Abstract
The interaction of dietary fats and carbohydrates on liver mitochondria were examined in male FBNF1 rats fed 20 different low-fat isocaloric diets. Animal growth rates and mitochondrial respiratory parameters were essentially unaffected, but mass spectrometry-based mitochondrial lipidomics profiling revealed increased levels of cardiolipins (CLs), a family of phospholipids essential for mitochondrial structure and function, in rats fed saturated or trans fat-based diets with a high glycemic index. These mitochondria showed elevated monolysocardiolipins (a CL precursor/product of CL degradation), elevated ratio of trans-phosphocholine (PC) (18:1/18:1) to cis-PC (18:1/18:1) (a marker of thiyl radical stress), and decreased ubiquinone Q9; the latter two of which imply a low-grade mitochondrial redox abnormality. Extended analysis demonstrated: i) dietary fats and, to a lesser extent, carbohydrates induce changes in the relative abundance of specific CL species; ii) fatty acid (FA) incorporation into mature CLs undergoes both positive (>400-fold) and negative (2.5-fold) regulation; and iii) dietary lipid abundance and incorporation of FAs into both the CL pool and specific mature tetra-acyl CLs are inversely related, suggesting previously unobserved compensatory regulation. This study reveals previously unobserved complexity/regulation of the central lipid in mitochondrial metabolism.
Keywords: mitochondria, ubiquinones, saturated fatty acids, trans fatty acids, monounsaturated fatty acids, polyunsaturated fatty acids, glycemic index, lipidomics
Cardiolipins (CLs) are a subclass of phospholipids unique to mitochondria. Each CL is a dimeric phospholipid consisting of two phosphatidyl head groups, connected on a glycerol backbone, and four fatty acyl chains (1–3). Unlike most membrane phospholipids, CLs are predominantly found in the mitochondrial inner membrane and at contact sites between the inner and outer mitochondrial membrane. CLs comprise ∼25% of total phospholipids in the mitochondrial inner membrane (1–3). Mammalian CL has been reported to contain primarily (∼85%) 18:2 (linoleic) acyl side chains (2, 4, 5) that mediate the high affinity of CL to inner mitochondrial membrane proteins (3, 6, 7).
CLs play multiple key roles in the regulation of mitochondrial metabolism (2, 3, 8–10). These functions include maintenance of proper mitochondrial quaternary structure, regulation of essential enzymatic activities involved in electron transport and oxidative phosphorylation, and assembly of respiratory supercomplexes. In particular, CL is required for the proper functioning of mitochondrial respiratory complexes I, III, IV (cytochrome oxidase), ATP synthase (6, 11–19), cardiolipin synthase (20), and several transporters in the inner mitochondrial membrane [e.g., the ADP/ATP translocator (10, 18, 21), phosphate transporter (22), pyruvate transporter (23), and carnitine/acylcarnitine carrier (24)]. In the intermembrane space, CL serves as a binding receptor for creatine kinase (24) and electrostatically anchors cytochrome c to the inner mitochondrial membrane (26). CL has been proposed to participate directly in proton conductance via cytochromes (14) and to prevent osmotic instability and uncoupling at high respiration rates (27). CL also regulates mitochondrial biogenesis (3, 10).
Changes both in absolute CL levels and, to a much lesser extent, in FA composition of the CL pool have been observed pathophysiologically. Specifically, decreases in overall CL content and/or CL oxidative damage have been implicated or observed in a range of overt pathological conditions associated with mitochondrial dysfunction, including Barth syndrome (28, 29), experimental brain and heart ischemic-reperfusion injury (3, 8, 30), experimental traumatic brain injury (31), heart failure (32), and experimental diabetes (33, 34). During aging in rats, the total CL concentration is reduced in the heart, skin, and liver, and linoleate (18:2) decreases while arachidonate (20:4) increases (35, 36). Analysis of CL in hearts explanted from patients with dilated cardiomyopathy revealed a loss of tetralinoleoyl CL species (32). Surprisingly, all physiologically modified scenarios appeared associated with decreased CL levels (3, 8, 32, 33, 37, 38). There seemed to be no active regulation of CLs to restore homeostasis or to counteract a biased diet.
Different dietary interventions affect CL acyl chain composition (39–42). Functionally, thirty days on a diet deficient in linoleic acid (18:2), an essential FA, significantly reduces tetralinoleoyl CL and affects mitochondrial oxygen consumption in the rat heart (43–45). Conversely, dietary supplementation with linoleic acid restores tetralinoleoyl CL in cultured fibroblasts from Barth syndrome patients and elevates CL level (46). Notably, the preferential accumulation of 18:2 side chains and the tetralinoleoyl-CL species was, until recently, one of the only recognized levels of CL regulation. These data speak to the need for an essential FA in the diet to provide the raw building blocks of the CL molecule, a finding consistent with stochastic incorporation of FAs in the CL pool and the ability of diet to modulate the pool. These findings do not, however, address CL dietary-mediated regulation and/or response to a nutritionally replete diet. The present report addresses these latter issues.
The data discussed above suggest that an in depth study of the CL pool composition is central to probing the roles of CL in mitochondrial biology. Detailed analysis of the CL profile is technically challenging, because it requires either multiple extraction and purification steps or the simultaneous analysis of the intact CL molecule and each of its side chains both qualitatively and quantitatively within the context of the overall mitochondrial lipid pool. We used a newly developed lipidomics platform to examine diet-mediated changes in CL composition in depth. We also specifically focused on a healthy rat model, Fisher 344 × Brown Norway F1 (FBNF1), maintained on low-fat isocaloric diets differing systematically in primary fat and glycemic index (GI). Our results suggest that CL regulation is unexpectedly active and compensatory, and provide a biological model to probe these levels of CL regulation further.
RESEARCH DESIGN AND METHODS
Chemicals
LC-MS grade acetonitrile, methanol, and isopropanol, as well as HPLC grade and dimethyl sulfoxide (DMSO), were purchased from Fisher Scientific (Pittsburg, PA). Ammonium formate was purchased from Sigma-Aldrich (St. Louis, MO). A detailed list all lipid standards purchased for LC-MS as well as their abbreviations and sources are in the supplemental information of our previously published work (47–49).
Rat husbandry
Healthy male FBNF1 rats that were fed ad libitum one of 20 isocaloric diets that differed in fat and carbohydrate composition (n = 8/diet, 160 total). Body weights and food intake were measured twice a week and rats were sacrificed after 8 weeks (age ∼4 months). The diets were based on the AIN 76A semi-purified diet and were comprised of five different fat groups with the major FA constituent of each being either saturated (SFA), trans (TFA), monounsaturated (MUFA), or polyunsaturated (PUFA) with either a 34:1 or a 6:1 ω-6/ω-3 ratio. Each fat group was combined with one of four carbohydrate groups that varied based on their level of sucrose (0, 22, 44, or 65%) and predicted GI [81, 76, 69, 62; based on considering sucrose at 62, corn starch (75% amylopectin, 25% amylose) at 72, and maltodextrin at 120]. These diets are referred to in the report on the basis of their predicted GI. The fat, carbohydrate, and protein percentages in all diets were held consistent at 5, 65, and 20 (%w/w), equivalent to 12, 68, and 21 (kcal %) respectively. The initial body weight for all rats on all diets was 188 ± 2 g, and at sacrifice was 287 ± 2 g (mean ± SD).
Greater details of the husbandry and diets are presented in the supplementary material, following the ARRIVE criteria for reporting animal studies (50). All animal use was Institutional Animal Care and Use Committee approved by the Office for Research Subject Protection at Harvard Medical School.
Mitochondrial isolation and respiratory assay
After the animals were sacrificed, each rat liver was harvested and the mitochondria isolated by the standard differential centrifugation method, using sucrose-based buffers, as described and as used previously in our laboratory (51, 52). Liver isolation buffer contained 240 mM sucrose, 10 mM HEPES, pH 7.4, 1 mM EGTA, and 0.5% BSA. Mitochondrial protein concentration was determined by the Lowry method using BSA as a standard. Secondary gradient purification was not performed, so some contamination with nonmitochondrial lipids is likely, probably <5% (52). For lipid analysis, sample aliquots containing 1 mg of protein from mitochondria were washed in 160 mM KCl and frozen as dry pellets at −80°C before analysis.
Mitochondrial respiration was measured with the Oxygraph 2k electrode system from Oroboros Instruments (http://www.oroboros.at/). Incubation buffer contained 240 mM sucrose, 10 mM HEPES, pH 7.4, 3 μM EDTA, 1 mM KH2PO4, and 5 mM succinate. Respiration in the presence of substrates corresponds only to state 2 respiration (V2). Addition of 200 μM ADP initiates ATP synthesis coupled to proton reentry across the membrane, which corresponds to state 3 (V3). ADP exhaustion leads to a reduction in the respiratory rate and corresponds to state 4 (V4). The respiratory control ratio (RCR) was calculated as the ratio between the rates of respiration in state 3 and 2 (by Lardy, RCR = V3/V2).
