Abstract
Functional characterization of the rat elongases, Elovl5 and Elovl2, has identified that Elovl2 is crucial for omega-3 docosahexaenoic acid (DHA) (22:6n-3) synthesis. While the substrate specificities of the rat elongases had some overlap, only Elovl2 can convert the C22 omega-3 PUFA docosapentaenoic acid (DPA) (22:5n-3) to 24:5n-3, which is the penultimate precursor of DHA. In order to better understand the potential for these elongases to be involved in DHA synthesis, we have examined the molecular reasons for the differences between Elovl5 and Elovl2 in their ability to elongate DPA to 24:5n-3. We identified a region of heterogeneity between Elovl5 and Elovl2 spanning transmembrane domains 6 and 7. Using a yeast expression system, we examined a series of Elovl2/Elovl5 chimeras and point mutations to identify Elovl2 residues within this region which are responsible for DPA substrate specificity. The results indicate that the cysteine at position 217 in Elovl2 and a tryptophan at the equivalent position in Elovl5 explain their differing abilities to elongate DPA to 24:5n-3. Further studies confirmed that Elovl2 C217 is a critical residue for elongation of DPA at the level observed in the native protein. Understanding the ability of elongases to synthesize 24:5n-3 may provide a basis for using sequence data to predict their ability to ultimately support DHA synthesis.
Keywords: chimera, desaturase, docosahexaenoic acid, eicosapentaenoic acid, elongase
Synthesis of the omega-3 C22 PUFA docosahexaenoic acid (DHA) (22:6n-3) from the C18 PUFA α-linolenic acid (ALA) (18:3n-3) requires a series of desaturation and elongation reactions. Although there is evidence that Δ6desaturase is rate-limiting for conversion of ALA to the C20 PUFA eicosapentaenoic acid (EPA) (20:5n-3), it is not rate-limiting for overall DHA synthesis because the downstream products of Δ6desaturase, stearidonic acid (SDA) (18:4n-3) and EPA, are poorly converted to DHA (1–4). Therefore, we examined the two elongases, Elovl5 and Elovl2, which have been overlooked as regulators of DHA synthesis. Using a yeast expression system, it was apparent that the substrate specificities of the two rat elongases had some overlap, but that only Elovl2 could convert endogenously formed C22 PUFA docosapentaenoic acid (DPA) (22:5n-3) to 24:5n-3, which is the penultimate precursor of DHA (5). Elovl2 performs the sequential elongation of EPA to DPA followed by further elongation to 24:5n-3.
Thus, Elovl2 is crucial for DHA synthesis at least in the rat where Elovl5 cannot elongate DPA to 24:5n-3 (5). This probably explains the poor or absent ability to produce DHA in species that do not have detectable Elovl2 such as barramundi (6, 7) or in species such as the rat in which Elovl2 is expressed at low levels (5). However, there is not an absolute Elovl2 dependence for DHA synthesis in all species because the sea bream, cobia, Atlantic bluefin tuna, and chicken Elovl5 have a small but measurable ability to elongate DPA (8–11). In order to better understand the potential for these elongases to be involved in DHA synthesis, we have sought the molecular reasons for the differences between Elovl5 and Elovl2 in their ability to elongate DPA to 24:5n-3.
Purification of membrane-bound elongases to determine the substrate binding pocket has proven to be unsuccessful (12). However, chimeric elongase proteins from yeast (13), the moss Physcomitrella patens (14), and the fungi Pythium irregulare and Phytophthore infestans (15) have been used to investigate the regions involved in C18 and C20 PUFA substrate specificity and product chain length determination. Therefore, we have constructed a series of rat Elovl2/Elovl5 chimeras and point mutations to examine the Elovl2 residues responsible for DPA substrate specificity using a yeast expression system.
