Abstract
Since spheroidal HDL particles (sHDL) are highly dynamic, molecular dynamics (MD) simulations are useful for obtaining structural models. Here we use MD to simulate sHDL with stoichiometries of reconstituted and circulating particles. The hydrophobic effect during simulations rapidly remodels discoidal HDL containing mixed lipids to sHDL containing a cholesteryl ester/triglyceride (CE/TG) core. We compare the results of simulations of previously characterized reconstituted sHDL particles containing two or three apoA-I created in the absence of phospholipid transfer protein (PLTP) with simulations of circulating human HDL containing two or three apoA-I without apoA-II. We find that circulating sHDL compared with reconstituted sHDL with the same number of apoA-I per particle contain approximately equal volumes of core lipid but significantly less surface lipid monolayers. We conclude that in vitro reconstituted sHDL particles contain kinetically trapped excess phospholipid and are less than ideal models for circulating sHDL particles. In the circulation, phospholipid transfer via PLTP decreases the ratio of phospholipid to apolipoprotein for all sHDL particles. Further, sHDL containing two or three apoA-I adapt to changes in surface area by condensation of common conformational motifs. These results represent an important step toward resolving the complicated issue of the protein and lipid stoichiometry of circulating HDL.
Keywords: apolipoprotein A-I, spheroidal HDL, molecular dynamics simulation, high density lipoproteins subspecies
High density lipoproteins (HDLs) are supramolecular assemblies of lipid and protein that represent a nanoscale “soft” form of condensed matter easily deformable by thermal fluctuations (1). Because the “soft” lipid environment in HDL modulates apoA-I conformation in dynamic ways, a detailed understanding of HDL structure-function requires new approaches.
The common lipid-associating motif in apoA-I is the amphipathic α helix (2, 3). The first tangible experimental evidence for the conformation of apoA-I on the edge of discoidal HDL (dHDL) was determination of a 4 Å resolution solution phase X-ray structure for residues 44–243 of lipid-free apoA-I reported by Borhani et al. in 1997 (4) that suggested an antiparallel double-belt model. In the first experimental test of the belt model, Axelsen et al. (5) used polarized attenuated total internal reflection Fourier-transform infrared spectroscopy to conclude that the result unambiguously supported a belt model. Theoretical considerations of the geometric and physical chemical nature of the apoA-I double-belt model resulted in the publication by our lab of an atomic resolution, antiparallel, double-belt, amphipathic helical model for dHDL (6) with a helix-helix registry termed LL5/5 oriented by salt bridges (6) surrounding the edge of a bilayer disc. Five laboratories subsequently have studied reconstituted dHDL using a variety of physical chemical methods, and the results have been consistent with our double-belt model (7–11). This model provided sound theoretical rationale for use of the lipid-free X-ray crystal structure as a valid model for molecular dynamics (MD) simulations of lipid-associated apoA-I. Comparison of the results of our MD simulations of the antiparallel, full-length apoA-I structure from an ensemble of 16 MD simulations of 105 Å dHDL (12–15) with the high-resolution C-truncated crystal structure of apoA-I by Mei and Atkinson (16) validates many of our key published MD findings (17).
We previously published MD simulations of sHDL using N-terminally truncated (Δ43)apoA-I (18). We also investigated the dynamics of the activation of LCAT by apoA-I using both all-atom (AA) and coarse-grained (CG) MD simulations of dHDL and sHDL (12). More recently, we developed methodology termed particle shrinkage that allows simulation of dHDL containing full-length apoA-I (12–15).
One of the major conundrums in performing MD simulations of sHDL particles is deciding upon the surface lipid concentration. One can use experimentally determined stoichiometries for reconstituted sHDL particles (19) or develop methods to calculate surface compositions of circulating sHDL particles. In this article, we use both approaches to perform MD simulations on sHDL particles containing two or three full-length apoA-I by combining the particle shrinkage methodology with a technique we call surface loading (12). We show that circulating and reconstituted sHDL particles with the same number of apoA-I appear to contain a constant molar ratio of core lipid to apoA-I, while circulating sHDL with the same number of apoA-I have a lower ratio of surface lipid to apoA-I than comparable reconstituted particles. The likely explanation is that in vitro reconstituted particles, in the absence of phospholipid transfer protein (PLTP), contain kinetically trapped phospholipid (PL). Further, we find that sHDL containing two or three apoA-I adapt to changes in surface area by condensation of common conformational motifs.
METHODS
Rationale for protein and lipid compositions of the simulated sHDL particles
Three basic categories of starting sHDL models were simulated: i) two newly designed particles based loosely upon published protein and lipid compositional data from sHDL particles reconstituted by sonication, S2-57 (2 apoA-I, 57 POPC, 16 CE, and 6 UC) and S3-155 (3 apoA-I, 155 POPC, 58 CE, 45 UC, and 14 TG) taken from a publication by Sparks et al. (19); ii) two particles whose stoichiometries were taken directly from protein and lipid compositional data on sHDL particles reconstituted in vitro using LCAT: S2-86 [2 apoA-I, 86 POPC, 12 CE, 6 UC, and 10 TG (20)]; and S3-94 [3 apoA-I, 94 POPC, 41 CE, and 6 UC (Silva, personal communication)]; iii) a series of particles whose stoichiometries were taken directly from compositional data of isolated native circulating sHDL particles by Huang et al. (21): S2-18, S2-30a, S2-30b, and S2-34 (2 apoA-I and progressively increasing POPC derived from the LpA-I particle, HDL[1]) and S3-41 (3 apoA-I, 41 POPC, 34 CE, 9 UC, and 7 TG derived from the LpA-I particle, HDL[4]).
The initial stoichiometry of the S2-18 particle is derived from the compositional data of isolated native circulating sHDL particles of Huang et al. (21) using data from their HDL[2]/HDL3C. However, since HDL[1] is beyond the range of their density isolates, the stoichiometry of the S2-18 particle is incorrect; MD simulation produces a particle in which the core lipid is insufficiently covered by the surface protein and lipid. As discussed in the Results section, since the initial particle was obviously incorrect, we used a theoretical approach to modify the composition of the initial HDL[1] model until the final particle in the series, S2-34, fell precisely on the linear regression line.
AA force field
AA simulations were performed using NAMD 2.6–2.9 (22) as described by Jones et al. (13). Each system was ionized and charge-neutralized with NaCl to 0.15M with the ‘‘Add Ions’’ plug-in of VMD (23). The TIP3P water model was used (24). The CHARMM 22 (25, 26) and 27 (27, 28) force fields were used for protein and lipid molecules, respectively.
AA MD simulations
Our AA simulations began with a discoidal particle with a POPC:CE:UC:TG:apoA-I molar ratio of 160:0:24:0:2 that we described in Jones et al. (15). Through a series of centrally removed POPC, random deletion of UC, and random mutation of UC to CE, all followed by 5 ns of 310 K and 1 atm equilibrations, we produced a spheroidal particle with a POPC:CE:UC:TG:apoA-I molar ratio of 57:16:6:0:2.
We then equilibrated the 57:16:6:0:2 particle for an additional 5 ns of 310 K and 1 atm, and we continued for another 5 ns at the same temperature and pressure after resolvation and reionization. These two particles, at 5 ns and 10 ns, respectively, were each subjected four times to the MD-simulated annealing (MDSA) 30 ns protocol, involving an initial temperature jump to 500 K as described by Jones et al. (12) to produce an ensemble of eight particles. The first set of four particles was reported previously, but the second set is new to this article (see flow diagram in supplementary Fig. I).
Spherical particles are named by the number of apoA-I and the number of POPC molecules they contain; e.g., S2-57 (also called 2-57) contains 2 apoA-I and 57 POPC; S3-155 (also called 3-155) contains 3 apoA-I and 155 POPC. They may also contain other lipids.
CG force field
All eight final structures of the AA 30 ns simulations described in the previous section were converted to CG using the MARTINI force field for lipid (29) and protein (30) molecules, respectively. Further details about CG parameters can also be found in our previous work (18). The protein secondary structure was assigned using as a reference the one generated by AA simulations (13). This approach was employed to give more flexibility to the apoA-I chains (12, 18) of the previously described CG simulations for particles of identical molar ratios. However, even after this change, the majority of protein residues still had an α-helical conformation (80%), and all the proline residues were in a β-turn conformation.
CG MD simulations
Creation of particles S2-57.
After mapping the starting AA sHDL particles with CG beads, all the particles were solvated with CG water and ionized with CG ions to get neutral systems with a physiological ionic strength of 0.15 M. These structures were subjected to 2,000 steps of conjugate gradient energy minimization, first without and then with CG water and ions. Then CGMD simulations of 19.6 µs were performed at 310 K and 1 atm. All CGMD simulations were performed using GROMACS 4.0.7 (31). Nonbonded van der Waals and electrostatic interactions were truncated using a cutoff distance of 12 Å. Coordinate trajectories were updated every 10 ps of simulation. The pressure was held constant at 1 atm for all MD simulations using the Berendsen barostat (32). In all CGMD simulations, the effective time, which is the time of simulation multiplied by four (29, 33), is used.
Creation of particles S3-155.
The starting structure for all six particles with a POPC:CE:UC:TG:apoA-I molar ratio of 155:58:45:14:3 was an AA disk with 566 POPC and both an apoA-I double-belt and an apoA-I hairpin on the edge. After converting to CG, two similar separate reduction, modification, and CGMD sequences were performed to explore conformational space (labeled D11 to S11 and D21 to S21 in supplementary Fig. I-B). Then, two different branching simulation protocols including 4 μs T-jumps in which protein was heated to 380 K were performed to obtain the six particles S12, S13, S14, S23, S25, and S26. See the flow diagram in supplementary Fig. I-B.
