154. [Google Scholar]
- 7.Chen XR, Ding XB, Zheng ZH, Peng YX. Macromol Biosci. 2004;5:157–163. doi: 10.1002/mabi.200400150. [DOI] [PubMed] [Google Scholar]
- 8.Choucair A, Lavigueur C, Eisenberg A. Langmuir. 2004;20:3894–3900. doi: 10.1021/la035924p. [DOI] [PubMed] [Google Scholar]
- 9.Bhargava P, Zheng JX, Li P, Quirk RP, Harris FW, Cheng SZD. Macromolecules. 2006;39:4880–4888. [Google Scholar]
- 10.Discher DE, Ahmed F. Annu Rev Biomed Eng. 2006;8:323–341. doi: 10.1146/annurev.bioeng.8.061505.095838. [DOI] [PubMed] [Google Scholar]
- 11.Liu SY, Billingham NC, Armes SP. Angew Chem, Int Ed. 2001;40:2328–2331. doi: 10.1002/1521-3773(20010618)40:12<2328::AID-ANIE2328>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Pispas S, Hadjichristidis N. Langmuir. 2003;19:48–54. [Google Scholar]
- 13.Bhargava P, Tu YF, Zheng JX, Xiong HM, Quirk RP, Cheng SZD. J Am Chem Soc. 2007;129:1113–1121. doi: 10.1021/ja0653019. [DOI] [PubMed] [Google Scholar]
- 14.Rao J, Luo Z, Ge Z, Liu H, Liu S. Biomacromolecules. 2007;8:3871–3878. doi: 10.1021/bm700830b. [DOI] [PubMed] [Google Scholar]
- 15.Ueki T, Watanabe M, Lodge TP. Macromolecules. 2009;42:1315–1320. [Google Scholar]
- 16.Schacher F, Walther A, Muller AHE. Langmuir. 2009;25:10962–10969. doi: 10.1021/la901182c. [DOI] [PubMed] [Google Scholar]
- 17.Li ZB, Hillmyer MA, Lodge TP. Macromolecules. 2006;39:765–771. [Google Scholar]
- 18.Zhang LF, Eisenberg A. Macromolecules. 1999;32:2239–2249. [Google Scholar]
- 19.Johnson BK, Prud'homme RK. Phys Rev Lett. 2003;91:118302. doi: 10.1103/PhysRevLett.91.118302. [DOI] [PubMed] [Google Scholar]
- 20.Battaglia G, Ryan AJ. J Phys Chem B. 2006;110:10272–10279. doi: 10.1021/jp060728n. [DOI] [PubMed] [Google Scholar]
- 21.Cui HG, Chen ZY, Zhong S, Wooley KL, Pochan DJ. Science. 2007;317:647–650. doi: 10.1126/science.1141768. [DOI] [PubMed] [Google Scholar]
- 22.Hayward RC, Pochan DJ. Macromolecules. 2010;43:3577–3584. [Google Scholar]
- 23.Meli L, Santiago JM, Lodge TP. Macromolecules. 2010;43:2018–2027. [Google Scholar]
- 24.Wang XS, Guerin G, Wang H, Wang YS, Manners I, Winnik MA. Science. 2007;317:644–647. doi: 10.1126/science.1141382. [DOI] [PubMed] [Google Scholar]
- 25.Berthier DL, Schmidt I, Fieber W, Schatz C, Furrer A, Wong K, Lecommandoux S. Langmuir. 2010;26:7953–7961. doi: 10.1021/la904832d. [DOI] [PubMed] [Google Scholar]
- 26.Yang PD, Zhao DY, Margolese DI, Chmelka BF, Stucky GD. Chem Mater. 1999;11:2813–2826. [Google Scholar]
- 27.Underhill RS, Liu GJ. Chem Mater. 2000;12:2082–2091. [Google Scholar]
- 28.Chen J, Zeng F, Wu SZ. ChemPhysChem. 2010;11:1036–1043. doi: 10.1002/cphc.200900999. [DOI] [PubMed] [Google Scholar]
- 29.Allen C, Maysinger D, Eisenberg A. Colloids Surf, B. 1999;16:3–27. [Google Scholar]
- 30.Kataoka K, Harada A, Nagasaki Y. Adv Drug Delivery Rev. 2001;47:113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 31.Torchilin VP. J Controlled Release. 2001;73:137–172. doi: 10.1016/s0168-3659(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 32.Gaucher G, Dufresne MH, Sant VP, Kang N, Maysinger D, Leroux JC. J Controlled Release. 2005;109:169–188. doi: 10.1016/j.jconrel.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 33.Edlund U, Albertsson AC. Degradable Aliphatic Polyesters. 2002;157:67–112. [Google Scholar]