EXPERIMENTAL PROCEDURES
Construction of the chimeric elongase protein
Amplification of the 600 bp rat 5′-Elovl2 (position 1-600 bp), 171 bp rat 3′-Elovl2 (position 670-840 bp), and 69 bp (position 643-711 bp) rat Elovl5 fragments was performed using the chimera 1 primers in supplementary Table I, template pYES2-Elovl2 or pYES2-Elovl5, respectively (5), and Finnzymes Phusion Hot Start High-Fidelity DNA Polymerase (New England BioLabs Inc., Arundel, Qld, Australia). Cycling conditions were as follows: initial denaturation step at 98°C for 30 s, followed by 25 cycles of denaturation at 98°C for 10 s, annealing at 72/70°C (Elovl2/Elovl5) for 20 s, and extension at 72°C for 15 s, followed by a final extension at 72°C for 5 min. The Elovl2 PCR products were gel purified and DpnI digested to remove template, while the 69 bp Elovl5 PCR product was used directly for subsequent amplifications. Chimera 1 was formed using two steps. Amplification of the 5′-Elovl2+Elovl5 and Elovl5+3′-Elovl2 fragments was initially performed with template 5′-Elovl2 and Elovl5 or Elovl5 and 3′-Elovl2, respectively, using the Elovl2 cycling conditions above. Chimera 1 was then amplified using the Elovl2 primers containing restriction enzyme sites flanking the open reading frame (supplementary Table I, chimera 1) and template 5′-Elovl2+Elovl5 and Elovl5+3′-Elovl2. Cycling conditions were as follows: initial denaturation step at 98°C for 30 s, followed by 10 cycles of denaturation at 98°C for 10 s, annealing at 65–10°C (with each cycle the temperature was reduced by 1°C) for 20 s, and extension at 72°C for 15 s, followed by 20 cycles of denaturation at 98°C for 10 s, annealing at 56°C for 20 s, and extension at 72°C for 15 s, followed by a final extension at 72°C for 5 min. The chimera 1 cDNA and the expression vector pYES2 (Invitrogen Australia Pty. Ltd., Mount Waverley, Vic, Australia) were restriction enzyme treated and ligated using T4 DNA ligase (1.5 Weiss units) (Promega, WI). Transformation of the resulting construct, pYES2-chimera1 into MAX Efficiency® DH5a™ Competent Escherichia coli cells (Invitrogen Australia Pty. Ltd.) was performed using heat-shock. Putative transformants were selected using 100 mg ml−1 ampicillin and PCR screening. Recombinant plasmids were purified and sequenced at the Institute of Medical and Veterinary Science (Adelaide, Australia).
Site directed mutagenesis of the chimeric elongase, Elovl5, or Elovl2
Site directed mutagenesis (SDM) was used to change Elovl5 amino acids in chimeric protein 1 back to the equivalent Elovl2 amino acids. A series of SDM resulted in the construction of chimeric proteins 2–5. Individual amino acid changes were also made in Elovl5 or Elovl2 using SDM. Complementary primers with a minimum of twelve base pairs on either side of the introduced mutation were designed (supplementary Table I). PCR amplification was performed using the primers and corresponding template outlined in supplementary Table I and Finnzymes Phusion Hot Start High-Fidelity DNA Polymerase (New England BioLabs Inc.). Cycling conditions were as follows: initial denaturation step at 98°C for 30 s, followed by 25 cycles of denaturation at 98°C for 10 s and annealing/extension at 72°C for 4 min, followed by a final extension at 72°C for 5 min. SDM products were cleaned, DpnI digested, and transformed into E. coli as previously described.
Functional characterization of the chimeric elongase proteins in Saccharomyces cerevisiae
S. cerevisiae strain INVSc1 was transformed with each chimeric construct for the production of recombinant protein as previously described (5). Recombinant yeast expressing chimeric elongase protein was supplemented with 100–200 μM of 20:5n-3 (EPA) (Sapphire Bioscience, Waterloo, NSW, Australia) for 24 h. Each chimeric protein was functionally characterized before subsequent constructs were made. Data are expressed as the mean ± SD of incubations from three independent samples.
Fatty acid analysis
Total lipid was extracted from yeast cells and analyzed by gas chromatography as previously described (16). The amount of each fatty acid was expressed as a percentage of the total amount of all fatty acids. This was done by expressing the peak area for an individual fatty acid as a percentage of the total peak area for all fatty acids.
Statistical analysis
One-way ANOVA with Tukey's post hoc test was performed using Graphpad Prism version 5.03 for Windows (Graphpad Software, San Diego, CA). Statistical significance was set at P < 0.05.
RESULTS
Sequence analysis of rat Elovl5 and Elovl2
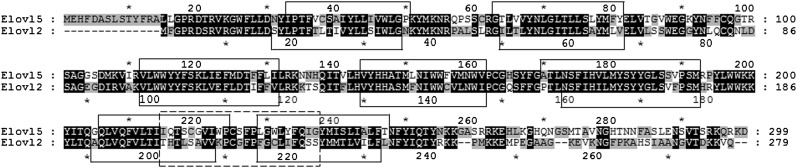
The rat Elovl5 and Elovl2 proteins share 56% identity and include the structural features characteristic of microsomal fatty acyl elongases including seven transmembrane domains (TMDs). Twenty-three residues spanning TMD6 and TMD7 in Elovl5 and Elovl2 were identified to have lower identity, 43%, compared with any similar region between TMD1 and TMD5 (Fig. 1).
Fig. 1.
