Abstract
Several studies have shown that housing conditions and environmental exposure to a series of stimuli lead to behavior improvement in several species. While more works have been focused on illustrating changes of the proteome and transcriptome following enriched environment exposure in mice, little has been done to understand changes in the brain metabolome in this paradigm due to the complexity of this type of analysis. In this paper, lipidomics focused on phospholipids and gangliosides were conducted for brain tissues of mice exposed to enriched or impoverished conditions. We optimized previously reported method and established a reliable relative comparison method for phospholipids and gangliosides in brain tissue using prefractionation with weak anion exchange cartridge. We used liquid chromatography mass spectrometry to explore metabolic signatures of the cerebral cortex and hippocampus after confirming the animals had significant memory differences using the fear conditioning paradigm and brain immunohistochemistry. Although both cerebral cortex and hippocampus regions did not show major alterations in ganglioside composition, we found significant differences in a series of phospholipids containing 22:6 fatty acid in the prefrontal cortex, indicating that environmental enrichment and impoverished housing conditions might be a relevant paradigm to study aberrant lipid metabolism of docosahexaenoic acid consumption. Our study highlights the hypothesis-generating potential of lipidomics and identifies novel region-specific lipid changes possibly linked not only to change of memory function in these models, but also to help us better understand how lipid changes may contribute to memory disorders.
Keywords: liquid chromatography, mass spectrometry, phospholipid, ganglioside, docosahexaenoic acid, phospho-cAMP response element binding protein
Enriched environment is a paradigm where animals are introduced to novel, complex, and stimulating surroundings that have been known to promote structural changes in the brain and to enhance learning and memory performance in rodents (1). Recent studies have confirmed that exposure to enriched conditions (ECs) results in altered expression of various neurochemical markers (2, 3) as well as neurophysiological changes such as enhanced synaptic plasticity (4). For instance, environmental enrichment is reported to modify the cAMP-dependent protein kinase (PKA)-dependence of long-term potentiation as well as improving hippocampal-dependent memory (5). However, the exact cellular and molecular basis for enrichment-induced structural or neurophysiological modifications remains unknown. Altered intracellular signaling and modified synaptic strength may support enrichment-induced cognitive changes. Microarray studies following short- and long-term environmental enrichment exposure have identified genes involved in DNA/RNA synthesis, neuronal signaling, neuronal growth/structure, cell death, and protein processing (6) as possible targets for neuronal adaptations in response to environmental enrichment. In addition, proteomic studies following environmental enrichment exposure have identified many regulated proteins that contribute to energy metabolism, cytoplasmic organization/biogenesis, and signal transduction processes (7). However, a global assessment of alterations of brain metabolite expression remains to be undertaken.
Metabolomics has particular relevance to drug discovery and development, as metabolites often mirror the end result of genomic and protein perturbations in various diseases and are most closely associated with phenotypic changes. Current metabolomics research involves the identification and quantification of hundreds to thousands of small molecular mass metabolites (<1,500 Da) in cells, tissues, or biological fluids. The aim of such studies is typically to identify disease pathways, to discover new diagnosis biomarkers, to understand the mechanism of action of therapeutic compounds, and to uncover the pharmacodynamics and kinetic markers of drugs in patients and in preclinical in vivo and in vitro models (8).
Lipidomics is a lipid-targeted metabolomics approach aiming at comprehensive analysis of lipids in biological systems (9–11). Recently, lipid profiling, or lipidomic research, has captured increased attention due to the well-recognized roles of lipids in numerous human diseases such as diabetes, obesity, atherosclerosis, and Alzheimer's disease (AD) (12–18). Investigating lipidomic biochemistry will not only provide insights into the specific roles of lipid molecular species in health and disease, but will also assist in identifying potential biomarkers for establishing preventive or therapeutic approaches for human health (9, 12, 19, 20). Recent technological advancements in mass spectrometry and improvements in chromatographic techniques have led to the rapid expansion of this research field (10, 11, 21).
Hence, to better understand the degree and patterns of altered metabolites changed by environmental enrichment, we have optimized the previous methodology and applied reliable and sensitive lipidomic methods to analyze the impact of housing environment on the cortical and hippocampal lipidome.
EXPERIMENTAL PROCEDURES
Chemicals and reagents
Most of the reagents used in the experiments were of analytical grade and were purchased from Wako Pure Chemicals Co. (Osaka, Japan). Sulfatides from bovine brain, ethylenediaminetetraacetic acid (EDTA), and 28% aqueous ammonia were purchased from Sigma-Aldrich (Dorset, UK). Deionized water was obtained from a Milli-Q water system (Millipore, Milford, MA). Ganglioside mixture ammonium salt was purchased from Wako Pure Chemicals Co.
Animals
Mice were treated humanely and the experiment received prior ethical approval in accordance with Eisai company policy. All procedures were performed in the animal facility accredited by the Center for Accreditation of Laboratory Animal Care and Use Japan Health Science Foundation. C57Bl6 mice (female, 5 weeks old) were obtained from Charles River, Japan and initially housed in groups of eight for 1 week in standard cages (300 × 195 × 135 mm) prior to the beginning of the study. Mice were given ad libitum access to food and water, and were kept on a 12 h light/dark cycle. Upon commencement of the enrichment study, the enriched group was placed in a large (900 × 900 × 400 mm) cage containing regular food pellets spread randomly in the bedding, two suspended drinking bottles, and a sawdust base in addition to several novel objects including tubes, running wheels, platforms, beams, ladders, etc. These enrichment objects were rearranged, and some were removed and replaced by novel items on a weekly basis for a 5 week period. This approach was to maintain a degree of novelty in order to stimulate learning about objects and their relationships. The nonenriched groups were placed in a cage (275 × 125 × 145 mm), with a food dispenser, suspended drinking bottle, and sawdust base bedding with no additional stimuli. The cage covers were made of plexiglass with perforated holes for aeration. Both EC and impoverished condition (IC) groups were placed in their corresponding environments for a total experimental period of 5 weeks (see supplementary Fig. I for enriched environment cage view) and cages were cleaned on a regular basis.
Fear conditioning
Fear conditioning is a behavioral paradigm in which mice learn to predict aversive events and is used to evaluate memory function in rodents. It is thought to be a form of learning in which an aversive stimulus (e.g., an electrical shock) is associated with a particular neutral context (e.g., a cage). The association of the cage with the electric shock results in the expression of a fear response to the originally neutral stimulus that manifests itself with increased freezing or immobility that is associated with memory. Fear conditioning is thought to depend upon an area of the brain called the amygdala, but it also involves the hippocampus for the reception of affective impulses and the cortex for memory retention (22).
The fear conditioning apparatus consisted of a transparent square cage [300 × 240 × 210 mm (from grid to ceiling)] with a grid floor wired to a shock generator. Mice were placed individually in the cage for 120 s and then three 0.5 mA electric shocks were given separated by a 60 s interval followed by a period of 120 s. Mice were then returned to their respective housing environment. Twenty-four hours later, mice were put in the same apparatus again (without any electric shock) and freezing time was automatically recorded with a computerized data collection system that can detect animal movements.
Tissue harvest
At the end of the 5 weeks of training and behavioral testing, EC and IC mice were decapitated with cervical dislocation in accordance with local ethical guidelines. All efforts were made to minimize the suffering and number of animals used in each experiment. Brains were rapidly dipped into ice-cold normal saline. The hippocampus and cortex were dissected free from surrounding regions, quickly snap-frozen in liquid nitrogen, and stored at −80°C until use.
