Abstract
It has been proposed that bile acid suppression of CYP7A1 gene expression is mediated through a gut-liver signaling pathway fibroblast growth factor (FGF)15/19-fibroblast growth factor receptor 4 which is initiated by activation of farnesoid X receptor in the ileum but not in the liver. This study evaluated whether FGF15/19 protein levels in the portal blood reflected changes in FGF15/19 mRNA in the ileum. Studies were conducted in Sprague Dawley rats and New Zealand white rabbits fed regular chow (controls), supplemented with cholesterol (Ch) or cholic acid (CA). After feeding CA, ileal FGF15 mRNA increased 8.5-fold in rats and FGF19 rose 16-fold in rabbits associated with 62 and 75% reduction of CYP7A1 mRNA, respectively. Neither FGF15 nor FGF19 protein levels changed in the portal blood to correspond with the marked increase of FGF15/19 mRNA levels in the ileum or inhibited CYP7A1 expression in the liver. Further, in Ch-fed rats, CYP7A1 mRNA increased 1.9-fold (P < 0.001) although FGF15 mRNA levels in the ileum and portal blood FGF15 protein levels were not decreased. In Ch-fed rabbits, although FGF19 mRNA levels in the ileum and liver did not increase significantly, CYP7A1 mRNA declined 49% (P < 0.05). We were unable to find corresponding changes of FGF15/19 protein levels in the portal blood in rats and rabbits where the mRNA levels of FGF15/19 in the ileum and CYP7A1 in the liver change significantly.
Keywords: fibroblast growth factor 15, fibroblast growth factor 19, regulation, bile acid
Farnesoid X receptor (FXR) is the bile acid sensor that has been identified as the negative regulator for CYP7A1 transcription (1–3). Hydrophobic bile acids, chenodeoxycholic acid (CDCA), lithocholic acid, and deoxycholic acid (DCA) are potent ligands that activate FXR. It was proposed that activated FXR enhances SHP expression in the liver, which in turn inhibits LRH-1-mediated transcriptional activation of the CYP7A1 gene (4, 5). This hypothesis, involving a cascade reaction in the liver, provides an explanation at the molecular level for the indirect repression of CYP7A1 by bile acid-activated FXR. Our results from in vivo studies in rabbits (6–8) and rats (9) agree with the hypothesis that activation of hepatic FXR by the increased circulating enterohepatic bile acid pool plays a crucial role in the downregulation of CYP7A1 expression.
Pandak et al. (10) reported that intravenous perfusion of taurocholate via the internal jugular vein failed to downregulate CYP7A1 expression in rats with bile fistula and proposed that there should be an “intestinal factor” which is necessary for the downregulation of CYP7A1. When the bile duct was ligated in rats (11) or mice (12), where bile acids accumulated in the liver and did not reach the intestine, CYP7A1 expression unexpectedly increased indicating a role for the intestine in regulation of CYP7A1. Furthermore, it was suggested that bile acids infused through peripheral (10, 13, 14) and portal venous systems (15) did not exert “feedback regulation” to downregulate CYP7A1/bile acid synthesis in rats.
Recently, an alternative hypothesis was proposed that CYP7A1 gene expression was controlled by the gut-hepatic signal fibroblast growth factor (FGF)15 expressed in the intestine via the ileal FXR-FGF15-FGF4 receptor pathway (12). FGF15 in mice (12), or its ortholog FGF19 in humans (16), is the target gene of FXR. In mice, FGF15 is mainly expressed in the ileum but not in the liver (12), while FGF19 also is expressed in the hepatocytes (17, 18). Thus, it has been proposed that FGF15 is selectively induced by bile acids-FXR in the ileum that acts as an intestinal-hepatic signal through FGF receptor 4 (FGFR4) interacting with SHP to repress CYP7A1 expression in the liver (12, 16). Furthermore, it was shown that activation of the FXR-FGF15 pathway in the intestine, but not activation of FXR-SHP in the liver, suppresses CYP7A1 gene expression (19). More recently, Kong et al. (20) reported that both FGFR4 and SHP are responsible for suppressing Cyp7a1 expression in the liver of mice after activation of FXR, but FGFR4 seems more important.
Despite the above findings, it is not known whether FGF15/19 protein levels in the portal blood reflect changes of FGF15/19 gene expression in the ileum to reach the liver and control the gene expression of CYP7A1. In the present study we investigated this relationship.
MATERIALS AND METHODS
Animal experiments
In this study, rats and rabbits were fed cholesterol (Ch)- or cholic acid (CA)-containing chow respectively, as these treatments have been shown to cause significant changes in activation of FXR in the ileum and liver and hepatic gene expression of CYP7A1 in these animals.
Sprague Dawley rats (n = 24), males weighing around 250 g were divided into three different groups (n = 8): rats fed regular chow (control), 2% Ch, and 1% CA respectively for 7 days. On the seventh feeding day, rats were euthanized around 12:00 PM, 4 h after the dark cycle was started (8:00 AM–8:00 PM). Portal and peripheral blood, ileal mucosa, and liver tissues were collected immediately under anesthesia [ketamine (60 mg/kg) + xylazine (6 mg/kg) im] right before euthanasia.
New Zealand white rabbits (n = 24) were assigned into three experimental groups (n = 8) and fed chow (control), 2% Ch, and 0.3% CA for 7 days. The rabbits were fed 4 ounces of chow every day at 8:00 AM under light cycle (7:00 AM–7:00 PM). After completing the treatments, the rabbits were euthanized under anesthesia [ketamine (40 mg/kg) + xylazine (4 mg/kg) iv] approximately 4 h after the last feeding at 8:00 AM. Portal and peripheral blood, ileal mucosa, and liver tissues were collected immediately before euthanasia.
Rats and rabbits were chosen as animal models in this study because we have studied extensively the regulation of CYP7A1 in Ch- and CA-fed rats and rabbits. We know that CYP7A1 in the liver and FXR in the ileum and liver is up- and downregulated in these models (8, 9). Thus, these models are appropriate for studying the relationship between the expression of ileal FGF15/19 and the protein levels of the gut-hepatic signal in the portal blood, as well as the expression of CYP7A1 in the liver.
The animal protocol was approved by the Institutional Animal Care and Use Committee at the Veterans Affairs Medical Center, East Orange, NJ.
Measurements of plasma FGF15 and FGF19 protein concentrations
FGF15 protein levels in the rat plasma were measured with the ELISA method using the Enzyme-linked Immunosorbent Assay kit for FGF15 (USCN Life Science Inc., Wuhan, China). This product has been assessed and certified as meeting the requirements of the International Organization for Standardization, ISO 13485:2003 and EN ISO 13485:2012 (certificate CN09/21756) and ISO 9001:2008 (certificate CN09/21757) by SGS United Kingdom Ltd, UK. According to the instruction manual, we added 100 μl standard or rat plasma samples to coated plates and incubated for 2 h at 37°C. Aspirated and added 100 μl detection reagent A and then incubated for 1 h at 37°C. Aspirated and washed five times with wash buffer. Added 100 μl detection reagent B and incubated for 30 min at 37°C. Aspirated and washed five times. Added 90 μl substrate solution and incubated 15–25 min at 37°C. The reaction was stopped by adding 50 μl of the stop solution. The result of each well was read out at 450 nm immediately using a DTX880 plate reader (Beckman Coulter, Inc., Fullerton, CA).