LC-MS conditions and experiments
Details of the LC-MS method, data analysis, and CL identification have been described (47–49). The approach detects lipids from six of the LIPID MAPS defined biological lipid categories including, FAs, sterols, glycerophospholipids, sphingolipids, glycerolipids, and prenols (PRs) as well as many lipid subclasses within these broad categories.
Immediately before lipid extraction, each aliquot of mitochondria (containing 1 mg of protein) was dissolved in 40 μl DMSO and the membranes were disrupted by water bath sonication for 1 h. A mitochondrial pool sample was created by combining 8 μl from the sonicated mitochondria of each rat. Lipids were extracted according to the method of (53), substituting dichloromethane for chloroform (54) and then injecting 10 μl of sample onto the LC-MS system.
Lipid extracts were separated on an Ascentis Express C18 2.1 × 150 mm 2.7 μm column (Sigma-Aldrich) connected to a Thermo Fisher Scientific autosampler, an Accela quaternary HPLC pump, and an Exactive benchtop orbitrap mass spectrometer (Thermo Fisher, San Jose, CA) equipped with a heated electrospray ionization (HESI) probe. All LC-MS files from the mitochondria isolated from rats in the diet studies were analyzed using the MS label-free differential analysis software package SIEVE v1.3 (Vast Scientific, Cambridge, MA).
Each sample was normalized for the amount of protein prior to lipid extraction, as 1 mg protein aliquots of mitochondria were used. Changes to the total CL amount per milligram of liver mitochondrial protein in each animal were calculated by taking the sum of the total MS signal for all 26 identified CL species.
We used both the Human Metabolome Database and the Metlin Database for exact mass searches of less than 0.010 ppm. The details of this identification scheme have been previously published (47).
Data analysis
Statistical analysis, simulations, and graphics were generally conducted/created using in-house code written in R (available upon request). Chi-square analysis with Yates correction performed on quantpsy.org (55).
Simulation-calculated probabilities of CLs
CLs are made up of four FAs. We identified 26 CLs in our study that have combinations of 10 distinct FAs. We used the multinomial distribution to calculate the probabilities of each of the 26 CLs. The multinomial formula is given by:
where: i) n is number of trials, in our case this would be four (number of FAs/CL); ii) n1, n2, … nk are numbers of each FA in the CL; iii) k is 10, as there are 10 unique FAs that make up the CLs identified in this study; and iv) P1, P2, … Pk are the probabilities of each of the 10 FAs being chosen at random, i.e., the probabilities are given by the frequency of their occurrence across all samples (the frequency was counted over all the diets and animals). We calculated the probabilities for each of the 26 CLs across all the diets and compared it to the observed frequencies.
Dendrograms and heatmaps
Data were preprocessed as follows: i) normalized the data for each rat by dividing the intensity by the total CL in that rat; ii) took the median value for each CL for each diet; iii) scaled the data for the heatmaps for each CL (row scaling); and iv) generated the dendrograms using hierarchical clustering with complete linkage. The distance matrix was based on Pearson correlation given by the formula as.dist[1 − cor(t(x))], where x is the data with CLs along the rows, and diets along the columns. This was transposed to carry out the correlation between the CLs and converted to a distance matrix. All calculations for dendrograms and heatmaps were done in R, and the dendrograms and heatmaps visualized using the heatmap.2 function from the gplots package.
RESULTS
There were no significant differences in food consumption or body weight gain across all diets, although rats on TFA diets, regardless of carbohydrate source, exhibited a trend of having the highest body weight at the time of sacrifice (I. Stavrovskaya, S. Bird, V. Marur, M. Sniatynski, S. Baranov, H. Greenberg, C. Porter, and B. Kristal, unpublished observations).
Mitochondrial CL levels are increased in high GI diets containing TFA or SFA
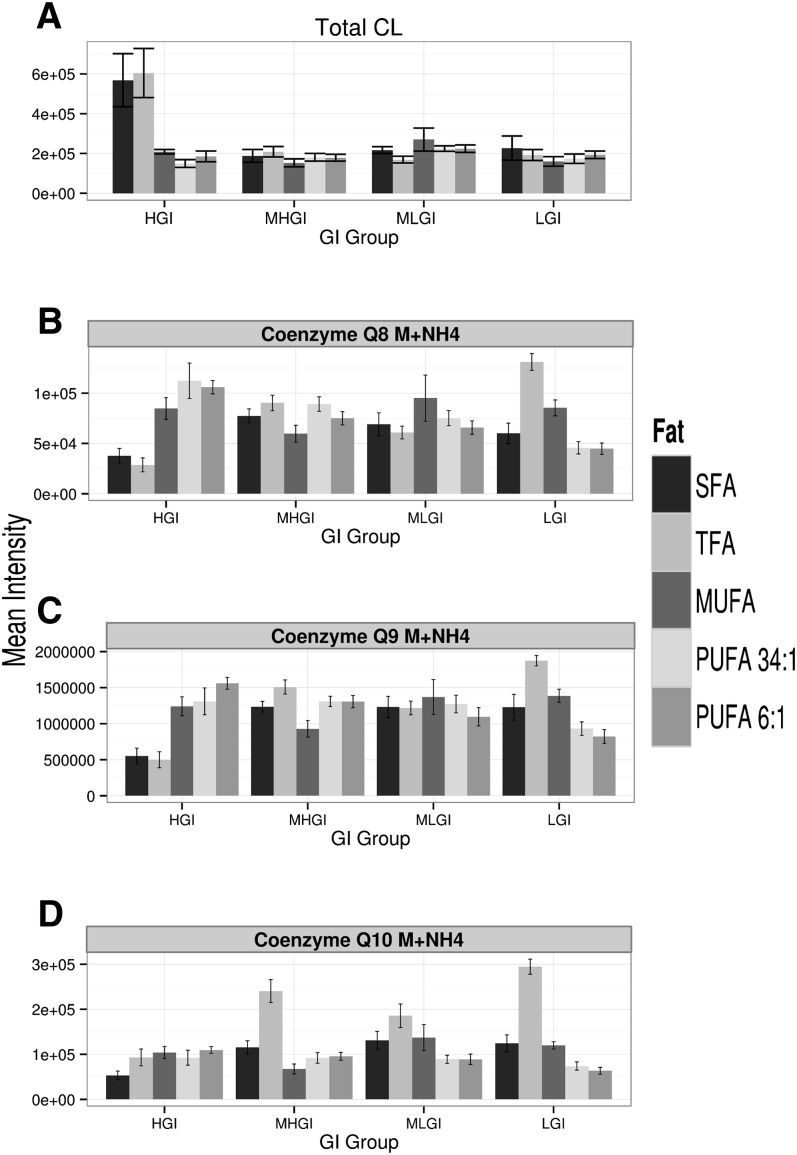
The insights gained into the linkages between diet and mitochondrial biology begin with a paradoxical observation: Rats on two “unhealthy” diets, which were expected to have decreased CL levels, actually had significantly elevated levels. Diets enriched in either SFA or TFA and, to a lesser extent, diets having high GI (HGI), are known to contribute to metabolic diseases, including diabetes. Similarly, diabetes itself is associated with abnormal mitochondrial functions. Decreases in mitochondrial CLs are a consequence of a suite of biochemical stressors and pathophysiological circumstances, and such decreases themselves can cause secondary mitochondrial dysfunction. Together, these conceptual arguments led to a working hypothesis that CLs would be decreased by these diets. This hypothesis was tested by assessing CL levels in the 160 liver mitochondrial samples isolated from the rats described above. CL levels were assessed using a mass spectrometry-based method recently developed in our laboratory that enables identification, structural characterization, and quantitative profiling of over 300 lipids in mitochondria (47, 48). In contrast to the working hypothesis, the mean total CL levels were increased ∼50% by HGI diets containing TFA or SFA relative to the level associated with other diets (Fig. 1A, P < 10−15 by ANOVA, supplementary Fig. IA for these data broken down by fat). CL levels appeared to have greater inter-animal variability in rats fed these diets, suggesting a potential secondary factor/cause for this unexpected increase.
Fig. 1.
Diets coupling high GI with SFA or TFA are associated with elevations in mitochondrial CLs and decreased mitochondrial PRs. A: The relative levels of total CLs per milligram of liver mitochondrial protein in each animal (mean ± SEM) was calculated by taking the sum of the total MS signal for all 26 identified CL species. Relative intensity is presented in arbitrary units; B–D: Mitochondrial levels (per mg mitochondrial protein) of Q8, Q9, and Q10. Within a panel, diets are broken down by GI and then, within each level of GI by dietary fat group [N = 8 for all bars (N = 160 total), mean ± SEM]. Again, relative intensity is presented in arbitrary units and normalized per mg mitochondrial protein. See text for ANOVA P values and supplemental material for post hoc tests. See supplementary Fig. I for these data broken down by fat. MHGI, moderately high glycemic index; MLGI, moderately low glycemic index; LGI, low glycemic index.