From a topological standpoint, at least one hairpin is required in a disc trimer. Cross-linking data by Silva et al. (34) suggests the presence of both a double belt and a hairpin structure in the trimer. The only other possibility for the trimer is the presence of three hairpins in various head-to-tail and head-to-head orientations. We simulated such starting structures, and the results were uninteresting. Some have suggested that the timer could be a triple belt. Besides possibly being too thick for a bilayer, such a trimer would be unable to make a disc larger than 120 Å trimer discs are in the 150 Å range (14, 15, 35).
Creation of particles S3-94, S3-41a, S3-41b, and S2-86.
Similar steps were performed to create four particles with POPC:CE:UC:TG:apoA-I molar ratios of 94:41:6:0:3, 41:34:9:7:3, 41:36:9:8:3, and 86:12:6:10:2, respectively. All particles except S2-86 were subjected to an initial 2 μs simulation followed by a 4 μs T-jump with protein heated to 380 K. Then all four were simulated for 10 μs. See flow diagrams in supplementary Fig. I-A.
Creation of particles S2-34, S2-30b, and S2-30a.
The starting structure for all three particles was an AA sHDL particle with a POPC:CE:UC:TG:apoA-I molar ratio of 57:16:6:0:2 from the set of S2-57. To obtain S2-34, this structure was subjected to the mutation of 5 CE molecules to TG to reach a POPC:CE:UC:TG:apoA-I molar ratio of 57:11:6:5:2. The energy-minimized structure of this latter model was converted to CG and subjected to the manual insertion of 1 CE and 5 TG molecules, followed by the deletion of 23 POPC molecules to obtain a POPC:CE:UC:TG:apoA-I molar ratio of 34:12:6:10:2, and then it was solvated with CG polarizable water and ions and simulated for 10.8 μs (supplementary Fig. I-A). To obtain S2-30b, an AA sHDL particle from the set of S2-57 was subjected to the deletion of 22 POPC and 1 CE molecule, and to the mutation of 5 POPC molecules to TG molecules, in order to reach a POPC:CE:UC:TG:apoA-I molar ratio of 30:15:6:5:2. Then, the energy-minimized structure of the latter model was converted to CG and simulated for 22.4 μs. To obtain S2-30a, from the energy-minimized structure of particle S2-30b, 3 TG molecules were deleted to reach a POPC:CE:UC:TG:apoA-I molar ratio of 30:15:6:2:2. Then the structure was solvated with polarizable water and ions and simulated for 25.6 μs. (supplementary Fig. I-A).
Root mean-square deviation
The root mean-square deviation (RMSD) of protein α carbon atoms was calculated for all 57:16:6:2 particles over the entire duration of the eight AA and CG simulations, and then an average RMSD was calculated for each ensemble.
Fraction of total α-helicity
The fraction of all protein residues that were α-helical were measured every 20 ps over the course of all eight 30 ns AA trajectories by using VMD's protein secondary structure determination program, STRIDE (36). Then an average was calculated for all the simulated AA particles. The fraction of the time that each protein residue was α-helical (fractional α-helicity per residue) was calculated over the last 20% of each 30 ns trajectory and averaged over the eight simulated structures.
Radial density distributions
The radial density distribution plots describe the number densities of the atoms and CG beads for each component of the system (37). Two different distributions were calculated with respect to the center of mass of the particle for both the AA and CG simulations. The first measures the mole fraction of each molecular component (what fraction of the atoms or CG beads found at that distance belong to each component), and the second measures the distribution of each molecular component (the fraction of the total atoms or CG beads of each component that are found at that distance). To calculate the mole fractions, six to eleven frames over the last 20% of the simulation were selected, and each atom or CG bead of each molecular component was measured from the center of mass. To calculate the distributions, all frames over the last 20% of the simulation were selected, and each atom or CG bead of each molecular component was measured from the center of mass.
Average Cα coordinate structures
Structures created by averaging the Cα coordinates over all final structures were calculated for the AA and CG ensembles by using the “measure avpos” command in VMD after aligning over the Cα atoms containing the less solvent-accessible salt bridges (residues 77–188).
RESULTS
Simulations of reconstituted sHDL particles containing two apoA-I
Particle S2-57.
Particle S2-57, the first particle containing two apoA-I that we simulated, consisted of two apoA-I, 57 POPC, 16 CE, and six UC. This stoichiometry (with the addition of six UC) was derived from work by Sparks et al. (19), in which sHDL was produced by sonication. We monitored the structural and dynamic behavior of each model sHDL particle using two different MD methodologies: i) MDSA allows the exploration of an increased conformational space via AA simulations and an initial temperature jump; and ii) CGMD allows the exploration of a longer time scale by using CG simulations to decrease computational costs. The equilibration of these systems was monitored by measuring the RMSD of each system as described in the Methods section. The average RMSD of the ensemble of simulated particles over the entire time of simulation reaches a plateau at about 16 ns and 19.6 µs for AA and CG simulations, respectively (see supplementary Fig. II-A, B), suggesting equilibration.
Changes in total α helicity.
The fraction of all protein residues that were α-helical over the course of all the eight 30 ns trajectories of AA simulations are plotted in supplementary Fig. II-C. The α-helicity percentage plateaus at approximately 72%, comparable to the experimental value reported by Rye et al. (38) for reconstituted sHDL particles in vitro (69%).
Structural features of AA and CG ensembles.
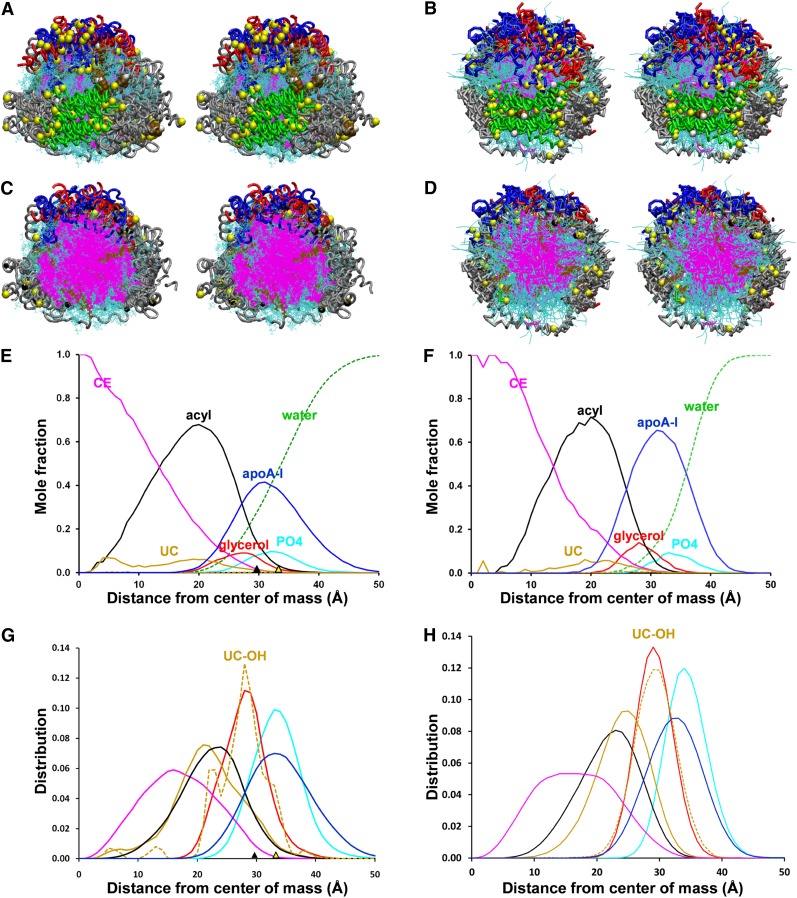
During our MD simulations, driven by the hydrophobic effect, the CE on the surface of the original discs form a separate phase or CE droplet, converting the discs to sHDL of approximately 68 Å in unhydrated diameter. Fig. 1 shows various views of the eight AA and CG sHDL ensembles in which apoA-I has been aligned in its central domain, residues 78–188; this domain represents the location of the six less solvent-accessible salt bridges involved in registration of the LL5/5 antiparallel double-belt conformation (6, 13–15, 18, 39, 40).
Fig. 1.
Properties of aligned ensembles of S2-57 sHDL particles formed after AA and CG simulations. The protein chains were aligned between residues 78 and 188, the domain containing the interhelical, less solvent-accessible salt bridges. A, B. Cross-eyed stereo views from helix 5 side (green) of AA and CG ensembles, respectively. The protein is in licorice ribbon representation: prolines, yellow space filling; helix 5, green; helix 10, red; residues 1–43, blue; remainder of protein, silver; CE, magenta lines; POPC, cyan lines; UC, brown space filling; UC-OH, white space filling. C, D. Cross-eyed stereo views of cross-sections of aligned AA and CG ensembles, respectively. Graphic representation is the same as (A, B) except UC is in line format. E, F. Plots of radial mole fractions of components of MDSA and CGMD simulations, respectively, averaged over the last 20% of their trajectories. G, H. Plots of radial distributions of components of MDSA and CGMD, respectively, averaged over the last 20% of their trajectories. CE, magenta; UC, gold; POPC acyl chains, black; UC-OH, dashed gold; glycerol moiety of POPC, red; apoA-I, blue; POPC headgroups, cyan; solvent, dashed green. The black triangles on the x axis represent the mean radius of gyration; the yellow triangles on the x axis represent the radius calculated from molecular volumes of the individual components.