USCN Life Science Inc. has tested the specificity of this rat FGF15 ELISA kit. Recombinant rat proteins of FGF18, FGF16, FGF10, TNH-α, and IL-6, and recombinant mouse proteins of FGF15, FGF21, FGF10, TNH-α, and IL-4 were prepared at 10 ng/ml for a mid-range recombinant rat FGF15 control. When these recombinant proteins were assayed, no significant cross-reactivity with the above prepared nonrat FGF15 recombinant proteins was observed.
A dilution test was performed to test whether this ELISA kit for rat FGF15 could distinguish changes of FGF15 concentrations corresponding to the applied amounts of rat plasma samples in a linear correlation. Because 100 μl of rat portal plasma were used for routine FGF15 protein measurements, the dilution test was performed in each set of 100, 75, 50, 25, and 12.5 μl of plasma from the same rat portal sample. A total of five sets of the above diluted plasma samples from five different rats were tested. The tested plasma samples were all diluted to 100 μl using the Plasma Sample Diluent (ImmunoChemistry Technologies, LLC, Bloomington, MN) and then were measured using the rat ELISA kit (USCN Life Science Inc.). The results of portal FGF15 concentrations (pg/ml) in the 75, 50, 25, and 12.5 μl plasma samples were divided respectively by the concentration in the corresponding 100 μl plasma sample to obtain relative concentrations (%). In this way, the data from five sets of diluted plasma samples could be pooled together for analysis of correlation coefficient (r) and evaluation of the significance of the linear relationship between the applied amounts of plasma and the corresponding concentrations of FGF15 protein using GraphPad software (GraphPad InStat, San Diego, CA).
The dilution test was also performed using the recombinant rat FGF15 protein solution. The recombinant rat FGF15 protein was synthesized by GenScript USA Inc., Piscataway, NJ. Combined with Western blot analysis, this dilution test was used to demonstrate that the FGF15 ELISA kit used in the present study could identify quantitatively changes of rat FGF15 protein levels in our samples. The test method was the same as used above except that instead of portal blood samples, two sets of the solution were used that contained 100, 50, 25, and 12.5 ng/ml of the recombinant FGF15 protein. It should be noted that the ELISA kit employed only a key truncated peptide of the whole rat FGF15 protein and thus, the molecular weight of rat FGF15 did not match that of the calibration peptide. Therefore, in this test, we continued to use relative FGF15 concentrations (concentrations relative to the concentration in 100 ng/ml solution) to quantitatively represent corresponding FGF15 concentrations in the recombinant protein samples. To obtain relative concentrations (%), the results of FGF15 concentrations (pg/ml) in the 50, 25, and 12.5 ng/ml recombinant rat FGF15 protein sample were divided respectively by the FGF15 concentration in the 100 ng/ml recombinant FGF15 sample.
To test the specificity of this commercial ELISA kit for detecting rat FGF15 protein, an immune-depletion test was performed in 10 paired rat portal plasma samples, 100 μl each + 100 μl Plasma Sample Diluent (ImmunoChemistry Technologies, LLC), with or without adding commercial FGF15 antibody (D-19), a goat polyclonal IgG (Santa Cruz Biotechnology Inc., Santa Cruz, CA). Each paired plasma sample was from the same plasma specimen. This FGF15 antibody is recommended for detection of FGF15 of mouse or rat. In each paired plasma sample (100 μl each + 100 μl Plasma Sample Diluent), one sample was added 10 μg of the FGF15 antibody (D-19) as the positive sample and another sample was added no FGF15 antibody but 10 μg of anti-insulin antibody (mouse monoclonal; Abcam, San Francisco, CA) used as controls. After overnight reaction at 4°C with shaking, 20 μl of protein A/G plus-Agarose (Santa Cruz Biotechnology, Inc., Dallas, TX) were added to both the positive and control samples and kept at 4°C for 4 h with shaking. Then the supernatant was collected after spinning with centrifugation for 1 min at 1,000 g. FGF15 protein concentrations in the supernatant from these paired samples were determined using the ELISA kit from USCN Life Science Inc.
FGF19 protein levels in the rabbit plasma samples were measured using an “in-house” designed indirect ELISA method. Rabbit FGF19 standard protein (Everest Biotech Ltd., Upper Heyford, UK) was diluted 1× with coating buffer (CB2; ImmunoChemistry Technologies, LLC) to make a series of standards (1,000 pg to 15.6 pg/ml). Two hundred microliters of the standards and rabbit (portal or peripheral) plasma samples were added to each well in the plate. After incubation overnight at 4°C, block agent (300 μl of 1% BSA in PBS) (BSA; Fisher Scientific, Fair Lawn, NJ) was added to each well to block unoccupied binding sites. After overnight incubation at 4°C, the plate was washed three times. Then, 1:200 diluted goat anti-rabbit FGF19 antibody, custom-made by Everest Biotech Ltd., in 1% BSA block buffer (BSA; Fisher Scientific) was added (100 μl per well). After incubation for 2 h at room temperature, the wells were washed three times with wash buffer. Then donkey anti-goat IgG-HRP (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) 1:200 diluted with block buffer was added to each well and incubated for 30 min at room temperature. The plate was then was washed three times and 100 μl of tetramethylbenzidine in mildly acidic buffer (SUB2; ImmunoChemistry Technologies) was added to the wells. After incubation for 10–30 min at room temperature, stop buffer (STOP1; ImmunoChemistry Technologies) was added to each well. The absorbance of each well was read by microplate reader DTX880 (Beckman Coulter, Inc.).
The peptide (TSAHGVSHCFLRIR) to produce rabbit FGF19 antibody was chosen by Everest Biotech Ltd. using generally accepted algorithms to identify a suitable part of the protein that was most antigenic and most likely be at the surface of the naturally folded FGF19. The candidate peptide was probed against the rabbit and hare protein database in NCBI (National Center for Biotechnology Information) in order to verify that there was no overlap of the amino acid sequence of the chosen peptide with any other known proteins in these species. In this way, it rules out possible cross-reactivity of the antibody once it is affinity purified using the immunizing peptide and used in immune assays on rabbit tissues and matrices.
Dilution tests were also performed to determine whether the indirect ELISA method could distinguish changes of FGF19 concentrations that corresponded to the applied amounts of rabbit portal plasma samples in a linear fashion. Because 200 μl of rabbit portal plasma were used for routine FGF19 protein measurements, the dilution tests were performed in each set of 225, 200, 175, 150, and 125 μl of portal plasma prepared from the same rabbit sample. A total of 10 sets (each set had five samples) of the above diluted plasma samples from 10 different rabbits were tested. The tested plasma samples were all diluted to 225 μl using the Plasma Sample Diluent (ImmunoChemistry Technologies, LLC) and then were measured using the above described indirect ELISA method. The results of portal FGF19 concentrations (pg/ml) in the 200, 175, 150, and 125 μl plasma samples were divided respectively by the concentration in the 225 μl plasma sample to obtain relative concentrations (%). In this way, the data from 10 sets of diluted plasma samples could be pooled together for statistical analysis.