Separate sets of mice of the same strain and age and housed the exact same way were used to determine the housing effects on the proliferation of neural progenitor cells. Fifty milligrams per kilogram of 5′-bromo-2′-deoxyuridine (BrdU) (Sigma, St. Louis, MO), a thymidine analog that is used to label proliferating cells in brain, was intraperitoneally administered three times at 4 h intervals on the last day of the 5 week housing period. After BrdU injection, the animals were euthanized, under deep anesthesia with 1 ml of sodium-pentobarbital (Abbott Laboratories, Chicago, IL), by transcardial perfusion with saline followed by 4% paraformaldehyde in phosphate-buffered saline. The brain was removed, placed in 4% paraformaldehyde in phosphate-buffered saline overnight at 4°C, cryoprotected in 25% sucrose, and coronally sectioned at 40 μm for immunohistochemical (IHC) analysis.
Immunohistochemistry
Free-floating sections of hippocampus were stained by BrdU for determination. Sections were incubated in 2% H2O2 for 30 min to eliminate endogenous peroxidase. DNA denaturation was conducted by incubation for 3 h in 50% formamide/2× saline sodium citrate (SSC) at 60°C, followed by 2× SSC rinse. Sections were incubated for 30 min in 2 N HCl at 37°C, and then for 10 min in borate buffer (pH 8.5). They were blocked with 3% normal rabbit serum (Vector Laboratories, Burlingame, CA) in 0.2% Triton X-100, and incubated with rat anti-BrdU antibody (1:500; Accurate Chemical and Scientific Corporation, Westbury, NY) overnight at 4°C. They were incubated for 1 h with biotinylated anti-rat IgG antibody (1:200; Vector Laboratories), followed by incubation with avidin-biotin complex (ABC Elite kit, Vector Laboratories). Cells were visualized with diaminobenzidine (DAB) using a DAB substrate kit (Vector Laboratories).
For phospho-cAMP response element binding protein (p-CREB) staining, sections were incubated in 2% H2O2 for 30 min, blocked with 3% normal goat serum (Vector Laboratories), and incubated with rabbit anti p-CREB antibody (1:200; Cell Signaling Technology, Beverly, MA) overnight at 4°C. They were further incubated for 1 h with biotinylated anti-rabbit IgG antibody (1:200; Vector Laboratories) followed by the incubation with avidin-biotin complex. And cells were visualized with DAB.
Sample preparation for phospholipidomics
The deep-frozen tissue samples (approximately 30 mg wet weight hippocampus and 15 mg wet weight cerebral cortex) were thawed on ice and were homogenized in 10 times volume of sodium phosphate buffer containing 0.01% butylated hydroxytoluene (BHT) and protease inhibitors, Roche complete protease inhibitor cocktail (Roche Diagnostics Ltd., Mannheim, Germany), and stored at −80°C until use.
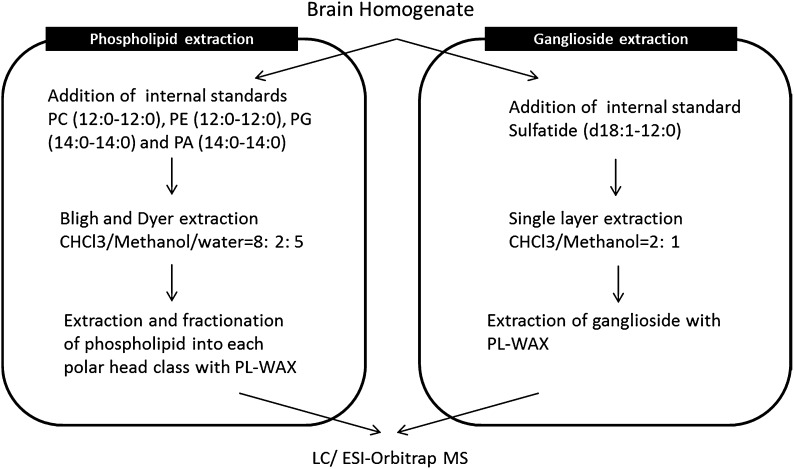
Phospholipid measurement was performed as described previously (16). In brief (see Fig. 1 for the protocol), approximately 25 μl of homogenate (corresponding to 34 μg total protein) was spiked with 200 μl of internal standard (IS) solution (100 ng/ml of phosphatidylcholine (PC) (12:0-12:0), phosphatidylethanolamine (PE) (12:0-12:0), phosphatidylglycerol (PG) (14:0-14:0), and phosphatidic acid (PA) (14:0-14:0) in methanol). Chloroform (CHCl3)/methanol (8:1) (1.8 ml) containing 10% BHT as anti-oxidant and 1.0 ml of sodium phosphate buffer containing 18% sodium chloride (NaCl) aqueous solution were added to the sample, and mixed well. The solution was centrifuged for 10 min at 2,000 g. The lower organic phase was transferred to a new tube and extracted again with CHCl3. The combined organic solution was evaporated to dryness under a nitrogen gas stream at 40°C, the residue was reconstituted in CHCl3 and the solution was used as a sample for fractionation of lipid classes. A 140 mg bed of PL-WAX (Agilent Technologies, Palo Alto, CA) packed in a Pasteur pipette, was sequentially prewashed with water, 1 M hydrochloride (HCl), water, 0.1 M sodium hydroxide (NaOH), water, methanol, acetic acid, methanol, and CHCl3. The sample was applied to the PL-WAX column and stepwise elution was begun with CHCl3 (3 × 1 ml) to wash out neutral lipids. Among the bound lipids, PC and lyso-phosphatidylcholine were eluted with CHCl3/methanol (95:5; 3 × 1 ml), followed by the elution of PE with CHCl3/methanol (1:1) (3 × 1 ml). The acidic lipids [e.g., PA, phosphatidylserine (PS), PG, phosphatidylinostol (PI), and cardiolipin] were then eluted with CHCl3/methanol/28% aqueous ammonia/acetic acid (50:25:1.17:0.35) (3 × 1 ml). Each eluate was dried under nitrogen gas and the fractionated samples were stored at −30°C until use. When required, each fraction was redissolved in 100 μl of methanol for LC/ESI-MS analysis.
Fig. 1.
Workflow for extraction and fractionation of phospholipids and gangliosides from a mouse brain.
Sample preparation for ganglioside measurement
Approximately 20 μl of homogenate (corresponding to 27 μg protein) was spiked with 30 μl of internal control solution [500 nM of sulfatide (d18:1-12:0) in methanol] and 1 μl of BHT as anti-oxidant reagent. The gangliosides were extracted by means of the optimized previous method (23). Briefly (see Fig. 1 for the protocol), gangliosides were extracted with single layer extraction using 1 ml of CHCl3 /methanol (2:1) and 0.8 ml of CHCl3 :methanol:water containing 1 ml of 10% BHT. The combined organic solution was evaporated to dryness under a nitrogen gas stream at 40°C, the residue was reconstituted in methanol and the solution was used as a sample for fractionation of lipid classes. The sample was applied to the PL-WAX column and stepwise elution was begun with methanol (3 × 1 ml) to wash out neutral lipids and phospholipids containing plus-charged nitrogen, such as PC and PE. Among the bound lipids, acidic glycolipid mixtures containing gangliosides and sulfatides were eluted with 5% ammonium hydroxide in methanol (3 × 1 ml). Ganglioside fraction was dried under nitrogen gas and the fractionated samples were stored at −30°C until use. When required, each fraction was redissolved in 100 μl of methanol for LC/ESI-MS analysis.