Fig. 1A, B are cross-eyed stereo images of the aligned sHDL particles produced by AA and CG, respectively. It is obvious that, although produced by two very different MD methods, both ensembles of particles have similar, though dynamic, protein conformations. Cross-sectional views of the two aligned sHDL ensembles in Fig. 1C, D show clearly how the hydrophobic effect generated by MD simulation drives the CE (magenta) to the particle center of each type of ensemble, while the POPC (cyan) and UC (brown) are driven to form surface monolayers.
The radial distributions of molecular components of the AA and CG ensembles of sHDL particles averaged over the last 20% of their trajectories are plotted as mole fractions and individual distributions of each component in Fig. 1E, H. The most important observation from these plots is that both methods of MD simulation produce similar molecular distributions. Both ensembles contain only CE in their centers (Fig. 1E, F); in both ensembles the POPC glycerol and the UC hydroxyl moieties have overlapping radial distributions (Fig. 1G, H), results that have been reported for membrane bilayers but never, to the best of our knowledge, for lipoprotein particles. In both ensembles, protein and POPC headgroups have overlapping radial distribution, and the acyl chains, as well as UC, have similar distribution curves.
Particle S2-86.
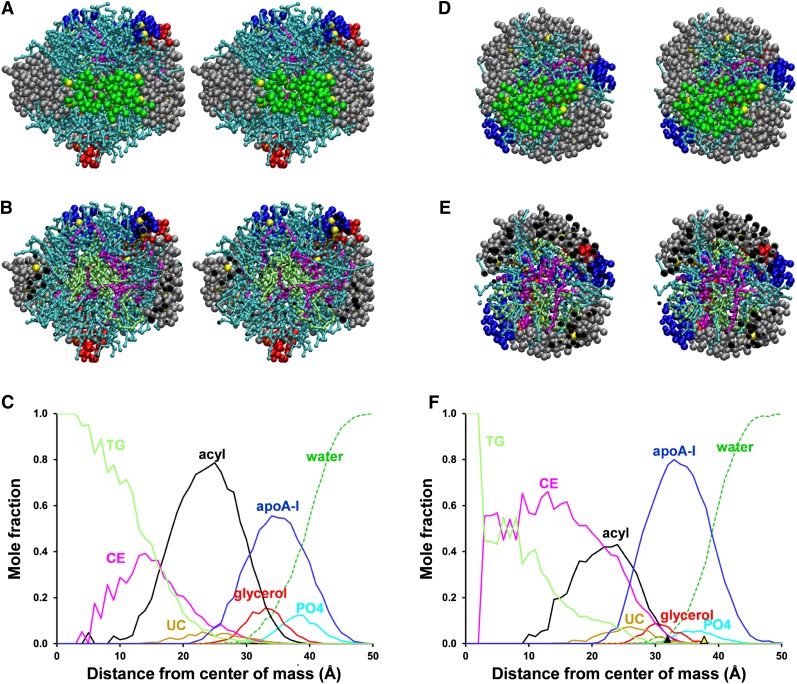
Particle S2-86, the second sHDL containing two apoA-I simulated by CGMD (Fig. 2A–C), was composed of two apoA-I, 86 POPC, 12 CE, six UC, and 10 TG. This stoichiometry was derived from work by Silva et al. (20) in which a reconstituted apoA-I:POPC disc was incubated with human LCAT and LDL as a phospholipid donor, resulting in a large spherical particle, that in turn was converted to a smaller sphere, S2-86, by incubating with CETP and Intralipid. Fig. 2A, B illustrate cross-eyed stereo images of the intact particle and the particle in cross-section, respectively. Fig. 2C represents the radial distributions of molecular components of the S2-86 particle averaged over the last 20% of their trajectory plotted as mole fractions; the radial distributions of protein and the different lipid moieties are similar to those seen for the S2-57 particles in Fig. 1E, F, with one important exception. The core lipid, TG, present in this particle, occupies the very center of the particle, displacing CE. Central TG can also be seen in the cross-sectional view (Fig. 2B).
Fig. 2.
Properties of S2-86 and S2-34 sHDL particles formed after 10 μs CGMD simulations. A, B. Cross-eyed stereo images from helix 5 side of the S2-86 particle intact and in cross-section, respectively. The protein is in space-filling mode with colors used in Fig. 1A. POPC, cyan CPK; CE, magenta CPK; UC, brown CPK; UC-OH, white CPK; TG, lime CPK. C. Plot of radial mole fractions of components of S2-86 particle, averaged over the last 20% of its trajectory. CE, magenta; UC, gold; POPC acyl chains, black; UC-OH, dashed gold; glycerol moiety of POPC, red; apoA-I, blue; POPC headgroups, cyan; solvent, dashed green. D, E. Cross-eyed stereo images from helix 5 side of S2-34 particle intact and in cross-section, respectively. The prolines are in space-filling mode with colors used in Fig. 1A. POPC, cyan CPK; CE, magenta CPK; UC, brown CPK; UC-OH, white CPK; TG, lime CPK. F. Plot of radial mole fractions of components of S2-34 particle, averaged over the last 20% of its trajectory. Color code as in (C).
Simulations of reconstituted sHDL particles containing three apoA-I
Particle S3-155.
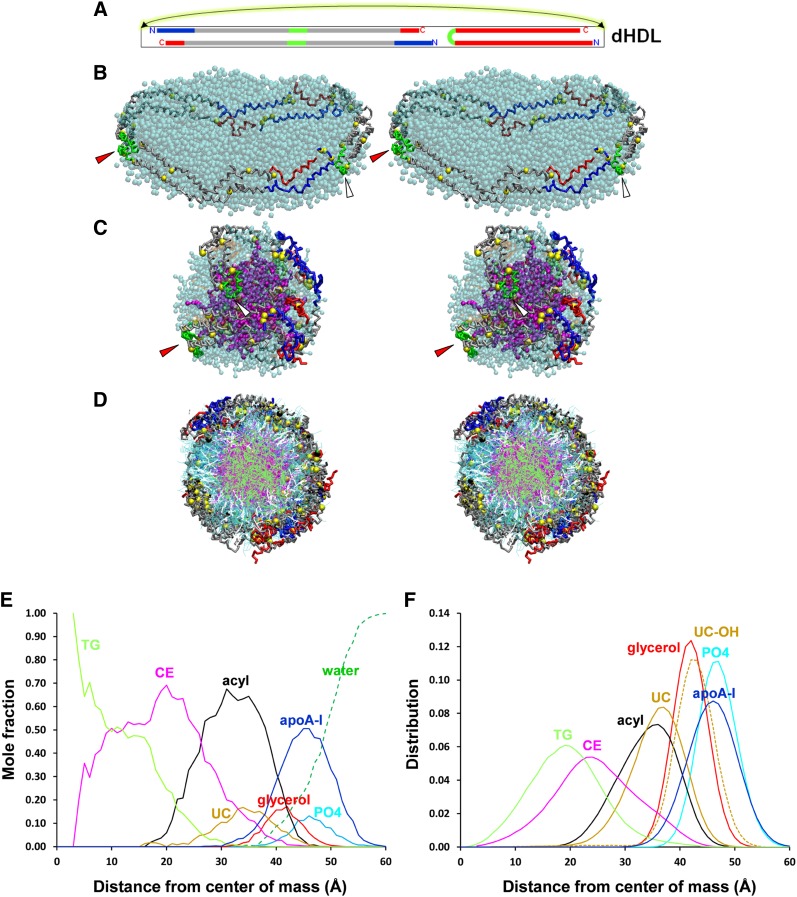
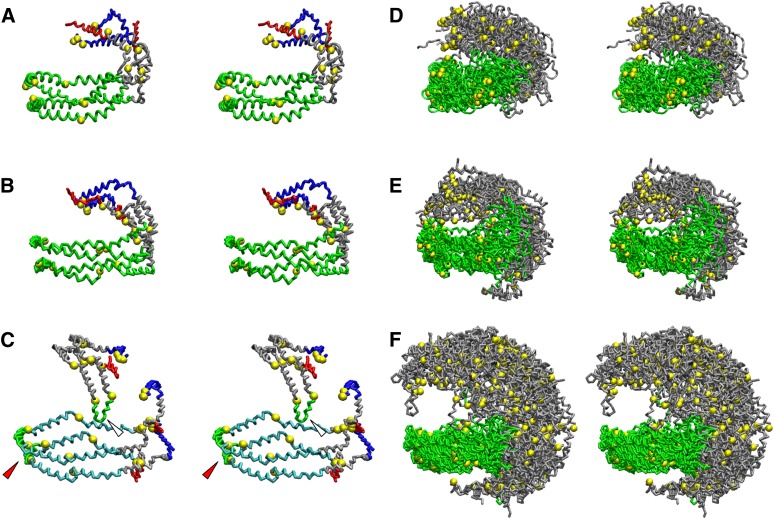
Particle S3-155, the first particle containing three apoA-I simulated by CGMD, was composed of three apoA-I, 155 POPC, 58 CE, 45 UC, and 14 TG. This stoichiometry represents a modification of a particle described by Sparks et al. (19) in which sHDL was produced by sonication by, among other changes, the addition of UC and TG to the particle. Fig. 3A shows a schematic diagram of the starting structure, a large discoidal POPC particle whose edge is lined by three apoA-I arranged as a double-belt dimer adjacent to a hairpin monomer. Fig. 3B shows a cross-eyed stereo image of the discoidal particle after an initial removal of 60 central POPC followed by 5 ns MD simulation at 310 K. In this view, the POPC surface monolayer has been rendered transparent to allow visualization of the entire apoA-I double-belt hairpin. An ensemble of six particles, approximately 80.9 Å in unhydrated diameter, was produced by a network of T-jumps (see the flow diagram in supplementary Fig. I-B). Fig. 3C is a cross-eyed stereo image of one representative example of the ensemble of S3-155 particles in which the POPC surface monolayer has been rendered transparent to allow visualization of the CE/TG core. Fig. 3D is a cross-eyed stereo image of a cross-section of an aligned ensemble of the six particles - the double-belt region of apoA-I has been aligned in its central domain, residues 78–188; this domain represents the location of the six less solvent-accessible salt bridges involved in registration of the LL5/5 antiparallel double-belt conformation (6, 13–15, 18, 39, 40). This image highlights the distribution of surface and core lipids. The radial distributions of molecular components of the S3-155 particle were averaged over the last 20% of their trajectories and plotted as mole fractions or individual distributions of each component in Fig. 3E, F. The radial distributions of protein and lipid are similar to that seen in the previous simulations; again, note the central displacement of CE by TG (Fig. 3D, E).