- 34.Torchilin VP. Pharm Res. 2007;24:1–16. doi: 10.1007/s11095-006-9132-0. [DOI] [PubMed] [Google Scholar]
- 35.Otsuka H, Nagasaki Y, Kataoka K. Curr Opin Colloid Interface Sci. 2001;6:3–10. [Google Scholar]
- 36.Riley T, Govender T, Stolnik S, Xiong CD, Garnett MC, Illum L, Davis SS. Colloids Surf, B. 1999;16:147–159. [Google Scholar]
- 37.Gong CY, Wei XW, Wang XH, Wang YJ, Guo G, Mao YQ, Luo F, Qian ZY. Nanotechnology. 2010;21:215103. doi: 10.1088/0957-4484/21/21/215103. [DOI] [PubMed] [Google Scholar]
- 38.Bae Y, Jang WD, Nishiyama N, Fukushima S, Kataoka K. Mol BioSyst. 2005;1:242–250. doi: 10.1039/b500266d. [DOI] [PubMed] [Google Scholar]
- 39.Bae Y, Kataoka K. Adv Drug Delivery Rev. 2009;61:768–784. doi: 10.1016/j.addr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Lavasanifar A, Samuel J, Kwon GS. Adv Drug Delivery Rev. 2002;54:169–190. doi: 10.1016/s0169-409x(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 41.Osada K, Christie RJ, Kataoka K. J R Soc Interface. 2009;6:S325–S339. doi: 10.1098/rsif.2008.0547.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae Y, Fukushima S, Harada A, Kataoka K. Angew Chem, Int Ed. 2003;42:4640–4643. doi: 10.1002/anie.200250653. [DOI] [PubMed] [Google Scholar]
- 43.Top A, Kiick KL. Adv Drug Delivery Rev. 2010;62:1530–1540. doi: 10.1016/j.addr.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han KK, Richard C, Biserte G. Int J Biochem. 1983;15:875–884. [Google Scholar]
- 45.Lee TAT, Cooper A, Apkarian RP, Conticello VP. Adv Mater. 2000;12:1105–1110. [Google Scholar]
- 46.Dreher MR, Raucher D, Balu N, Colvin OM, Ludeman SM, Chilkoti A. J Controlled Release. 2003;91:31–43. doi: 10.1016/s0168-3659(03)00216-5. [DOI] [PubMed] [Google Scholar]
- 47.Furgeson DY, Dreher MR, Chilkoti A. J Controlled Release. 2006;110:362–369. doi: 10.1016/j.jconrel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Meyer DE, Shin BC, Kong GA, Dewhirst MW, Chilkoti A. J Controlled Release. 2001;74:213–224. doi: 10.1016/s0168-3659(01)00319-4. [DOI] [PubMed] [Google Scholar]
- 49.Farmer RS, Argust LM, Sharp JD, Kiick KL. Macromolecules. 2006;39:162–170. doi: 10.1021/ma051534t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S, Kiick KL. Macromolecules. 2008;41:764–772. doi: 10.1021/ma702128a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Top A, Kiick KL, Roberts CJ. Biomacromolecules. 2008;9:1595–1603. doi: 10.1021/bm800056r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farmer RS, Top A, Argust LM, Liu S, Kiick KL. Pharm Res. 2008;25:700–708. doi: 10.1007/s11095-007-9344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Top A, Roberts CJ, Kiick KL. Biomacromolecules. 2011;12:2184–2192. doi: 10.1021/bm200272w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kline SR. J Appl Crystallogr. 2006;39:895–900. [Google Scholar]
- 55.Abramoff MD, Magelhaes PJ, Ram SJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 56.Huang H, Chung B, Jung J, Park HW, Chang T. Angew Chem, Int Ed. 2009;48:4594–4597. doi: 10.1002/anie.200900533. [DOI] [PubMed] [Google Scholar]