Comprehensive analysis of phospholipids, gangliosides, and sulfatides with LC/MS
Phospholipids, gangliosides, and sulfatides were measured with a Shimadzu 20AD system with a SIL-20AC auto-sampler, a CTO-20A column oven, and an LTQ Orbitrap mass spectrometer (ThermoFisher, San Jose, CA) with an ESI probe. For phospholipid analysis, a total of 2.5 μl of fraction 2 or 10 μl of other fractions was injected onto a 1.0 mm internal diameter × 150 mm length Capcell Pack C18 column (Shiseido, Tokyo, Japan), prewashed with 2 mM EDTA solution at a flow rate of 50 μl/min, with a total run time of 80 min. The gradient used consisted of solvent A (water:acetonitrile:methanol = 4:4:2 containing 0.1% formic acid and 0.028% aqueous ammonia) and solvent B (isopropanol:methanol = 8:2 containing 0.1% formic acid and 0.028% aqueous ammonia). For ganglioside and sulfatide analysis, a total of 10 μl of sample was injected onto a 2.0 mm internal diameter × 50 mm length Capcell Pack C18 column at a flow rate of 150 μl/min with a total run time of 35 min. In each measurement, gradient elution was applied; the detail gradient programs and other LC conditions are listed in supplementary Table I. The mass spectrometer was operated in the negative ion mode. The spray voltage was set at −4,500 kV. A cycle of one full Fourier transform scan mass spectrum (m/z 400–2,000, resolution of 30,000) followed by three data-dependent MS/MS and MS/MS/MS acquired in the linear ion trap or C-trap with normalized collision energy (setting of 35%) was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the XCalibur data system.
Standard sample preparation
Ganglioside and sulfatide mix standard stock solutions (1 mg/ml) were prepared in methanol, and were used for the development of prefractionation protocol, the optimization of chromatographic separation in LC/MS, and the identification of gangliosides and sulfatides.
Data analysis and statistical analysis
The mass spectrometric data were acquired using Xcalibur, and the initial metabolomics profiling was performed using in-house-developed Mass ++ data analysis software to obtain a peak list, align retention times, and obtain peak areas normalized with IS or total ion counts. The obtained data matrix was used for statistical analysis. The P values and fold changes of lipids were compared between groups using the R software package or a Student's t-test was performed to evaluate group differences. A difference of P < 0.05 was regarded as significant.
RESULTS AND DISCUSSION
Behavior analysis
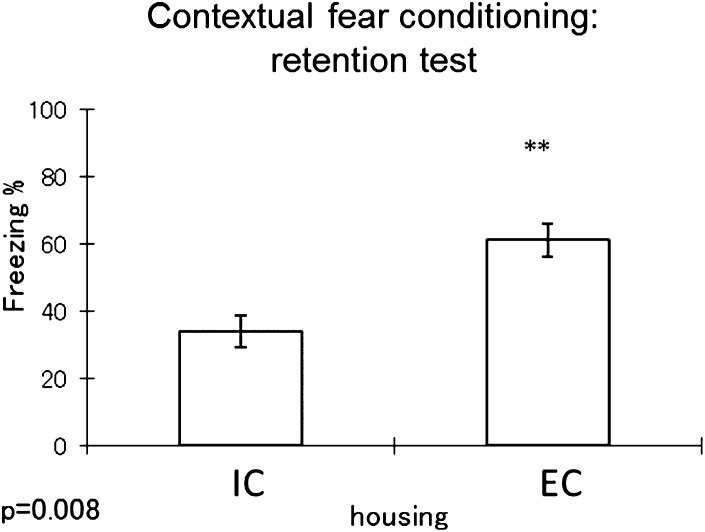
Prior to euthanizing mice and to conduct immunochemistry and metabolomic analysis of changes in the brain induced by the housing environment, we performed a series of experiments to confirm that the environmental paradigm the mice were put in was sufficient to cause changes in various types of behavior that relate to depression and anxiety, and most importantly to confirm that the model induced significant changes in short-term memory as reported in previous studies (24). One well-established model we used to confirm differences in memory acquisition was the fear conditioning paradigm (5, 24). We chose this test because several brain regions are involved in the learning process that includes the amygdala and the hippocampus, as well as the cortex (25). As shown in Fig. 2, mice that were housed in ICs showed a significant reduction in freezing time, hinting at memory deficiency compared with mice housed in ECs. This told us that the 5 week housing condition was sufficient to induce changes in brain regions involved in memory retention. After getting this confirmation, mice were euthanized as indicated, and brain regions collected for IHC and metabolomic analysis.
Fig. 2.
Contextual fear conditioning test result (n = 8 per group). The y axis represents percent of freezing time. Higher percent is associated with better memory retention. Student t -test result is shown on the figure. ** P < 0.01.
Immunohistochemistry
It is suggested that BrdU has been a principal marker for mitotic cells in studies of adult neurogenesis (26). Endogenous cAMP response element binding protein (CREB) activation has been shown to promote neuronal survival in the dentate gyrus (DG) (27). Several reports implicate CREB activation in the formation of long-term memory in mice (28). In addition, it is reported that environmental enrichment affects spatial memory and hippocampal CREB immune reactivity (29).
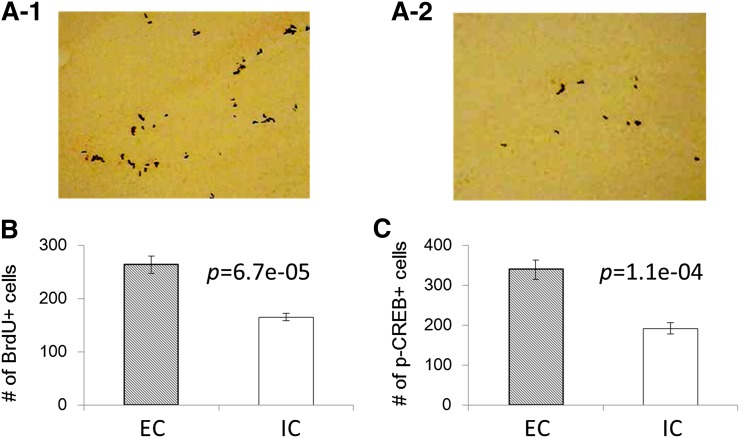
To elucidate the change of cell proliferation and memory formation in the brain after different housing in this study, we performed BrdU and p-CREB staining of the DG in the dorsal hippocampus (Fig. 3). Figure 3A shows the representative staining and Fig. 3B shows the summary of IHC data for BrdU. As a result, we found the increase of BrdU staining, which is linked to neuronal progenitor cell proliferation, in EC mouse brain compared with IC (P = 6.7e-5, Fig. 3A, B). In addition, it was revealed that p-CREB, which is linked to memory formation, was increased in EC mouse brain compared with IC (P = 1.1e-4, Fig. 3C). These results also suggested that the 5 week housing condition was sufficient to induce changes in brain regions involved in memory retention. Then we performed metabolomics analysis using the hippocampus and cortex of mice to understand any changes associated with these different housing conditions.
Fig. 3.
Immunohistochemistry (BrdU staining) of DG. The upper two pictures [ECs (A-1), ICs (A-2)] show the representative IHC photos and the lower graph shows the summary of IHC data (B). Increased BrdU staining is linked to neuronal progenitor cell differentiation. Summary of IHC data (p-CREB staining) of DG (C). Increased p-CREB staining in the DG as reported in the literature is linked to increased memory formation.