Fig. 3.
Properties of S3-155 sHDL particles created by CGMD simulations. A. Schematic of the starting double-belt hairpin disc. B. Cross-eyed stereo image of the discoidal particle after an initial removal of 60 central POPC followed by 5 ns MD simulation at 310 K. In this and in (C), the POPC and UC surface monolayer are in cyan and brown, respectively, and are transparent to allow visualization of the entire apoA-I double-belt hairpin, the red arrowheads denote the helix 5 antiparallel pair, and the white arrowheads denote the helix 5 hairpin. C. Cross-eyed stereo image of a representative example of the ensemble of six S3-155 particles. CE, magenta space filling; UC, brown space filling; TG, lime space filling. D. Cross-eyed stereo image of a cross-section of the aligned ensemble of six particles - the double-belt region of apoA-I has been aligned in its central domain, residues 78–188. POPC, cyan stick; UC, white stick; CE, magenta stick; TG, lime stick. E, F. Plots of radial mole fractions and radial distributions of components of the ensemble of CGMD simulations, respectively, averaged over the last 20% of their trajectories. TG, lime; CE, magenta; UC, gold; POPC acyl chains, black; UC-OH, dashed gold; glycerol moiety of POPC, red; apoA-I, blue; POPC headgroups, cyan; solvent, dashed green.
Particle S3-94.
Particle S3-94, the second sHDL containing three apoA-I simulated by CGMD (Fig. 4A–C), was composed of three apoA-I, 94 POPC, 41 CE, six UC and no TG. This stoichiometry is derived from work by Silva (Silva, personal communication) in which a reconstituted apoA-I:POPC disc was incubated with human LCAT and LDL as a phospholipid donor, resulting in a large spherical particle, S3-94. Fig. 4A, B illustrate cross-eyed stereo images of the intact particle and the particle in cross-section, respectively. The particle contains no core TG. The radial distributions of molecular components of the S3-94 particle were averaged over the last 20% of their trajectories and plotted as mole fractions in Fig. 4C. Once again, the radial distributions of protein and lipid are similar to that seen in the previous simulations.
Fig. 4.
Properties of S3-94 and S3-41 sHDL particles formed after 10 μs CGMD simulations. A, B. Cross-eyed stereo images from helix 5 side of the S3-94 particle intact and in cross-section, respectively. The protein is in space-filling mode with colors used in Fig. 1A. POPC, CE, UC, UC-OH, and TG are in CPK mode and in colors used in Fig. 2. C. Plot of radial mole fractions of components of S3-94 particle, averaged over the last 20% of its trajectory. CE, magenta; UC, gold; POPC acyl chains, black; UC-OH, dashed gold; glycerol moiety of POPC, red; apoA-I, blue; POPC headgroups, cyan; solvent, dashed green. D, E. Cross-eyed stereo images from helix 5 side of S3-41 particle intact and in cross-section, respectively. The protein is in space-filling mode with colors used in Fig. 1A. POPC, CE, UC, UC-OH, and TG are in CPK mode and in colors used in Fig. 2. F. Plot of radial mole fractions of components of S3-41 particle, averaged over the last 20% of its trajectory. Color code as in (C).
Simulations of native circulating sHDL particles containing two and three apoA-I
Best-fit volumetric analyses of the stoichiometry of native sHDL particles.
As described Ref. 41, we developed a volumetric analysis procedure for selecting native sHDL protein and lipid stoichiometries (21) for best fits to dimensions determined by nondenaturing gradient gel electrophoresis (NDGGE) for HDL isolated by immunoaffinity chromatography (42) Table 2 in Ref. 41 provides an example of this volumetric method applied to the well-characterized S2-86 (20) and S3-94 particles (Silva, personal communication). Seven particles HDL[1]–HDL[7] are described in Table 3 of Ref. 41, with HDL[1], HDL[4], and HDL[7] being predominantly LpA-I (particles with apoA-I but without apoA-II).
MD simulations of circulating sHDL particles.
On the assumption that HDL[4] is the best characterized of the seven particles with a stoichiometry of 3 apoA-I: 41 PL: 34 CE: 9 UC: 7 TG (Table 3 in Ref. 41), we created a sHDL particle with this composition (named S3-41), simulated it by CGMD for 10 μs, and compared the resulting particle to S3-94 (supplementary Fig. II-D is an RMSD plot showing that the simulation reaches equilibrium at 5 μs). The final S3-41 particle has an unhydrated diameter of 74 Å, significantly smaller than the 80.9 Å unhydrated diameter of the S3-94 particle. Fig. 4D, E illustrate cross-eyed stereo images of the intact particle and the particle in cross-section, respectively. Although S3-41 has less surface lipid (PL + UC) than S3-94 (compare Fig. 4D, E with Fig. 4A, B), importantly it has the same core volume (48,339 Å3 versus 47,946 Å3, respectively), resulting in a much thinner layer of surface lipid (compare Fig. 4E with Fig. 4B). The radial distributions of molecular components of the S3-41 particle were averaged over the last 20% of their trajectories and plotted as mole fractions in Fig. 4F; the radial distributions of protein and lipid are similar to those found in the other simulations. In spite of the small particle size, TG displaces CE from the core center.
HDL[4] compositionally falls in the middle of the five particles that Huang et al. (21) isolated for their analyses. This fact and the apoA-I-alone nature of the particle make it the best characterized of the seven circulating particles. In contradistinction, HDL[1], the smallest of the circulating apoA-I-alone particles with a diameter of 76.4 Å and a low concentration, falls outside the density range of the five particles isolated by Huang et al. (21) for their analyses. When subjected to best-fit volumetric analysis, its inaccurately calculated stoichiometry (Table 3 in Ref. 41) places it well outside the composition and size of circulating sHDL (see Fig. 5A).
Fig. 5.
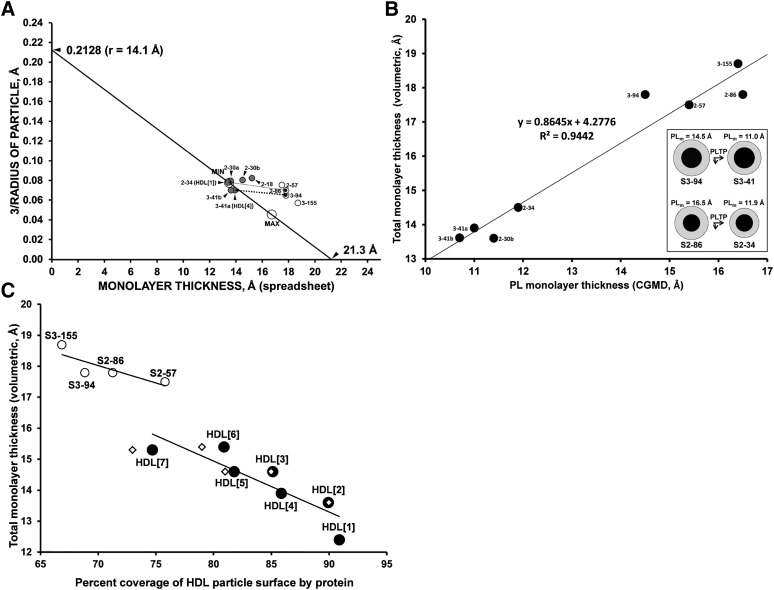
Linear regression plots of monolayer thickness measured by volumetric analysis versus surface-to-volume ratios for reconstituted sHDL particles. A. Plot of monolayer thickness measured by volumetric analysis based upon molecular volumes versus surface-to-volume ratios assuming spherical shapes for four reconstituted sHDL particles, S2-57, S2-86, S3-155 and S3-94 (open circles); six CGMD-simulated models of circulating sHDL (gray-filled circles); and upper (d = 131.8 Å) and lower (r = 13.5 Å) limits (open circles labeled MAX and MIN, respectively) of monolayer thickness plotted along the linear regression line (solid diagonal) taken from Fig. 6 in Ref. 41. The x and y intercepts of the linear regression line are indicated. The surface-to-volume ratio (S/V = (4πr2) / (4/3πr3) = 3 / radius) varies inversely with particle diameter and is a measure of the ratio of surface moieties (protein and polar lipid) to core lipid. The thick dotted arrow indicates the pathway of conversion of S3-94 to S3-41a. The thin dotted arrow indicates the pathway of conversion of S2-86 to S2-34. B. Plot of PL thickness measured by simulations versus total monolayer thickness measured by volumetric analysis for eight CGMD particles, S2-30b, S2-34, S2-57, S2-86, S3-41a, S3-41b, S3-94, and S3-155. The linear regression line y = 0.8645x + 4.2776 is shown. Insert: Cross-sectional schematic diagram to scale showing changes of cores (black) and surface monolayers (gray) during conversion of S3-94 to S3-41 and S2-86 to S2-34. presumably driven by PLTP. C. Plot of percentage coverage of HDL particle surface area versus total monolayer thickness measured by volumetric analysis for four reconstituted CGMD particles S2-57, S2-86, S3-94, and S3-155 (open circles) and seven native particles HDL[1]–HDL[7] (filled circles). Two linear regression lines are shown for each group. Measure of percentage surface area occupied by protein for HDL[2], HDL[3], and HDL[5]–HDL[7] from Huang et al. (21) (small open diamonds).
