Abstract
Adipose tissue lipogenesis is paradoxically impaired in human obesity, promoting ectopic triglyceride (TG) deposition, lipotoxicity, and insulin resistance. We previously identified mitogen-activated protein kinase kinase kinase kinase 4 (Map4k4), a sterile 20 protein kinase reported to be upstream of c-Jun NH2-terminal kinase (JNK) signaling, as a novel negative regulator of insulin-stimulated glucose transport in adipocytes. Using full-genome microarray analysis we uncovered a novel role for Map4k4 as a suppressor of lipid synthesis. We further report here the surprising finding that Map4k4 suppresses adipocyte lipogenesis independently of JNK. Thus, while Map4k4 silencing in adipocytes enhances the expression of lipogenic enzymes, concomitant with increased conversion of 14C-glucose and 14C-acetate into TGs and fatty acids, JNK1 and JNK2 depletion causes the opposite effects. Furthermore, high expression of Map4k4 fails to activate endogenous JNK, while Map4k4 depletion does not attenuate JNK activation by tumor necrosis factor α. Map4k4 silencing in cultured adipocytes elevates both the total protein expression and cleavage of sterol-regulated element binding protein-1 (Srebp-1) in a rapamycin-sensitive manner, consistent with Map4k4 signaling via mechanistic target of rapamycin complex 1 (mTORC1). We show Map4k4 depletion requires Srebp-1 upregulation to increase lipogenesis and further show that Map4k4 promotes AMP-protein kinase (AMPK) signaling and the phosphorylation of mTORC1 binding partner raptor (Ser792) to inhibit mTORC1. Our results indicate that Map4k4 inhibits adipose lipogenesis by suppression of Srebp-1 in an AMPK- and mTOR-dependent but JNK-independent mechanism.
Keywords: lipid synthesis, insulin, mitogen-activated protein kinase kinase kinase kinase 4, sterol-regulated element binding protein-1, c-Jun NH2-terminal kinase, mechanistic target of rapamycin, AMP-protein kinase
Extreme human obesity triggers adipose dysfunction, paradoxically resulting in decreased capacity for lipid synthesis and contributing to ectopic lipid deposition, lipotoxicity, and whole-body insulin resistance (1). Lipodystrophic human subjects and mouse models demonstrate that deficits in adipose lipogenesis correlate with excess lipid deposition in liver and muscle as well as whole-body insulin resistance, highlighting the importance of adipose lipogenesis in metabolic dysfunction (1–4). Adipose tissue expression and activity of the lipogenic transcription factors peroxisome proliferator-activated receptor γ (PPARγ), sterol-regulated element binding protein-1 (Srebp-1), and carbohydrate-responsive element binding protein (Chrebp, also known as Mlxipl) are suppressed in obese humans, leading to decreased expression of lipogenic enzymes and decreased adipose fatty acid (FA) and triglyceride (TG) synthesis; however, the mechanisms that lead to this inhibition are unclear (2, 3, 5–10). Therefore, understanding this deregulation is fundamental in developing strategies that promote adipose lipogenesis, to increase lipid sequestration in adipocytes and prevent ectopic fat deposition during obesity.
Srebp-1 and Srebp-2 are transcription factors important for regulating the expression of genes involved in synthesis and uptake of cholesterol, FAs, TGs, and phospholipids (11). These factors reside in the endoplasmic reticulum and upon demand for lipid synthesis are transported to the Golgi apparatus where they are cleaved, allowing the functional N-terminal fragment to translocate into the nucleus and regulate lipogenic gene expression (11). Srebp transcription factors have distinctive specificities: Srebp-2 mainly controls expression of genes involved in the cholesterol biosynthetic pathway while Srebp-1 controls lipogenic gene expression; however, overlap in function has been reported (11, 12). Srebp-1 is an important regulator of lipogenesis because it controls the expression of solute carrier family 2 (facilitated glucose transporter) member 4 (Slc2a4 or Glut4) (13), ATP citrate lyase (Acly) (14), acetyl-CoA carboxylase α (Acaca) (15), FA synthase (Fasn) (16, 17), stearoyl-CoA desaturase 1 (Scd-1) (18), glycerol-3-phosphate acyltransferase (Gpat) (19), and PPARγ (20, 21). Srebp-1, in turn, is positively regulated by mechanistic target of rapamycin complex 1 (mTORC1) (22–25) and negatively regulated by AMP-protein kinase (AMPK) (26–28). Although the mechanism used by mechanistic target of rapamycin (mTOR) to promote the activity of Srebp-1 is not fully understood, increased PPARγ function in response to mTORC1 activation may contribute to increased Srebp-mediated adipose lipogenesis (22, 29–34).
Mitogen-activated protein kinase kinase kinase kinase 4 (Map4k4) is a serine/threonine protein kinase related to Saccharomyces cerevisiae sterile 20 (Ste20) protein kinases. Previous work has suggested that Map4k4 is a pro-inflammatory kinase that activates c-Jun NH2-terminal kinase (JNK) protein kinase (35–37). This would be in keeping with a role for Map4k4 as a Ste20-like protein kinase, upstream of the terminal MAP kinases (38–40). We identified Map4k4 in an RNA interference (RNAi) screen as a protein kinase that regulates insulin-stimulated glucose transport in cultured adipocytes (41). Map4k4 has also been reported to inhibit mTORC1, resulting in decreased PPARγ protein levels (29). Because Map4k4 expression increases in adipose tissue in obese subjects while adipose lipogenesis decreases (42), and Map4k4 negatively regulates mTOR (29), we aimed to test the role of Map4k4 in adipose lipogenesis and whether its actions require mTOR or the JNK protein kinase pathway. These studies extend our previous understanding of Map4k4 as a potential regulator of adipocyte lipid synthesis by demonstrating that Map4k4 suppresses lipogenesis in an mTOR-dependent and JNK-independent manner.
MATERIALS AND METHODS
Materials and chemicals
Bovine insulin, FA-free BSA, d-glucose, sodium pyruvate, and sodium acetate were purchased from Sigma. Tumor necrosis factor α (TNFα) was purchased from Calbiochem. 14C-U-glucose (250 μCi/ml) and 14C-acetate (250 μCi/ml) were purchased from Perkin Elmer. Flag-JNK1 and Flag-JNK2 were developed by R. Davis (University of Massachusetts Medical School) (Addgene plasmid 13798 and 13801, respectively) (43, 44).
Cell culture
3T3-L1 fibroblasts were grown and differentiated into adipocytes as previously described (45). Briefly, 3T3-L1 fibroblasts were grown to confluence in complete medium [high glucose (25 mM) DMEM containing 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml of streptomycin]. Two days after confluence, differentiation medium (0.25 μM dexamethasone, 0.5 mM 1-methyl-3-isobutylxanthine, and 10−7 M insulin) was added. On the fourth day after differentiation medium was added, adipocytes were either infected with 40 μl of 1.43 × 1012 particles/ml of adenovirus (AdCMV-HA-Control or AdCMV-HA-Map4k4, University of Massachusetts Medical School Viral Vector Core Facility, the cytomegalovirus (CMV) driven adenoviral vector expresses Map4k4 with an N-terminal triple hemagglutinin (HA) epitope tag) or washed with PBS, trypsinized, and transfected by electroporation (Bio-Rad Gene Pulser II; 0.18 kV, 960 microfarads) with small interfering RNA (siRNA) (scrambled, Map4k4, JNK1/2, and Srebp1/2 from Dharmacon). A green fluorescent protein-expressing control virus or Map4k4 D152N-expressing virus was also used and added at a dose of 40 μl of 1.43 × 1012 particles/ml for 72 h. These adenoviruses were gifts from Diane L. Barber (Department of Cell and Tissue Biology, University of California, San Francisco, CA). Transfection of HEK 293T cells was achieved using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Briefly, cells were plated at a density of 2.5 × 105 cells per 6-well plate a day before transfection. One microgram of DNA was used for each transfection and empty vector was used to achieve equal amounts of DNA between conditions. Knockdown experiments in HEK 293T cells were achieved using Lipofectamine RNAi Max (Invitrogen) following the manufacturer's protocol.