To test the specificity of the indirect ELISA method for detecting rabbit FGF19 protein, an immune-depletion test was performed in 10 paired rabbit portal plasma samples using anti-FGF19 mAb, a monoclonal anti-human FGF19 antibody produced by Abnova (Taipei, Taiwan). This FGF19 antibody was chosen because we found that it showed some cross-reaction to rabbit FGF19 protein. In each paired plasma sample (200 μl/each), one sample was added 10 μg of the FGF19 antibody as the positive sample and another sample was added no FGF19 antibody but 10 μg of anti-insulin antibody (mouse monoclonal; Abcam, San Francisco, CA) as controls. After overnight reaction at 4°C with shaking, 20 μl of protein A/G plus-Agarose (Santa Cruz Biotechnology, Inc., Dallas, TX) were added to both the positive and control samples and kept at 4°C for 4 h with shaking. Then the supernatant was collected after spinning with centrifugation for 1 min at 1,000 g. FGF19 protein concentrations in the supernatant from these paired samples were determined using the indirect ELISA method.
Western blot analysis
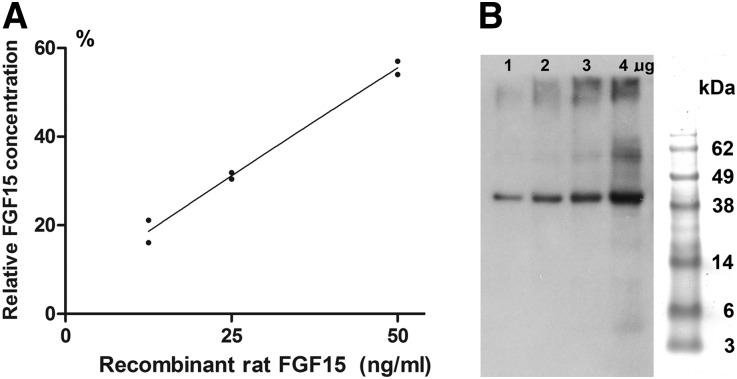
To confirm the recombinant rat FGF15 protein (41.67 kDa, synthesized by GenScript USA Inc.), Western blot analysis was performed using FGF15 goat polyclonal IgG (Santa Cruz Biotechnology, Santa Cruz, CA), an antibody used to identify both mouse and rat FGF15 protein. The recombinant rat FGF15 protein (41.67 kDa, synthesized by GenScript USA Inc.) was loaded 1, 2, 3, and 4 μg per well respectively in Tris-acetate Mini Gels (Invitrogen, Carlsbad, CA). After denaturing the protein samples, the gel was run for 40 min. Then the gel was transferred to a polyvinylidene difluoride membrane using iBlot dry blotting system (Invitrogen). After washing three times with TBS (Thermo Scientific, Rockford, IL), the membrane was incubated in blocking buffer (Thermo Scientific) for 1 h at room temperature with shaking. The blot was incubated with 1:100 FGF15 goat polyclonal IgG (Santa Cruz Biotechnology) at 4°C overnight. Then the blot was washed with TBS and 0.05% Tween 2 (TBST) and incubated with diluted (1:5,000) HRP-conjugate (Santa Cruz Biotechnology) at room temperature for 45 min with shaking. After washing with TBS, substrate working solution (Thermo Scientific) was added. The blot was exposed to X-ray film (Kodak, Rochester, NY).
Assays for mRNA expression
The mRNA expression was determined quantitatively by standard real-time PCR method. Total RNA was isolated using TRIzol reagent (Sigma, St. Louis, MO) according to the manufacturer's instructions. For quantitative RT-PCR analysis, 1 μg total RNA was transcribed to cDNA using the high cDNA capacity reverse transcription kit (Applied Biosystems, Foster City, CA). The quantitative RT-PCR was performed using TaqMan® Expression Assays (containing primer and probe) designed by Applied Biosystems for rat and rabbit CYP7A1, SHP, FGF15, and FGF19, respectively. Custom TaqMan gene expression assays for rabbit FGF19 were: forward primer, TCAGCTCCGGCTACAACGT; reverse primer, TGCTTGGCACTGCTCAGAGA; and middle FAM (a dye-labeled probe), TACCGCTCCACGACGC. All reactions were performed in triplicate in a 7500 Real Time PCR System (Applied Biosystems) with thermal cycling conditions as follows: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative mRNA levels were calculated by the comparative threshold cycle method using GAPDH as the internal standard. Each gene expression was expressed as a ratio between the controls (regular chow-fed) and treated animals.
Measurements of portal blood bile acid concentrations
The bile acid concentrations were measured by capillary gas liquid chromatography. Ten micrograms of glycoursocholic acid (internal standard) was added to 1 ml plasma from portal blood sample that was then deproteinized using C18 Sep-Pak (Waters, Milford, MA). The bile acids were deconjugated with 0.5 ml 4N NaOH at 115°C for 4 h, acidified with 0.5 ml 50% HCl, and extracted with water-saturated ethyl acetate. After the sample was methylated overnight with 300 μl methanolic hydrochloric acid, trimethylsilyl ether derivatives were prepared by adding 100 μl Sylon HTP (Supelco, Bellefonte, PA) and incubated at 55°C for 30 min. The bile acid methyl esters were applied to a capillary gas chromatograph (Hewlett-Packard, Palo Alto, CA) equipped with a 25 m fused silica CP-Sil 5-CB capillary column. The bile acids were quantitated by comparison to the known quantity (10 μg) of the internal standard.
Statistical analysis
Data in the text and figures are shown as mean ± SD. Statistical difference among multi-study groups was tested using ANOVA followed by the Bonferroni multiple comparisons test except for Fig. 1 and Fig. 2 where the data were analyzed for correlation coefficient (r) and linear relationship. The Student's t-test was used when only two groups were compared. GraphPad InStat, Instant Biostatistics Version 3.0 (GraphPad Software) was used for all statistical evaluations.