A deduced amino acid sequence alignment of Elovl5 and Elovl2 from rat. Elovl5 amino acid numbering is shown above the alignment and Elovl2 numbering shown below the alignment. An asterisk indicates every ten amino acids. Identity/similarity shading was based on the Gonnet series matrix produced by ClustalX where primary black shading indicates identical residues and secondary and tertiary gray shading indicates similar residues with an 80% and 60% cut off, respectively. Seven predicted transmembrane spanning domains were predicted using the HMMTOP transmembrane topology prediction server version 2.0 and are shown with a solid lined box and a region of lower identity between transmembrane domains 6 and 7 is shown with a dashed lined box.
Native Elovl5 and Elovl2 activity
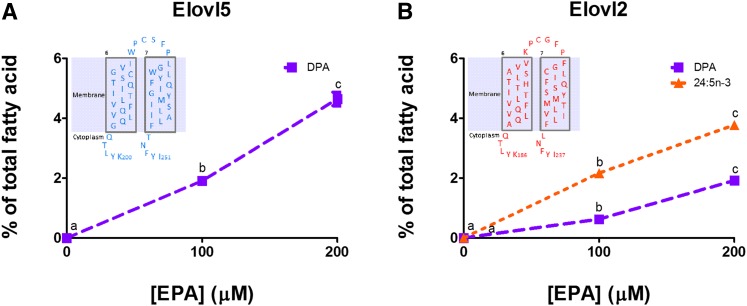
Although the topic of this investigation is DPA elongation, EPA was used as the substrate because it is common to both enzymes and the DPA that is elongated by Elovl2 is endogenously formed from EPA. EPA accumulation in the yeast cells expressing the rat Elovl5 or Elovl2 was proportional to the concentration of EPA added to the medium (data not shown). Elovl5 and Elovl2 synthesis of DPA increased proportionally with EPA substrate concentration (Fig. 2A, B). However, Elovl2 further elongated the newly synthesized DPA to 24:5n-3 (Fig. 2B), whereas Elovl5 did not (Fig. 2A).
Fig. 2.
Elongation of EPA by rat Elovl5 (A) and Elovl2 (B). Values represent the means ± SD of triplicate incubations. Values with different symbols are significantly different from each other. The inserts show the Elovl5 or Elovl2 amino acid sequence of TMD6, TMD7, and the extracellular loop.
Identification of Elovl2 residues involved in DPA substrate specificity
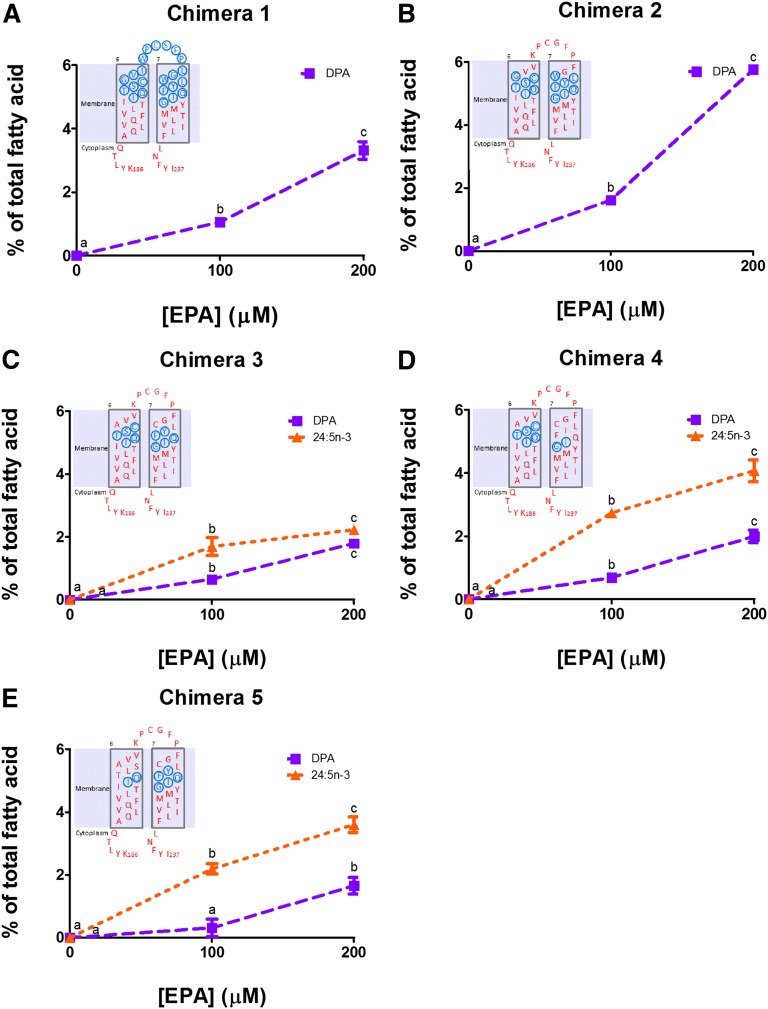
Twenty-three residues from T201 to S223 that span Elovl2 TMD6 and TMD7 were replaced with the equivalent residues from Elovl5 to form chimera 1 (Fig. 3). This resulted in a loss of the unique Elovl2 conversion of DPA to 24:5n-3, but retention of the ability to convert EPA to DPA (Fig. 4A).