RNA isolation, real-time PCR, Affymetrix gene chip analysis
Total RNA was extracted from adipocytes using TriPure isolation reagent (Roche) following the manufacturer's instructions. cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad) and quantitative Real-time PCR was performed using iQ SybrGreen supermix and analyzed as previously described (46, 47). 36B4 (Rplp0) and Hprt served as housekeeping internal controls.
For Affymetrix GeneChip analysis, RNA was isolated from three independent experiments and hybridized to three different murine genome MOE430-2 microarrays. RNA quality was measured using the Agilent 2100 bioanalyzer. Differentially expressed mRNAs were identified using a random variance t-test.
siRNA duplexes
siRNA sequences were purchased from Dharmacon RNAi Technologies (Thermo Scientific). Scramble control (CAGUCGCGUUUGCGACUGGUU), Map4k4 (GACCAACUCUGGCUUGUUAUU), JNK1 (GGAGUUAGAUCAUGAAAGAUU), JNK2 (GGAAAGAGCUAAUUUACAAUU), Srebp1 (Srebf1) SMARTpool (GGGCAGCUCUGUACUCCUU, GUACACUUCUGGAGACAUC, CAAACAAGCUGACCUGGAU, GCAAGGCCAUCGACUACAU), Srebp2 (Srebf2) SMARTpool (GAGGAAGGCCAUUGAUUAC, GGUGCAACCUCAGAUCAUU, GAAAGUUCCUAUCAAGCAA, CCAGUGCUCUAGAGUAUUU).
Immunoblotting
For studies on JNK signaling, cells were treated with 0.01% BSA or 50 ng/ml TNFα for 15 min. Cells were washed twice in cold PBS and collected in radio-imminoprecipitation assay buffer (RIPA) buffer supplemented with protease and phosphatase inhibitors (Thermo Scientific). Total cell lysates were resolved by SDS-PAGE and electrotransferred to nitrocellulose membranes. Membranes were incubated with indicated antibodies overnight at 4°C. Blots were washed with TBST (0.1% Tween 20 in Tris-buffered saline), incubated with horseradish peroxidase anti-mouse or anti-rabbit secondary antibody and visualized using an enhanced chemiluminescent substrate kit (Perkin Elmer). Densitometry analyses were performed using ImageJ. For experiments on insulin signaling, 3T3-L1 adipocytes were washed twice with PBS and 2 ml of low glucose DMEM [2.5% FA-free BSA, 1% (v/v) Pen/Strep, 0.5 mM d-glucose, 0.5 mM sodium acetate, and 2 mM sodium pyruvate) were added in all wells. Insulin (1 μM) was added to corresponding wells for 1.5 h. Phospho-JNK and JNK, DYKDDDDDK Tag, HA, phospho-4EBP and total 4EBP, phospho-Raptor, phospho-AMPK, total AMPK, phospho-ACC, phospho-S6, and total S6 antibodies were purchased from Cell Signaling. Phospho-cJun and cJun antibodies were purchased from Santa Cruz. Actin and Flag M2 antibodies were purchased from Sigma and Map4k4 antibody was purchased from Bethyl (#A301-503A).
Srebp immunoblotting
Srebp-1 antibody was purchased from Millipore (clone 2121) and Srebp-2 was purchased from Cayman Chemical (#10007663). A minimum of 30 μg of protein lysates (2% SDS, 150 mM NaCl, and 5 mM EDTA) were resolved by SDS-PAGE (8%) and electrotransferred to nitrocellulose membranes. Membranes were incubated with indicated antibodies overnight at 4°C (1:1,000). Blots were washed with TBST (0.1% Tween 20 in Tris-buffered saline), incubated with horseradish peroxidase anti-mouse (for Srebp-1) or anti-rabbit (for Srebp-2) secondary antibody (1 h) and visualized using an enhanced chemiluminescent substrate kit (Perkin Elmer). It is important to note that a nonspecific band is recognized by Srebp-2 that could be mistaken as the cleaved Srebp-2 protein. Srebp-1 antibody occasionally recognizes a nonspecific band circa 75 KDa.
In vitro protein kinase assay
Map4k4 kinase assays were performed by lysing cells (20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 1 μg/ml leupeptin) and precipitating Map4k4 with HA antibody (Cell Signaling) and protein A agarose beads. Precipitates were washed four times and mixed with 40 μl kinase buffer [20 mM HEPES (pH 7.4), 10 mM MgCl2, 20 mM β-glycerophosphate, 10 mM NaF, 0.2 mM sodium orthovanadate, and 1 mM DTT], 5 μg unphosphorylated Myelin basic protein (MBP) (Millipore), 250 μM ATP, and 1μCi/reaction ATP. Kinase reactions were performed at 30°C for 30 min and terminated with addition of SDS sample buffer and heating the samples to 95°C for 5 min. Reaction mixtures were resolved by SDS-PAGE analysis and transferred to a nitrocellulose membrane for autoradiogram and Western blot analysis.
Radiolabeled glucose and acetate conversion to TG and FA
The incorporation of the various radioactive substrates into TG and FA was measured as previously described (48). In brief, adipocytes were washed twice with PBS and 1 ml of labeling medium [2.5% FA-free BSA, 1% (v/v) Pen/Strep, 0.5 mM d-glucose, 0.5 mM sodium acetate, 2 mM sodium pyruvate, and 2 μCi/ml 14C-U-glucose or 14C-acetate] was added. Final concentration of 1 μM insulin was added to insulin-stimulated conditions. Cells were incubated at 37°C in a humidified incubator (5% CO2) for 4.5 h before lipid extraction. All metabolic processes were stopped by washing cells twice with cold PBS and lysing cells by the addition of modified Dole's extraction mixture (80 ml isopropanol, 20 ml hexane, and 2 ml of 1N H2SO4) (49). TGs were extracted with hexane, washed, and the solvent was evaporated. Incorporation of 14C-glucose or 14C-acetate into FAs of TGs was determined by evaporating the solvent from the neutral lipids, adding 1 ml KOH-ethanol (20 ml of 95% ethanol, 1 ml water, 1 ml saturated KOH), and heating samples to 80°C for 1 h. Sulfuric acid was added to the mixture to ensure complete saponification. Addition of hexane allowed hydrophobic separation, which was evaporated and counted by liquid scintillation. Incorporation data were normalized to cell number.
TLC
TGs and FAs were separated by TLC using hexane:ethyl ether:acetic acid (200:50:4). Autoradiography was performed with TG and FA standards. The silica gel from each radiolabeled spot was subsequently scraped and quantified by liquid scintillation.