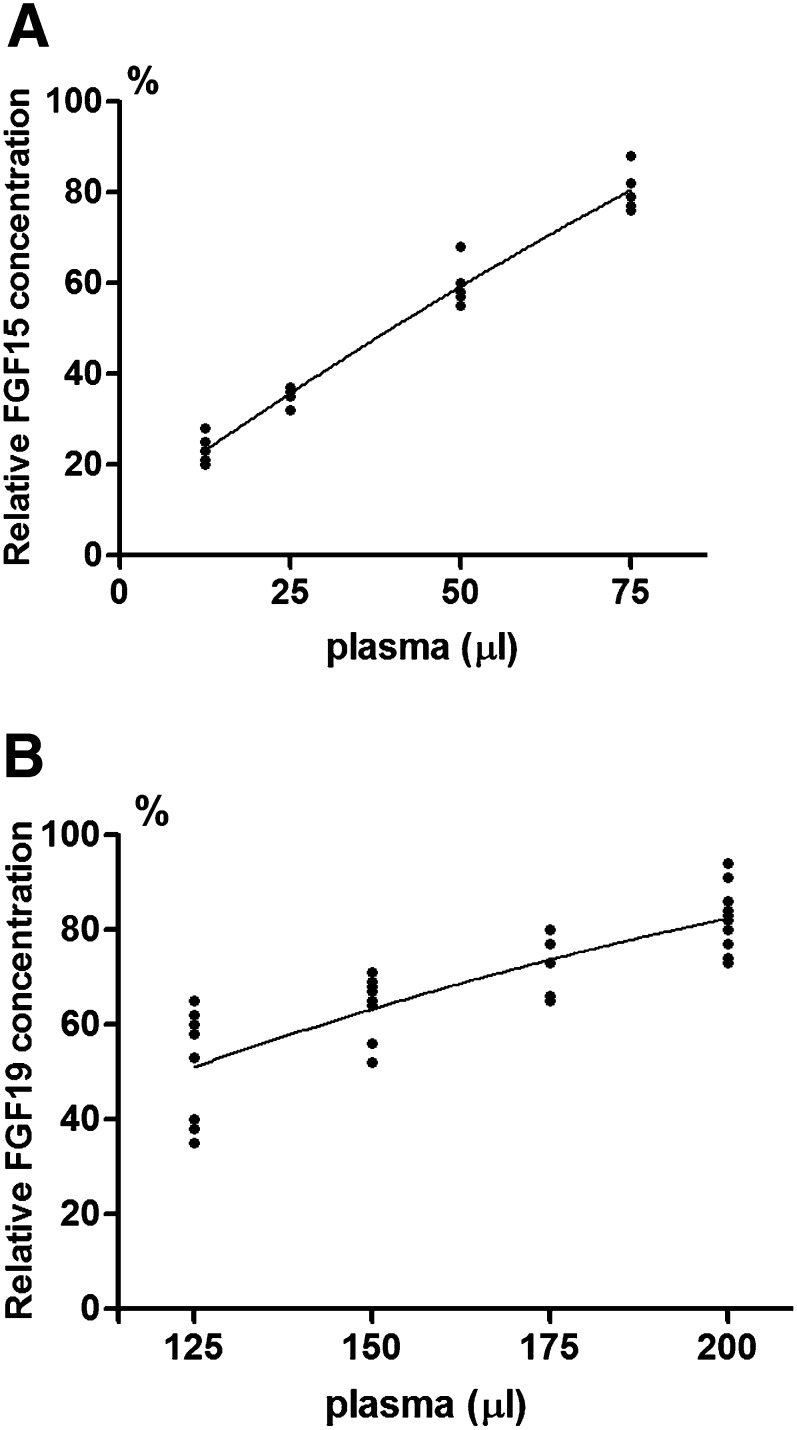
Fig. 1.
Dilution test for rat FGF15 ELISA kit (A) and rabbit FGF19 indirect ELISA (B). A: FGF15 relative concentrations versus diluted amounts of rat portal plasma were analyzed for regression and correlation. Data were obtained from five sets of tested rat portal plasma samples. Each set consisted of four plasma samples (75, 50, 25, and 12.5 μl). The relative FGF15 concentrations (%) are plotted as the Y axis and the corresponding amounts of applied plasma as the X axis. The correlation coefficient (r) = 0.9867 for the portal concentrations of FGF15 protein versus the diluted amounts of portal plasma. B: FGF19 relative concentrations versus diluted amounts of rabbit portal plasma are analyzed for regression and correlation. Data were obtained from 10 sets of tested rabbit portal plasma samples. Each set consisted of four plasma samples (200, 175, 150, and 125 μl). The relative FGF19 concentrations (%) are plotted as the Y axis and the corresponding amounts of applied plasma as the X axis. The correlation coefficient (r) = 0.8275 for the portal concentrations of FGF19 protein versus the diluted amounts of portal plasma.
Fig. 2.
Dilution test for rat FGF15 ELISA kit using recombinant rat FGF15 protein solution (A) and confirmation of the recombinant rat FGF15 protein by Western blot analysis (B). A: FGF15 relative concentrations versus diluted amounts of the recombinant rat FGF15 protein were analyzed for regression and correlation. Data were obtained from two sets of diluted recombinant FGF15 protein solutions. Each set consisted of four recombinant rat FGF15 solutions (100, 50, 25, and 12.5 ng/ml). The relative FGF15 concentrations (%) (concentrations relative to the concentration in 100 ng/ml solution) are plotted as the Y axis and the corresponding amounts of applied recombinant protein as the X axis. The correlation coefficient (r) was 0.9871 for the concentrations of FGF15 protein versus the diluted amounts of the recombinant rat FGF15 protein. B: Western blot analysis. Recombinant rat FGF15 protein (1, 2, 3, and 4 μg) were applied to the blot. Antibody FGF15 goat polyclonal IgG (Santa Cruz Biotechnology Inc.) was used to identify FGF15 protein at 41.67 kDa. On the right is SeeBlue Plus 2 prestained protein standard (Invitrogen).
RESULTS
A dilution test was performed to evaluate whether the ELISA kit for rat FGF15 could distinguish changes of FGF15 concentrations corresponding to the applied amounts of rat plasma samples in a linear correlation. The results from the dilution test in rat portal plasma are summarized in Fig. 1A, which demonstrates that the concentrations of FGF15 protein show a linear relationship with corresponding diluted amounts of portal plasma. The correlation coefficient (r) was 0.9867 indicating the linear correlation is highly significant (P < 0.001). The dilution test was also performed on the recombinant rat FGF15 protein (synthesized by GenScript USA Inc.). The measured relative FGF15 concentrations determined by the rat FGF15 ELISA kit also showed linear correlation (r = 0.9871, P < 0.001) with the given recombinant rat FGF15 protein (Fig. 2A). Because the recombinant rat FGF15 protein (41.67 kDa) was confirmed by Western blot analysis (Fig. 2B) using FGF15 antibody (Santa Cruz Biotechnology Inc.), this dilution test demonstrated that the FGF15 ELISA kit used in the present study could measure changes of rat FGF15 protein levels quantitatively.
The results from the dilution test in the rabbit portal plasma are summarized in Fig. 1B, which demonstrates that the portal concentrations of FGF19 protein show a linear correlation with the corresponding diluted amounts of portal plasma (r = 0.8275, P < 0.001).
The immune-depletion test in rat portal plasma showed that in samples (n = 10) where the FGF15 antibody was added, FGF15 protein levels (142 ± 35 pg/ml) were reduced 48 ± 16% (P < 0.001) as compared with those samples (n = 10) without added FGF15 antibody (280 ± 48 pg/ml). After addition of anti-human FGF19 antibody, FGF19 protein levels (211 ± 66 pg/ml) in the rabbit portal plasma samples (n = 10) detected by the indirect ELISA method were reduced 32 ± 19% (P < 0.01) as compared with those samples (n = 10) without the added FGF19 antibody (326 ± 103 pg/ml).
Hepatic CYP7A1 mRNA levels (n = 8) increased 87% (2.08 ± 0.56 relative units, P < 0.001) in the Ch-fed rats but decreased 62% (0.42 ± 0.16 relative units, P < 0.01) in rats fed CA as compared with the control group (1.11 ± 0.27 relative units) (Fig. 3). In rabbits, hepatic CYP7A1 mRNA levels decreased 49% (0.52 ± 0.30 relative units, P < 0.05) when fed Ch and declined 75% (0.26 ± 0.21 relative units, P < 0.001) in those fed CA as compared with the control group (1.02 ± 0.42 relative units) (Fig. 4). Similar to the expression of CYP7A1, CYP8B1 mRNA levels (n = 8) were suppressed 79% in the rats fed CA (0.34 ± 0.20 relative units vs. 1.64 ± 0.83 relative units, P < 0.01) and induced 78% in rats fed Ch (2.92 ± 0.98 relative units, P < 0.01). In rabbits, CYP8B1 expression was suppressed after feeding either CA (−0.44%, 2.09 ± 0.75 relative units, P < 0.01) or Ch (−46%, 1.13 ± 0.58 relative units, P < 0.01) as compared with the controls (1.16 ± 0.20 relative units).