Fig. 3.
Rat Elovl2/Elovl5 chimeric proteins. Amino acid numbering in the chimeras is according to the Elovl2 residue position. The table shows the amount of DPA and 24:5n-3 in yeast expressing chimeric proteins after incubation with 200 μM EPA.
Fig. 4.
Elongation of EPA by chimeric rat elongase proteins. Chimera 1 (A), chimera 2 (B), chimera 3 (C), chimera 4 (D), and chimera 5 (E). Values represent the means ± SD of triplicate incubations. Values with different symbols are significantly different from each other. The inserts show the Elovl5 residues in blue and Elovl2 residues in red within each chimeric construct across TMD6, TMD7, and the extracellular loop.
To investigate which combinations of residues were responsible for the DPA to 24:5n-3 function, residues were progressively changed back to the original Elovl2 sequence. Initially, changes were made to leave Elovl5 residues in each TMD with the rationale that these would be adjacent if TMD6 and TMD7 formed a channel for fatty acid substrates. Ten of the 23 Elovl5 residues that spanned the extracellular loop and the two adjacent membrane residues in each TMD were changed back to Elovl2 residues V207-G216 in chimera 2 (Fig. 3). Chimeric protein 2 displayed Elovl5-like activity with elongation of EPA to DPA, but no further (Fig. 4B). The restoration of a further three Elovl2 residues A206, C217, and L218 in chimeric protein 3 resulted in 24:5n-3 synthesis being partially returned (Figs. 3, 4C). Levels of 24:5n-3 reached 2.2% of total fatty acids after 200 μM EPA supplementation (Fig. 4C) compared with 3.8% 24:5n-3 with native Elovl2 (Fig. 3). In chimeras 4 and 5 a further three Elovl2 residues I219-Q221 or T203-S205 were restored in TMD7 or TMD6, respectively (Fig. 3). In both cases, this restored full Elovl2 DPA to 24:5n-3 activity with 24:5n-3 synthesis at 200 μM EPA supplementation reaching 4.1% and 3.6% of total fatty acids, respectively (Figs. 3, 4D, E).
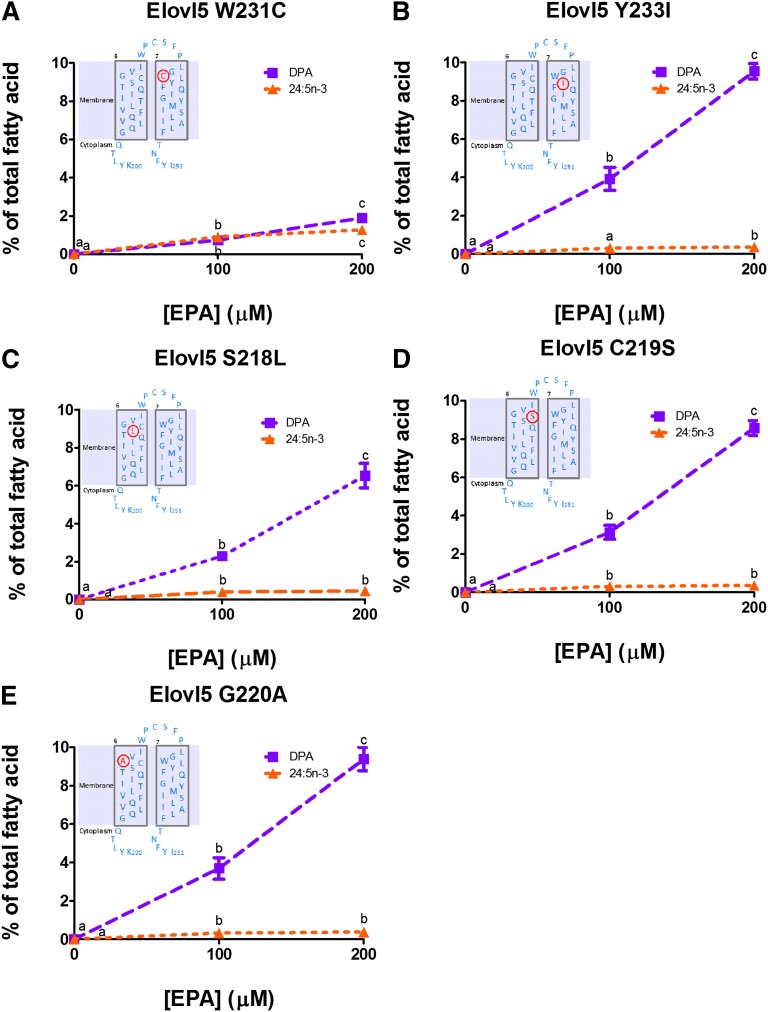
The effect of Elovl5 point mutations on EPA activity