RESULTS
Map4k4 depletion enhances metabolic gene expression
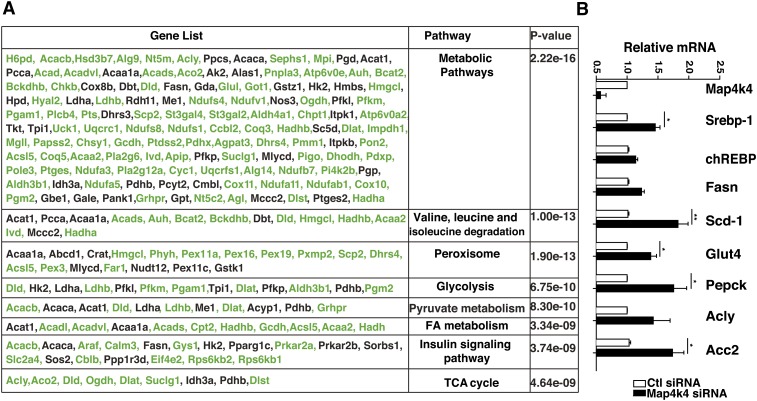
We have previously shown that Map4k4 depletion enhances PPARγ protein expression and TG accumulation in cultured adipocytes (29, 41). To further investigate the effects of Map4k4 depletion, we used Affymetrix mouse MOE430-2 microarrays to compare gene expression between scrambled siRNA- and Map4k4 siRNA-treated mature 3T3-L1 adipocytes. Kyoto encyclopedia of genes and genomes (KEGG) gene set enrichment profiles using scrambled siRNA- versus Map4k4 siRNA-treated adipocytes indicated that the most upregulated genes upon Map4k4 depletion are enriched in metabolic pathways (pathway 01100), valine, leucine, and isoleucine degradation (pathway 00280), peroxisome biosynthesis (pathway 04146), glycolysis (pathway 00010), pyruvate metabolism (pathway 00620), FA metabolism (pathway 00071), insulin signaling pathway (pathway 04910), and the TCA cycle (pathway 00020) (Fig. 1A). We further validated the mRNA expression of several metabolic genes upon Map4k4 depletion in cultured adipocytes (Fig. 1B). These analyses suggest that Map4k4 suppresses lipid metabolism-related processes in cultured adipocytes.
Fig. 1.
Map4k4 depletion enhances metabolic gene expression. A: KEGG gene set enrichment profile of differentially expressed genes upon Map4k4 depletion (P < 0.05, fold change 1.3) in murine 3T3-L1 mature adipocytes. The table shows the pathways that were most significantly upregulated by Map4k4 depletion. The gene list contains genes involved in each pathway and the genes in green further represent genes that were upregulated within the specific pathway upon Map4k4 depletion. B: Quantitative real-time PCR analysis of metabolic genes upon Map4k4 depletion in mature cultured adipocytes.
Map4k4 functions as a repressor of TG synthesis in cultured adipocytes
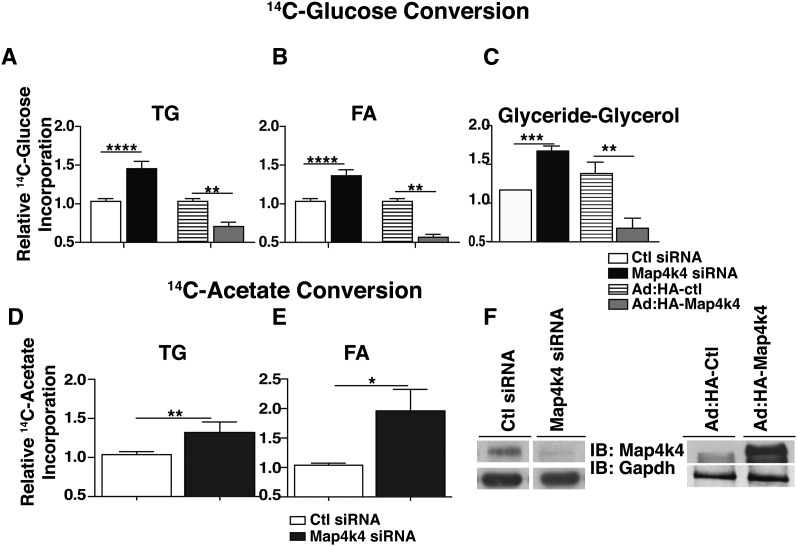
To further assess the role of Map4k4 as a negative regulator of TG synthesis in adipocytes, we depleted Map4k4 in mature 3T3-L1 adipocytes using siRNA or elicited increased Map4k4 expression using adenovirus vectors, and measured the conversion of 14C-glucose into neutral lipids. Consistent with the hypothesis that Map4k4 inhibits lipid synthesis, Map4k4 knockdown resulted in approximately 50% increased incorporation of the 14C-radiolabel into neutral lipids compared with the scrambled control (Fig. 2A). Conversely, Map4k4 overexpression significantly reduced 14C-radiolabel incorporation into TGs by ∼29% compared with the adenovirus HA-control (Ad: HA-control) (Fig. 2A). To determine whether Map4k4 was suppressing glycerol-3-phosphate formation or de novo lipogenesis, we measured 14C-radiolabel incorporation into both esterified FA on TG molecules (Fig. 2B) and glyceride-glycerol (Fig. 2C). We determined that Map4k4 inhibited both glyceride-glycerol formation and de novo lipid synthesis in mature adipocytes as seen by increased incorporation of the radiolabel into both molecules upon Map4k4 depletion and decreased incorporation upon Map4k4 overexpression (Fig. 2B, C). We also confirmed that these observed changes in neutral lipid synthesis and de novo lipogenesis reflect changes in TGs and FAs, respectively, by TLC analysis (data not shown).
Fig. 2.
Map4k4 represses TG synthesis in cultured adipocytes. A–C: 14C-glucose conversion into TGs (A), FAs (B), and glyceride-glycerol (C) is shown. Cells were transfected by electroporation with scrambled (7.5 μM) or Map4k4 (7.5 μM) siRNA (N = 9) or were infected with control (Ctl) virus (Ad: HA-Ctl) or HA-Map4k4 adenovirus (Ad: HA-Map4k4) (N = 4). Seventy-two hours posttransfection, cells were incubated with 14C-glucose for 4 h and TGs were extracted. D, E: 14C-acetate conversion into TGs (D) and FAs (E) is shown; lipids were extracted as in (A). F: Representative immunoblots (IB) depicting Map4k4 protein expression upon siRNA transfection or adenoviral overexpression. Samples were noncontiguous on the same gel. The data are represented as the average ± SE and were compared with appropriate controls by Student's t-test. ****P < 0.0001, ***P <.001, **P < 0.01, *P < .05.
We have previously reported that Map4k4 silencing increases Glut4 expression and glucose uptake (41), possibly providing the substrate for both glyceride-glycerol and FA synthesis. To distinguish the contribution of Map4k4 regulation to these processes, we used either control or Map4k4 siRNA-transfected adipocytes and measured 14C-acetate incorporation into TGs, a measure of de novo lipogenesis, as the radiolabel bypasses glucose transport and metabolism to provide substrate for FA synthesis. Consistent with the results from Fig. 2A, B, Map4k4 depletion significantly increased 14C-acetate incorporation into TGs and its constituent FAs (Fig. 2D, E). Representative protein immunoblots confirmed Map4k4 knockdown or overexpression at the protein level (Fig. 2F). These results indicate that Map4k4 is a negative regulator of glucose flux, de novo FA synthesis, and TG synthesis in cultured adipocytes.