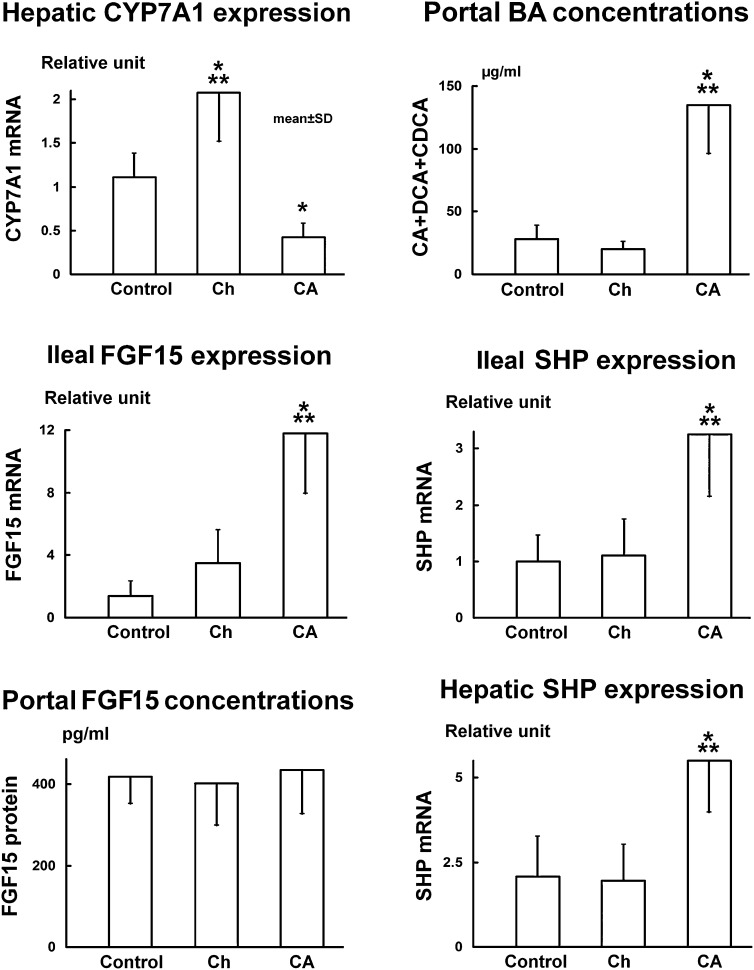
Fig. 3.
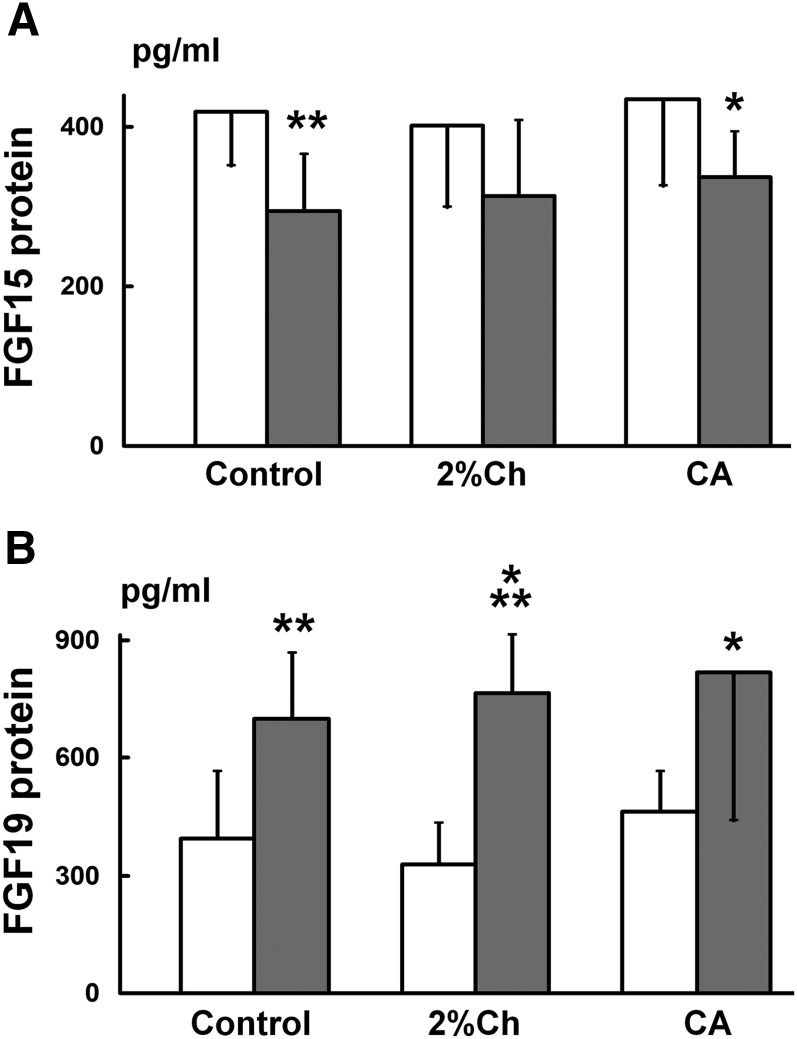
Results from the rat experiment show that FGF15 protein levels in the portal blood do not reflect the changes of FGF15 expression in the ileum or control the expression of CYP7A1 in the liver. Rats were fed with 1% CA or 2% Ch for 7 days. Data (n = 8) are represented as mean ± SD. *P < 0.05 and ***P < 0.001 when compared with the control group. Note that portal bile acid flux is represented by only the portion of CA plus DCA and CDCA in the bile, as these bile acids are potent FXR ligands which activate FXR in the ileum as well as in the liver. BA, bile acid.