Ubiquinone Q9, the central electron carrier in the rat mitochondrial electron transport chain, is decreased and inversely related to CL levels
Because CL plays multiple roles in mitochondria, we hypothesized that any secondary factor/cause that influenced CL levels would also play a central role. And thus, we examined the PR lipids, particularly ubiquinone Q9 (also known as coenzyme Q9, hereafter Q9). In general, the ubiquinones act as electron carriers, and their reduced forms are potent antioxidants. In mitochondria, these molecules are also the primary carriers in the electron transport chain. In humans, this function is primarily mediated by ubiquinone Q10, but in rodents the dominant species is Q9 [80–90% of the total CoQ] (56). Ubiquinone Q9 was therefore identified and quantitated within the previous lipidomics profiles (45, 46), and it displayed 10 times higher signal than other ubiquinone species (c.f., Fig. 1C vs. Fig. 1B, D, supplementary Fig. IC vs. Fig. IB, D for these data broken down by fat). Consistent with our working hypothesis, Q9 was reduced by HGI diets containing TFA or SFA (Fig. 1C, P < 10−13 by ANOVA; supplementary Fig. IC for these data broken down by fat; see also supplementary material for extended statistical analysis vs. all diets). Similar reductions were noted in the ubiquinone Q8 and the Q10 pool (Fig. 1B, D; supplementary Fig. IB, D, P < 10−13 for Q8, P < 10−15 for Q10 by ANOVA; see supplementary material for extended statistical analysis vs. all diets). The latter was generally elevated by TFA diets. Levels of the three ubiquinones were correlated (r2 = 0.6–0.9).
Mitochondrial respiration is essentially unaffected by manipulations used in this model system
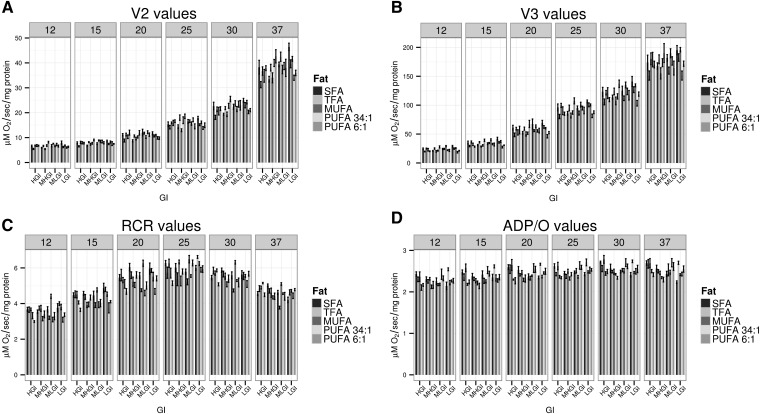
To address the possibility of overt functional impairment of mitochondria (and hence, overt effects of the biochemical changes noted), we tested whether the values for any of the respiratory states or ratios differed significantly across the diets. The assessment of mitochondrial respiratory parameters under different temperature conditions was originally aimed at evaluating potential changes in the membrane fluidity that we hypothesized would be observed in the different diets. Although small differences were observed (Fig. 2, supplementary Fig. II for these data broken down by fat), i) they did not exceed ∼20% of the median for all diets, ii) only a few of the 16,560 two-way comparisons were significant at an adjusted P < 0.05 using a Tukey test (not shown), iii) none were consistently observed for a given fat or GI group within a given temperature condition, and iv) none were consistently observed in the HGI diets containing TFA or SFA within a given temperature condition. Additional studies, e.g., with different substrates in different cohorts, are broadly consistent with these data (data not shown).
Fig. 2.
Respiration parameters are essentially unchanged across the diets. A: Oxygen consumption in V2 (5 mM succinate, no ADP). B: Oxygen consumption in V3 (with substrate and 200 µM ADP). C: RCR defined as V3/V2. D: ADP/O ratio. All studies were conducted at six temperatures as noted. Within a panel, diets are broken down by GI and then, within each level of GI by dietary fat group (N = 8 for all studies, mean ± SEM). See text for statistical discussion. See supplementary Fig. II for these data broken down by fat. MHGI, moderately high glycemic index; MLGI, moderately low glycemic index; LGI, low glycemic index.
The shifts in CL/Q9 populations are associated with endogenous markers of CL synthesis/turnover and oxidative stress
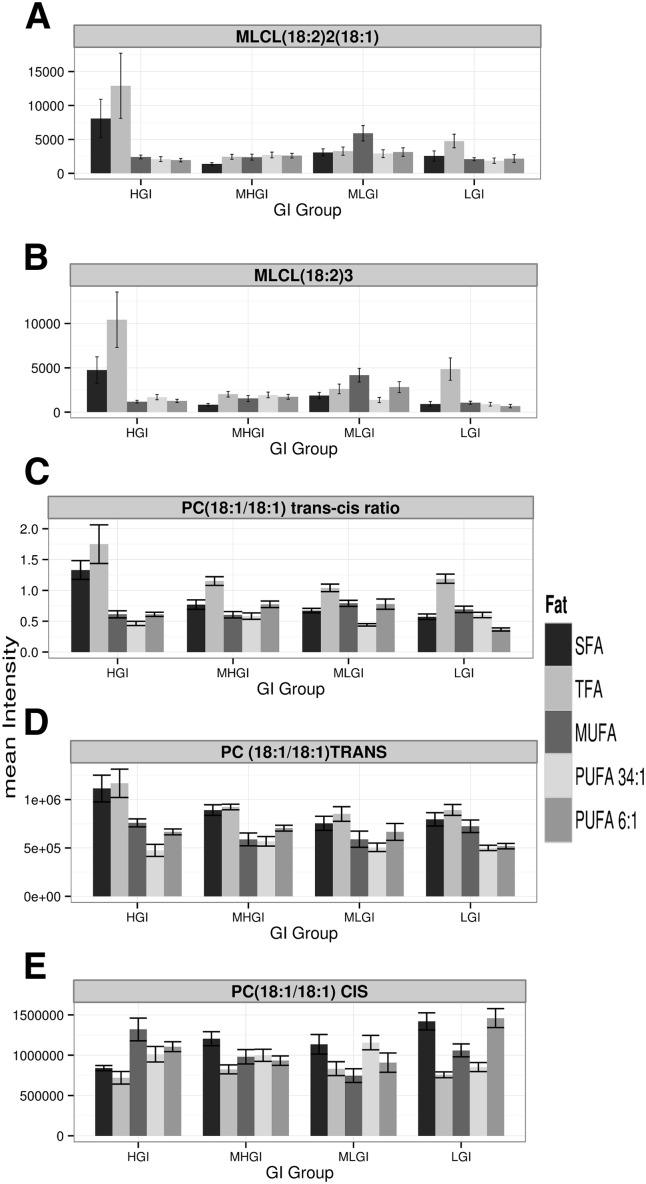
The most parsimonious interpretation of the above results is that the HGI diets containing TFA or SFA induce generalized oxidative stress in the mitochondria. We tested this hypothesis by examining two endogenous markers, the monolysocardiolipins (MLCLs) and the ratio of cis and trans isomers of endogenous phosphocholine (PC) (18:1/18:1). MLCLs are primarily known as intermediaries in CL biosynthesis, but they are also byproducts of the phospholipase A2 (PLA2)-mediated degradation of CLs, which could occur in the context of oxidative stress. Both MLCL species monitored were increased in the HGI diets containing TFA or SFA [Fig. 3A, B, by ANOVA in comparison to the remaining 18 diets: for MLCL (18:2)2(18:1) P < 10−6 comparing SFA/HGI and P < 10−8 for TFA/HGI; for MLCL (18:2)3 P < 10−10 comparing SFA/HGI and P < 10−14 for TFA/HGI, supplementary Fig. IIIA, B for these data broken down by fat] (see supplementary material for extended statistical analysis vs. all diets). Thiyl radicals, which can form during oxidative stress, isomerize the double bond of cis-PC (18:1/18:1) to trans-PC (18:1/18:1), allowing a readout of endogenous thiyl radical stress (49, 57). The trans-cis ratio of endogenous PC (18:1/18:1) is specifically elevated in the mitochondria from rats fed the HGI diets containing TFA or SFA (Fig. 3C, P < 10−15 by ANOVA). This change appears to result from both increased trans-PC (18:1/18:1) and decreased cis-PC (18:1/18:1) [Fig. 3D, E, P < 10−12 for trans-PC (18:1/18:1) and for cis-PC (18:1/18:1) by ANOVA, see supplementary Fig. IIID, E for these data broken down by fat] (see supplementary material for extended statistical analysis vs. all diets). Data are consistent with the hypothesized increase in oxidative stress.
Fig. 3.
Despite respiratory stability, diets coupling high GI with SFA or TFA are associated with elevated mitochondrial oxidative stress. A, B: Levels of MLCL (18:2)2(18:1) and MLCL (18:2)3. C–E: trans/cis ratio (C) of mitochondrial PC (18:1/18:1) trans (D) relative to PC (18:1/18:1) cis (E). Within a panel, diets are broken down by dietary fat and then, within each dietary fat group by GI [N = 8 for all bars (N = 160 total), mean ± SEM]. See text for ANOVA P values and supplementary material for post hoc tests. See supplementary Fig. III for these data broken down by fat. MHGI, moderately high glycemic index; MLGI, moderately low glycemic index; LGI, low glycemic index.