Map4k4 silencing does not enhance lipid synthesis via JNK signaling
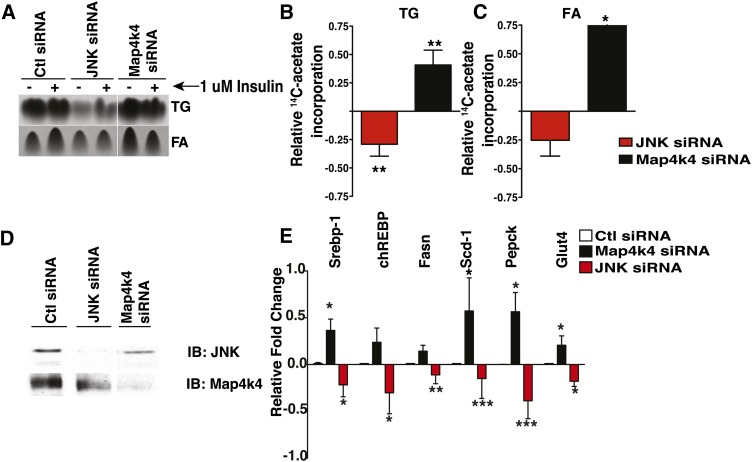
Map4k4 has been previously proposed to be a pro-inflammatory kinase and upstream activator of the JNK protein kinase cascade (35–37). Because the role of adipose JNK in lipid synthesis has not been clearly established, we tested whether Map4k4 repressed lipogenesis via JNK. We hypothesized that if Map4k4 was an upstream activator of JNK, then JNK depletion would result in enhanced lipid synthesis compared with scrambled control, similar to Map4k4 depletion. Thus, 3T3-L1 adipocytes were electroporated with siRNA against Map4k4 or JNK1 and JNK2, and 14C-acetate incorporation into FAs and TGs was used to assess the role of these kinases in TG synthesis and de novo lipogenesis. Consistent with results shown in Fig. 2, Map4k4 depletion significantly increased 14C-acetate incorporation into TGs (Fig. 3A, B). Surprisingly, depletion of the two major isoforms of JNK, JNK1 and JNK2, significantly reduced TG synthesis by 56%, opposite to the Map4k4 knockdown-induced increase in TG synthesis (Fig. 3A, B). Furthermore, 14C-acetate incorporation into FAs, a measure of de novo lipogenesis, was significantly increased upon Map4k4 silencing (∼47%) and decreased upon JNK depletion (∼20%) (Fig. 3C). Efficient Map4k4 and JNK protein depletion was confirmed by protein immunoblots (Fig. 3D). These data demonstrate that while Map4k4 downregulates lipogenesis, JNK1 and JNK2 are required for this process. We next examined whether JNK and Map4k4 depletion affected lipogenic gene expression in a dissimilar fashion using real-time PCR (Fig. 3C). Gene expression was normalized to 36B4, and the gene expression in Map4k4- or JNK-depleted cells was compared with that in scrambled siRNA-treated control cells. Comparison of the relative mRNA expression of Srebp-1, Chrebp, Fasn, Scd-1, Pepck, and Glut4 shows that while Map4k4 depletion increased the mRNA expression of these enzymes, JNK depletion decreased the mRNA expression of these genes (Fig. 3D). These results indicate that Map4k4 is a repressor of adipose lipogenesis while JNK is an unexpected positive regulator of this metabolic process. Thus, Map4k4 does not repress lipogenesis in a JNK-dependent manner.
Fig. 3.
Map4k4 does not require JNK to regulate lipid synthesis. A: Autoradiogram of TLC analysis of TGs and FAs extracted from adipocytes that were transfected with scrambled siRNA (14 μM), JNK1 and JNK2 siRNA (14 μM), or Map4k4 siRNA (14 μM). Seventy-two hours posttransfection, adipocytes were incubated with 14C-acetate and incorporation into TGs and FAs was determined. Samples were noncontiguous on the same TLC plate. B: Changes in 14C-acetate incorporation into TGs and FAs is shown in JNK- and Map4k4-depleted mature adipocytes relative to scrambled control (Ctl). Representative immunoblots (IB) for Map4k4 and JNK protein expression in control, Map4k4-, and JNK-depleted cells. C: Quantitative real-time PCR analysis of lipogenic gene expression in control, Map4k4-, and JNK-depleted adipocytes relative to scrambled control siRNA-treated cells (N = 5). Data are presented as average ± SE and were compared between groups by Student's t-test. *P < 0.01, **P < 0.001, ***P < 0.0001.
Map4k4 does not activate the JNK signaling pathway in adipocytes
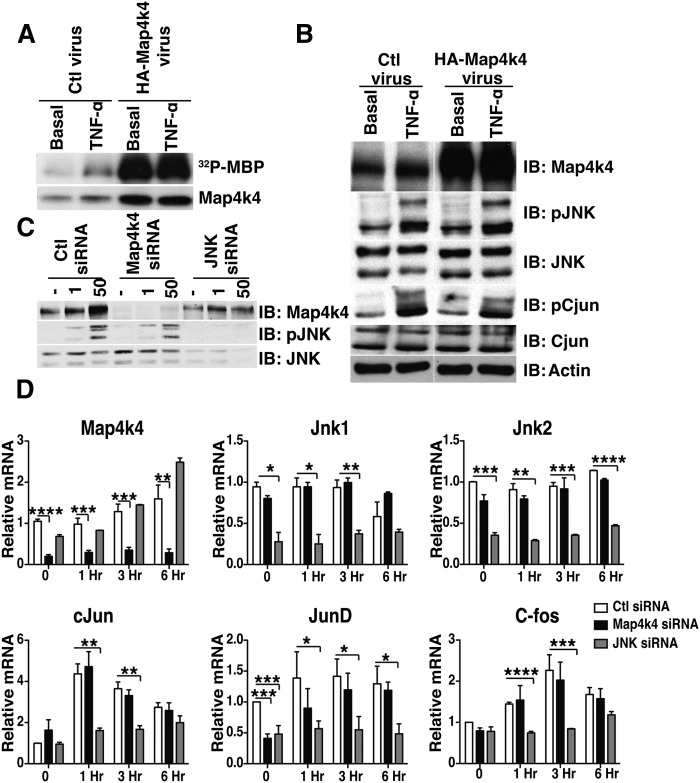
These divergent effects of Map4k4 and JNK were unexpected in light of previous reports that Map4k4 functions as an upstream activator of JNK in various cell models (35–37). To further investigate whether increased Map4k4 expression and activity would be sufficient to increase endogenous JNK signaling in adipocytes, we used adenoviral-mediated Map4k4 overexpression followed by stimulation with TNFα, a potent JNK activator (Fig. 4). Despite increased expression and activity of Map4k4 (Fig. 4A), phosphorylation of JNK and its downstream substrate cJun did not differ between empty virus (Ad: HA-control) and Map4k4 adenoviral (Ad: HA-Map4k4)-treated adipocytes in both basal and TNFα-treated conditions (Fig. 4B). Thus, while increased Map4k4 expression inhibited TG synthesis, it was not sufficient to activate endogenous JNK signaling in adipocytes. Furthermore, if Map4k4 is a required upstream activator of JNK, Map4k4 depletion should attenuate JNK activation in response to treatment with TNFα. Map4k4 depletion, however, did not blunt JNK activation (Fig. 4C) and also did not affect the expression of activator protein-1 (AP-1) transcriptional factors, cJun, C-fos, and JunD, in response to TNFα (Fig. 4D). As expected, and consistent with previous results (50), JNK depletion attenuated the maximal response of these AP-1 transcriptional factors in response to TNFα (Fig. 4D). These results indicate that Map4k4 is neither sufficient nor required to induce JNK activation in adipocytes. Furthermore, using gain- and loss-of-function approaches in HEK 293T cells, we show that cotransfection of both Map4k4 and JNK results in increased JNK activity (supplementary Fig. IVA); however, depletion of endogenous Map4k4 did not diminish JNK activity (supplementary Fig. IVB), suggesting that Map4k4 is not an endogenous modulator of JNK activity. These results support the notion that endogenous Map4k4 is not required for optimal endogenous JNK activation in adipocytes or HEK 293T cells, but that artificially high levels of exogenously expressed Map4k4 can induce JNK activation when JNK is also overexpressed.
Fig. 4.
Map4k4 is neither necessary nor sufficient for canonical JNK signaling. A: Map4k4 kinase activity in control (Ctl) and Map4k4-overexpressing adipocytes stimulated with TNFα for 30 min. B: Protein immunoblots (IB) of control and Map4k4-overexpressing adipocytes treated with 50 ng/ml TNFα for 15 min (N = 4). Samples were noncontiguous on the same gel. C: Protein immunoblot of electroporated adipocytes treated with control, Map4k4, or JNK1 and JNK2 siRNA and stimulated with 50 ng/ml TNFα for 15 min (N = 4). D: Quantitative real-time PCR analysis of AP-1 transcription factors in response to 50 ng/ml TNFα treatment in control, Map4k4-depleted, or JNK-depleted adipocytes (N = 3). Data are presented as average ± SE and were compared between groups by Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P<.0001.