Results with chimeric proteins 3, 4, and 5 demonstrate that the residues important for native Elovl2 DPA activity include L204-A206 in TMD6 and C217-I219 in TMD7. These residues correspond with Elovl5 S218-G220 in TMD6 and W231-Y233 in TMD7. Within these six residues, only Elovl2 L218/Elovl5 L232 is conserved. The effect of individual substitutions of the other five Elovl5 residues with the corresponding Elovl2 residue was investigated (Fig. 5). The Elovl5 W231C mutant showed a gain of Elovl2-like DPA to 24:5n-3 function (Fig. 5A) unlike the Elovl5 Y233I (Fig. 5B), S218L (Fig. 5C), C219S (Fig. 5D), and G220A (Fig. 5E) mutants which retained Elovl5-like EPA to DPA activity, with an insignificant amount of 24:5n-3 produced.
Fig. 5.
Elongation of EPA by rat Elovl5 containing amino acid point mutations Elovl5 W231C (A), Elovl5 Y233I (B), Elovl5 S218L (C), Elovl5 C219S (D), or Elovl5 G220A (E). Values represent the means ± SD of triplicate incubations. Values with different symbols are significantly different from each other. The inserts show the Elovl5 amino acid sequence of TMD6, TMD7, and the extracellular loop in blue with the Elovl2 amino acid point mutation in red.
The effect of Elovl2 point mutations on EPA activity
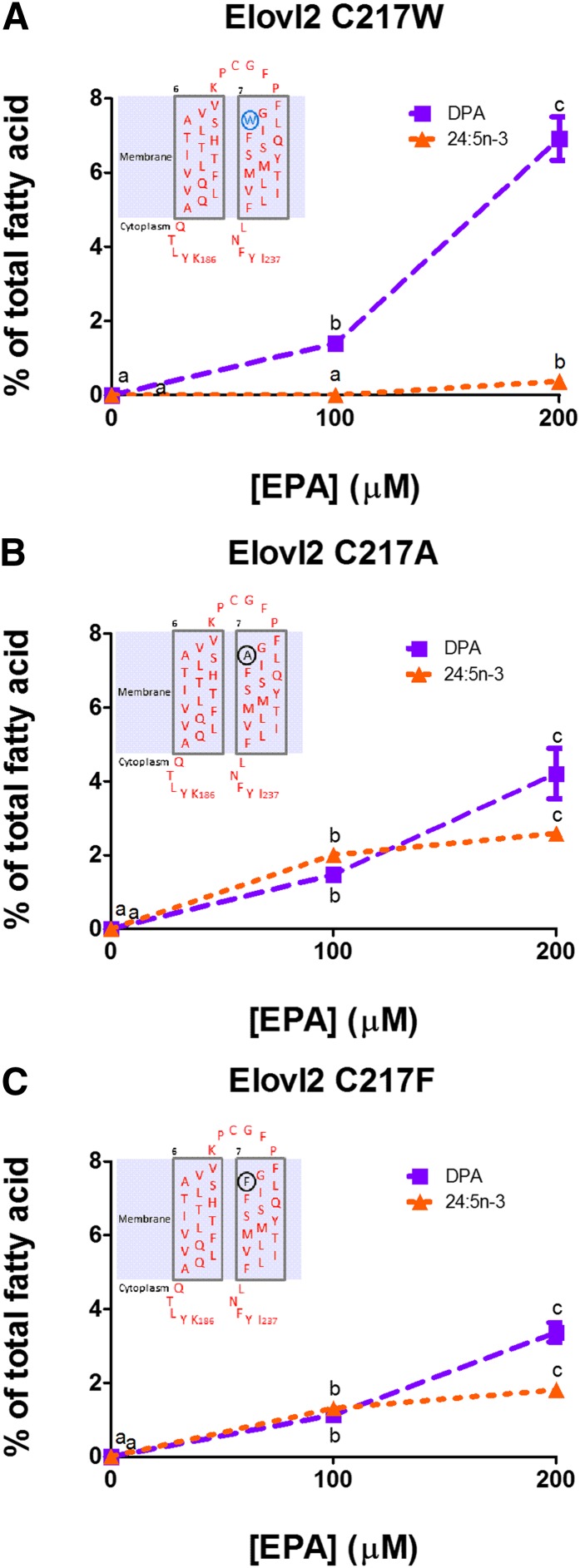
The substitution of cysteine for tryptophan in the Elovl5 W231C mutant showed the importance of Elovl2 cysteine at position 217 for elongation of DPA to 24:5n-3. When Elovl2 C217 was substituted into the equivalent position in Elovl5, there was a restoration of 24:5n-3 synthesis (Fig. 5A). In the reverse mutant, where Elovl5 W231 was substituted into the equivalent position in Elovl2, the ability to convert DPA to 24:5n-3 was lost but EPA to DPA synthetic capability was retained (Fig. 6A). To further examine the role of tryptophan in the loss of Elovl2 DPA elongation, Elovl2 point mutations were made with either the less space-filling residue alanine or another bulky residue phenylalanine. The Elovl2 C217A (Fig. 6B) and Elovl2 C217F (Fig. 6C) mutants retained DPA to 24:5n-3 activity, although at reduced levels with 24:5n-3 reaching 2.6 and 1.8%, respectively, after 200 μM EPA supplementation compared with 3.8% with native Elovl2 (Fig. 2B).