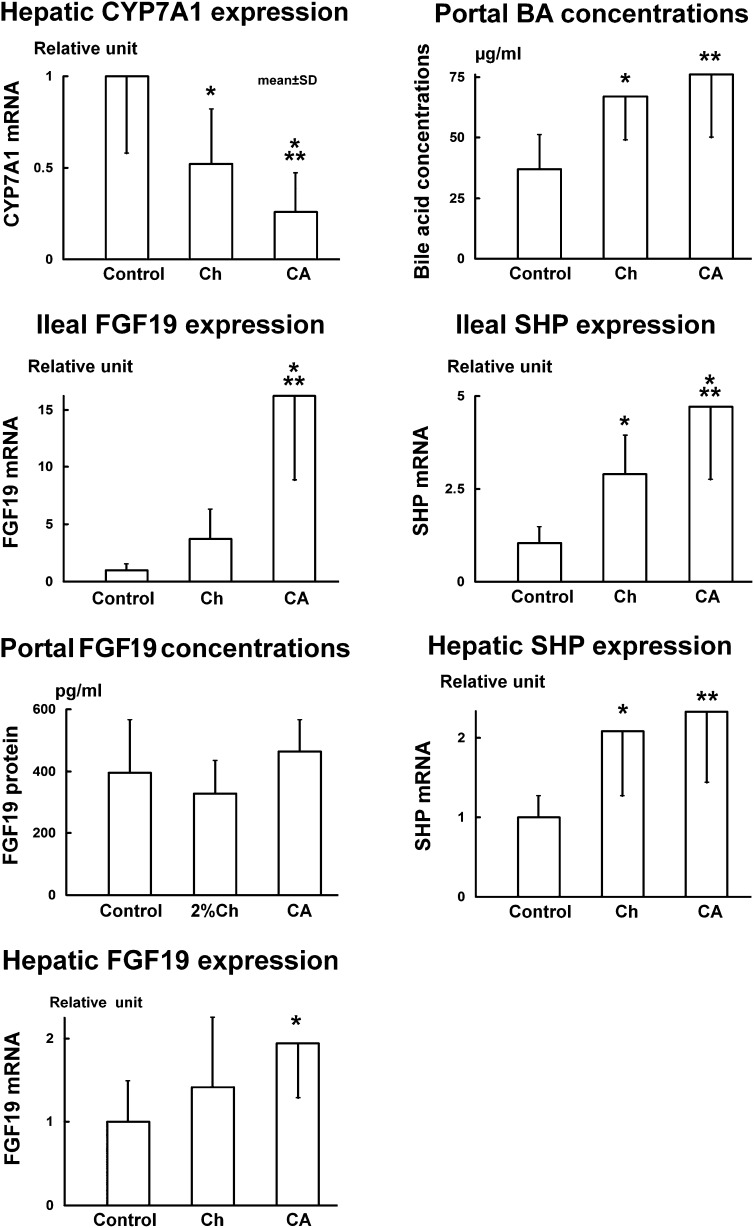
Fig. 4.
Results from the rabbit experiment show that FGF19 protein levels in the portal blood do not reflect the changes of FGF19 expression in the ileum or control the expression of CYP7A1 in the liver. Rabbits were fed with 0.3% CA or 2% Ch for 7 days. Data (n = 8) are represented as mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 when compared with the control group. BA, bile acid.
FGF15 mRNA levels in the ileum increased 8.5-fold (11.8 ± 3.8 relative units, P < 0.001) in CA-fed rats but not in Ch-fed rats (3.5 ± 2.1 relative units) as compared with the controls (1.4 ± 0.9 relative units). FGF19 mRNA levels increased 16-fold in the CA-fed rabbits (16.2 ± 7.3 relative units, P < 0.001) but not in the rabbits fed Ch (3.7 ± 2.6 relative units) as compared with the control group (1.0 ± 0.5 relative units).
In rabbits, FGF19 expression in the liver was measured by a routine real-time PCR method. Hepatic FGF19 mRNA levels increased 94% (1.94 ± 0.65 relative units, P < 0.05) in the CA-fed rabbits but not in the Ch group (1.42 ± 0.83 relative units) as compared with control rabbits (1.00 ± 0.49 relative units). However, the expression of FGF19 in the liver was much lower than in the ileum. The ratio of hepatic mRNA versus ileal mRNA was approximately 1:12 (0.10 ± 0.06 relative units vs. 1.00 ± 0.97 relative units).
Rat FGF15 protein levels in the portal blood did not change in either the CA-fed (436 ± 109 pg/ml) or the Ch-fed rats (402 ± 103 pg/ml) as compared with the control rats (419 ± 67 pg/ml). FGF15 protein levels in the peripheral blood also did not change in the CA-fed (338 ± 57 pg/ml) and Ch-fed rats (314 ± 95 pg/ml) as compared with control rats (295 ± 71 pg/ml). The FGF15 protein levels in the peripheral blood were 30% lower (P < 0.01 using t-test) than those in the portal blood in the control group, 22% lower in the Ch group and 24% lower (P < 0.05) in the CA group (Fig. 5A).
Fig. 5.
Comparison of FGF15/19 protein concentrations in the portal and peripheral blood. A: FGF15 protein levels in the rat study. Rats were fed with regular chow (Control), 1% CA, or 2% Ch for 7 days. Data (n = 8) are represented as mean ± SD. *P < 0.05 and **P < 0.01 when compared with that in the portal blood using the t-test. The open bar represents FGF15 levels in the portal blood while the gray bar represents FGF15 levels in the peripheral blood. B: FGF19 protein levels in the rabbit study. Rabbits were fed with regular chow (Control), 0.3% CA, or 2% Ch for 7 days. Data (n = 8) are represented as mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 when compared with that in the portal blood using the t-test. The open bar represents FGF19 levels in the portal blood while the gray bar represents FGF19 levels in the peripheral blood.
In the rabbit studies, portal blood FGF19 protein levels in either the CA (464 ± 102 pg/ml) or the Ch groups (329 ± 104 pg/ml) were not changed as compared with the control group (394 ± 172 pg/ml). The paired data (portal FGF19 protein levels vs. ileal FGF19 mRNA, n = 8) were statistically analyzed using GraphPad software to determine whether there was a positive correlation between the portal blood FGF19 protein levels and the ileal FGF19 expression in the rabbits fed CA. The correlation coefficient (r) was 0.2848 (P = 0.4941) indicating that FGF19 protein concentrations in the portal blood were not correlated with the FGF19 mRNA levels in the ileum in the CA-fed rabbits. In addition, FGF19 protein levels in rabbit peripheral blood also did not change significantly in the CA (817 ± 379 pg/ml) and the Ch (764 ± 148 pg/ml) groups as compared with the control rabbits (699 ± 167 pg/ml). The paired data (peripheral FGF19 protein levels vs. the corresponding ileal FGF19 mRNA, n = 8) were analyzed statistically using GraphPad software to determine whether there was a positive correlation between these two parameters in the rabbits fed CA. The correlation coefficient (r) was 0.5567 (P = 0.1518) indicating that peripheral FGF19 protein concentrations were not significantly correlated with FGF19 mRNA levels in the ileum in the CA-fed rabbits. In contrast to FGF15 in rats, FGF19 protein levels in rabbit peripheral blood were significantly higher than those in the portal blood (using Student's t-test): +77% in the control, P < 0.01; +76% in the CA group, P < 0.05; and +2.3-fold higher in the Ch group, P < 0.001 (Fig. 5B).
SHP mRNA levels in the rat ileum increased 3.3-fold (3.25 ± 1.11 relative units, P < 0.001) in the CA-fed rats but not in the Ch-fed rats (1.10 ± 0.64 relative units) as compared with the control group (1.00 ± 0.46 relative units). In rabbits, SHP mRNA levels in the ileum increased 4.5-fold (4.71 ± 1.96 relative units, P < 0.001) in the CA and 2.8-fold (2.90 ± 1.03 relative units, P < 0.05) in the Ch group as compared with the control rabbits (1.05 ± 0.43 relative units).
SHP mRNA levels in the liver increased 2.6-fold (5.50 ± 1.52 relative units, P < 0.001) in the CA-fed rats but remained unchanged (1.96 ± 1.06 relative units) in the Ch-fed rats as compared with the control rats (2.08 ± 1.19 relative units). In rabbits, hepatic SHP mRNA levels increased 2.3-fold (2.33 ± 0.89 relative units, P < 0.01) and 2.1-fold (2.09 ± 0.83 relative units, P < 0.05) in the CA and Ch groups respectively as compared with the control rabbits (1.00 ± 0.27 relative units).