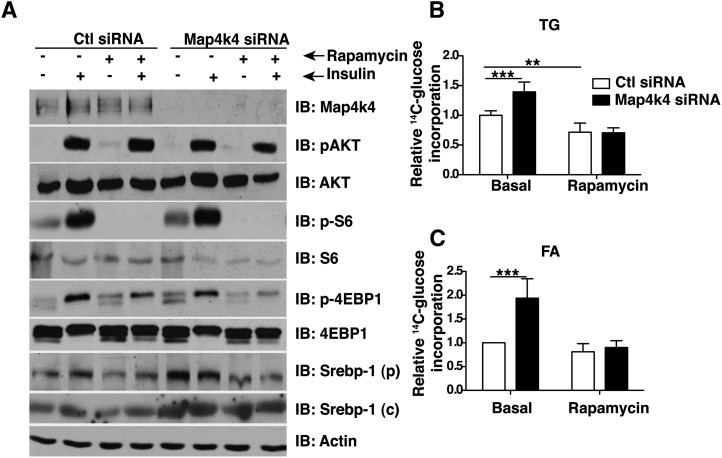
Map4k4 inhibits lipid synthesis in an mTOR-dependent manner
We have previously demonstrated that Map4k4 impairs mTORC1 signaling (29) and because mTORC1 signaling enhances lipid synthesis (22, 24, 34, 51), we tested whether Map4k4 required mTORC1 to inhibit lipid synthesis. We depleted Map4k4 using siRNA and treated the cells with rapamycin, an mTORC1 inhibitor, to repress mTORC1 signaling. Consistent with previous reports, Map4k4 depletion enhanced mTORC1 signaling as demonstrated by increased ribosomal protein S6 (S6) and eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1/Eif4ebp1) protein phosphorylation (29) (Fig. 5A). Interestingly, Map4k4 depletion also increased Srebp-1 protein levels, an important lipogenic transcription factor (Fig. 5A). Rapamycin treatment inhibited mTORC1 signaling, as evidenced by a lack of S6 and 4E-BP1 protein phosphorylation, and abolished the increase in Srebp-1 protein expression due to Map4k4 silencing (Fig. 5A). Rapamycin treatment also diminished the Map4k4 silencing-induced increase in 14C-glucose incorporation into TGs (Fig. 5B) and FAs (Fig. 5C). These results indicate that mTORC1 function is necessary for Srebp-1 expression and optimal lipid synthesis in cultured adipocytes and support the notion that Map4k4 represses the mTORC1/Srebp pathway to decrease adipose lipogenesis.
Fig. 5.
Map4k4 regulates mTOR to inhibit Srebp-1 expression and TG synthesis. A: 3T3-L1 adipocytes were electroporated with control siRNA or Map4k4 siRNA and treated with 100 nM rapamycin for 48 h and/or insulin for 1.5 h. Protein immunoblots (IB) depicting insulin-induced Akt and mTOR activation and rapamycin-induced inhibition in control and Map4k4-depleted adipocytes. B, C: Incorporation of 14C-glucose into TGs (B) and saponifiable FAs (C) in control siRNA-treated or Map4k4-depleted adipocytes treated with rapamycin. The data are represented as the average ± SE and were compared between groups by Student's t-test. N = 4, ***P < .001, **P < .01.
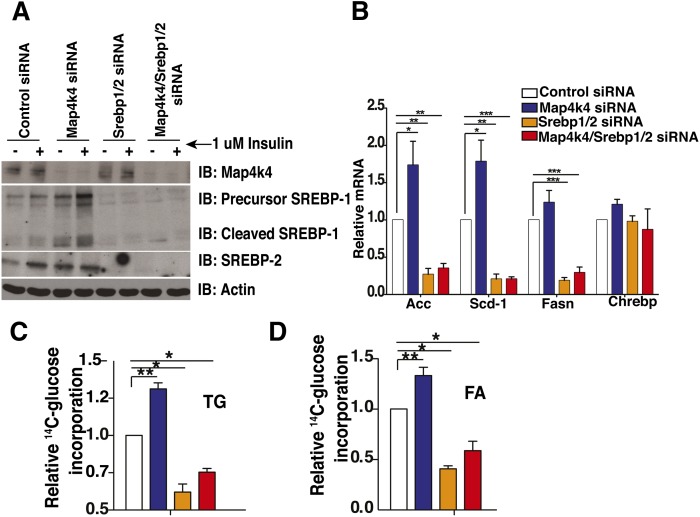
Map4k4 depletion requires Srebp expression to enhance lipid synthesis
We demonstrated that Map4k4 depletion enhances mTORC1 signaling and increases Srebp-1 protein expression (Fig. 5A). As an alternative approach to assess the effect of impaired Map4k4 function, we used a kinase-inactive mutant of Map4k4 (AdMap4k4 D152N) (52), previously shown to function as a dominant-negative inhibitor (53). Consistent with knockdown experiments (Fig. 5A), Map4k4 D152N overexpression resulted in a significant increase of Srebp-1 protein levels (supplementary Fig. II). To determine whether Map4k4 required this increase in Srebp-1 protein to enhance lipid synthesis, we performed knockdown experiments to suppress Map4k4, Srebp-1, and Srebp-2 expression in mature cultured adipocytes and examined conversion of 14C-glucose into TGs and FAs. Depletion of both Srebp-1 and Srebp-2 was required in these experiments because depletion of either transcription factor alone resulted in a compensatory increase of the other and downstream target genes (data not shown). Protein immunoblots demonstrated that Map4k4 knockdown resulted in a significant increase in precursor and cleaved Srebp-1 protein levels as well as an increase in total Srebp-2 protein levels (Fig. 6A). Furthermore, Map4k4 silencing significantly increased the mRNA expression of known Srebp-1 lipogenic target genes Acaca (∼73%), Scd-1 (∼78%), and Fasn (∼23%) (15, 17, 18) and depletion of Srebp severely blunted the enhanced expression of these genes in response to Map4k4 silencing (Fig. 6B). Importantly, Map4k4 depletion significantly increased 14C-glucose conversion into both TGs (Fig. 6C) and FAs (Fig. 6D), as demonstrated in previous figures, and depletion of both Srebp proteins significantly reduced the increase of radiolabel incorporation into TGs and FAs in response to Map4k4 silencing (Fig. 6C, D). These results indicate that Srebp proteins are necessary for optimal adipose lipogenesis and are also required for the Map4k4 knockdown-induced enhancement of lipid synthesis.
Fig. 6.
Map4k4 regulates lipogenesis in a Srebp-dependent manner. A: Mature 3T3-L1 adipocytes were transfected with control (Ctl) siRNA, Map4k4 siRNA, Srebp-1 and Srebp-2 siRNA, or Map4k4/Srebp-1/Srebp-2 siRNA and treated with 1 μM insulin for 1.5 h. Cells were harvested and Map4k4, Srebp-1, and Srebp-2 protein expression was analyzed by protein immunoblots (IB). B: Quantitative real-time PCR analysis of lipogenic gene expression in control, Map4k4, Srebp1, and Srebp2, and Srebp1-, Srebp2-, and Map4k4-depleted adipocytes. C, D: Incorporation of 14C-glucose into TGs (C) and FAs (D) in control, Map4k4-depleted, Srebp-1- and Srebp-2-depleted, or Map4k4/Srebp-1/Srebp-2-depleted adipocytes was measured. The data are represented as the average ± SE and were compared between groups by Student's t-test. N = 4, *P < 0.01, **P < 0.001, ***P < 0.0001.