Fig. 6.
Elongation of EPA by rat Elovl2 containing amino acid position 217 point mutations Elovl2 C217W (A), Elovl2 C217A (B), or Elovl2 C217F (C). Values represent the means ± SD of triplicate incubations. Values with different symbols are significantly different from each other. The inserts show the Elovl2 amino acid sequence of TMD6, TMD7, and the extracellular loop in red, the Elovl5 amino acid point mutation in blue, and alternative amino acid point mutations in black.
Comparing functionally characterized fish and mammalian Elovl5 and Elovl2
The region of 23 residues examined in chimera 1 was compared with the deduced Elovl5 and Elovl2 protein sequences across other mammals and fish (Fig. 7). There is only one position where a residue is conserved across Elovl5 from all species which is different to the conserved residue across Elovl2 from all species, and this is the tryptophan at position 231 in rat Elovl5 which is equivalent to the cysteine at position 217 in rat Elovl2 (Fig. 7).
Fig. 7.
A deduced amino acid sequence alignment of mammalian and fish Elovl5 and Elovl2. Identity/similarity shading was based on the Gonnet series matrix produced by ClustalX where primary black shading indicates identical residues and secondary and tertiary gray shading indicates similar residues with an 80% and 60% cut off, respectively. The conserved tryptophan in all Elovl5 sequences and cysteine in all Elovl2 sequences is indicated with an arrow.
DISCUSSION
The initial reason for examining the TMD6 and TMD7 region of the rat Elovl5 and Elovl2 arose from a report that the TMD6 and TMD7 region of the yeast elongase, Sur4p, was responsible for the elongation of C18 substrates to C26 and the determination of the chain length (13). If this region is important for elongation activity in the rat enzymes, including the different substrate specificities between Elovl5 and Elovl2, it is expected that within the two sequences there must be regions of homology which enable both proteins to elongate EPA and other regions of heterogeneity which enable Elovl5 and Elovl2 to elongate SDA or DPA, respectively. An alignment of the rat Elovl5 and Elovl2 proteins highlighted a 23 residue region of heterogeneity spanning TMD6 and TMD7. This region was targeted for involvement in DPA substrate specificity due to its lower sequence identity compared with the overall sequence.
Confirmation that the TMD6 and TMD7 region was important for the differing substrate specificities between Elovl5 and Elovl2 was provided by chimera 1 in which 23 residues that span Elovl2 TMD6 and TMD7 were replaced with the equivalent residues from Elovl5. This resulted in a loss of the unique Elovl2 conversion of DPA to 24:5n-3, but with retention of the ability to convert EPA to DPA. This success provided the platform for the sequential changes in the Elovl5 insert to determine which residues were important in restoring the Elovl2 functionality of converting EPA to DPA and then to 24:5n-3. Chimeras 4 and 5 demonstrated that five residues were potentially important, three in TMD6 and two in TMD7. Of these five residues, the point mutations revealed that it was the C217 residue of Elovl2 that was critical for elongation of DPA at the level observed in the native protein. A further finding was that the loss of DPA to 24:5n-3 activity in Elovl2 C217W appeared to be caused at least in part by the inclusion of a tryptophan residue, which is at the equivalent position in Elovl5, and not simply due to the loss of the cysteine residue at this position. Cysteine is a less space-filling residue than tryptophan, which may facilitate the entry of DPA further into the transmembrane channel. However, the same substitution with another hydrophobic residue such as phenylalanine, which contains a benzyl side chain similar to tryptophan or the structurally simple alanine, retained DPA to 24:5n-3 activity, although at reduced levels. The ability of Elovl2 to elongate DPA may be due to the effect of C217 in TMD7 on the structure of Elovl2.