- 57.Hussain H, Tan BH, Gudipati CS, He CB, Liu Y, Davis TP. Langmuir. 2009;25:5557–5564. doi: 10.1021/la8042389. [DOI] [PubMed] [Google Scholar]
- 58.Sommer C, Pedersen JS, Stein PC. J Phys Chem B. 2004;108:6242–6249. doi: 10.1021/jp036963c. [DOI] [PubMed] [Google Scholar]
- 59.Rubinson KA, Stanley C, Krueger S. J Appl Crystallogr. 2008;41:456–465. [Google Scholar]
- 60.Arleth L, Bauer R, Ogendal LH, Egelhaaf SU, Schurtenberger P, Pedersen JS. Langmuir. 2003;19:4096–4104. [Google Scholar]
- 61.Lee AS, Gast AP, Butun V, Armes SP. Macromolecules. 1999;32:4302–4310. [Google Scholar]
- 62.Bloomfield VA. Biopolymers. 2000;54:168–172. doi: 10.1002/1097-0282(200009)54:3<168::AID-BIP20>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 63.Weiss WF, Hodgdon TK, Kaler EW, Lenhoff AM, Roberts CJ. Biophys J. 2007;93:4392–4403. doi: 10.1529/biophysj.107.112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LaRue I, Adam M, Pitsikalis M, Hadjichristidis N, Rubinstein M, Sheiko SS. Macromolecules. 2006;39:309–314. [Google Scholar]
- 65.Jada A, Hurtrez G, Siffert B, Riess G. Macromol Chem Phys. 1996;197:3697–3710. [Google Scholar]
- 66.Derici L, Ledger S, Mai SM, Booth C, Hamley IW, Pedersen JS. Phys Chem Chem Phys. 1999;1:2773–2785. [Google Scholar]
- 67.Cogan KA, Gast AP, Capel M. Macromolecules. 1991;24:6512–6520. [Google Scholar]
- 68.Doniach S. Chem Rev. 2001;101:1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- 69.Kamatari YO, Konno T, Kataoka M, Akasaka K. J Mol Biol. 1996;259:512–523. doi: 10.1006/jmbi.1996.0336. [DOI] [PubMed] [Google Scholar]
- 70.Prosa TJ, Bauer BJ, Amis EJ. Macromolecules. 2001;34:4897–4906. [Google Scholar]
- 71.Willner L, Jucknischke O, Richter D, Roovers J, Zhou LL, Toporowski PM, Fetters LJ, Huang JS, Lin MY, Hadjichristidis N. Macromolecules. 1994;27:3821–3829. [Google Scholar]
- 72.Moffitt M, Yu YS, Nguyen D, Graziano V, Schneider DK, Eisenberg A. Macromolecules. 1998;31:2190–2197. [Google Scholar]
- 73.Richter D, Schneiders D, Monkenbusch M, Willner L, Fetters LJ, Huang JS, Lin M, Mortensen K, Farago B. Macromolecules. 1997;30:1053–1068. [Google Scholar]
- 74.Mortensen K. J Phys: Condens Matter. 1996;8:A103–A124. [Google Scholar]
- 75.Hamley IW, Castelletto V, Fundin J, Yang Z, Crothers M, Attwood D, Talmon Y. Colloid Polym Sci. 2004;282:514–517. [Google Scholar]
- 76.Castelletto V, Newby GE, Zhu Z, Hamley IW, Noirez L. Langmuir. 2010;26:9986–9996. doi: 10.1021/la100110f. [DOI] [PubMed] [Google Scholar]
- 77.Jain S, Bates FS. Science. 2003;300:460–464. doi: 10.1126/science.1082193. [DOI] [PubMed] [Google Scholar]
- 78.Zheng Y, Won YY, Bates FS, Davis HT, Scriven LE, Talmon Y. J Phys Chem B. 1999;103:10331–10334. [Google Scholar]
- 79.Srinivas G, Discher DE, Klein ML. Nat Mater. 2004;3:638–644. doi: 10.1038/nmat1185. [DOI] [PubMed] [Google Scholar]
- 80.Sheng YJ, Wang TY, Chen WM, Tsao HK. J Phys Chem B. 2007;111:10938–10945. doi: 10.1021/jp073408s. [DOI] [PubMed] [Google Scholar]
- 81.Sharma N, Top A, Kiick KL, Pochan DJ. Angew Chem, Int Ed. 2009;48:7078–7082. doi: 10.1002/anie.200901621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.