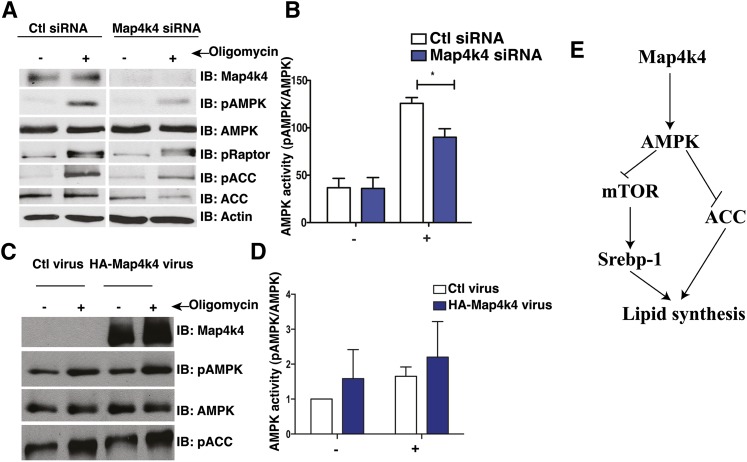
Map4k4 is a positive regulator of AMPK
Map4k4 has been implicated as a possible positive regulator of AMPK phosphorylation (54), and AMPK is a negative regulator of mTOR, suggesting a plausible mechanism by which Map4k4 may regulate mTOR and inhibit lipogenesis. We verified that Map4k4 regulates AMPK phosphorylation and activity in adipocytes by treating Map4k4-depleted adipocytes with oligomycin, an inhibitor of ATP synthase and potent activator of AMPK. As expected, treatment of adipocytes with oligomycin (500 nM for 30 min) increased AMPK phosphorylation. This response was significantly blunted in Map4k4-depleted adipocytes (Fig. 7A, B). Furthermore, decreased AMPK activation results in increased mTOR signaling, as assessed by decreased raptor phosphorylation (Ser792) and increased lipogenesis, as assessed by Acetyl-CoA carboxylase (ACC/Acaca) phosphorylation (Ser79) (Fig. 7A). On the other hand, increased Map4k4 expression and activity results in increased AMPK signaling as seen in Map4k4-overexpressing adipocytes treated with oligomycin (500 nM for 30 min) (Fig. 7C, D). These results suggest Map4k4 is necessary for optimal AMPK activation and provide insight into the mechanism by which Map4k4 inhibits mTOR and lipid synthesis in mature adipocytes.
Fig. 7.
Map4k4 regulates AMPK signaling. A: Protein immunoblot (IB) of 3T3-L1 adipocytes transfected with control (Ctl) siRNA or Map4k4 siRNA and treated with 500 nM oligomycin for 30 min (N = 5). B: Quantification of (A). C: Protein immunoblot of 3T3-L1 adipocytes infected with control virus (empty adenovirus) or HA-Map4k4 adenovirus and treated with 500 nM oligomycin for 30 min (N = 4). D: Quantification of (C). Samples were noncontiguous on the same gel. E: Schematic model coupling Map4k4 to AMPK and mTOR to inhibit lipid synthesis. The data are represented as the average ± SE and were compared between groups by Student's t-test. N = 4, *P < 0.05.
DISCUSSION
Here we show that adipose Map4k4 represses glucose incorporation into FAs and TGs, at least in part via upregulation of AMPK signaling which results in downregulation of mTOR function and Srebp-1 expression. We found that Map4k4 depletion enhances the conversion of both 14C-glucose and 14C-acetate into TGs and FAs, suggesting Map4k4 inhibits both de novo FA synthesis and FA esterification. Conversely, increased Map4k4 expression decreases conversion of 14C-glucose into TGs. We provide strong evidence that the ability of Map4k4 to repress adipose lipogenesis is independent of the JNK pathway, which we unexpectedly found to be required for optimal TG synthesis in cultured adipocytes. We believe these findings have direct relevance to adipose dysfunction including chronic inflammation and impaired lipid storage capacity that occurs with obesity because Map4k4 expression is elevated with increasing body mass index (42). This increased Map4k4 expression could contribute to impaired TG synthesis and storage in obese adipose tissue. Upon evaluating underlying mechanisms of Map4k4 action, we show here that Map4k4 is required for activation of AMPK signaling by oligomycin, which in turn is expected to modulate mTOR and Srebp activity as well as adipose lipogenesis. Thus, our results provide evidence for a model in which Map4k4 modulates AMPK, suppressing mTOR function and Srebp-1 expression to depress lipogenesis (Fig. 7E).
Our results are consistent with previous studies that describe a positive role of JNK in adipose lipogenesis (55–57). JNK1 depletion using anti-sense oligonucleotides in adipose tissue attenuates the expression of lipogenic enzymes Acly, Acaca, Fasn, and Scd-1 (55) and JNK2 suppression in human adipocytes attenuates Srebp-1 expression and activity resulting in decreased target lipid enzymes (57). Mice expressing a mature transcriptionally active Srebp-1 variant lacking all of the JNK phosphorylation sites are protected from hepatic steatosis and weight gain, thus demonstrating that JNK positively regulates Srebp-1 function (58). Furthermore, JNK is required for optimal Srebp-1 activation and the subsequent increase in Scd-1 and Fasn in response to keratinocyte growth factor treatment (56). We have therefore confirmed the positive role of JNK in adipose lipogenesis and have further established that Map4k4 does not require JNK to inhibit TG synthesis. We also provide evidence that Map4k4 is neither required nor sufficient to activate endogenous JNK signaling. Consistent with previous reports, we find that cotransfection of JNK and Map4k4 enhanced JNK signaling (35–37); however, increased Map4k4 activity and expression were not sufficient to activate endogenous JNK, and Map4k4 depletion did not attenuate TNFα-induced JNK activation. Therefore, our results suggest that Map4k4 is not an upstream activator of JNK in cultured adipose cells or HEK 293T cells, and that previous results placing Map4k4 in the JNK pathway may be due to artificially high ectopic expression of both of these protein kinases.
These results extend our previous findings that Map4k4 represses mTOR signaling in adipocytes (29) and shed insight into the mechanism by which Map4k4 represses lipid synthesis. The role of Srebp-1 in adipose lipogenesis has been largely dismissed because Srebp-1-null animals form functional fat depots (59, 60). However, the loss of Srebp-1 increases Srebp-2 expression and activity, compensating for the loss of Srebp-1 (59, 60). Furthermore, while overexpression of the nuclear fragment of Srebp-1c causes lipodystrophy (61), overexpression of a different isoform, Srebp-1a, increases the expression of downstream lipogenic target genes such as Fasn and Scd-1 (62). Because we observed compensation of Srebp isoforms in cultured adipocytes (data not shown), we targeted both Srebp-1 and Srebp-2 to deplete lipogenesis (Fig. 6). Interestingly, liver-specific depletion of SCAP, a regulatory protein that is required for Srebp protein proteolytic cleavage, decreased Srebp-1 and Srebp-2 expression and reduced obesity-induced hepatic steatosis, consistent with the notion that depletion of both Srebp proteins are required to diminish lipogenesis (63, 64). Our results support an active role for Srebp-1 and Srebp-2 in adipose lipogenesis and demonstrate that Map4k4 inhibits lipid synthesis via suppression of Srebp proteins.