An alignment of the deduced Elovl5 and Elovl2 protein sequences from other functionally characterized mammals and fish supports the essentiality of cysteine at the equivalent position across all Elovl2 proteins (Fig. 7). Likewise a tryptophan is found at the equivalent position across all 16 of the Elovl5 sequences used in the alignment (Fig. 7).
Although chimera 1 and Elovl2 C231W resulted in a loss of DPA activity making the enzymatic activity of these proteins more Elovl5-like, these proteins did not gain significant Elovl5-like SDA activity (data not shown). Similarly, the gain of DPA activity by Elovl5 W231C did not result in a loss of Elovl5-like SDA activity (data not shown). These findings suggest that the residues responsible for SDA substrate specificity are not within TMD6 and TMD7, but instead in a separate region of Elovl5.
The opposite chimeric construct was made by replacing the 23 residues from I215 to G237 that span Elovl5 TMD6 and TMD7 with the equivalent from Elovl2. Interestingly, this chimeric protein was inactive when expressed in yeast and no longer able to convert SDA or EPA (data not shown). A similar finding was reported in the fungi elongases when chimeric proteins of PirELO and PinELO were made. The inclusion of a region of PirELO residues in PinELO resulted in a gain of EPA substrate specificity, whereas the reciprocal chimera resulted in an inactive PirELO which was no longer able to convert GLA or EPA (15).
We have reported that the chicken Elovl5 has some ability to elongate DPA (9). This is unlike the Elovl5 enzymes of rat (5, 17), human (18), and most, but not all, fish (6, 8, 16, 19, 20). The current study does not identify sequence differences between the chicken and rat Elovl5, which may explain their different abilities to elongate DPA. Also, it does not identify sequence variability which could explain the higher activity of the chicken Elovl5 which converts 20% DPA to 24:5n-3 compared with Elovl5 DPA conversion activities of 5–9% in sea bream, zebrafish, cobia, and Atlantic bluefin tuna (8, 10, 11, 21). The sites within these Elovl5 enzymes that confer DPA elongation ability may not be within the transmembrane regions examined in this study with rat enzymes.
The results of this study provide a starting point for further examination of the differing abilities of Elovl5 and Elovl2 to elongate DPA and the differing abilities of Elovl5 enzymes in different species to elongate DPA. A comprehensive understanding of the molecular differences responsible for these differing activities could allow sequence data to be used to assess the ability of a species or different breeds of a domestic species to elongate DPA to 24:5n-3, a critical and perhaps rate-limiting reaction for DHA synthesis.
Supplementary Material
Footnotes
Abbreviations:
- ALA
- α-linolenic acid
- DHA
- docosahexaenoic acid
- DPA
- docosapentaenoic acid
- EPA
- eicosapentaenoic acid
- SDA
- stearidonic acid
- SDM
- site directed mutagenesis
- TMD
- transmembrane domain
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
REFERENCES
- 1.Burdge G. C., Jones A. E., Wootton S. A. 2002. Eicosapentaenoic and docosapentaenoic acids are the principal products of alpha-linolenic acid metabolism in young men. Br. J. Nutr. 88: 355–363 [DOI] [PubMed] [Google Scholar]
- 2.James M. J., Ursin V. M., Cleland L. G. 2003. Metabolism of stearidonic acid in human subjects: comparison with the metabolism of other n-3 fatty acids. Am. J. Clin. Nutr. 77: 1140–1145 [DOI] [PubMed] [Google Scholar]
- 3.Fu Z., Sinclair A. J. 2000. Increased alpha-linolenic acid intake increases tissue alpha-linolenic acid content and apparent oxidation with little effect on tissue docosahexaenoic acid in the guinea pig. Lipids. 35: 395–400 [DOI] [PubMed] [Google Scholar]
- 4.Bowen R. A., Clandinin M. T. 2000. High dietary 18:3n-3 increases the 18:3n-3 but not the 22:6n-3 content in the whole body, brain, skin, epididymal fat pads, and muscles of suckling rat pups. Lipids. 35: 389–394 [DOI] [PubMed] [Google Scholar]