Extraction and LC/MS analysis of phospholipids from tissue samples
Depending on the lipids of interest, normal-phase liquid chromatography (NPLC) or reverse-phase liquid chromatography (RPLC) is usually applied to phospholipid measurement. NPLC and RPLC columns provide characteristic lipid separations. For instance, the separation of PLs on a NPLC column is mainly achieved based on the difference in the polar head-group of the PLs, whereas separation on a RPLC column relies on the difference of chain length or of the number of double bonds of fatty acids (i.e., essentially on the lipophilicity) (16). Aiming at comprehensive and relative comparison of multiple PL classes via two-dimensional (2D) separation prior to analysis, we developed a procedure for 2D phospholipidomics that would separate low-abundance acidic PLs from high-abundance neutral lipids and cationic PLs, such as triacylglycerols (TAGs) and PCs with high throughput. In this study, we applied this highly comprehensive method to identify altered metabolites underlying different environment. EDTA solution was used as a column prewashing solution in LC/MS to prevent the peak tailing before the analysis of PLs, as previously reported (16).
To clarify the reproducibility of our 2D-phospholipidomic methodology in biological samples, we conducted a phospholipidomic analysis in triplicate using a mouse brain (see supplementary Figs. II and III for representative LC-MS profiles and MS/MS spectra). The recovery and repeatability are summarized in supplementary Table II. Four ISs eluted in different fractions were chosen and they were never found in biological samples: PC (12:0-12:0) was used as IS of fraction 2, PE (12:0-12:0) was for fraction 3, and PG (14:0-14:0) and PA (14: 0-14: 0) were selected as ISs of fraction 4, and many phospholipid peaks were detected in mouse brain samples. These ISs were reproducibly recovered over 80% at better than 15% of the coefficient of variation (CV). Moreover, fourteen brain-endogenous PLs among six different polar head-groups in the range of fractions 2–4 were evaluated about repeatability of measurements with four ISs. Major brain phospholipids could be separated into three fractions based on polar head-groups and detected with good reproducibility [CV(%): better than 12%] after the IS normalization. Because these results were reproducible, this reliable relative comparison method was used for comprehensive 2D-phospholipidomic analysis of brain lipids.
Extraction and LC/MS analysis of gangliosides from tissue samples
It was supposed that other highly abundant gangliosides can mask many low-abundant gangliosides of interest. Therefore, lipid class separation after extraction can be useful for isolation of targeted lipids. Several research groups have utilized a procedure that can separate low-abundant gangliosides from neutral lipids with off-line sample preparation, followed by RPLC-ESI MS analysis. However, there is no quantitative and comprehensive method in the literature using 2D ganglioside separation prior to MS analysis.
The recovery of gangliosides was very low in the liquid-liquid extraction, which was used for phospholipids. The ganglioside analysis was conducted separately from phospholipid analysis. As reported previously (23, 30), anionic extraction cartridges are probably suitable for the separation of ganglioside classes containing sialic acid from neutral and basic compounds, as well as the prefractionation of PLs. We applied the PL-WAX method, which was used as PL class separation, to the prefractionation of gangliosides and sulfatides. PL-WAX gave the reproducible elution of ganglioside and sulfatide classes with 5% ammonium hydroxide in methanol. Sulfatide (d18:1, 12:0) was chosen as the internal control, and the recovery and repeatability are summarized in supplementary Table III. Peak area ratio of each class of gangliosides was very reproducible. All ganglioside standards were recovered over 50% at better than 10% of CV (%). The gangliosides have multiple carbohydrates with different numbers of carboxylic acid moieties, which may require use of multiple ISs. However, the availability of commercial isotope-labeled standards is limited and the costs can be prohibitive to large scale use. In addition, preparing multiple isotope-labeled standards for large scale lipidomics is challenging, given the diverse and complex structures of the lipids. Therefore, from the practical point of view, sulfatide (d18:1-12:0) was selected as the internal control for all gangliosides. Then, gangliosides were analyzed in triplicates using a mouse brain to confirm the reproducibility of real biological samples (see supplementary Figs. IV and V for representative LC-MS profiles and MS/MS spectra). Table 1 shows the reproducibility result of each class of gangliosides, which were better than 13% of CV (%). Because these results are reproducible for comprehensive ganglioside measurement, we applied this method to brain ganglioside analysis.
TABLE 1.
Reproducibility of endogenous gangliosides from mouse brains (n = 3)
| Compound | RT (min) | Theoretical Value |
Measured Value (m/z) | Internal Control Ratio (mean ± SD, n = 3) | CV (%) | |
| Exact Mass | m/z | |||||
| GM1, d18:1-18:0 | 4.9 | 1545.8767 | 1544.8694 | 1544.8676 | 11.2 ± 0.2 | 2.0 |
| GM1, d18:1-20:0 (d20:1-18:0) | 6.7 | 1573.908 | 1572.9007 | 1572.8988 | 2.1 ± 0.0 | 1.7 |
| GM2, d18:1-18:0 | 5.2 | 1383.8238 | 1382.8166 | 1382.8147 | 0.56 ± 0.03 | 5.8 |
| GM2, d18:1-20:0 (d20:1-18:0) | 7 | 1411.8551 | 1410.8479 | 1410.8451 | 0.085 ± 0.009 | 11.0 |
| GM3, d18:1-18:0 | 5.4 | 1180.7445 | 1179.7372 | 1179.735 | 0.96 ± 0.02 | 1.7 |
| GM3, d18:1-20:0 (d20:1-18:0) | 7.3 | 1208.7758 | 1207.7685 | 1207.7667 | 0.11 ± 0.01 | 7.9 |
| GD1, d18:1-18:0 | 4.3 | 1836.9721 | 917.4788 | 917.4773 | 38.6 ± 2.0 | 5.2 |
| GD1, d18:1-20:0 (d20:1-18:0) | 5.8 | 1865.0034 | 931.4944 | 931.4928 | 9.9 ± 0.4 | 4.1 |
| GD2, d18:1-18:0 | 4.4 | 1674.9193 | 836.4523 | 836.4528 | 0.029 ± 0.003 | 12.1 |
| GD3, d18:1-18:0 | 4.9 | 1471.8399 | 734.9127 | 734.9118 | 0.18 ± 0.01 | 7.5 |
| GD3, d18:1-20:0 (d20:1-18:0) | 6.7 | 1499.8712 | 748.9283 | 748.9272 | 0.024 ± 0.003 | 11.9 |
| GT1, d18:1-18:0 | 3.9 | 2128.0675 | 1063.0265 | 1063.0243 | 11.3 ± 0.3 | 2.7 |
| Sulfatide, d18:1-22:0 (d20:1-20:0) | 10.6 | 863.6156 | 862.6084 | 862.6067 | 6.0 ± 0.3 | 4.5 |
| Sulfatide, d18:1-24:0 (d20:1-22:0) | 14.1 | 891.6469 | 890.6397 | 890.6385 | 15.7 ± 0.4 | 2.4 |
Lipid measurement of mouse brain tissues
Lipids have multiple roles, functioning as membrane components, energy stores, cell-structural components, and regulators of complex signaling pathways as well as serving as ligands and mediators of various protein-protein interactions and cell-cell interactions. Therefore, defects in lipid metabolism have been linked to numerous human diseases that include AD and Parkinson's disease (9, 12).
Gangliosides are sialic acid-containing glycosphingolipids that are expressed in the outer leaflet of the plasma membrane of all vertebrate cells (31). Gangliosides are involved in a variety of functions, including serving as antigens, receptors for bacterial toxins, mediators of cell adhesion, and mediators and modulators of signal transduction (32). Gangliosides are most abundant in the nervous system. Their expression in the nervous system is cell specific and developmentally regulated, and their abundance and species undergo dramatic changes during differentiation of the cell (33–35).