We modified the HDL[1] particle by creating two sHDL particles with increased surface lipid and decreasing core lipid, named S2-30b (30 POPC: 15 CE: 6 UC: 5 TG) and S2-30a (30 POPC: 15 CE: 6 UC: 2 TG), respectively, simulated them for more than 20 μs by CGMD, and analyzed the properties of each particle.
We then created a third particle, S2-34 (34 POPC: 12 CE: 6 UC: 10 TG), in which the core composition (with a volume of 29,898 Å3) was identical to S2-86, and simulated it for 10 μs by CGMD. The resulting particle, shown in Fig. 2D, E, has an unhydrated diameter of 65.3 Å, smaller than the 74.2 Å unhydrated diameter of the S2-86 particle. The radial distributions of molecular components of the S2-34 particle were averaged over the last 20% of their trajectories and plotted as mole fractions of each component in Fig. 2F; the radial distributions of protein and lipid are similar to that found in the other simulations; e. g., even in this small particle, TG displaces CE from the core center.
Comparison of general properties of reconstituted versus native sHDL particles.
Using the volumetric analysis, we calculated the thickness of the surface lipid monolayer for the reconstituted and native sHDL particles. In Fig. 5A, we have plotted monolayer thickness versus particle surface-to-volume ratio (S/V = 3/radius) for the four reconstituted sHDL particles, S2-57, S2-86, S3-155, and S3-94, and for the six circulating sHDL-like particles subjected to CGMD simulations, S2-18, S2-30b, S2-30a, S2-34, S3-41a, and S3-41b. The solid line is the linear regression line for the six native sHDL particles (see Fig. 6 in Ref. 41) whose equation is y = −0.01x + 0.2128. We have plotted the PL thickness measured by CGMD simulations versus the total monolayer thickness measured by volumetric analysis in Fig. 5B and the percentage coverage of HDL particle surface by protein versus total monolayer thickness measured by volumetric analysis in Fig. 5C. When the four reconstituted sHDL particles, S2-57, S2-86, S3-155 and S3-94, are plotted in Fig. 5A–C, they fall sufficiently far away from the plots of the native particles to show that all four reconstituted particles have surface lipid thicknesses greater than the largest of the native sHDL particles (large, open circle marked MAX). For example, for S3-94 to be converted to S3-41a (HDL[4]) and S2-86 to be converted to S2-34 (HDL[1]), the core volumes would not change, but the number of surface PL would decrease from 94 to 41 and 86 to 34, for S3 and S2 particles, respectively. This reduction in surface PL reduces the monolayer thicknesses from 14.5 Å to 11.0 Å for S3, and from 16.5 Å to 11.9 Å for S2 (Table 1 and Fig. 5A, thick and thin dotted arrows, respectively). The fact that the monolayer thicknesses of sHDL particles S2-86 and S3-94 reconstituted by the LCAT reaction in the absence of PLTP are identical despite the difference in particle diameters (80 Å versus 93 Å) suggests disequilibrium between the two particles. In the insert to Fig. 5B, the changes in PL monolayer thickness (PLm) of reconstituted particles S3-94 and S2-86, and of corresponding native particles S3-41 and S2-34, respectively, are shown schematically.
Fig. 6.
Cross-eyed stereo licorice images of the ensembles of the AA and CG simulations of S2-57 and the CG simulations of S3-155. A, B. Average structures of the AA and CG ensembles of S2-57. Aligned residues 78–188, green; residues 1–43, blue; helix 10, red; prolines, yellow space filling; remainder, silver. C. Average structure of the CG ensemble of six S3-155 simulations. Aligned residues 78–188, cyan; helix 5, green; residues 1–43, blue; helix 10, red; prolines, yellow space filling; remainder, silver. Red arrowhead, antiparallel helix 5 pair; white arrowhead, helix 5 hairpin. D, F. Aligned structures of the protein backbones of AA and CG ensembles for the S2-57 and the S3-155 ensemble, respectively. Aligned residues 78–188 in the double belt, green; prolines, yellow space filling; remainder of the double belt and all of the hairpins, silver.
TABLE 1.
Best-fit volumetric analysis of simulated sHDL particles
| Computational Models | |||||||
| Object | Origin | Stoichiometry |
Diameter |
Monolayer Thickness |
|||
| Lipid (PC:CE:UC:TG) | Apolipoprotein (A-I:A-II) | NDGGE (Å) | Calculated Diameter (Å) | Volumetric Analysis | MD Simulation | ||
| 2-57 | Model | 57:16:6:0 | 2:0 | — | 73.6–79.2 | 17.5 | 15.4 |
| 3-155 | Model | 155:58:45:14 | 3:0 | — | 98.7–104.3 | 18.7 | 16.4 |
| S80 (2-86) | Ref. (20) | 86:12:6:10 | 2:0 | 80 Å | 79.8–85.4 | 17.8 | 16.5 |
| S93 (3-94) | (Silva, personal communication) | 94:41:6:0 | 3:0 | 93 Å | 86.5–92.1 | 17.8 | 14.5 |
| 2-34 a | HDL[1] | 34:12:6:10 | 2:0 | — | 71.3–77.3 | 14.5 | 11.9 |
| 3-41a b | HDL[4] | 41:34:9:7 | 3:0 | 85.0 ± 1.2 | 79.6–85.2 | 13.9 | 11.0 |
Modification of composition of HDL[1] to place it on the regression line in Fig. 5A.
Two S3-41 particles with differing lipid compositions were simulated. S3-41a is equivalent to HDL[4]; S3-41b had a slightly greater lipid composition.
Our results suggest that sHDL particles such as S2-86 and S3-94 reconstituted by LCAT and CETP are not optimal models for circulating sHDL particles since they possess more surface lipids than do the circulating particles exposed to PLTP (and perhaps other proteins that remodel HDL). We suggest, instead, that sHDL particles reconstituted in the absence of PLTP are metastable; i. e. the surface PL is kinetically trapped. In the absence of PLTP it is impossible for excess surface PL, due to an almost infinitesimally small critical micelle concentration, to be transferred between particles in any biologically relevant period of time.
At least three factors influence the PL monolayer thickness in HDL subspecies: i) acyl chain tilt increases with decreasing radius and results in decreasing monolayer thickness; ii) the wedge effect of amphipathic helixes on monolayers induces increased monolayer curvature (43); and iii) the wedge effect also induces monolayer thinning (44). The wedge effects likely predominate.
The effect of the thinning of the polar lipid monolayer as native circulating HDL subspecies decrease in size has an uncertain effect on PL packing: As we demonstrated previously, LDL appears to have structural mechanisms built into apoB that dampen changes in surface pressure between different-sized subspecies (44). Similarly, because of the tendency for different domains of apoA-I to pop off and on the HDL surface, packing density might increase, decrease, or remain constant between HDL subspecies.
Further, there are a number of reasons that it is difficult to accurately predict the packing (or surface area per PL) for the different HDL subspecies. First, as we have pointed out elsewhere (13), it is very difficult to measure surface area per PL molecule on MD-simulated spheroidal structures, such as HDL. Second, calculations of surface area per PL using the volumetric method of analysis of sHDL particles is an even greater challenge, predominantly because apoA-I in particular possesses regions with variable lipid affinity. Furthermore, UC has a condensing effect on the surface area of PL monolayers (45).
A useful parameter for distinguishing between the structures of reconstituted and circulating sHDL is quantification of the percentage of the particle surface covered by protein. We have used an approximate method to calculate surface area coverage with protein with our volumetric calculations. In these calculations, we assume that each PL has a surface area of 65 Å2 that is condensed by 1% for each 2 mol % of UC and that the surface area per UC is constant at 27 Å2 (45). This approximation allows an estimate of the percentage surface area occupied by protein, and these calculations are plotted against total monolayer thickness (volumetric) in Fig. 5C for both native (filled circles) and reconstituted (open circles). While HDL[4] and S2-86 each have the same unhydrated diameters, 74 Å and 74.2 Å, respectively, protein covers 86% of the surface of HDL[4] but only71% of the surface of S2-86. HDL[7] and S3-94 have unhydrated diameters of 98 Å and 81Å but very similar percentages of protein surface coverage, 75% and 69%, respectively. Comparison of reconstituted with circulating sHDL particles with equal numbers of apoA-I clearly illustrates the structural difference between the two forms of HDL. Circulating HDL[4] and reconstituted S3-94 both contain three apoA-I per particle, but HDL[4] has greater protein surface coverage, 86% versus 69%. This difference reflects significant differences in condensation of apoA-I folding.
On the basis of two different assumptions from the ones we have used (i.e., a fixed area of 55 Å2 per PL and a surface area equivalent to a sphere with the hydrated, not the volumetric, HDL radii), Huang et al. (21) also calculated the percentage surface area occupied by protein for their isolated HDL subspecies equivalent to our HDL[2], HDL[3], HDL[5], HDL[6], and HDL[7]; their numbers are plotted onto Fig. 5C (small, open diamonds). In spite of significant differences in assumptions, both our calculations and those of Huang et al. (21) of the percentage of area occupied by protein suggests very protein-rich particle surfaces for native HDL subspecies.