The observed elevation in CL levels is broadly, but not always, observed across different CL species
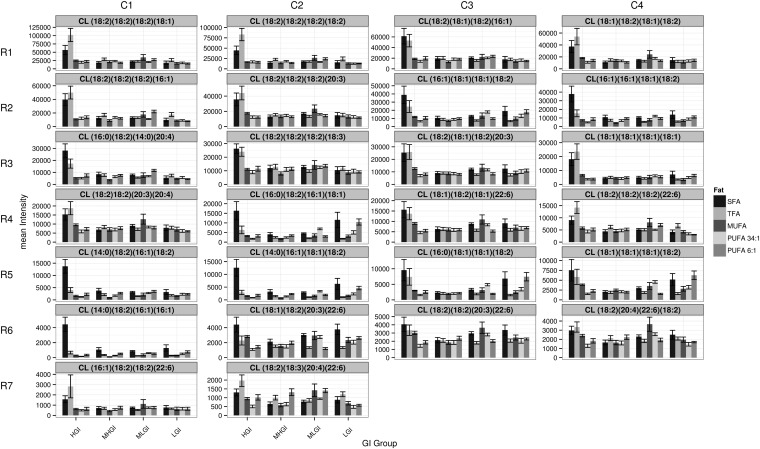
We next tested the hypothesis that the increase in CL was generalized, i.e., the changes were driven by an increase in overall CL synthesis or a decrease in CL degradation versus a counter-hypothesis that the difference would be localized to only a subset of the CL species. Our previous work described 26 CL species in a pooled sample drawn from this cohort of rats (47). We now determined that all of these CL species are present in all mitochondria assayed (Fig. 4, supplementary Fig. IV for these data broken down by fat). The data are presented in Fig. 4, ranging from the most abundant to the least abundant CL species (read left to right across and then down). These data provide partial, but not complete, support for the generalized CL elevation hypothesis. Specifically, most of the more abundant CL species show increased levels similar to those observed in the overall population, but some CL species are increased by other diets. For example, the level of CL (16:0)(18:2)(16:1)(18:1) (located at position row 4 column 2, hereafter R4-C2) is increased by two diets with low GI, and some CL species [e.g., CL (14:0)(18:2)(16:1)(18:2), R5-C1] are not elevated by one of the diets in question (HGI, with TFA or SFA). Therefore, we examined diet-specific effects on mitochondrial CL acyl chain composition further.
Fig. 4.
The effects of macronutrient shifts on CL levels are broad but not uniform. The mass spectrometry signal of each CL is presented in each of 26 panels. The Y-axis scale is varied to highlight the inter-CL species changes across the diets; thus, each panel has a different Y-axis scaling. Abundance is highest in the upper left corner and decreases moving right and then down. Each panel's title includes the CL FA makeup, with the first two and last two located on the same phosphatidylglycerol moiety. Within a panel, diets are broken down by dietary fat and then, within each dietary fat group by GI (mean ± SEM, N = 8/diet, 160 total). See supplementary Fig. IV for these data broken down by fat. MHGI, moderately high glycemic index; MLGI, moderately low glycemic index; LGI, low glycemic index.
FA chain composition of mature CLs is both unexpectedly complex and nonprobabilistic
We next addressed the relative occupancy of the four acyl components of the mature CL. For these analyses, we normalized the absolute abundance of each of 26 structurally unique CL species by the total CL amount in a given sample, i.e., each CL species was expressed as a fraction of total CL in mitochondrial samples from that rat (Fig. 5). As noted, there is a long history suggesting that there is a preferential retention of linoleic acid (18:2) in mature CLs, which has been reported to run to ∼85% (5). Together, the preferential accumulation of 18:2 side chains and an apparent preference for identical side chain accumulation (3, 5) suggested that the tetralinoleoyl CL species would dominate the CL pool composition.
Fig. 5.
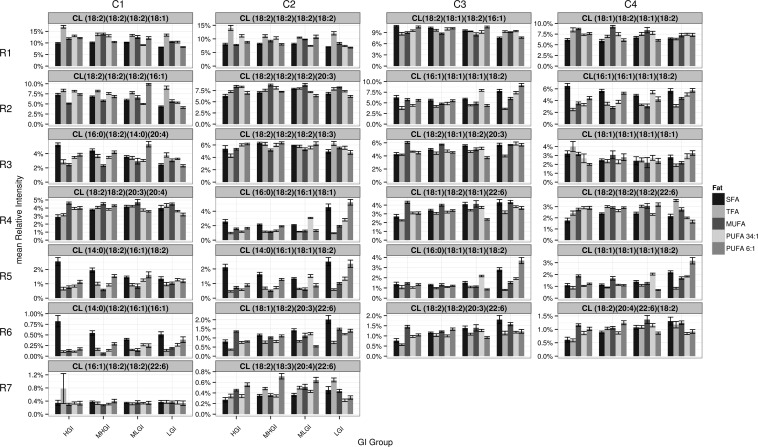
Changes in the relative abundance of each CL. The abundance of each CL as a percentage of the total CLs (normalized at the level of the individual rats) is presented in each of 26 panels (mean ± SEM, N = 8/diet, 160 total). Specifics are as in Fig. 4. See supplementary Fig. V for these data broken down by fat. MHGI, moderately high glycemic index; MLGI, moderately low glycemic index; LGI, low glycemic index.
The detailed profiling method shows the story is much more complicated, and is consistent with growing evidence of non-tetralinolyl species in mature CLs (3, 5, 7, 8, 44, 58, 59). The data show a spectrum of abundance from 0.34% of total CLs {CL (18:2)(18:3)(20:4)(22:6) [R7-C2]} to 11.9% of total CLs {CL (18:2)3(18:1) [R1-C1]}. We are aware that CLs with frequencies of consistently <0.1% would not be followed in our mass spectrometry quantitation approach. We would similarly not expect our approach to detect oxidized CL species, as, under the basal conditions studied, we would expect: i) comparatively few oxidation events relative to those occurring in more commonly studied pathological conditions; and ii) any generated oxidized CLs to be short-lived due to the rapid deacylation induced by PLA2. Therefore, we would expect any oxidized CL species to be below our limit of detection.
More strikingly, the CL (18:2)4 [R1-C2] is only 8.5% of the total CL in our mitochondrial samples; 38% of the total CL pool of the study has at least three 18:2 chains (median percentage of all 20 diets). Depending on the diet, the five most abundant CLs (i.e., approximately top fifth by abundance), each of which has at least two 18:2 FA chains, comprise ∼44% of the total CL pool (median percentage of all 20 diets). The five least abundant CLs observed (i.e., approximately bottom fifth in abundance) make up only 2.3% of the total pool (median percentage of all 20 diets).
Closer examination of the data showed both expected and counter-intuitive aspects. When the data for each diet are initially grouped by fat classes (as they are in supplementary Fig. V), we observe an elevation of CL species containing SFA chains in mitochondria both from rats maintained on diets enriched in SFA, and, unexpectedly, in rats maintained on diets enriched in PUFA. These phenomena can be seen for CL (14:0)(16:1)(18:1)(18:2) [R5-C2], CL (16:0)(16:1)(18:1)(18:2) [R4-C2], CL (14:0)(16:1)2(18:2) [R6-C1], CL (14:0)(16:1)(18:2)2 [R5-C1], and CL (14:0)(16:0)(18:2)(20:4) [R3-C1], but not consistently for CL (16:0)(18:1)(18:1)(18:2) [R5-C3], the only other CL detected that has a saturated side chain.
Consistent with this counter-intuitive relationship between the behavior caused by SFA-enriched diets, the behavior of CL (18:1)(18:2)(20:3)(22:6) [R6-C2] is counter-intuitive; this CL is increased by both MUFA- and SFA-enriched diets as much or more than the more ω-3-enriched diets. Also unexpectedly, CL (18:2)4 [R1-C2] is increased by TFA diets, and CL (18:2)2(20:3)(22:6) [R6-C3] by several of the SFA diets.
CL (18:1)(18:2)(20:3)(22:6) [R6-C2] exemplifies another observed trait: within diets containing a given fat, the predicted GI also influences CL makeup. For example, CL (18:1)(18:2)(20:3)(22:6) [R6-C2] is progressively increased by SFA-enriched diets, and as predicted, GI decreases. Focusing, for simplicity, just on GI-mediated effects in the SFA diets, we note similar behaviors in CL (18:2)2(20:3)(22:6) [R6-C3], CL (18:1)2(18:2)(22:6) [R4-C3], and CL (18:2)(20:4)(22:6)(18:2) [R6-C4]. Notably, these other CLs also each lack SFA chains and contain a 22:6 side chain. In contrast, we note the opposite effects (decreasing CL levels with decreasing GI in SFA diets) on CL (16:0)(18:2)(14:0)(20:4) [R3-C1], CL (14:0)(18:2)(16:1)(18:2) [R5–C1], and CL (14:0)(18:2)(16:1)2 [R6-C1]. Each of these CLs has a SFA and two shorter chain FAs. There are, however, exceptions to the “saturated versus nonsaturated” relationships {e.g., CL (18:2)(18:1)(18:2)(16:1) [R1-C3] and CL(18:2)3(16:1) [R2-C1] decrease with decreasing predicted GI}, indicating that these relationships, although potential themes, are neither necessary nor sufficient to determine the effects on individual CLs. Other CLs seem to respond more abruptly to changes in GI (or its relative sucrose makeup), including CL (16:0)(18:2)(16:1)(18:1) [R4-C2] and CL (16:0)(18:1)2(18:2) [R5-C3].