MS/MS spectra in positive mode provide good information to identify ceramide core. In this experiment, we measured lipids using negative ionization mode due to the lack of sensitivity of positive ionization mode for sphingolipids in Orbitrap. So we could not determine the exact structural information of ceramide core from MS/MS and MS/MS/MS spectrum. We only semi-identified all gangliosides using theoretical m/z, retention time of mixed ganglioside standards, and limited MS/MS information (supplementary Figs. II and III). To elucidate if ICs or ECs affect ganglioside metabolism in mouse brain, we compared gangliosides in mice exposed to each condition. A total of 20 gangliosides (GM1, 4; GM2, 2; GM3, 4; GD1, 5; GD1, 1; GD3, 2; and GT1, 2) and fourteen sulfatides were detected in mouse brain and compared between ICs and ECs (supplementary Table IV). Although ganglioside metabolism is highly related to neuronal functions (33–35), we could not see any difference between mouse brains of ICs and ECs, suggesting that housing condition does not affect brain gangliosides, and that the behavior change seen in this study does not correlate with ganglioside metabolism.
Phospholipids not only contribute to the structural and physical properties of biological membranes but participate actively in cell signaling. There are also several reports showing that brain phospholipids are highly linked to brain function, such as blood-brain barrier (36), neurotransmission (37–39), or ion channels (40, 41). These functions are related to the relative concentrations of various phospholipid classes and the degree of unsaturation of their fatty acids, which varies greatly among different cell types and their subfractions (42). 2D phospholipid separation was applied to identify if there are any alterations of PLs following exposure to different environments. Thousands of peaks were detected from the hippocampus and cerebral cortex. The number of detected peaks was approximately 2-fold more than conventional one-dimensional analysis (one-dimensional, 1,073 peaks; 2D, 2,037 peaks), suggesting that the combination of NPLC and RPLC was more suitable to find lower abundant molecules in mouse brain, because high-abundance phospholipids may mask many low-abundance lipids of interest.
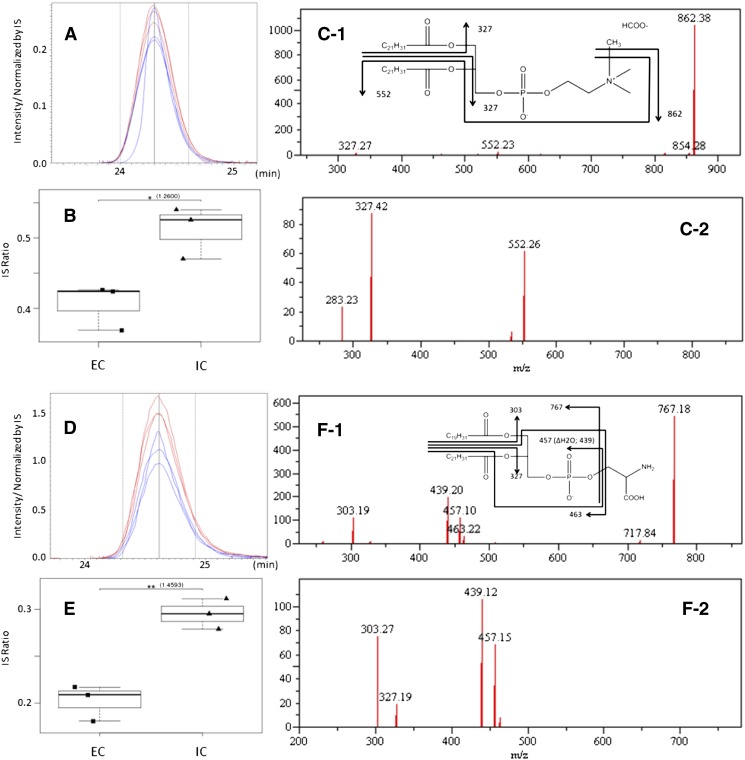
Although we could not see any significant changes in ganglioside analysis (supplementary Table IV), we could detect changes in a total of 38 phospholipids in the hippocampus and cerebral cortex [hippocampus, five phospholipids in fraction 2 (probably PCs); cerebral cortex, seven phospholipids in fraction 2, thirteen in fraction 3 (probably PEs), and thirteen in fraction 4 (probably PAs, PGs, Pis, or PSs); see supplementary Table V]. We tried to identify those peaks using the information of retention time, exact mass, MS/MS, and MS/MS/MS spectrum. We were able to identify six phospholipid candidate structures which were significantly changed after environmental enrichment based on t-test and rate of change (rate of change > 25% and P < 0.05) (Table 2). Figure 4 shows the examples for the chromatograms, box plots of peaks comparison between both housing conditions, and MS/MS spectra with lipid chemical structures. Interestingly, all phospholipids, which significantly changed and were identified as candidate structures with MS/MS and MS/MS/MS spectra, contained C22:6 fatty acid, such as PC (22:6-22:6), PE (22:6-22:6), PE (20:4-22:6), PS (22:6-22:6), PS (20:4-22:6), and PS (18:1-22:6). All of them were significantly decreased in ECs compared with ICs. In addition, these phospholipids were changed in the cerebral cortex but not in the hippocampus.
TABLE 2.
Statistical results of phospholipidomics analyzed by R software
| Sample Type | Measured |
Theoretical (m/z) | ECs (mean ± SD) | ICs (mean ± SD) | Rate of Change (ICs vs. ECs) | P | ||
| RT (min) | m/z | Identification by MS/MS | ||||||
| Cerebral cortex | 24.3 | 922.570 | PC (1-acyl-2-acyl 22:6-22:6) | 922.560 | 0.41 ± 0.03 | 0.51 ± 0.04 | 26% up | <0.05 |
| Cerebral cortex | 25.6 | 810.516 | PE (1-acyl-2-acyl 20:4-22:6) | 810.507 | 0.59 ± 0.03 | 0.83 ± 0.08 | 42% up | <0.01 |
| Cerebral cortex | 24.9 | 834.507 | PE (1-acyl-2-acyl 22:6-22:6) | 834.507 | 1.00 ± 0.05 | 1.30 ± 0.13 | 31% up | <0.05 |
| Cerebral cortex | 31.7 | 818.535 | PS (1-alk-2-acyl 18:1-22:6) | 818.534 | 0.16 ± 0.01 | 0.23 ± 0.01 | 42% up | <0.001 |
| Cerebral cortex | 24.6 | 854.501 | PS (1-acyl-2-acyl 20:4-22:6) | 854.497 | 0.20 ± 0.02 | 0.30 ± 0.02 | 46% up | <0.01 |
| Cerebral cortex | 24.0 | 878.499 | PS (1-acyl-2-acyl 22:6-22:6) | 878.497 | 0.87 ± 0.09 | 1.15 ± 0.05 | 32% up | <0.01 |
Showing the peak lists which were significantly changed between ICs and ECs, and were identified candidate structures with MS/MS and MS/MS/MS spectra, based on Student's t-test P value and rate of change (rate of change > 25% and P < 0.05).
Fig. 4.
A: Chromatogram of m/z 922.570 at 24.3 min [PC (22:6-22:6)]. B: Categorized box and whisker plot of PC (22:6-22:6). C-1: MS/MS spectrum of m/z 922.570 at 24.3 min and fragmentation pattern with chemical structure of PC (22:6-22:6). C-2: MS/MS/MS spectrum of m/z 862.38 of (C-1). D: Chromatogram of m/z 854.501 at 24.6 min [PS (20:4-22:6)]. E: Categorized box and whisker plot of PS (20:4-22:6). F-1: MS/MS spectrum of m/z 854.501 at 24.6 min and fragmentation pattern with chemical structure of PS (20:4-22:6). F-2: MS/MS/MS spectrum of m/z 767.18 of (F-1).