Our results suggest that sHDL particles, such as S2-86 and S3-94, reconstituted by LCAT and CETP are not optimal models for circulating sHDL particles since they possess more surface lipids than do the circulating particles exposed to PLTP (and perhaps other proteins that remodel HDL). We suggest, instead, that sHDL particles reconstituted in the absence of PLTP are metastable (i.e., the surface PL is kinetically trapped). In the absence of PLTP, it is impossible for excess surface PL, due to an almost infinitesimally small critical micelle concentration, to be transferred between particles in any biologically relevant period of time.
Changes in protein conformation in reconstituted and circulating sHDL particles containing two and three apoA-I
Comparison of the average protein structures of the S2-57 and S3-155 sHDL particles.
The protein backbone in the final frames of each AA and CG ensemble of S2-57 were aligned between residues 78 and 188 (green in Fig. 6D, E), the location of the less solvent-accessible salt bridges. The remarkable similarity between the protein structures in the AA and CG simulations is dramatically illustrated by the strong similarity between the two average structures created from the aligned ensembles (Fig. 6A, B). Overall, both average structures display clam-like conformations. In both average structures, the double belts hinge at approximately the same region, the “sticky” N-terminal domains (blue) self-associate (15), and the C-terminal helix 10 domains (red) are independent or “promiscuous” (15). The double belt in this average structure (red arrowhead) forms a clam-like structure similar to that created by S2-57 but, given its larger unhydrated diameter of 80.9 Å versus 68 Å, one that is more open. The surface above the open double belt is bridged by the hairpin monomer of apoA-I (white arrowhead). The overall coverage of the small and large sHDL particles is similar. Each resembles a football helmet with a green face guard (aligned domain).
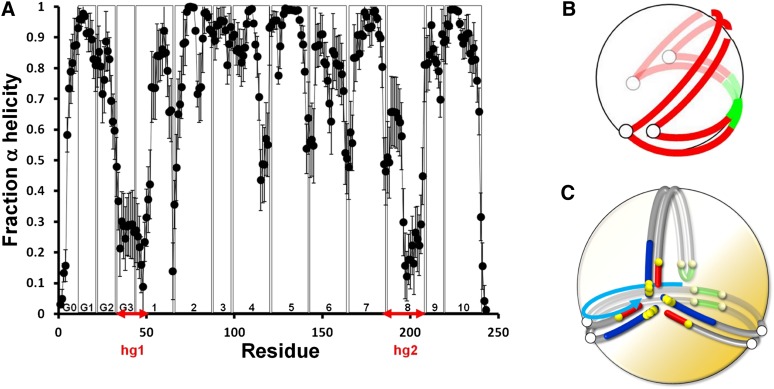
A plot of the fraction helicity of each residue averaged over the last 20% of all eight AA simulations of S2-57 is shown in Fig. 7A. This plot shows two dramatic and symmetric decreases in helicity: hg1: residues 33–52 (tandem repeat G3 and almost half of helix 1); hg2: residues 185–207 (starts at the GGA sequence at the junction of helix 7 and 8 and ends at the end of helix 8). Validation of the hg1 domain, essentially a loop-helix-loop motif, is provided by the presence of a similar motif located at residues 35–43 in the C-truncated apoA-I crystal structure (16). Interestingly, the dip in helicity at hg2 partially spares residues 189–194, an exception that includes the completely conserved residue pair E191-Y192 (46) and the residue Y192 that Shao et al. (47) suggest acts as a preferred atherogenic oxidation target in apoA-I.
Fig. 7.
Relationship of secondary and tertiary conformational features of the MD simulations. A. Fraction helicity for each residue averaged over the last 20% of the ensemble of eight AA S2-57 simulations. B. Schematic illustration of folding of discoidal double belt around hinges (open circles) to cover surface of sHDL. The pairwise helix 5 domain is green. C. Schematic illustration of folding of discoidal double belt and hairpin to cover surface of sHDL. The pairwise helix 5 domain and the helix 5 hairpin are green. The light blue arrow illustrates the dynamics of motion of the hairpin that swings in a right-handed spiral from the helix 5 pair to the N-terminal domains with an increase in simulation time (see supplementary Fig. III).
From these results, we derive the clam-fold model shown in Fig. 7B for the conformational adaptation of apoA-I from a largely planar double belt on the edge of dHDL particles to a clam-like structure that distributes itself around the surface of an sHDL particle. During this conformational change, the double belt hinges (the open circles in Fig. 7B) at the domains noted in Fig. 7A: hg1, residues 33–52, and hg2, residues 185–207. These are approximately the same residues that create the more flattened hinges in S3-155 (open circles in Fig. 7C).
Changes in protein conformation of sHDL particles during transition between large and small particles.
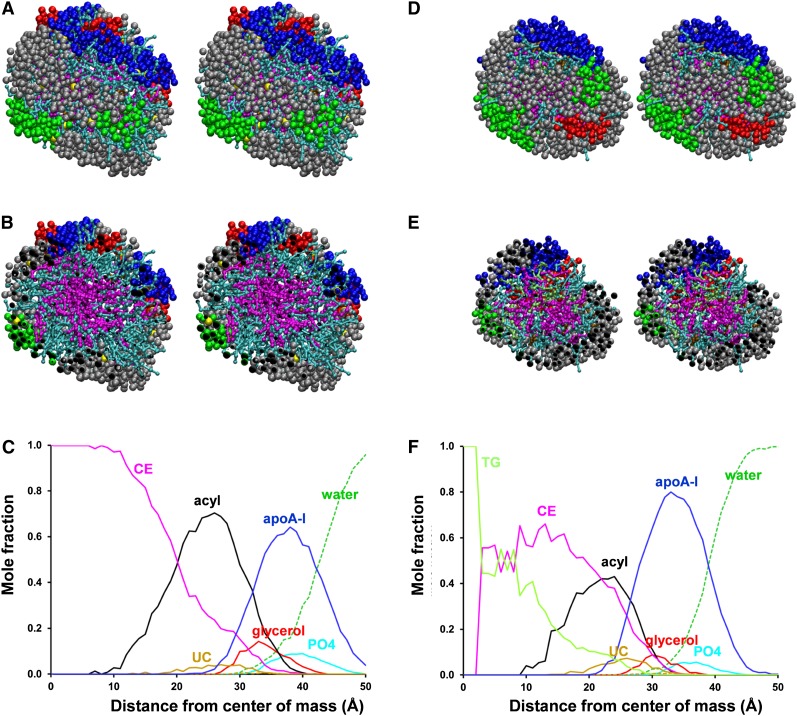
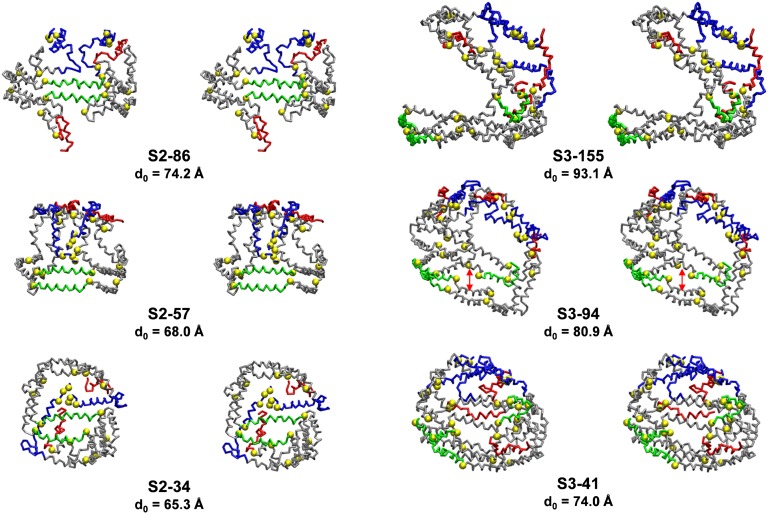
Fig. 8 compares cross-eyed stereo licorice representations of the protein conformations of the three simulated sHDL particles containing two apoA-I (S2, left-hand column) with the three containing three apoA-I (S3, right-hand column). The three S2 particles, S2-86, S2-57, and S2-34, have an unhydrated diameter (d0) of 74.2, 68.0, and 65.3 Å, respectively. The conformation of apoA-I in each particle represent variations on the clam-fold double-belt conformation noted for S2-57 (Fig. 8, middle-left panel). The largest, S2-86, represents an expanded version of the clam-fold structure in which the sticky N-terminal domains (blue) interact but in a more open fashion (Fig. 8, upper-left panel). The smallest of the S2 particles, S2-34, forms a clam-fold structure condensed by twisting upon itself so that the sticky N-terminal pair (blue) end up on the opposite side of the particle from the helix 5 antiparallel pair (Fig. 8, lower-left panel).
Fig. 8.
Cross-eyed stereo licorice images of protein conformations of the S2 and S3 particles after CGMD simulations. The three S2 particles (left-hand column) contain two apoA-I in a double belt, and the three S3 particles (right-hand column) contain three apoA-I in a double belt plus hairpin. From top to bottom, the S2 and S3 particles show the effects of core and surface lipid reductions on protein conformation. Residues 1–43, blue: helix 5, green; helix 10, red; prolines, yellow space filling. The unhydrated diameters calculated from volumes of lipid and protein components assuming a perfectly spherical shape is designated d0.