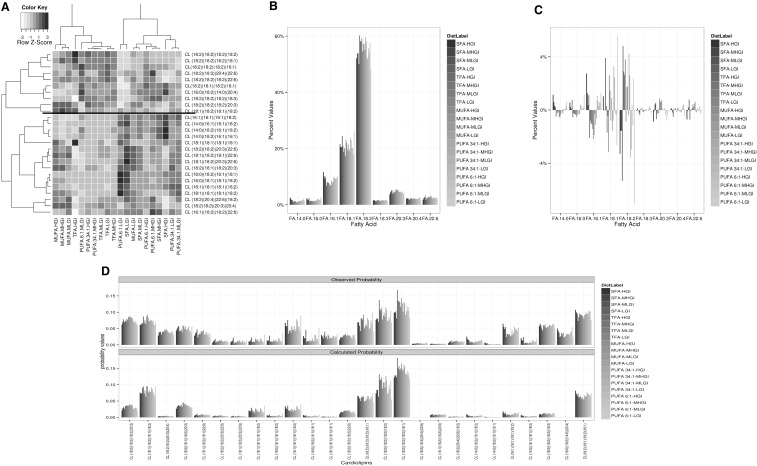
We next sought higher level relationships between the diets and between the CL species by using two-dimensional hierarchical clustering analysis (Fig. 6A). As implemented, the clustering algorithm determines natural de novo groups of data by looking at the relative correlation of the changes in relative occupancy of the CL pool across the different diets and the relative similarity of the diets across the individual CLs. The dendrograms are similar to evolutionary trees, with the most similar members having the shortest distance to a common node (branch point). Nodes are rotationally symmetrical, so absolute distance apart is not significant. We will not discuss the diet-specific clustering aspect.
Fig. 6.
FA incorporation into CLs reveals natural groups as well as positive and negative regulation. A: Two-way hierarchical clustering analysis was performed as described in Research Design and Methods. Clusters were built based on a distance metric assigned by the Pearson correlation of the median data for each CL within each diet, and then normalized as percent of total CL per rat. Coloring is by Z-scores of median intensity of CLs by diet. B: Percentage of each FA in the CL pool from the rats in this study, with the bars ordered and colored by fat, with secondary order by GI. C: Difference from the median representation for that FA in the CL pool, with the bars ordered and colored by fat, with secondary order by GI [note scale difference with panel (B)]. D: Comparison of the level of each CL observed (top) and expected (based on simulations, see Research Design and Methods). MHGI, moderately high glycemic index; MLGI, moderately low glycemic index; LGI, low glycemic index.
We predicted that the clustering by CLs would recapitulate known aspects of biochemistry, but this was not consistently observed. Specifically, the synthesis of CLs involves the condensation of two diacylglycerides followed by remodeling. As such, we would predict that the CLs with three 18:1 or three 18:2 chains would cluster tightly with those having four 18:1 or four 18:2 chains, respectively. Consistent with this hypothesis, the top cluster contains all CL molecules containing 3 or 4 (18:2) chains, whereas the bottom cluster has none (6 of 10 vs. 0 of 16, P < 0.005 by Chi-square with Yates correction) (55). Conversely, within the bottom cluster, the CL (18:1)4 and the only CL with three 18:1 chains are separated. Thus, the established biochemistry does not completely define the apparent coordinate regulation of the CL groups.
The FA composition of the CL pool is not directly related to the presence of a given FA in the diet
The above result suggests a level of coordinate regulation beyond that predicted by the likely precursors and their products; we therefore further examined the relationship between total FAs in the CL population and the makeup of the CL pools. Overall, the individual FA groups that comprised the CL pools were similar across all 20 diets (Fig. 6B, C). We observed that the 18:2 chains comprised a mean of 55 ± 3.3% (mean ± SD) of the total acyl chains across all diets. This is slightly less than the lower end of the scale commonly reported (60–90%), but we note that most of our diets have lower (18:2) content than typical rodent diets, both in absolute amount and in percentage of the dietary FAs. Overall SFA composition (means of 1.7% for 16:0 and 1.4% for 14:0) were consistent with previous reports (60) that saturated species do not exceed 6–8% of total FAs. We note that the shorter chain FAs (14:0, 16:0, and 16:1) show a trend toward higher concentration in the saturated and PUFA diets. In contrast, levels of 18:1, 18:3, 20:4, and 22:6 had minimal or no consistent change across all diets.
Consistent with the data shown in Figs. 4 and 5, the data about the overall FA composition of the CL pool support the view that there is little direct relationship between the presence of a lipid in the diet and the presence of that lipid in the CL pool; indeed, the relationship often seems counter-intuitive. These data support the hypothesis that there is selection (both positive and negative) for individual FA chains to be incorporated into the CL pool.
Composition of the mature CLs is species specific and can be driven by mass action or by positive or negative selection
We next asked whether the relationship existed one level lower. Would the presence of a FA in the CL pool lead to essentially concentration-driven incorporation into mature CL species? This is functionally a test of the hypothesis that the above selection for individual FAs into the CL pool is the only level of selection, after which individual CLs are driven by mass effects. The data we have disprove this working hypothesis (Fig. 6D) and show that the picture is, in fact, species specific. Consider CL (18:2)4: the four FA groups that comprise this CL are present at 55.4% of the FA pool, so it would be expected to occur (by random assortment) at 0.554 × 0.554 × 0.554 × 0.554 = 9.4%, which is close to that observed (means of diet medians = 9.1%). In contrast, consider CL (16:0)(18:2)(14:0)(20:4) [R3-C1] (Fig. 5). The estimated predicted abundance of this molecule is 0.017 × 0.554 × 0.014 × 0.021 × 24 = 0.0066%, or 446-fold less than is observed (3.4%). Thus, for some CLs there is an added level of positive selection. The reverse situation occurs for CL (16:1)(18:2)2(22:6) [R7-C1] (Fig. 5). The estimated predicted abundance of this molecule is 0.087 × 0.554 × 0.554 × 0.025 × 12 = 0.80%, or 2.5-fold greater than is observed (0.31%). Thus, for some CLs, there is an added level of negative selection.
It is also noteworthy that we do not detect many expected CLs. As one example, we never observe a CL having both a 16:1 (8.8% of all FAs) and a 20:3 FA group (4.8% of all FAs), but we would expect the hypothetical CL (16:1)(18:2)2(20:3) at 0.088 × 0.046 × 0.554 × 0.554 × 12 = ∼1.5% of the total pool, or ∼5–15 times our limit of detection, suggesting that such species are either never formed or are preferentially destroyed. Thus, there also exists a level of exclusion of potential CL species or, at the very least, they are reduced below the level of detection (estimated at ∼0.1% of the CL pool).
DISCUSSION
Our data qualitatively and quantitatively alter the previous conceptual view of CL regulation. They provide a biological/experimental model for the systematic investigation of the levels of regulation identified in this report. These data also provide insight into the early consequences of physiological elevations of oxidative stress in mitochondria. The extensive work on CL demonstrates the importance of CL from both the bottom up (biochemistry) and from the top down (disease) (2, 3, 6, 8–34, 37, 38). Until recently, the known regulation of CL could be summarized by two broad concepts: i) the over-representation of 18:2 FAs; and ii) the almost ubiquitous linkage between morbidity/pathology, decreases in CL/reduction in CL total content, loss of 18:2 FAs, and loss/failure of mitochondrial respiratory capacity. The literature shows that abnormalities and deficiencies in the CL pool exert a penetrant effect on mitochondrial physiology. The aforementioned concepts paint the CL pool itself as a passive player, subject to dietary lipid availability and susceptible to oxidative damage. From a theoretical standpoint, however, it is logical that CL content and FA composition should be regulated because: i) CL is the most important lipid in eukaryotic energy production; and ii) it is synthesized endogenously.
More than a target-regulated response to environmental influences
Our insights into the connections between diet and mitochondrial biology began with a paradoxical observation: Two “unhealthy” diets that were expected to, if anything, decrease CL levels, were instead associated with elevated levels of CL. Our follow-up analyses of these results revealed five additional levels of CL regulation, detailed below.