The brain is particularly enriched with the PUFA docosahexaenoic acid (DHA) (22:6n-3) (43), but the brain does not have de novo synthesis systems for n-3 PUFAs, indicating that it must be taken up from preformed DHA present in plasma (44) through the blood-brain barrier to enter the brain (45–47). Upon its entry, DHA is activated by an acyl-CoA synthetase (48, 49) and is esterified via an acyl-CoA transferase to the sn-2 position of phospholipids. Phospholipid DHA (sn-2 esterified) is the main storage form of DHA in the brain and is released by phospholipase A2 when needed by brain cells (44). In addition, DHA helps maintain membrane fluidity in the brain (50), promotes cell survival (51–53), acts as a secondary messenger via coupling to neuroreceptors (54–56), and is converted, via oxygenation, to a variety of signaling molecules, some of which have potent anti-inflammatory properties (57, 58). Thus, it is not surprising that 22:6n-3 is important in neural development (59, 60) and the alterations in DHA could reflect underlying metabolic changes taking place during a variety of neurological disorders, including stroke (61), AD (62), and major depression (63). It is also reported that insufficiency of DHA could impair neuronal plasticity and repair of the brain following injuries. Indeed, DHA deficiency in the brains of patients with AD has been associated with impaired learning and memory (64).
Recently, Holguin, Huang, and Wurtman (65) reported that although chronic DHA administration did not affect the performance of EC rats in the Morris water maze, its administration in IC rats restored their impaired memory, indicating that lipid metabolism related with DHA is linked to memory dysfunction in ICs. It has been suggested that fear conditioning involves long-term memory formation, a process that involves the cerebral cortex (22). In our study, mice housed in ICs showed a significant reduction in freezing time, hinting at memory deficiency compared with mice housed in ECs. In addition, our lipidomic studies using mouse brain exposed to ECs and ICs revealed that the amount of phospholipid species containing DHA was significantly different in the cerebral cortex region. Although our studies could not directly link the change of PLs containing DHA level and the change of memory function, these findings might be consistent with previous reports that the impaired memory of IC rats was highly related with DHA metabolism (65).
Our data and previous studies suggest that DHA metabolism might be changed in the IC brain and that the consumption of DHA could be decreased in the IC brain compared with the EC brain, which in return negatively impacts some DHA-dependent memory function in ICs. All information considered together, our phospholipidomic study highlights the hypothesis-generating potential of lipidomics for better understanding diseases implicating memory dysfunction. Moreover our findings suggest that a significant change of a signaling pool of phospholipids containing DHA in the prefrontal cortex of mice exposed to ICs or ECs may indicate that these mice are relevant tools to study aberrant lipid metabolism of DHA consumption.
To our knowledge, there is no previous report for the comparison of phospholipids and gangliosides in brain between the EC and the IC group, and any clear report of brain phospholipids containing DHA change among both groups. Our results confirm the value of our refined method for tissue analysis using prefractionation. On the other hand, poor fragmentation in MS/MS, MS/MS/MS, and the lipid database prevented us from identifying lipid structures and fully understanding comprehensively all metabolic changes happening in ICs and ECs. Further work will also be required to address the relevance of the difference of phospholipids containing DHA in the cerebral cortex. In addition, we need to elucidate how those lipid metabolic changes in mouse brain lead to change of memory function between ICs and ECs.
CONCLUSION
We confirmed that the 2D-phospholipidomic method we optimized could be applied for biological tissue sample analysis. We also optimized a 2D-phospholipidomic method and established a reliable relative comparison method for gangliosides in tissue samples. Acidic phospholipids are coeluted with the gangliosides from a WAX column. In addition, acidic phospholipids containing ester bonds are not stable in alkaline solution during the WAX column process, and some endogenous metabolites interfere with the retention of basic phospholipids in the WAX column. These are reasons why liquid-liquid extraction was used for phospholipids before fractionation, and gangliosides were directly applied to the WAX column without liquid-liquid extraction. PL-WAX, packed in a Pasteur pipette for PL sample prefractionation, also provided good separation of gangliosides from high-abundance lipid classes, like neutral lipid and PC. Both analytical methods for PLs and gangliosides performed sufficiently well to permit detection of subtle changes in metabolite patterns in biological tissue samples, because we chose several ISs corresponding to different polar head-groups. Application of our analytical method highlighted significant differences in phospholipid peaks between brain samples of ECs and ICs, although no significant change was observed for gangliosides. Interestingly, phospholipids containing DHA were significantly changed and a difference of DHA consumption in brain in each housing condition might affect memory performance. It has been reported very recently (66) that male rats switched from a DHA-enriched diet to a high-fat diet showed increased anxiety accompanied by a decrease of p-CREB and other proteins linked to neuronal plasticity in the hippocampus. We have also observed similar anxiety induction using the Light-Dark test in the same mice used in this paper (supplementary Fig. VI) which corroborates the open field data described by Sharma, Zhuang, and Gomez-Pinilla (66). We also observed the decrease of p-CREB in poor housing conditions. The fact that a decrease in phospholipid-containing DHA levels is induced by housing conditions independently of diet change suggests the importance of this metabolite in memory and also anxiety-related behaviors, and further expands the knowledge regarding how maintaining not only a healthy diet but also a healthy lifestyle affects brain lipid environment. The 2D lipidomic measurement approaches we described are expected to be useful for obtaining new biological insight on the importance of lipid metabolism for brain functions.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Tatsuji Nakamura and Dr. Ken Aoshima who provided carefully considered feedback and valuable comments. Special thanks also go to Mr. Mitsuru Fukuda and Dr. Shigeru Koikegami whose support has helped us very much throughout the production of this study. Finally, the authors would like to thank Core Research for Evolutional Science and Technology (CREST) for a grant that made this study possible.
Footnotes
Abbreviations:
- AD
- Alzheimer's disease
- BHT
- butylated hydroxytoluene
- BrdU
- 5′-bromo-2′-deoxyuridine
- CREB
- cAMP response element binding protein
- 2D
- two-dimensional
- CV
- coefficient of variation
- DAB
- diaminobenzidine
- DG
- dentate gyrus
- DHA
- docosahexaenoic acid
- EC
- enriched condition
- IC
- impoverished condition
- IHC
- immunohistochemical
- IS
- internal standard
- NPLC
- normal-phase liquid chromatography
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- p-CREB
- phospho-cAMP response element binding protein
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinostol
- PKA
- cAMP-dependent protein kinase
- PS
- phosphatidylserine
- RPLC
- reverse-phase liquid chromatography
- SSC
- saline sodium citrate TAG, triacylglycerol
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and five tables.