The three S3 particles S3-155, S3-94, and S3-41 have a d0 of 93.1, 80.9, and 74.0 Å, respectively. Like the three S2 particles, the three S3 particles each represent variations on a common apoA-I conformational motif, the double-belt hairpin noted for the largest, S3-155 (Fig. 8, upper-right panel). The middle S3 particle, S3-94, represents a condensed version of the double-belt hairpin (Fig. 8, middle-right panel), and the smallest, S3-41, represents a further condensation of the same basic double-belt hairpin structure (Fig. 8, lower-right panel). With particle shrinkage, the double-belt portion of the structures approaches the clam-fold conformation, and as the surface PL decreases, various portions of the double belts approach one another (red double-arrow in S3-94 model).
DISCUSSION
Role of hydrophobic effect in MD simulations
The concept of using a discoidal particle in which core lipids have been inserted by mutation into the disc bilayer is based upon the ability of MD simulations to simulate the hydrophobic effect. For CGMD, the hydrophobic effect is incorporated into the force fields for individual amino acid and lipid moieties. For example, a CG representation of water has a repulsive force toward hydrophobic moieties, such as CE. So as discs containing CE and TG are simulated by CGMD, these hydrophobic lipids are forced out of the polar monolayer by the force field, and by phase separation, they form a CE/TG core. Simulations by AAMD require no modified force field; rather, the statistical thermodynamic process that leads to the hydrophobic effect comes directly into play. Water contact with CE/TG and other hydrophobic moieties (a low probability event by statistical thermodynamics) is minimized by phase separation of the CE/TG or, for hydrophobic amino acids, by interaction with the acyl chains of surface PL, resulting in water entering a higher probability state.
Existing structural models of sHDL
Two basic structural topologies have been proposed for discoidal HDL: the picket fence and the double-belt models. On the basis of numerous physical chemical studies, the double-belt arrangement is now widely accepted as generally correct. Several variations on the double-belt model have been proposed, including the solar flare, the double superhelix, the belt buckle, and the looped belt models. None of these models is directly relevant to the structure of sHDL.
The models for sHDL can be divided into two basic types: particles containing dimeric apoA-I and those containing three or more apoA-I. The major issue so far for dimeric apoA-I has been whether the double-belt model remains intact or separates into individual helical segments during conversion of discs to spheres; all experimental evidence so far suggests that the double belt remains intact. Two models have been proposed for sHDL containing three or more apoA-I, the symmetric trefoil model and the asymmetric double-belt hairpin model.
Simulations of sHDL containing two apoA-I
Reconstituted sHDL particles.
The striking similarity of the sHDL structures for S2-57, a particle whose composition is derived from a sonication form of reconstitution, from two dramatically different MD simulation methods, AA and CG, supports in a reasonably compelling way the conclusion that MD simulations provide robust virtual experimental imaging of the structure and dynamics of sHDL. Both MD methods have strengths and weaknesses but not the same ones. The strength of AA is that it is all-atom; the principle weaknesses are a failure to robustly sample long simulation times and the use of high temperature jumps to overcome that limitation. The strength of CGMD is that it samples longer simulation times; the principle weakness is the use of pseudoatoms to represent clusters of heavy atoms.
Our results reported here support the concept first suggested computationally (18) and then experimentally (20) that the intramolecular interactions between individual apoA-I chains do not change substantially from the LL5/5 double-belt registry upon conversion from dHDL to sHDL. Rather, the double belt seems to adapt its conformation to associate with a curved spheroidal surface. For the double-belt dimer, there are essentially no degrees of freedom in conformational space not explored using this technique. If the sticky N-termini remain in contact, there are only two possibilities: the double belt can either fold back upon itself to form the clam-fold structure or twist. Our MD simulations suggest that both types of double-belt conformational changes occur, with twisting following the initial clam-fold structure during particle shrinkage. Although there are conflicting reports as to how often dimeric apoA-I occurs in sHDL (21, 48), a recent study by Gauthamadasa et al. (49) suggests that the dimer may be a significant oligomeric form of apoA-I in circulating HDL. Our volumetric analyses support this possibility (41).
The hinging of apoA-I double belts at or near hg1 (residues 33–52) and hg2 (residues 185–207) have also been observed in dHDL particles of different sizes (14), and it appears to be a general property of apoA-I double belts. Further, residues 185–207 in the middle of hg2, partially helical in our AA sHDL simulations, were also shown to be part of a loop-helix-loop motif in hg2 during MD simulations of dHDL (14).
Circulating sHDL particles.
Our MD simulation studies of the S2-86 sHDL particle, derived compositionally from published data on the composition of the S80 particle described by Silva et al. (20), along with our simulations of the native S2-34 particle and the S2-57 particle reconstituted by sonication, suggest that the clam-fold model (Fig. 7B) represents a general structural paradigm for double-belt apoA-I on the surface of both S2 and S3 forms of sHDL. Conversely, in the smallest of the S2 particles, S2-34, whose hydrated diameter is equal to that of the circulating HDL[1] particle, the clam-fold structure has twisted upon itself, so that, unlike the intermediate-sized particle S2-57, the N-N interactions have moved to the other side of the particle from the helix 5 antiparallel pairing. This twisting manner of folding or unfolding is virtually identical to the mechanism we proposed, whereby dHDL adjusts to a varying PL content by incremental winding or unwinding of the twisted apoA-I double-belt saddle shape on the discoidal particle (14).
Simulations of sHDL containing three apoA-I
The results of our MD simulations and analyses of sHDL particles containing three apoA-I show a similar pattern of folding or unfolding of the apoA-I in the transitions from larger to smaller particles as the particles containing two apoA-I. Our simulated structures are quite similar to the structure proposed by Wu et al. (50) called the helical dimer-hairpin. In the absence of PLTP, however, the major caveat with any reconstitution method producing sHDL particles is the issue of kinetically trapped excess surface lipid highlighted here; sHDL particles, such as S3-94, containing three apoA-I per particle reconstituted by LCAT alone also possess more surface lipids than do the circulating particles exposed to PLTP because the surface PL of S3-94 is kinetically trapped. Our S3-41 model, on the other hand, more accurately reflects the lipid structure of the circulating apoA-I-alone sHDL particle HDL[4] that is 85 Å in diameter (42).
The major uncertainty with our studies of sHDL containing three apoA-I is the starting model and the resulting apoA-I conformation on the sHDL surface; a disc containing a double-belt hairpin perimeter subjected to our method that combines the particle shrinkage methodology (12–14) with surface loading (12) will likely always produce the same general type of surface conformation changes in apoA-I. For apoA-I trimers, not only can a starting discoidal model contain several different conformations, a double-belt hairpin or several combinations of three hairpins, but the method is incapable of producing a final trefoil structure as proposed by Davidson's lab (20, 21). One argument for the trefoil model has been that all three (or four) apoA-I must by necessity be in equilibrium. The trefoil model requires that the double belt separate into single strands and reassemble. If the trefoil model is correct, some additional metabolic process, such as the action of LCAT or the presence of apoA-II, must be involved in overcoming the energy barrier required to split the double belt into single strands.
TG favors the core center
Finally, our MD simulation studies yield one consistent finding regardless of the particle studied: TG displaces CE from sHDL core centers. This may be due to the slight increase in polarity of CE versus TG; the low dielectric of the particle core would result in a slight increased attraction of CE for the charged particle surface compared with TG, resulting in an equilibrium that favors TG in the particle center. Another factor favoring TG over CE in the core center might be an increased flexibility of TG versus CE.
CONCLUSIONS
Our MD simulation models provide high-resolution information about the probable structure and dynamics of circulating HDL, suggesting, in particular, that a large percentage of the particle surface area, up to 85%, is covered by apolipoprotein. The high percentage of surface area coverage by apolipoproteins can be tested by experimental methods, such as cryo-electron microscopy (51–53) and chemical cross-linking (21). NMR can be used to test the low percentage of surface area coverage by surface lipid predicted by our models. Measurement of the chemical shift of the choline N-methyl region of PL with high field 1H NMR can distinguish between annular PL (within 7Å of the protein) and bulk PL (14, 54, 55). The recombinant sHDL should have a greater percentage of surface lipid in direct contact with the protein than circulating HDL particles (compare structures of recombinant sHDL with circulating HDL in Figs. 2 and 4). These models also can serve as blueprints for comprehensively understanding the mechanisms involved in the biological functions of HDL; for example, the role of apoA-I as a platform for attachment of the HDL proteome (56, 57), including apoA-II.
Supplementary Material
Acknowledgments
The authors thank UAB Information Technology and the Department of Mechanical Engineering, University of Alabama at Birmingham, for use of the commodity cluster Cheaha that they jointly maintain.
Footnotes
Abbreviations:
- AA
- all atom
- CE
- cholesteryl ester
- CG
- coarse grained
- CGMD
- coarse-grained molecular dynamics
- dHDL
- discoidal HDL
- MD
- molecular dynamics
- MDSA
- molecular dynamics simulated annealing
- NDGGE
- nondenaturing gradient gel electrophoresis
- PL
- phospholipid
- PLTP
- phospholipid transfer protein
- POPC
- palmitoyloleoylphosphatidylcholine
- RMSD
- root mean-square deviation
- sHDL
- spheroidal HDL
- TG
- triglyceride
- UC
- unesterified cholesterol
This work was supported by National Institutes of Health Grants HL-34343 and HL-102515.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures.