First, CL levels can increase in the context of an apparent mitochondrial oxidative stress (e.g., decreased ubiquinones, increased trans/cis lipids). Notably, these changes occur in a nonmorbid condition, e.g., with no obvious pathology and in the context of functional mitochondria. Thus, these changes may represent one aspect of an efficacious compensatory change in CL in response to the mitochondrial damage. Indirect evidence from pathological studies suggests that this compensatory capacity is limited, e.g., diabetes is associated with abnormal mitochondrial function. Consistent with this idea, multiple reports support a model in which, when compensatory capacity is overwhelmed, mitochondrial CLs decrease in the context of a suite of biochemical stressors and/or pathophysiological circumstances that appear to cause secondary mitochondrial dysfunction (2, 3, 8–27). Notably, under these circumstances, we find that mitochondrial respiration is robust against the loss of Q9. This indicates that the biochemical changes observed in CL are premorbid, nonstochastic, and likely in response to the subthreshold mitochondrial biochemical changes (coincident oxidative stress and loss of Q9). This idea is supported by the data from Lenaz (61) showing that the size of the ubiquinone pool is a limiting factor for mitochondrial respiratory rate. The observation of substantially reduced levels of Q9 in the presence of normal respiration is, to our understanding, potentially unique in the literature, as usually such drops are associated with considerable morbidity/loss of respiratory capacity (e.g., in heart failure), consistent with Lenaz's work. These data would suggest that, at least in the context of our study, Q9 has a very low control coefficient.
Second, CL levels (overall or individual species) are not directly correlated with any mitochondrial parameter. This observation complements the well-established data linking morbidity and reduced CL by showing that there exists a range of mitochondrial changes in which CL undergoes a potentially compensatory response and a range in which this potential response capacity is lost and/or overwhelmed.
Third, our data extend links between diet and CL regulation, and more specifically, our understanding of the regulation of FA incorporation into CL pools. Previous studies focused on evidence that limiting an animal's 18:2 FA intake was associated with decreasing CL levels, mitochondrial dysfunction, and morbidity (3, 8, 38, 45). These studies also showed that many FAs are not observed in CL pools (i.e., active exclusion). Such experiments showed that, in the face of nutritional deficiency, CL pools could take up relevant FAs. Here we show a qualitatively different picture. Analysis of individual CL species provides additional support for compensatory changes. Although none of the diets fed were deficient in 18:2, the diets that increase CL levels have less 18:2 than the 34:1 and 6:1 ω-6/ω-3 diets and the same 18:2 as the non-HGI versions of the SFA and TFA diets. Thus, the observed increase must, at some level, be regulated rather than stochastic. Our data provide evidence that the CL pool composition is regulated by both dietary fat and dietary carbohydrate. The response to dietary fat is subtle and often inverted. For example, 6-fold increases in dietary ω-3 FAs are associated with minimal or no change in the overall 22:6 FA incorporation into CL pools (Fig. 6B, C) and even decreases of some 22:6-containing species, e.g., CL (18:1)(18:2)(18:1)(22:6) (Fig. 4, R4-C3; supplementary Fig. IV). In contrast, these same diets are associated with increases in the overall 14:0, 16:0, and 16:1 FA incorporation into CL pools (Fig. 6B, C). The combination of nonproportional and often counter-proportional results implies active selection (both positive and negative) for incorporation/maintenance of individual FA chains into the CL pool. These data expand the point, which is implied in the literature, that higher levels of 18:2 occur in CLs than in the diets themselves. Similarly, incorporation of 18:2 is 54.8 ± 3.5% (mean ± SD), despite diets varying ∼4-fold (from 0.95% to 3.8% of the diet) in this essential FA. Our data and interpretations are also generally consistent with data presented in a recent study from Aoun et al. (62). Aoun et al. (62) suggest that dietary fats modulate the total CL content and FA composition at the level of CL synthase. They also do not see a fish oil-related increase in ω-3 FAs in CLs in their 5% diet. Their data also suggested a linkage between CL levels and ATP synthase. The Aoun et al. (62) study, however, used mitochondria isolated from frozen tissue and assessed only CL population level FAs. As such, they would have been unable to observe any effects at the level of intact CLs, and they were unable to determine whether the ATP synthase effect that they observed had any functional consequences. Functional consequences would not be predicted a priori, as ATP synthase generally does not have a high control coefficient relative to ADP translocase. It is also difficult to predict how the freeze-thaw damage would have affected subsequent purification (e.g., loss of mitochondria, mitochondrial outer membranes, or cross-contamination with other organelles). Recovered CL is notably increased in a high-fat diet-induced steatosis component of their study, but the appropriate normalization relative to ours and other studies with functional mitochondria is unclear. Thus, direct comparison with our data is not possible.
Fourth, both positive and negative selection occur at the level of individual mature CLs. We observed positive selection of ∼400-fold and negative selection of ≥2.4-fold. Furthermore, some FAs, e.g., 20:4, are only present in four CLs, suggesting that only a select set of mature molecules is positively regulated, and at least the potential CL (16:1)(18:2)2(20:3) is actively negatively regulated (it is predicted to be abundant enough at ∼1.4% to be observable). We also note that, in this animal model, CLs that contain two identical chains are always split between the phosphatidyl head groups on the backbone (which was directly observable using our LC-MS method (47, 48), these data differ from previous observations in other models (4, 5, 8, 34, 38, 58, 59).
Fifth, we detected groups of CLs, as seen in Fig. 6A. The most parsimonious explanation of the observed results is that CLs divide into two broad groups, primarily characterized by the presence or absence of three or four 18:2 chains. Although this idea is possible, we think it is more likely that the concept of functional groupings of CLs is valid, but the specifics are condition dependent. If so, there would be situations (genetic and/or environmental) in which the dominant factors are, for example, the presence or absence of arachidonic acid (20:4) or docosahexaenoic acid (22:6). We can see, for example, partial clustering of the smaller, more saturated fatty acyl chain containing CLs in the lower half of the dendrogram in Fig. 6A. In either case, the result is suggestive of functional differences of these classes, and is consistent with literature that specific CL subgroups/acyl chains might have differential biochemical roles in mitochondrial biology.
Parsimony suggests that regulatory levels four and five might be linked, as both concern mature CL amounts. The controls exerted at these two levels, however, appear to be at least semi-independent, as they do not consistently covary across the identified clusters.
What mechanistically controls CL composition?
The diversity of CL molecules is known to be the end result of two processes: de novo synthesis and remodeling. De novo CL synthesis occurs by a mechanism universal for all phospholipids via a precursor phosphatidic acid (3, 5, 8, 38, 63). The acyl specificity of CL is imparted by the remodeling phase of CL biosynthesis, which takes place via two known mechanisms: i) conventional CL remodeling, which occurs by repeated cycles of MLCL deacylation and subsequent reacylation either by an acyl-CoA-dependent or an acyl-CoA-independent pathway; and ii) CL remodeling that occurs after a transacylation reaction in which acyl chains are transferred from a variety of phospholipids (e.g., phosphatidylethanolamine and phosphatidylcholine) to MLCL via a phospholipid-lysophospholipid transacylase termed tafazzin (64–70).
The mechanism by which tafazzin generates CL acyl specificity is also unknown. It is known that tafazzin is required to maintain structural uniformity and molecular symmetry (e.g., the formation of identical 1,2-diacylglycerol moieties) among CL species, and might be selective for 18:2 acyl chains (59). In contrast, in vitro studies on the purified enzyme showed that tafazzin itself does not possess much specificity for acyl groups, carbon positions, or double bond localization. Tafazzin mutations also lead to the deficiency of only one CL species, tetralinoleoyl (18:2)4. Schlame (69) proposed a mathematical model of CL pattern formation suggesting that the abundance of CL (18:2)4 and CL diversity might result from transacylation equilibrium of multiple molecular species rather than tafazzin substrate specificity. Zhang et al. (71) employed a computational modeling approach to help elucidate the CL remodeling mechanism. Their model, based on a suite of data from different animal tissues, suggested that proportional incorporation and nonconstrained distribution of available FAs underlies the composition of mature CL populations. Their model suggested that, in the majority of tissues, the headgroups on acyl donors are the main determinants of contribution to CL, rather than selectivity on individual acyl chains. Our data showing nonproportional incorporation of FAs from the CL pool into mature CLs (Fig. 6C, D) indicates a role for the acyl chains in determining mature CL composition and suggests that the Zhang et al. (71) model is incomplete. The data provided here, which biologically complements the tissue-specific data available (71), may help increase the overall predictive accuracy of this model.
The diet/animal model
The biological model (diets + rat strain) we used in the present study has several important features. Most studies that look at the effects of CL changes and/or diet on mitochondrial function focus on animals with known disease susceptibilities or animals already showing overt abnormalities. For example, in the recent study from Aoun et al. (62), the effect of dietary fats on CL content and composition was examined in obese Wistar rats that were about 200 g heavier than our rats. In contrast, we fed robust FBNF1 rats low-fat diets and tested systematic variation across both the fat and carbohydrate components of the diet. We observed no problems with weight gain, which, along with food consumption, was conserved across the study (I. Stavrovskaya, S. Bird, V. Marur, M. Sniatynski, S. Baranov, H. Greenberg, C. Porter, and B. Kristal, unpublished observations).
CONCLUSION
In conclusion, a combination of a unique animal model with an in-depth lipidomics analysis of the rat liver mitochondrial CL pools has revealed unexpected complexity in the regulation of one of the most important lipids in eukaryotic energy production. This revelation occurs in the context of early stage oxidative challenge to mitochondria that is not associated with overt loss of function. The data suggest that the CL pool is regulated in the following ways: i) dietary fats and, to a lesser extent, carbohydrates induce changes in the relative abundance of specific CL species; ii) FA incorporation into mature CLs undergoes both positive (∼400-fold) and negative (≥2.4-fold) regulation; iii) subclasses of CL species are coordinately regulated; and iv) dietary lipid abundance and incorporation of FAs into both the CL pool and specific mature tetra-acyl CLs are inversely related, suggesting compensatory regulation. The basic model, a simple two-way dietary cross, provides a diverse custom expandable system for the investigation of the underlying mechanism and implications of these results for both CL and physiological levels of oxidative stress in mitochondria.