REFERENCES
- 1.Hebb D. O. 1947. The effects of early experience on problem solving at maturity. Am. Psychol. 2: 306–307 [Google Scholar]
- 2.Huang F. L., Huang K. P., Wu J., Boucheron C. 2006. Environmental enrichment enhances neurogranin expression and hippocampal learning and memory but fails to rescue the impairments of neurogranin null mutant mice. J. Neurosci. 26: 6230–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naka F., Narita N., Okado N., Narita M. 2005. Modification of AMPA receptor properties following environmental enrichment. Brain Dev. 27: 275–278 [DOI] [PubMed] [Google Scholar]
- 4.Irvine G. I., Abraham W. C. 2005. Enriched environment exposure alters the input-output dynamics of synaptic transmission in area CA1 of freely moving rats. Neurosci. Lett. 391: 32–37 [DOI] [PubMed] [Google Scholar]
- 5.Duffy S. N., Craddock K. J., Abel T., Nguyen P. V. 2001. Environmental enrichment modifies the PKA-dependence of hippocampal LTP and improves hippocampus-dependent memory. Learn. Mem. 8: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rampon C., Jiang C. H., Dong H., Tang Y. P., Lockhart D. J., Schultz P. G., Tsien J. Z., Hu Y. 2000. Effects of environmental enrichment on gene expression in the brain. Proc. Natl. Acad. Sci. USA. 97: 12880–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNair K., Broad J., Riedel G., Davies C. H., Cobb S. R. 2007. Global changes in the hippocampal proteome following exposure to an enriched environment. Neuroscience. 145: 413–422 [DOI] [PubMed] [Google Scholar]
- 8.Wilcoxen K. M., Uehara T., Myint K. T., Sato Y., Oda Y. 2010. Practical metabolomics in drug discovery. Expert Opin. Drug Discov. 5: 249–263 [DOI] [PubMed] [Google Scholar]
- 9.Hu C., van der Heijden R., Wang M., van der Greef J., Hankemeier T., Xu G. 2009. Analytical strategies in lipidomics and applications in disease biomarker discovery. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2836–2846 [DOI] [PubMed] [Google Scholar]
- 10.Han X., Gross R. W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412 [DOI] [PubMed] [Google Scholar]
- 11.Han X., Gross R. W. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44: 1071–1079 [DOI] [PubMed] [Google Scholar]
- 12.Wenk M. R. 2005. The emerging field of lipidomics. Nat. Rev. Drug Discov. 4: 594–610 [DOI] [PubMed] [Google Scholar]
- 13.Watson A. D. 2006. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J. Lipid Res. 47: 2101–2111 [DOI] [PubMed] [Google Scholar]
- 14.Steinberg D. 2005. Thematic review series: the pathogenesis of atherosclerosis. An interpretive history of the cholesterol controversy: part II: the early evidence linking hypercholesterolemia to coronary disease in humans. J. Lipid Res. 46: 179–190 [DOI] [PubMed] [Google Scholar]
- 15.Sato Y., Suzuki I., Nakamura T., Bernier F., Aoshima K., Oda Y. 2012. Identification of a new plasma biomarker of Alzheimer's disease using metabolomics technology. J. Lipid Res. 53: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y., Nakamura T., Aoshima K., Oda Y. 2010. Quantitative and wide-ranging profiling of phospholipids in human plasma by two-dimensional liquid chromatography/mass spectrometry. Anal. Chem. 82: 9858–9864 [DOI] [PubMed] [Google Scholar]
- 17.Han X. 2010. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer's disease. Biochim. Biophys. Acta. 1801: 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han X. 2005. Lipid alterations in the earliest clinically recognizable stage of Alzheimer's disease: implication of the role of lipids in the pathogenesis of Alzheimer's disease. Curr. Alzheimer Res. 2: 65–77 [DOI] [PubMed] [Google Scholar]
- 19.Rosenson R. S. 2010. New technologies personalize diagnostics and therapeutics. Curr. Atheroscler. Rep. 12: 184–186 [DOI] [PubMed] [Google Scholar]
- 20.Kochanek P. M., Berger R. P., Bayir H., Wagner A. K., Jenkins L. W., Clark R. S. 2008. Biomarkers of primary and evolving damage in traumatic and ischemic brain injury: diagnosis, prognosis, probing mechanisms, and therapeutic decision making. Curr. Opin. Crit. Care. 14: 135–141 [DOI] [PubMed] [Google Scholar]
- 21.Taguchi R., Houjou T., Nakanishi H., Yamazaki T., Ishida M., Imagawa M., Shimizu T. 2005. Focused lipidomics by tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 823: 26–36 [DOI] [PubMed] [Google Scholar]
- 22.Runyan J. D., Moore A. N., Dash P. K. 2004. A role for prefrontal cortex in memory storage for trace fear conditioning. J. Neurosci. 24: 1288–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda K., Shimizu T., Taguchi R. 2008. Targeted analysis of ganglioside and sulfatide molecular species by LC/ESI-MS/MS with theoretically expanded multiple reaction monitoring. J. Lipid Res. 49: 2678–2689 [DOI] [PubMed] [Google Scholar]
- 24.Briand L. A., Robinson T. E., Maren S. 2005. Enhancement of auditory fear conditioning after housing in a complex environment is attenuated by prior treatment with amphetamine. Learn. Mem. 12: 553–556 [DOI] [PubMed] [Google Scholar]
- 25.Alvarez R. P., Biggs A., Chen G., Pine D. S., Grillon C. 2008. Contextual fear conditioning in humans: cortical-hippocampal and amygdala contributions. J. Neurosci. 28: 6211–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gratzner H. G. 1982. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: a new reagent for detection of DNA replication. Science. 218: 474–475 [DOI] [PubMed] [Google Scholar]
- 27.Walton M., Connor B., Lawlor P., Young D., Sirimanne E., Gluckman P., Cole G., Dragunow M. 1999. Neuronal death and survival in two models of hypoxic-ischemic brain damage. Brain Res. Brain Res. Rev. 29: 137–168 [DOI] [PubMed] [Google Scholar]
- 28.Yin J. C., Tully T. 1996. CREB and the formation of long-term memory. Curr. Opin. Neurobiol. 6: 264–268 [DOI] [PubMed] [Google Scholar]
- 29.Williams B. M., Luo Y., Ward C., Redd K., Gibson R., Kuczaj S. A., McCoy J. G. 2001. Environmental enrichment: effects on spatial memory and hippocampal CREB immunoreactivity. Physiol. Behav. 73: 649–658 [DOI] [PubMed] [Google Scholar]
- 30.Lee H., An H. J., Lerno L. A., Jr, German J. B., Lebrilla C. B. •••. Rapid profiling of bovine and human milk gangliosides by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Int. J. Mass Spectrom. 305: 138–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haughey N. J., Bandaru V. V., Bae M., Mattson M. P. 2010. Roles for dysfunctional sphingolipid metabolism in Alzheimer's disease neuropathogenesis. Biochim. Biophys. Acta. 1801: 878–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakomori S. 2003. Structure, organization, and function of glycosphingolipids in membrane. Curr. Opin. Hematol. 10: 16–24 [DOI] [PubMed] [Google Scholar]
- 33.Ngamukote S., Yanagisawa M., Ariga T., Ando S., Yu R. K. 2007. Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains. J. Neurochem. 103: 2327–2341 [DOI] [PubMed] [Google Scholar]
- 34.Yanagisawa M., Yu R. K. 2007. The expression and functions of glycoconjugates in neural stem cells. Glycobiology. 17: 57R–74R [DOI] [PubMed] [Google Scholar]
- 35.Yu R. K. 1994. Development regulation of ganglioside metabolism. Prog. Brain Res. 101: 31–44 [DOI] [PubMed] [Google Scholar]