REFERENCES
- 1.Zlotnick A. 2004. Viruses and the physics of soft condensed matter. Proc. Natl. Acad. Sci. USA. 101: 15549–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segrest J. P., Jackson R. L., Morrisett J. D., Gotto A. M., Jr 1974. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 38: 247–258 [DOI] [PubMed] [Google Scholar]
- 3.Segrest J. P., Garber D. W., Brouillette C. G., Harvey S. C., Anantharamaiah G. M. 1994. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 45: 303–369 [DOI] [PubMed] [Google Scholar]
- 4.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koppaka V., Silvestro L., Engler J. A., Brouillette C. G., Axelsen P. H. 1999. The structure of human lipoprotein A-I. Evidence for the “belt” model. J. Biol. Chem. 274: 14541–14544 [DOI] [PubMed] [Google Scholar]
- 6.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758 [DOI] [PubMed] [Google Scholar]
- 7.Tricerri M. A., Behling Agree A. K., Sanchez S. A., Jonas A. 2000. Characterization of apolipoprotein A-I structure using a cysteine-specific fluorescence probe. Biochemistry. 39: 14682–14691 [DOI] [PubMed] [Google Scholar]
- 8.Davidson W. S., Hilliard G. M. 2003. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass specrometry study. J. Biol. Chem. 278: 27199–27207 [DOI] [PubMed] [Google Scholar]
- 9.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. 2006. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281: 20418–20426 [DOI] [PubMed] [Google Scholar]
- 10.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46: 7811–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. 2007. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14: 861–868 [DOI] [PubMed] [Google Scholar]
- 12.Jones M. K., Catte A., Li L., Segrest J. P. 2009. Dynamics of activation of lecithin:cholesterol acyltransferase by apolipoprotein A-I. Biochemistry. 48: 11196–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones M. K., Catte A., Patterson J. C., Gu F., Chen J., Li L., Segrest J. P. 2009. Thermal stability of apolipoprotein A-I in high-density lipoproteins by molecular dynamics. Biophys. J. 96: 354–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. 2010. Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285: 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones M. K., Gu F., Catte A., Li L., Segrest J. P. 2011. “Sticky” and “promiscuous”, the yin and yang of apolipoprotein A-I termini in discoidal high-density lipoproteins: a combined computational-experimental approach. Biochemistry. 50: 2249–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mei X., Atkinson D. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segrest J. P., Jones M. K., Catte A., Thirumuruganandham S. P. 2012. Validation of previous computer models and MD simulations of discoidal HDL by a recent crystal structure of apoA-I. J. Lipid Res. 53: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catte A., Patterson J. C., Bashtovyy D., Jones M. K., Gu F., Li L., Rampioni A., Sengupta D., Vuorela T., Niemela P., et al. 2008. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 94: 2306–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparks D. L., Lund-Katz S., Phillips M. C. 1992. The charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. J. Biol. Chem. 267: 25839–25847 [PubMed] [Google Scholar]
- 20.Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kale´ L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151: 283–312 [Google Scholar]
- 23.Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14: 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen W. L., Chandrasekhar J., Madura, R. W. Impey, and M. L. Klein J. D. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79: 926–935 [Google Scholar]
- 25.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4: 187–217 [Google Scholar]
- 26.MacKerell A. D., Jr, Bashford D., Bellot M., Dunbrack R. L., Jr, Evanseck J., Field M. J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102: 3586–3616 [DOI] [PubMed] [Google Scholar]
- 27.Feller S. E., Pastor R. W. 1997. Length scales of lipid dynamics and molecular dynamics. Pac. Symp. Biocomput. 1997: 142–150 [PubMed] [Google Scholar]
- 28.Schlenkrich M., Brickmann J., MacKerell A., Jr, Karplus M. 1996. An empirical potential energy function for phospholipids: criteria for parameter optimization and applications. In Biological Membranes: A Molecular Perspective from Computation and Experiment. K. M. Merz and B. Roux, editors. Birkhauser, Boston. 31–81. [Google Scholar]
- 29.Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H. 2007. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 111: 7812–7824 [DOI] [PubMed] [Google Scholar]
- 30.Monticelli L., Kandasamy S. K., Periole X., Larson R. G., Tieleman D. P., Marrink S. J. 2008. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 4: 819–834 [DOI] [PubMed] [Google Scholar]
- 31.Hess B., Kutzner C., van der Spoel D., Lindahl E. 2008. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4: 435–447 [DOI] [PubMed] [Google Scholar]
- 32.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81: 3684–3690 [Google Scholar]
- 33.Marrink S. J., de Vries A. H., Mark A. E. 2004. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 108: 750–760 [Google Scholar]
- 34.Silva R. A., Hilliard G. M., Li L., Segrest J. P., Davidson W. S. 2005. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 44: 8600–8607 [DOI] [PubMed] [Google Scholar]
- 35.Li L., Chen J., Mishra V. K., Kurtz J. A., Cao D., Klon A. E., Harvey S. C., Anantharamaiah G. M., Segrest J. P. 2004. Double belt structure of discoidal high density lipoproteins: molecular basis for size heterogeneity. J. Mol. Biol. 343: 1293–1311 [DOI] [PubMed] [Google Scholar]
- 36.Frishman D., Argos P. 1995. Knowledge-based protein secondary structure assignment. Proteins. 23: 566–579 [DOI] [PubMed] [Google Scholar]
- 37.Vuorela T., Catte A., Niemela P. S., Hall A., Hyvonen M. T., Marrink S. J., Karttunen M., Vattulainen I. 2010. Role of lipids in spheroidal high density lipoproteins. PLOS Comput. Biol. 6: e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rye K. A., Hime N. J., Barter P. J. 1995. The influence of cholesteryl ester transfer protein on the composition, size, and structure of spherical, reconstituted high density lipoproteins. J. Biol. Chem. 270: 189–196 [DOI] [PubMed] [Google Scholar]
- 39.Catte A., Patterson J. C., Jones M. K., Jerome W. G., Bashtovyy D., Su Z., Gu F., Chen J., Aliste M. P., Harvey S. C., et al. 2006. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys. J. 90: 4345–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Li S., Jones M. K., Segrest J. P. 2012. Rotational and hinge dynamics of discoidal high density lipoproteins probed by interchain disulfide bond formation. Biochim. Biophys. Acta. 1821: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segrest J. P., Cheung M. C., Jones M. K. 2013. Volumetric determination of apolipoprotein stoichiometry of circulating HDL subspecies. J. Lipid Res. 54: 2733–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung M. C., Segrest J. P., Albers J. J., Cone J. T., Brouillette C. G., Chung B. H., Kashyap M., Glasscock M. A., Anantharamaiah G. M. 1987. Characterization of high density lipoprotein subspecies: structural studies by single vertical spin ultracentrifugation and immunoaffinity chromatography. J. Lipid Res. 28: 913–929 [PubMed] [Google Scholar]
- 43.Campelo F., McMahon H. T., Kozlov M. M. 2008. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 95: 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hristova K., Wimley W. C., Mishra V. K., Anantharamiah G. M., Segrest J. P., White S. H. 1999. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J. Mol. Biol. 290: 99–117 [DOI] [PubMed] [Google Scholar]
- 45.Rog T., Pasenkiewicz-Gierula M., Vattulainen I., Karttunen M. 2009. Ordering effects of cholesterol and its analogues. Biochim. Biophys. Acta. 1788: 97–121 [DOI] [PubMed] [Google Scholar]
- 46.Bashtovyy D., Jones M. K., Anantharamaiah G. M., Segrest J. P. 2011. Sequence conservation of apolipoprotein A-I affords novel insights into HDL structure-function. J. Lipid Res. 52: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao B., Oda M. N., Bergt C., Fu X., Green P. S., Brot N., Oram J. F., Heinecke J. W. 2006. Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J. Biol. Chem. 281: 9001–9004 [DOI] [PubMed] [Google Scholar]
- 48.Colvin P. L., Moriguchi E., Barrett P. H., Parks J. S., Rudel L. L. 1999. Small HDL particles containing two apoA-I molecules are precursors in vivo to medium and large HDL particles containing three and four apoA-I molecules in nonhuman primates. J. Lipid Res. 40: 1782–1792 [PubMed] [Google Scholar]
- 49.Gauthamadasa K., Rosales C., Pownall H. J., Macha S., Jerome W. G., Huang R., Silva R. A. G. D. 2010. Speciated human high-density lipoprotein protein proximity profiles. Biochemistry. 49: 10656–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z., Gogonea V., Lee X., May R. P., Pipich V., Wagner M. A., Undurti A., Tallant T. C., Baleanu-Gogonea C., Charlton F., et al. 2011. The low resolution structure of ApoA1 in spherical high density lipoprotein revealed by small angle neutron scattering. J. Biol. Chem. 286: 12495–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones M. K., Zhang L., Catte A., Li L., Oda M. N., Ren G., Segrest J. P. 2010. Assessment of the validity of the double superhelix model for reconstituted high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285: 41161–41171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L., Yan F., Zhang S., Lei D., Charles M. A., Cavigiolio G., Oda M., Krauss R. M., Weisgraber K. H., Rye K. A., et al. 2012. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat. Chem. Biol. 8: 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L., Tong H., Garewal M., Ren G. 2013. Optimized negative-staining electron microscopy for lipoprotein studies. Biochim. Biophys. Acta. 1830: 2150–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brouillette C. G., Jones J. L., Ng T. C., Kercret H., Chung B. H., Segrest J. P. 1984. Structural studies of apolipoprotein A-I/phosphatidylcholine recombinants by high-field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry. 23: 359–367 [DOI] [PubMed] [Google Scholar]
- 55.Brouillette C. G., Segrest J. P., Ng T. C., Jones J. L. 1982. Minimal size phosphatidylcholine vesicles: effects of radius of curvature on head group packing and conformation. Biochemistry. 21: 4569–4575 [DOI] [PubMed] [Google Scholar]
- 56.Shiflett A. M., Bishop J. R., Pahwa A., Hajduk S. L. 2005. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J. Biol. Chem. 280: 32578–32585 [DOI] [PubMed] [Google Scholar]
- 57.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.