Note added in proof
During the publication process, the lipid and mitochondrial data reported herein became available: Stavrovskaya, I.G., S. S. Bird, V. R. Marur, M. J. Sniatynski, S. V. Baranov, H. K. Greenberg, C. L. Porter, and B. S. Kristal. 2013. Dietary macronutrients modulate the fatty acyl composition of rat liver mitochondrial cardiolipins. Dryad Digital Repository. doi:10.5061/dryad.1t93v
Supplementary Material
Acknowledgments
The authors thank ThermoFisher for the loan of an Exactive Benchtop orbitrap for demonstration testing and financial support for scientific meeting attendance.
Footnotes
Abbreviations:
- CL
- cardiolipin
- GI
- glycemic index
- HGI
- high glycemic index
- MLCL
- monolysocardiolipin
- PC
- phosphocholine: PLA2, phospholipase A2
- PR
- prenol
- RCR
- respiratory control ratio
- SFA
- saturated fatty acid
- TFA
- trans fatty acid
These studies were funded by National Institutes of Health Grants U01-ES-16048 (B.S.K.) and P30-DK-040561 (W.A.W., Principal Investigator). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. B.S.K. is the inventor on general metabolomics-related IP that has been licensed to Metabolon via Weill Medical College of Cornell University and for which he receives royalty payments via Weill Medical College of Cornell University. He also consults for and has a small equity interest in the company. Metabolon offers biochemical profiling services and is developing molecular diagnostic assays detecting and monitoring disease. Metabolon has no rights or proprietary access to the research results presented and/or new IP generated under these grants/studies. B.S.K.’s interests were reviewed by the Brigham and Women's Hospital and Partners Healthcare in accordance with their institutional policy. Accordingly, upon review, the institution determined that B.S.K.’s financial interest in Metabolon does not create a significant financial conflict of interest (FCOI) with this research. The addition of this statement where appropriate was explicitly requested and approved by Brigham and Women's Hospital.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures, 13 tables, and text.
REFERENCES
- 1.Ioannou P. V., Golding B. T. 1979. Cardiolipins: their chemistry and biochemistry. Prog. Lipid Res. 17: 279–318 [DOI] [PubMed] [Google Scholar]
- 2.Hoch F. L. 1992. Cardiolipins and biomembrane function. Biochim. Biophys. Acta. 1113: 71–133 [DOI] [PubMed] [Google Scholar]
- 3.Schlame M., Rua D., Greenberg M. L. 2000. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39: 257–288 [DOI] [PubMed] [Google Scholar]
- 4.Schlame M., Otten D. 1991. Analysis of cardiolipin molecular species by high-performance liquid chromatography of its derivative 1,3-bisphosphatidyl-2-benzoyl-sn-glycerol dimethyl ester. Anal. Biochem. 195: 290–295 [DOI] [PubMed] [Google Scholar]
- 5.Schlame M., Ren M., Xu Y., Greenberg M. L., Haller I. 2005. Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids. 138: 38–49 [DOI] [PubMed] [Google Scholar]
- 6.Robinson N. C. 1993. Functional binding of cardiolipin to cytochrome c oxidase. J. Bioenerg. Biomembr. 25: 153–163 [DOI] [PubMed] [Google Scholar]
- 7.Schlame M., Rustow W. 1990. Lysocardiolipin formation and reacylation in isolated rat liver mitochondria. Biochem. J. 272: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chicco A. J., Sparagna G. C. 2007. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292: C33–C44 [DOI] [PubMed] [Google Scholar]
- 9.Schlame M., Ren M. 2009. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta. 1788: 2080–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claypool S. M., Koehler C. M. 2012. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry M., Green D. E. 1980. Cardiolipin requirement by cytochrome oxidase and the catalytic role of phospholipid. Biochem. Biophys. Res. Commun. 93: 1238–1246 [DOI] [PubMed] [Google Scholar]
- 12.Fry M., Green D. E. 1981. Cardiolipin requirement for electron transfer in complex I and III of mitochondrial respiratory chain. J. Biol. Chem. 256: 1874–1880 [PubMed] [Google Scholar]
- 13.Eble K. S., Coleman W. B., Hantgan R. R., Cunningham C. C. 1990. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem. 265: 19434–19440 [PubMed] [Google Scholar]
- 14.Hoch F. L. 1998. Cardiolipins and mitochondrial proton-selective leakage. J. Bioenerg. Biomembr. 30: 511–532 [DOI] [PubMed] [Google Scholar]
- 15.Claypool S. M. 2009. Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim. Biophys. Acta. 1788: 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M., Mileykovskaya E., Dowhan W. 2002. Gluing the respiratory chain together. Cardiolipin is required for supercomplexes formation in the inner mitochondrial membrane. J. Biol. Chem. 277: 43553–43556 [DOI] [PubMed] [Google Scholar]
- 17.Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schagger H. 2003. Cardiolipin stabilizes respiratory chain supracomplexes. J. Biol. Chem. 278: 52873–52880 [DOI] [PubMed] [Google Scholar]
- 18.Klingenberg M. 2009. Cardiolipin and mitochondrial carriers. Biochim. Biophys. Acta. 1788: 2048–2058 [DOI] [PubMed] [Google Scholar]
- 19.Acehan D., Malhotra A., Xu Y., Ren M., Stokes D. L., Schlame M. 2011. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 100: 2184–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlame M., Hostetler K. Y. 1991. Solubilization, purification, and characterization of cardiolipin synthase from rat liver mitochondria. Demonstration of its phospholipid requirement. J. Biol. Chem. 266: 22398–22403 [PubMed] [Google Scholar]
- 21.Beyer K., Nuscher B. 1996. Specific cardiolipin binding interferes with labeling of sulfhydryl residues in the adenosine diphosphate/adenosine triphosphate carrier protein from beef heart mitochondria. Biochemistry. 35: 15784–15790 [DOI] [PubMed] [Google Scholar]
- 22.Kadenbach B., Mende P., Kolbe H. V., Stipani I., Palmieri F. 1982. The mitochondrial phosphate carrier has an essential requirement for cardiolipin. FEBS Lett. 139: 109–112 [DOI] [PubMed] [Google Scholar]
- 23.Nałecz K. A., Bolli R., Wojtczak L., Azzi A. 1986. The monocarboxylate carrier from bovine heart mitochondria: partial purification and its substrate-transporting properties in a reconstituted system. Biochim. Biophys. Acta. 851: 29–37 [DOI] [PubMed] [Google Scholar]
- 24.Noël H., Pande S. V. 1986. An essential requirement of cardiolipin for mitochondrial carnitine acylcarnitine translocase activity. Lipid requirement of carnitine acylcarnitine translocase. Eur. J. Biochem. 155: 99–102 [DOI] [PubMed] [Google Scholar]
- 25.Müller M., Cheneval D., Carafoli E. 1986. The mitochondrial creatine phosphokinase is associated with inner mitochondrial membrane cardiolipin. Adv. Exp. Med. Biol. 194: 151–156 [DOI] [PubMed] [Google Scholar]
- 26.Ott M., Robertson J. D., Gogvadze V., Zhivotovsky B., Orrenius S. 2002. Cytochrome c release from mitochondria proceeds by a two-step process. Proc. Natl. Acad. Sci. USA. 99: 1259–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koshkin V., Greenberg M. L. 2002. Cardiolipin prevents rate-dependent uncoupling and provides osmotic stability in yeast mitochondria. Biochem. J. 364: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauff K. D., Hatch G. M. 2006. Cardiolipin metabolism and Barth syndrome. Prog. Lipid Res. 45: 91–101 [DOI] [PubMed] [Google Scholar]
- 29.Schlame M., Ren M. 2006. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 580: 5450–5455 [DOI] [PubMed] [Google Scholar]
- 30.Lesnefsky E. J., Minkler P., Hoppel C. L. 2009. Enhanced modification of cardiolipin during ischemia in the aged heart. J. Mol. Cell. Cardiol. 46: 1008–1015 [DOI] [PubMed] [Google Scholar]
- 31.Ji J., Kline A. E., Amoscato A., Samhan-Arias A. K., Sparvero L. J., Tyurin V. A., Tyurina Y. Y., Fink B., Manole M. D., Puccio A. M., et al. 2012. Lipidomics identifies cardiolipin as a mitochondrial target for redox therapy of brain injury. Nat. Neurosci. 15: 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparagna G. C., Chicco A. J., Murphy R. C., Bristow M. R., Johnson C. A., Rees M. L., Maxey M. L., McCune S. A., Moore R. L. 2007. Loss of cardiac tetralinoleoyl cardiolipin in human and experimental heart failure. J. Lipid Res. 48: 1559–1570 [DOI] [PubMed] [Google Scholar]
- 33.Moreira P. I., Santos M. S., Moreno A. M., Proenca T., Seica R., Oliveira C. R. 2004. Effect of streptozotocin-induced diabetes on rat brain mitochondria. J. Neuroendocrinol. 16: 32–38 [PubMed] [Google Scholar]