- 36.Hussain S. T., Roots B. I. 1994. Effect of essential fatty acid deficiency & immunopathological stresses on blood brain barrier (B-BB) in Lewis rats: a biochemical study. Biochem. Soc. Trans. 22: 338S. [DOI] [PubMed] [Google Scholar]
- 37.Minami M., Kimura S., Endo T., Hamaue N., Hirafuji M., Togashi H., Matsumoto M., Yoshioka M., Saito H., Watanabe S., et al. 1997. Dietary docosahexaenoic acid increases cerebral acetylcholine levels and improves passive avoidance performance in stroke-prone spontaneously hypertensive rats. Pharmacol. Biochem. Behav. 58: 1123–1129 [DOI] [PubMed] [Google Scholar]
- 38.Kim H. Y., Edsall L. 1999. The role of docosahexaenoic acid (22:6n-3) in neuronal signaling. Lipids. 34(Suppl): S249–S250 [DOI] [PubMed] [Google Scholar]
- 39.Delion S., Chalon S., Guilloteau D., Besnard J. C., Durand G. 1996. alpha-Linolenic acid dietary deficiency alters age-related changes of dopaminergic and serotoninergic neurotransmission in the rat frontal cortex. J. Neurochem. 66: 1582–1591 [DOI] [PubMed] [Google Scholar]
- 40.Vreugdenhil M., Bruehl C., Voskuyl R. A., Kang J. X., Leaf A., Wadman W. J. 1996. Polyunsaturated fatty acids modulate sodium and calcium currents in CA1 neurons. Proc. Natl. Acad. Sci. USA. 93: 12559–12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishikawa M., Kimura S., Akaike N. 1994. Facilitatory effect of docosahexaenoic acid on N-methyl-D-aspartate response in pyramidal neurones of rat cerebral cortex. J. Physiol. 475: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mozzi R., Buratta S., Goracci G. 2003. Metabolism and functions of phosphatidylserine in mammalian brain. Neurochem. Res. 28: 195–214 [DOI] [PubMed] [Google Scholar]
- 43.Diau G. Y., Hsieh A. T., Sarkadi-Nagy E. A., Wijendran V., Nathanielsz P. W., Brenna J. T. 2005. The influence of long chain polyunsaturate supplementation on docosahexaenoic acid and arachidonic acid in baboon neonate central nervous system. BMC Med. 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green J. T., Orr S. K., Bazinet R. P. 2008. The emerging role of group VI calcium-independent phospholipase A2 in releasing docosahexaenoic acid from brain phospholipids. J. Lipid Res. 49: 939–944 [DOI] [PubMed] [Google Scholar]
- 45.Chen C. T., Ma D. W., Kim J. H., Mount H. T., Bazinet R. P. 2008. The low density lipoprotein receptor is not necessary for maintaining mouse brain polyunsaturated fatty acid concentrations. J. Lipid Res. 49: 147–152 [DOI] [PubMed] [Google Scholar]
- 46.Hamilton J. A., Brunaldi K. 2007. A model for fatty acid transport into the brain. J. Mol. Neurosci. 33: 12–17 [DOI] [PubMed] [Google Scholar]
- 47.Robinson P. J., Noronha J., DeGeorge J. J., Freed L. M., Nariai T., Rapoport S. I. 1992. A quantitative method for measuring regional in vivo fatty-acid incorporation into and turnover within brain phospholipids: review and critical analysis. Brain Res. Brain Res. Rev. 17: 187–214 [DOI] [PubMed] [Google Scholar]
- 48.Bazinet R. P., Weis M. T., Rapoport S. I., Rosenberger T. A. 2006. Valproic acid selectively inhibits conversion of arachidonic acid to arachidonoyl-CoA by brain microsomal long-chain fatty acyl-CoA synthetases: relevance to bipolar disorder. Psychopharmacology (Berl.). 184: 122–129 [DOI] [PubMed] [Google Scholar]
- 49.Mashek D. G., Li L. O., Coleman R. A. 2006. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 47: 2004–2010 [DOI] [PubMed] [Google Scholar]
- 50.Salem N., Jr, Litman B., Kim H. Y., Gawrisch K. 2001. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids. 36: 945–959 [DOI] [PubMed] [Google Scholar]
- 51.Bazan N. G. 2005. Lipid signaling in neural plasticity, brain repair, and neuroprotection. Mol. Neurobiol. 32: 89–103 [DOI] [PubMed] [Google Scholar]
- 52.Kim H. Y. 2007. Novel metabolism of docosahexaenoic acid in neural cells. J. Biol. Chem. 282: 18661–18665 [DOI] [PubMed] [Google Scholar]
- 53.Rao J. S., Ertley R. N., Lee H. J., DeMar J. C., Jr, Arnold J. T., Rapoport S. I., Bazinet R. P. 2007. n-3 polyunsaturated fatty acid deprivation in rats decreases frontal cortex BDNF via a p38 MAPK-dependent mechanism. Mol. Psychiatry. 12: 36–46 [DOI] [PubMed] [Google Scholar]
- 54.DeGeorge J. J., Nariai T., Yamazaki S., Williams W. M., Rapoport S. I. 1991. Arecoline-stimulated brain incorporation of intravenously administered fatty acids in unanesthetized rats. J. Neurochem. 56: 352–355 [DOI] [PubMed] [Google Scholar]
- 55.Garcia M. C., Kim H. Y. 1997. Mobilization of arachidonate and docosahexaenoate by stimulation of the 5–HT2A receptor in rat C6 glioma cells. Brain Res. 768: 43–48 [DOI] [PubMed] [Google Scholar]
- 56.Jones C. R., Arai T., Rapoport S. I. 1997. Evidence for the involvement of docosahexaenoic acid in cholinergic stimulated signal transduction at the synapse. Neurochem. Res. 22: 663–670 [DOI] [PubMed] [Google Scholar]
- 57.Ariel A., Serhan C. N. 2007. Resolvins and protectins in the termination program of acute inflammation. Trends Immunol. 28: 176–183 [DOI] [PubMed] [Google Scholar]
- 58.Schwab J. M., Chiang N., Arita M., Serhan C. N. 2007. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 447: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson R. A., Makrides M. 1998. The role of long chain polyunsaturated fatty acids (LCPUFA) in neonatal nutrition. Acta Paediatr. 87: 1017–1022 [DOI] [PubMed] [Google Scholar]
- 60.Neuringer M., Connor W. E., Lin D. S., Barstad L., Luck S. 1986. Biochemical and functional effects of prenatal and postnatal omega 3 fatty acid deficiency on retina and brain in rhesus monkeys. Proc. Natl. Acad. Sci. USA. 83: 4021–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marcheselli V. L., Hong S., Lukiw W. J., Tian X. H., Gronert K., Musto A., Hardy M., Gimenez J. M., Chiang N., Serhan C. N., et al. 2003. Novel docosanoids inhibit brain ischemia-reperfusion-mediated leukocyte infiltration and pro-inflammatory gene expression. J. Biol. Chem. 278: 43807–43817 [DOI] [PubMed] [Google Scholar]
- 62.Calon F., Lim G. P., Yang F., Morihara T., Teter B., Ubeda O., Rostaing P., Triller A., Salem N., Jr, Ashe K. H., et al. 2004. Docosahexaenoic acid protects from dendritic pathology in an Alzheimer's disease mouse model. Neuron. 43: 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinclair A. J., Begg D., Mathai M., Weisinger R. S. 2007. Omega 3 fatty acids and the brain: review of studies in depression. Asia Pac. J. Clin. Nutr. 16(Suppl. 1): 391–397 [PubMed] [Google Scholar]
- 64.Soderberg M., Edlund C., Kristensson K., Dallner G. 1991. Fatty acid composition of brain phospholipids in aging and in Alzheimer's disease. Lipids. 26: 421–425 [DOI] [PubMed] [Google Scholar]
- 65.Holguin S., Huang Y., Liu J., Wurtman R. 2008. Chronic administration of DHA and UMP improves the impaired memory of environmentally impoverished rats. Behav. Brain Res. 191: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma S., Zhuang Y., Gomez-Pinilla F. 2012. High-fat diet transition reduces brain DHA levels associated with altered brain plasticity and behaviour. Sci Rep. 2: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.