Mechanistically, it is unclear how mTOR positively regulates Srebp-1 function (22–25). Rapamycin, a general mTORC1 inhibitor, disrupts Srebp-1 expression and maturation, thus decreasing lipogenesis (22, 24, 31–33). We have previously shown that Map4k4 inhibits mTOR function and we further show here that this inhibition resulted in decreased Srebp-1 expression and adipose lipogenesis. Map4k4 depletion enhances protein translation in an mTOR-dependent manner (29) and this may explain the increased Srebp-1 protein expression (both precursor and cleaved product) upon Map4k4 depletion. Interestingly, Map4k4 depletion does not affect Srebp-1 protein stability (data not shown) as previously reported for PPARγ (29) and Myf5 (53). Furthermore, we provide data here that suggests Map4k4 inhibits mTOR function via AMPK modulation. Map4k4 depletion attenuates AMPK signaling, resulting in decreased ACC and raptor phosphorylation, translating into increased lipid synthesis.
Thus, Map4k4 represses lipid synthesis in an mTOR- and Srebp-1-dependent manner and unexpectedly this is mediated independent of JNK-signaling. Because adipose tissue Map4k4 expression is increased during obesity (42), Map4k4 may be an important negative regulator of adipose lipogenesis in metabolic disease.
Supplementary Material
Acknowledgments
The authors thank Diane L. Barber (Department of Cell and Tissue Biology, University of California, San Francisco, CA) for the Map4k4 adenovirus vectors, Joseph Virbasius for critical reading of the manuscript, and members of the Czech lab for helpful discussions.
Footnotes
Abbreviations:
- Acaca
- acetyl-CoA carboxylase α Ad, adenovirus
- Ad
- adenovirus
- Acly
- ATP citrate lyase
- AMPK
- AMP-protein kinase
- AP-1
- activator protein-1
- Chrebp
- carbohydrate-responsive element binding protein
- CMV
- cytomegalovirus
- JNK
- c-Jun NH2-terminal kinase
- HA
- hemagglutinin
- Map4k4
- mitogen-activated protein kinase kinase kinase kinase 4
- mTOR
- mechanistic target of rapamycin
- mTORC1
- mechanistic target of rapamycin complex 1
- MBP
- myelin basic protein
- PPARγ
- peroxisome proliferator-activated receptor γ RNAi, RNA interference
- Scd-1
- stearoyl-CoA desaturase 1
- siRNA
- small interfering RNA
- TG
- triglyceride
- TNFα
- tumor necrosis factor α Srebp-1, sterol-regulated element binding protein-1
- Srebp-1
- sterol-regulated element binding protein-1
This work was supported by National Institutes of Health Grant DK-030898 (to M.P.C.) and core facility support from the University of Massachusetts Medical School Diabetes and Endocrinology Research Center (DERC) Grant DK032520.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.Unger R. H., Scherer P. E. 2010. Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol. Metab. 21: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diraison F., Dusserre E., Vidal H., Sothier M., Beylot M. 2002. Increased hepatic lipogenesis but decreased expression of lipogenic gene in adipose tissue in human obesity. Am. J. Physiol. Endocrinol. Metab. 282: E46–E51 [DOI] [PubMed] [Google Scholar]
- 3.Roberts R., Hodson L., Dennis A. L., Neville M. J., Humphreys S. M., Harnden K. E., Micklem K. J., Frayn K. N. 2009. Markers of de novo lipogenesis in adipose tissue: associations with small adipocytes and insulin sensitivity in humans. Diabetologia. 52: 882–890 [DOI] [PubMed] [Google Scholar]
- 4.Huang-Doran I., Sleigh A., Rochford J. J., O'Rahilly S., Savage D. B. 2010. Lipodystrophy: metabolic insights from a rare disorder. J. Endocrinol. 207: 245–255 [DOI] [PubMed] [Google Scholar]
- 5.Ranganathan G., Unal R., Pokrovskaya I., Yao-Borengasser A., Phanavanh B., Lecka-Czernik B., Rasouli N., Kern P. A. 2006. The lipogenic enzymes DGAT1, FAS, and LPL in adipose tissue: effects of obesity, insulin resistance, and TZD treatment. J. Lipid Res. 47: 2444–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman M. A., Peroni O. D., Villoria J., Schon M. R., Abumrad N. A., Bluher M., Klein S., Kahn B. B. 2012. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 484: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurtado del Pozo C., Vesperinas-Garcia G., Rubio M. A., Corripio-Sanchez R., Torres-Garcia A. J., Obregon M. J., Calvo R. M. 2011. ChREBP expression in the liver, adipose tissue and differentiated preadipocytes in human obesity. Biochim. Biophys. Acta. 1811: 1194–1200 [DOI] [PubMed] [Google Scholar]
- 8.Richardson D. K., Czech M. P. 1978. Primary role of decreased fatty acid synthesis in insulin resistance of large rat adipocytes. Am. J. Physiol. 234: E182–E189 [DOI] [PubMed] [Google Scholar]
- 9.Czech M. P., Richardson D. K., Smith C. J. 1977. Biochemical basis of fat cell insulin resistance in obese rodents and man. Metabolism. 26: 1057–1078 [DOI] [PubMed] [Google Scholar]
- 10.Czech M. P. 1976. Cellular basis of insulin insensitivity in large rat adipocytes. J. Clin. Invest. 57: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon T. I., Osborne T. F. 2012. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im S. S., Kwon S. K., Kang S. Y., Kim T. H., Kim H. I., Hur M. W., Kim K. S., Ahn Y. H. 2006. Regulation of GLUT4 gene expression by SREBP-1c in adipocytes. Biochem. J. 399: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato R., Okamoto A., Inoue J., Miyamoto W., Sakai Y., Emoto N., Shimano H., Maeda M. 2000. Transcriptional regulation of the ATP citrate-lyase gene by sterol regulatory element-binding proteins. J. Biol. Chem. 275: 12497–12502 [DOI] [PubMed] [Google Scholar]
- 15.Lopez J. M., Bennett M. K., Sanchez H. B., Rosenfeld J. M., Osborne T. F. 1996. Sterol regulation of acetyl coenzyme A carboxylase: a mechanism for coordinate control of cellular lipid. Proc. Natl. Acad. Sci. USA. 93: 1049–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin M. J., Wong R. H., Pandya N., Sul H. S. 2007. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J. Biol. Chem. 282: 5453–5467 [DOI] [PubMed] [Google Scholar]
- 17.Boizard M., Le Liepvre X., Lemarchand P., Foufelle F., Ferre P., Dugail I. 1998. Obesity-related overexpression of fatty-acid synthase gene in adipose tissue involves sterol regulatory element-binding protein transcription factors. J. Biol. Chem. 273: 29164–29171 [DOI] [PubMed] [Google Scholar]
- 18.Mauvoisin D., Rocque G., Arfa O., Radenne A., Boissier P., Mounier C. 2007. Role of the PI3-kinase/mTor pathway in the regulation of the stearoyl CoA desaturase (SCD1) gene expression by insulin in liver. J. Cell Commun. Signal. 1: 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ericsson J., Jackson S. M., Kim J. B., Spiegelman B. M., Edwards P. A. 1997. Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1- and sterol regulatory element-binding protein-responsive gene. J. Biol. Chem. 272: 7298–7305 [DOI] [PubMed] [Google Scholar]
- 20.Kim J. B., Spiegelman B. M. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10: 1096–1107 [DOI] [PubMed] [Google Scholar]
- 21.Fajas L., Schoonjans K., Gelman L., Kim J. B., Najib J., Martin G., Fruchart J. C., Briggs M., Spiegelman B. M., Auwerx J. 1999. Regulation of peroxisome proliferator-activated receptor gamma expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol. Cell. Biol. 19: 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takashima M., Ogawa W., Emi A., Kasuga M. 2009. Regulation of SREBP1c expression by mTOR signaling in hepatocytes. Kobe J. Med. Sci. 55: E45–E52 [PubMed] [Google Scholar]
- 23.Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Brown M. S., Goldstein J. L. 2010. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. USA. 107: 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakan I., Laplante M. 2012. Connecting mTORC1 signaling to SREBP-1 activation. Curr. Opin. Lipidol. 23: 226–234 [DOI] [PubMed] [Google Scholar]