- 34.Han X., Yang J., Cheng H., Yang K., Abendschein D. R., Gross R. W. 2007. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: a shotgun lipidomics study. Biochemistry. 46: 6417–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepe S., Tsuchiya N., Lakatta E. G., Hansford R. G. 1999. PUFA and aging modulate cardiac membrane mitochondrial composition and Ca2+ activation of PDH. Am. J. Physiol. 276: H149–H158 [DOI] [PubMed] [Google Scholar]
- 36.Lee H. J., Mayette J., Rapoport S. I., Bazinet R. P. 2006. Selective remodeling of cardiolipin fatty acids in the aged rat heart. Lipids Health Dis. 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesnefsky E. J., Hoppel C. H. 2008. Cardiolipin as an oxidative target in cardiac mitochondria in the aged rat. Biochim. Biophys. Acta. 1777: 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparagna G. C., Lesnefsky E. J. 2009. Cardiolipin remodeling in the heart. J. Cardiovasc. Pharmacol. 53: 290–301 [DOI] [PubMed] [Google Scholar]
- 39.Blomstrand R., Svensson L. 1983. The effect of partially hydrogenated marine oils on the mitochondrial function and membrane phospholipid fatty acids in rat heart. Lipids. 18: 151–170 [DOI] [PubMed] [Google Scholar]
- 40.Berger A., German J. B. 1990. Phospholipid fatty acid composition of various mouse tissues after feeding α-linoleate or eicosatrienoate. Lipids. 25: 473–480 [DOI] [PubMed] [Google Scholar]
- 41.Høy C-E., Holmer G. 1990. Influence of dietary linoleic acid and trans fatty acids on the fatty acid profile of cardiolipins in rats. Lipids. 25: 455–459 [DOI] [PubMed] [Google Scholar]
- 42.Berger A., German J. B., Gershwin M. E. 1992. Effect of various dietary fats on cardiolipin acyl composition during ontogeny of mice. Lipids. 27: 605–612 [DOI] [PubMed] [Google Scholar]
- 43.Yamaoka S., Urade R., Kito M. 1988. Mitochondrial function in rats is affected by modification of membrane phospholipids with dietary sardine oil. J. Nutr. 118: 290–296 [DOI] [PubMed] [Google Scholar]
- 44.Yamaoka S., Urade R., Kito M. 1990. Cardiolipin molecular species in rat heart mitochondria are sensitive to essential fatty acid-deficient dietary lipids. J. Nutr. 120: 415–421 [DOI] [PubMed] [Google Scholar]
- 45.Yamaoka-Koseki S., Urade R., Kito M. 1991. Cardiolipins from rats fed different dietary lipids affect bovine heart cytochrome c oxidase activity. J. Nutr. 121: 956–958 [DOI] [PubMed] [Google Scholar]
- 46.Valianpour F., Wanders R. J., Overmars H., Vaz F. M., Barth P. G., van Gennip A. H. 2003. Linoleic acid supplementation of Barth syndrome fibroblasts restores cardiolipin levels: implication for treatment. J. Lipid Res. 44: 560–566 [DOI] [PubMed] [Google Scholar]
- 47.Bird S. S., Marur V. R., Sniatynski M. J., Greenberg H. K., Kristal B. S. 2011. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal. Chem. 83: 940–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bird S. S., Marur V. R., Stavrovskaya I. G., Kristal B. S. 2013. Qualitative characterization of the rat liver mitochondrial lipidome using LC-MS profiling and high energy collisional dissociation (HCD) all ion fragmentation. Metabolomics. 9 (1 Suppl.): 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bird S. S., Marur V. R., Stavrovskaya I. G., Kristal B. S. 2012. Separation of cis-trans phospholipid isomers using reverse phase LC with high resolution MC detection. Anal. Chem. 84: 5509–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kilkenny C., Browne W., Cuthill I. C., Emerson M., Altman D. G. 2010. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br. J. Pharmacol. 160: 1577–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stavrovskaya I. G., Narayanan M. V., Zhang W., Krasnikov B. F., Heemskerk J., Young S. S., Blass J. P., Brown A., Beal M. F., Friedlander R. M., et al. 2004. Clinically-approved heterocyclics act on a mitochondrial target and reduce stroke-induced pathology. J. Exp. Med. 200: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krasnikov B. F., Zorov D. B., Antonenko Y. N., Zaspa A. A., Kulikov I. V., Kristal B. S., Cooper A. J. L., Brown A. M. 2005. Comparative kinetic analysis reveals that inducer-specific ion release precedes the mitochondrial permeability transition. Biochim. Biophys. Acta. 1708: 375–392 [DOI] [PubMed] [Google Scholar]
- 53.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 54.Cequier-Sánchez E., Rodríguez C., Ravelo A. G., Zárate R. 2008. Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J. Agric. Food Chem. 56: 4297–4303 [DOI] [PubMed] [Google Scholar]
- 55.Preacher K. J. 2001. Calculation for the chi-square test: An interactive calculation tool for chi-square tests of goodness of fit and independence [Computer software]. Available from http://quantpsy.org
- 56.Turunen M., Olsson J., Dallner G. 2004. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 1660: 171–199 [DOI] [PubMed] [Google Scholar]
- 57.Chatgilialoglu C., Crich D., Komatsu M., Ryu I. 1999. Chemistry of acyl radicals. Chem. Rev. 99: 1991–2070 [DOI] [PubMed] [Google Scholar]
- 58.Ma B. J., Taylor W. A., Dolinsky V. W., Hatch G. M. 1999. Acylation of monolysocardiolipin in rat heart. J. Lipid Res. 40: 1837–1845 [PubMed] [Google Scholar]
- 59.Xu Y., Kelley R. I., Blanck J. J., Schlame M. 2003. Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 278: 51380–51385 [DOI] [PubMed] [Google Scholar]
- 60.Wolff L., Entressangles B. 1991. Compositional changes of fatty acids in the 1(1″)- and 2(2″)-positions of cardiolipin from liver, heart, and kidney mitochondria of rats fed low-fat diet. Biochim. Biophys. Acta. 1082: 136–142 [DOI] [PubMed] [Google Scholar]
- 61.Lenaz G. 2001. A critical appraisal for the mitochondrial coenzyme Q pool. FEBS Lett. 509: 151–155 [DOI] [PubMed] [Google Scholar]
- 62.Aoun M., Fouret G., Michel F., Bonafos B., Ramos J., Cristol J-P., Carbonneau M-A., Coudray C., Feillet-Coudray C. 2012. Dietary fatty acids modulate liver mitochondrial cardiolipin content and its fatty acid composition in rats with non alcoholic fatty liver disease. J. Bioenerg. Biomembr. 44: 439–452 [DOI] [PubMed] [Google Scholar]
- 63.Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., Langer T. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338: 815–818 [DOI] [PubMed] [Google Scholar]
- 64.Malhotra A., Xu Y., Ren M., Schlame M. 2009. Formation of molecular species of mitochondrial cardiolipin. 1. A novel transacylation mechanism to shuttle fatty acids between sn-1 and sn-2 positions of multiple phospholipid species. Biochim. Biophys. Acta. 1791: 321-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Y., Malhorta A., Ren M., Schlame M. 2006. The enzymatic function of tafazzin. J. Biol. Chem. 281: 39217–39224 [DOI] [PubMed] [Google Scholar]
- 66.Taylor W. A., Hatch G. M. 2009. Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1). J. Biol. Chem. 284: 30360–30371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiebish M. A., Yang K., Sims H. F., Jenkins C. M., Liu X., Mancuso D. J., Zhao Z., Guan S., Abendschein D. R., Han X., et al. 2012. Myocardial regulation of lipidomic flux by cardiolipin synthase: setting the beat for bioenergetic efficiency. J. Biol. Chem. 287: 25086–25097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J., Liu X., Wang H., Zhang W., Chan D. C., Shi Y. 2012. Lysocardiolipin acyltransferase 1 (ALCAT1) controls mitochondrial DNA fidelity and biogenesis through modulation of MFN2 expression. Proc. Natl. Acad. Sci. USA. 109: 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schlame M. 2009. Formation of molecular species of mitochondrial cardiolipin 2. A mathematical model of pattern formation by phospholipid transacylation. Biochim. Biophys. Acta. 1791: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sparagna G. C., Johnson C. A., McCune S. A., Moore R. L., Murphy R. C. 2005. Quantitation of cardiolipin molecular species in spontaneously hypertensive heart failure rats using electrospray ionization mass spectrometry. J. Lipid Res. 46: 1196–1204 [DOI] [PubMed] [Google Scholar]
- 71.Zhang L., Bell R. J., Kiebish M. A., Seyfried T. N., Han X., Gross R. W., Chuang J. H. 2011. A mathematical model for the determination of steady-state cardiolipin remodeling mechanisms using lipidomic data. PLoS ONE. 6: e21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.