Portal blood bile acid concentrations representing the total portal bile acid flux returning to the liver increased 3.6-fold (147 ± 39 μg/ml, P < 0.001) in the CA-fed rats but not in the Ch-fed rats (41 ± 12 μg/ml) as compared with control rats (41 ± 13 μg/ml). In rat bile, only CA, DCA, and CDCA are potent ligands of FXR which activate FXR in the ileum and liver. Because these bile acids in the portal flux regulate CYP7A1 activity (9, 21) in Fig. 3, this fraction of CA + DCA + CDCA in the bile acid flux was illustrated to represent the regulating portal bile acid flux. This portion of the bile acid flux in the CA-fed rats (135 ± 39 μg/ml) increased 4.8-fold (P < 0.001) as compared with the control group (28 ± 11 μg/ml). However, this portion of bile acid flux did not increase in the Ch-fed rats (20 ± 6 μg/ml). In rabbits, portal bile acid concentrations (95% DCA) increased both in those fed CA (+2-fold, 147 ± 39 μg/ml, P < 0.01) and Ch (+1.8-fold, 67 ± 18 μg/ml, P < 0.05) as compared with the control rabbits (37 ± 14 μg/ml).
DISCUSSION
Although for 9 years the intestinal FXR-FGF15/19-FGFR4 signaling pathway has been proposed to regulate hepatic CYP7A1, no data have been presented that measure signal protein concentrations in the portal blood of rats, mice, and rabbits. We recognized that it was crucial to determine signal (FGF15/19 protein) levels in the portal blood to provide evidence to ascertain the role of ileal FGF15 or FGF19 in regulation of CYP7A1 expression. If, as proposed, FGF15/19 acts as the gut-hepatic signal which is released from the ileum and brings messages to the liver that determine CYP7A1 expression, the signal FGF15/19 protein levels in the portal blood should reflect the changes of FGF15/19 mRNA levels in the ileum that correlate inversely with the expression of CYP7A1 in the liver. In the present study, we failed to find direct evidence that indicates that this proposed gut-hepatic signal carries the message from the ileum to the liver through the portal system. In rats and rabbits, neither FGF15 nor FGF19 protein levels within the portal blood reflected the changes of ileal FGF15/19 expression nor inversely correlated with CYP7A1 expression at either site in the portal system. As seen in the rat and rabbit CA-fed groups, the expression of FGF15/19 in the ileum was enhanced 8.5-fold and 16-fold, respectively, while the expression of CYP7A1 in the liver was suppressed 62 and 75%, respectively. However, we did not find corresponding changes in FGF15/19 protein levels in the portal blood (see Figs. 3, 4). Therefore, it appears that the signal portal plasma FGF15/19 levels are not related to the expression of FGF15/19 in the ileum or the inhibition of CYP7A1 in the liver, at least in rats and rabbits. It has been reported that ileal FGF15 mRNA expression undergoes a circadian rhythm (22) and that the peak time for postprandial enhancement of ileal FGF15 expression is 60 min in mice (23). Furthermore, studies in humans showed diurnal rhythm and postprandial increase of FGF19 protein levels in peripheral blood (24, 25). Because we only measured the FGF15/19 protein levels in the portal blood at a single time point, the significant response of the portal or peripheral FGF15/19 protein levels to the enhanced FGF15/19 expression in the ileum might be missed. However, in our study, the CA-fed rats and rabbits were sacrificed at the time point when ileal expression of FGF15/19 was enhanced and associated with significantly downregulated CYP7A1 expression. In this case, comparisons with the control group at the same time point should be informative and able to discern possible correlations between ileal FGF15/19 expression and changes in the portal FGF15/19 protein levels.
Although it is well known that CYP7A1 expression can be regulated by multiple pathways and factors (26), in the present study, we focused on whether the bile acid flux returning to the liver could also downregulate CYP7A1 expression. We expected that reabsorbing the increased bile acid flux would enhance ileal FGF15/19 expression as the bile acid pool is reabsorbed and passes through the ileal enterocytes where it activates ileal FXR and in turn stimulates the expression of its target gene FGF15/19. Because increase of the absorbed circulating bile acid pool (flux) and the induction of FGF15/19 expression overlap in the ileum, it was impossible to separate and identify whether the suppression of CYP7A1 in the CA-fed rats and rabbits or the Ch-fed rabbits is due solely to enhanced expression of ileal FGF15/19 or the increased portal bile acid flux returning to the liver, or possibly both. However, there were several examples where the changes in CYP7A1 expression were obviously separate and not related to FGF15/19 expression in the ileum. In Ch-fed rats, where ileal FGF15 mRNA levels did not decrease, the significant enhancement of hepatic CYP7A1 mRNA levels was not dependent on the FGF15 expression in the ileum. In this case, the returning bile acid flux would better explain the upregulation of CYP7A1. Because the portal bile acid flux was not increased in the Ch-fed rats, hepatic FXR was not activated and thus the activation of LXRα induced by dietary Ch stimulated the expression of CYP7A1 (9). In Ch-fed rabbits, where FGF19 mRNA levels in the ileum were not increased significantly, hepatic CYP7A1 mRNA levels were reduced. The above cases suggest that suppression of CYP7A1 expression in the liver is not solely determined by the ileal FGF19 expression, but rather can be regulated by changes in the portal bile acid flux returning to the liver. These data may also show species differences in regulating bile acid synthesis.
We also found that the expression of FGF19 in the liver of rabbits could be measured easily by standard real-time PCR methodology. It has not yet been ascertained whether hepatic FGF19 expression in rabbits plays a role to determine CYP7A1 expression, although in the CA-fed rabbits hepatic FGF19 mRNA levels were significantly increased and CYP7A1 mRNA levels were inhibited. However, we also noted that hepatic FGF19 mRNA levels were not enhanced in Ch-fed rabbits, whereas CYP7A1 expression was inhibited. As FGF19 is also expressed in the liver in humans (17, 18), the significance of the FGF19 protein produced from the liver to downregulate CYP7A1 remains to be clarified, as well as whether the expression of CYP7A1 could be estimated by measurements of FGF19 protein levels in the peripheral blood.
Furthermore, we also determined and compared FGF15/19 levels in the peripheral blood with the portal blood. This comparison provided more convincing evidence to show whether FGF15/19 protein levels in peripheral blood reflected the changes of FGF15/19 expression in the ileum and CYP7A1 mRNA in the liver, as some clinical investigators have suggested in humans (27). In other words, we tried to ascertain whether changes of FGF15/19 expression in the ileum could be detected by measuring the changes of FGF15/19 protein levels in the peripheral blood. The observations in this study suggested that similar to FGF15/19 levels in the portal blood, FGF15/19 levels in peripheral blood do not correspond significantly to the changes of FGF15/19 mRNA levels in the ileum, although in CA-fed rabbits FGF19 levels in the peripheral blood tended to rise. Because our study only observed the changes of FGF19 at one single time point, it remains to be seen whether FGF19 protein levels in the peripheral blood mirror the changes of FGF19 expression in the ileum at other time points. In contrast to rats, where FGF15 protein levels in the peripheral blood were lower than those in the portal blood, rabbit FGF19 protein levels in the peripheral blood were significantly higher than in the portal blood. This increment of FGF19 in the peripheral blood could be contributed by FGF19 secreted from the liver. Thus, this finding also suggests that the expression of FGF19 in the liver is considerable and may be important in rabbits.