- 26.Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Xu S., Mihaylova M. M., Zheng B., Hou X., Jiang B., Park O., Luo Z., Lefai E., Shyy J. Y., et al. 2011. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 13: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw R. J. 2009. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol. (Oxf.). 196: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guntur K. V., Guilherme A., Xue L., Chawla A., Czech M. P. 2010. Map4k4 negatively regulates peroxisome proliferator-activated receptor (PPAR) gamma protein translation by suppressing the mammalian target of rapamycin (mTOR) signaling pathway in cultured adipocytes. J. Biol. Chem. 285: 6595–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersten S. 2002. Peroxisome proliferator activated receptors and obesity. Eur. J. Pharmacol. 440: 223–234 [DOI] [PubMed] [Google Scholar]
- 31.Kim J. E., Chen J. 2004. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 53: 2748–2756 [DOI] [PubMed] [Google Scholar]
- 32.Houde V. P., Brule S., Festuccia W. T., Blanchard P. G., Bellmann K., Deshaies Y., Marette A. 2010. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 59: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakrabarti P., English T., Shi J., Smas C. M., Kandror K. V. 2010. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 59: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porstmann T., Santos C. R., Lewis C., Griffiths B., Schulze A. 2009. A new player in the orchestra of cell growth: SREBP activity is regulated by mTORC1 and contributes to the regulation of cell and organ size. Biochem. Soc. Trans. 37: 278–283 [DOI] [PubMed] [Google Scholar]
- 35.Yao Z., Zhou G., Wang X. S., Brown A., Diener K., Gan H., Tan T. H. 1999. A novel human STE20-related protein kinase, HGK, that specifically activates the c-Jun N-terminal kinase signaling pathway. J. Biol. Chem. 274: 2118–2125 [DOI] [PubMed] [Google Scholar]
- 36.Su Y. C., Han J., Xu S., Cobb M., Skolnik E. Y. 1997. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 16: 1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H., Su Y. C., Becker E., Treisman J., Skolnik E. Y. 1999. A Drosophila TNF-receptor-associated factor (TRAF) binds the ste20 kinase Misshapen and activates Jun kinase. Curr. Biol. 9: 101–104 [DOI] [PubMed] [Google Scholar]
- 38.Delpire E. 2009. The mammalian family of sterile 20p-like protein kinases. Pflugers Arch. 458: 953–967 [DOI] [PubMed] [Google Scholar]
- 39.Dan I., Watanabe N. M., Kusumi A. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11: 220–230 [DOI] [PubMed] [Google Scholar]
- 40.Cobb M. H., Xu S., Hepler J. E., Hutchison M., Frost J., Robbins D. J. 1994. Regulation of the MAP kinase cascade. Cell. Mol. Biol. Res. 40: 253–256 [PubMed] [Google Scholar]
- 41.Tang X., Guilherme A., Chakladar A., Powelka A. M., Konda S., Virbasius J. V., Nicoloro S. M., Straubhaar J., Czech M. P. 2006. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc. Natl. Acad. Sci. USA. 103: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isakson P., Hammarstedt A., Gustafson B., Smith U. 2009. Impaired preadipocyte differentiation in human abdominal obesity: role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes. 58: 1550–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dérijard B., Hibi M., Wu I. H., Barrett T., Su B., Deng T., Karin M., Davis R. J. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 76: 1025–1037 [DOI] [PubMed] [Google Scholar]
- 44.Sluss H. K., Barrett T., Derijard B., Davis R. J. 1994. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol. Cell. Biol. 14: 8376–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Z. Y., Zhou Q. L., Coleman K. A., Chouinard M., Boese Q., Czech M. P. 2003. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc. Natl. Acad. Sci. USA. 100: 7569–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- 47.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 48.Rodbell M. 1964. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J. Biol. Chem. 239: 375–380 [PubMed] [Google Scholar]
- 49.Dole V. P. 1956. A relation between non-esterified fatty acids in plasma and the metabolism of glucose. J. Clin. Invest. 35: 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seki E., Brenner D. A., Karin M. 2012. A liver full of JNK: signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology. 143: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown N. F., Stefanovic-Racic M., Sipula I. J., Perdomo G. 2007. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 56: 1500–1507 [DOI] [PubMed] [Google Scholar]
- 52.Baumgartner M., Sillman A. L., Blackwood E. M., Srivastava J., Madson N., Schilling J. W., Wright J. H., Barber D. L. 2006. The Nck-interacting kinase phosphorylates ERM proteins for formation of lamellipodium by growth factors. Proc. Natl. Acad. Sci. USA. 103: 13391–13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M., Amano S. U., Flach R. J., Chawla A., Aouadi M., Czech M. P. 2013. Identification of Map4k4 as a novel suppressor of skeletal muscle differentiation. Mol. Cell. Biol. 33: 678–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie M., Zhang D., Dyck J. R., Li Y., Zhang H., Morishima M., Mann D. L., Taffet G. E., Baldini A., Khoury D. S., et al. 2006. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc. Natl. Acad. Sci. USA. 103: 17378–17383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X. X., Murray S. F., Watts L., Booten S. L., Tokorcheck J., Monia B. P., Bhanot S. 2008. Reduction of JNK1 expression with antisense oligonucleotide improves adiposity in obese mice. Am. J. Physiol. Endocrinol. Metab. 295: E436–E445 [DOI] [PubMed] [Google Scholar]
- 56.Chang Y., Wang J., Lu X., Thewke D. P., Mason R. J. 2005. KGF induces lipogenic genes through a PI3K and JNK/SREBP-1 pathway in H292 cells. J. Lipid Res. 46: 2624–2635 [DOI] [PubMed] [Google Scholar]
- 57.Ito M., Nagasawa M., Omae N., Tsunoda M., Ishiyama J., Ide T., Akasaka Y., Murakami K. 2013. A novel JNK2/SREBP-1c pathway involved in insulin-induced fatty acid synthesis in human adipocytes. J. Lipid Res. 54: 1531–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotzka J., Knebel B., Haas J., Kremer L., Jacob S., Hartwig S., Nitzgen U., Muller-Wieland D. 2012. Preventing phosphorylation of sterol regulatory element-binding protein 1a by MAP-kinases protects mice from fatty liver and visceral obesity. PLoS ONE. 7: e32609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimano H., Yahagi N., Amemiya-Kudo M., Hasty A. H., Osuga J., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K., Harada K., et al. 1999. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J. Biol. Chem. 274: 35832–35839 [DOI] [PubMed] [Google Scholar]
- 60.Shimano H., Shimomura I., Hammer R. E., Herz J., Goldstein J. L., Brown M. S., Horton J. D. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimomura I., Hammer R. E., Richardson J. A., Ikemoto S., Bashmakov Y., Goldstein J. L., Brown M. S. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12: 3182–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton J. D., Shimomura I., Ikemoto S., Bashmakov Y., Hammer R. E. 2003. Overexpression of sterol regulatory element-binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J. Biol. Chem. 278: 36652–36660 [DOI] [PubMed] [Google Scholar]
- 63.Kuriyama H., Liang G., Engelking L. J., Horton J. D., Goldstein J. L., Brown M. S. 2005. Compensatory increase in fatty acid synthesis in adipose tissue of mice with conditional deficiency of SCAP in liver. Cell Metab. 1: 41–51 [DOI] [PubMed] [Google Scholar]
- 64.Moon Y. A., Liang G., Xie X., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Brown M. S., Goldstein J. L., Horton J. D. 2012. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab. 15: 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.