- 5.Gregory M. K., Gibson R. A., Cook-Johnson R. J., Cleland L. G., James M. J. 2011. Elongase reactions as control points in long-chain polyunsaturated fatty acid synthesis. PLoS ONE. 6: e29662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu W. C., Muhlhausler B. S., James M. J., Stone D. A., Gibson R. A. 2012. An alternative n-3 fatty acid elongation pathway utilising 18:3n-3 in barramundi (Lates calcarifer). Biochem. Biophys. Res. Commun. 423: 176–182 [DOI] [PubMed] [Google Scholar]
- 7.Tu W. C., Muhlhausler B. S., James M. J., Stone D. A., Gibson R. A. 2013. Dietary alpha-linolenic acid does not enhance accumulation of omega-3 long-chain polyunsaturated fatty acids in barramundi (Lates calcarifer). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 164: 29–37 [DOI] [PubMed] [Google Scholar]
- 8.Agaba M. K., Tocher D. R., Zheng X., Dickson C. A., Dick J. R., Teale A. J. 2005. Cloning and functional characterisation of polyunsaturated fatty acid elongases of marine and freshwater teleost fish. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 142: 342–352 [DOI] [PubMed] [Google Scholar]
- 9.Gregory M. K., Geier M. S., Gibson R. A., James M. J. 2013. Functional characterization of the chicken fatty acid elongases. J. Nutr. 143: 12–16 [DOI] [PubMed] [Google Scholar]
- 10.Morais S., Mourente G., Ortega A., Tocher J. A., Tocher D. R. 2011. Expression of fatty acyl desaturase and elongase genes, and evolution of DHA:EPA ratio during development of unfed larvae of Atlantic bluefin tuna (Thunnus thynnus L.). Aquaculture. 313: 129–139 [Google Scholar]
- 11.Zheng X., Ding Z., Xu Y., Monroig O., Morais S., Tocher D. R. 2009. Physiological roles of fatty acyl desaturases and elongases in marine fish: characterisation of cDNAs of fatty acyl Δ6 desaturase and elovl5 elongase of cobia (Rachycentron canadum). Aquaculture. 290: 122–131 [Google Scholar]
- 12.Guillou H., Zadravec D., Martin P. G., Jacobsson A. 2010. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49: 186–199 [DOI] [PubMed] [Google Scholar]
- 13.Denic V., Weissman J. S. 2007. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 130: 663–677 [DOI] [PubMed] [Google Scholar]
- 14.Eiamsa-Ard P., Kanjana-Opas A., Cahoon E. B., Chodok P., Kaewsuwan S. 2013. Two novel Physcomitrella patens fatty acid elongases (ELOs): identification and functional characterization. Appl. Microbiol. Biotechnol. 97: 3485–3497 [DOI] [PubMed] [Google Scholar]
- 15.Vrinten P. L., Hoffman T., Bauer J., Qiu X. 2010. Specific protein regions influence substrate specificity and product length in polyunsaturated fatty acid condensing enzymes. Biochemistry. 49: 3879–3886 [DOI] [PubMed] [Google Scholar]
- 16.Gregory M. K., See V. H., Gibson R. A., Schuller K. A. 2010. Cloning and functional characterisation of a fatty acyl elongase from southern bluefin tuna (Thunnus maccoyii). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155: 178–185 [DOI] [PubMed] [Google Scholar]
- 17.Inagaki K., Aki T., Fukuda Y., Kawamoto S., Shigeta S., Ono K., Suzuki O. 2002. Identification and expression of a rat fatty acid elongase involved in the biosynthesis of C18 fatty acids. Biosci. Biotechnol. Biochem. 66: 613–621 [DOI] [PubMed] [Google Scholar]
- 18.Leonard A. E., Bobik E. G., Dorado J., Kroeger P. E., Chuang L. T., Thurmond J. M., Parker-Barnes J. M., Das T., Huang Y. S., Mukerji P. 2000. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 350: 765–770 [PMC free article] [PubMed] [Google Scholar]
- 19.Hastings N., Agaba M. K., Tocher D. R., Zheng X., Dickson C. A., Dick J. R., Teale A. J. 2004. Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from alpha-linolenic acid in Atlantic salmon (Salmo salar). Mar. Biotechnol. (NY). 6: 463–474 [DOI] [PubMed] [Google Scholar]
- 20.Meyer A., Kirsch H., Domergue F., Abbadi A., Sperling P., Bauer J., Cirpus P., Zank T. K., Moreau H., Roscoe T. J., et al. 2004. Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J. Lipid Res. 45: 1899–1909 [DOI] [PubMed] [Google Scholar]
- 21.Agaba M., Tocher D. R., Dickson C. A., Dick J. R., Teale A. J. 2004. Zebrafish cDNA encoding multifunctional Fatty Acid elongase involved in production of eicosapentaenoic (20:5n-3) and docosahexaenoic (22:6n-3) acids. Mar. Biotechnol. (NY). 6: 251–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.