Although we have not definitely clarified yet whether FGF15/19 in the ileum or the portal bile acid flux is responsible for the downregulation of CYP7A1 expression, we noted that unlike FGF15/19 expression in the ileum, changes in the portal bile acid flux were consistent with alterations of CYP7A1 mRNA levels in the Ch-fed rats and rabbits. Furthermore, we also noted that SHP mRNA levels in the liver changed in line with the portal bile acid flux and inversely with the hepatic CYP7A1 expression. These findings were exemplified when the portal bile acid flux increased as seen in CA-fed rats and rabbits and Ch-fed rabbits, hepatic SHP mRNA levels rose correspondingly whereas portal blood FGF15/19 protein levels did not change or mirror the expression of FGF15/19 in the ileum. Thus, we believe that the returning portal bile acid flux also plays an important role in the regulation of CYP7A1 expression through the activation of the hepatic FXR-SHP pathway. Recently, Kir et al. (28) proposed that, in the liver, SHP repressed CYP7A1 transcription by interacting with HNF4/LRH-1 at their overlapping binding sites in the CYP7A1 promoter region. This report also supported the significance of hepatic bile acid-FXR-SHP cascade in the regulation of CYP7A1.
Acknowledgments
The authors would like to thank Bibi Pcolinsky for her excellent technical assistance in analysis of portal blood bile acid concentrations.
Footnotes
Abbreviations:
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- Ch
- cholesterol
- DCA
- deoxycholic acid
- FGF
- fibroblast growth factor
- FGFR4
- fibroblast growth factor receptor 4
- FXR
- farnesoid X receptor
- SHP
- short heterodimer partner
This study was supported by a Merit Review Grant from the Department of Veterans Affairs, Health Services Research, USA. All the authors have no potential conflicts of interest.
REFERENCES
- 1.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365 [DOI] [PubMed] [Google Scholar]
- 2.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368 [DOI] [PubMed] [Google Scholar]
- 3.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553 [DOI] [PubMed] [Google Scholar]
- 4.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526 [DOI] [PubMed] [Google Scholar]
- 5.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515 [DOI] [PubMed] [Google Scholar]
- 6.Shang Q., Pan L., Saumoy M., Chiang J. Y. L., Tint G. S., Salen G., Xu G. 2007. An overlapping binding site in the CYP7A1 promoter allows activation of FXR to override the stimulation by LXRα. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G817–G823 [DOI] [PubMed] [Google Scholar]
- 7.Xu G., Pan L., Li H., Forman B. M., Erickson S. K., Shefer S., Bolloneni J., Batta A. K., Christie J., Wang T., et al. 2002. Regulation of the farnesoid X receptor (FXR) by bile acid flux in rabbits. J. Biol. Chem. 277: 50491–50496 [DOI] [PubMed] [Google Scholar]
- 8.Xu G., Li H., Pan L., Shang Q., Honda A., Ananthanarayanan M., Erickson S. K., Shneider B. L., Shefer S., Bollineni J., et al. 2003. Farnesoid X receptor (FXR)-mediated down-regulation of CYP7A1 dominates LXRα in long term cholesterol fed NZW rabbits. J. Lipid Res. 44: 1956–1962 [DOI] [PubMed] [Google Scholar]
- 9.Xu G., Pan L., Li H., Shang Q., Honda A., Shefer S., Bollineni J., Matsuzaki Y., Tint G. S., Salen G. 2004. Dietary cholesterol stimulates CYP7A1 in rats because farnesoid X receptor (FXR) is not activated. Am. J. Physiol. Gastrointest. Liver Physiol. 286: G730–G735 [DOI] [PubMed] [Google Scholar]
- 10.Pandak W. M., Heuman D. M., Hylemon P. B., Chiang J. Y. L., Vlahcevic Z. R. 1995. Failure of intravenous infusion of taurocholate to down-regulate cholesterol 7 alpha-hydroxylase in rats with biliary fistula. Gastroenterology. 108: 533–544 [DOI] [PubMed] [Google Scholar]
- 11.Dueland S., Reichen J., Everson G. T., Davis R. A. 1991. Regulation of cholesterol and bile acid homoeostasis in bile-obstructed rats. Biochem. J. 280: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225 [DOI] [PubMed] [Google Scholar]
- 13.Davis R. A., Musso C. A., Malone-Mcneal M., Lattier G. R., Hyde P. M. 1988. Examination of bile acid negative feedback regulation in rats. J. Lipid Res. 29: 202–211 [PubMed] [Google Scholar]
- 14.Stange E. F., Scheibner J., Lutz C., Ditschuneit H. 1988. Feedback regulation of bile acid synthesis in the rat by dietary vs. intravenous cholate or taurocholate. Hepatology. 8: 879–886 [DOI] [PubMed] [Google Scholar]
- 15.Nagano M., Kuroki S., Mizuta A., Furukawa M., Noshiro M., Chijiiwa K., Tanaka M. 2004. Regulation of bile acid synthesis under reconstructed enterohepatic circulation in rats. Steroids. 69: 701–709 [DOI] [PubMed] [Google Scholar]
- 16.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaap F. G., van der Gaag N. A., Gouma D. J., Jansen P. L. M. 2009. High expression of the bile salt homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 49: 1228–1235 [DOI] [PubMed] [Google Scholar]
- 18.Song K. H., Li T., Owsley E., Strom S., Chiang J. Y. 2009. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 49: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim I., Ahn S., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672 [DOI] [PubMed] [Google Scholar]
- 20.Kong B., Wang L., Chiang J. Y., Zhang Y., Klaassen C. D., Guo G. L. 2012. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 56: 1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reihnér E., Björkhem I., Angelin B., Ewerth S., Einarsson K. 1989. Bile acid synthesis in humans: regulation of hepatic microsomal cholesterol 7 alpha-hydroxylase activity. Gastroenterology. 97: 1498–1505 [DOI] [PubMed] [Google Scholar]
- 22.Stroeve J. H. M., Brufau G., Stellaard F., Gonzalez F. J., Staels B., Kuipers F. 2010. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab. Invest. 90: 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potthoff M. J., Boney-Montoya J., Choi M., He T., Sunny N. E., Satapati S., Suino-Powell K., Xu H. E., Gerard R. D., Finck B. N., et al. 2011. FGF15/19 Regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab. 13: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angelin B., Larsson T. E., Rudling M. 2012. Circulating fibroblast growth factors as metabolic regulators–a critical appraisal. Cell Metab. 16: 693–705 [DOI] [PubMed] [Google Scholar]
- 25.Lundåsen T., Gälman C., Angelin B., Rudling M. 2006. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J. Intern. Med. 260: 530–536 [DOI] [PubMed] [Google Scholar]
- 26.Chiang J. Y. L. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walters J. R. F., Tasleem A. M., Omer O. S., Brydon W. G., Dew T., Roux C. W. L. 2009. A new mechanism for bile acid diarrhea: defective feedback inhibition of bile acid biosynthesis. Clin. Gastroenterol. Hepatol. 7: 1189–1194 [DOI] [PubMed] [Google Scholar]
- 28.Kir S., Zhang Y., Gerard R. D., Kliewer S. A., Magelsdorf D. J. 2012. Nuclear receptors HNF4α and LRH-1 cooperate in regulating Cyp7a1 in vivo. J. Biol. Chem. 287: 41334–41341 [DOI] [PMC free article] [PubMed] [Google Scholar]