Abstract
We have performed plasma lipid profiling using liquid chromatography electrospray ionization tandem mass spectrometry on a population cohort of more than 1,000 individuals. From 10 μl of plasma we were able to acquire comparative measures of 312 lipids across 23 lipid classes and subclasses including sphingolipids, phospholipids, glycerolipids, and cholesterol esters (CEs) in 20 min. Using linear and logistic regression, we identified statistically significant associations of lipid classes, subclasses, and individual lipid species with anthropometric and physiological measures. In addition to the expected associations of CEs and triacylglycerol with age, sex, and body mass index (BMI), ceramide was significantly higher in males and was independently associated with age and BMI. Associations were also observed for sphingomyelin with age but this lipid subclass was lower in males. Lysophospholipids were associated with age and higher in males, but showed a strong negative association with BMI. Many of these lipids have previously been associated with chronic diseases including cardiovascular disease and may mediate the interactions of age, sex, and obesity with disease risk.
Keywords: lipidomics, mass spectrometry, obesity, cardiovascular disease, disease risk
Circulating lipids, their biosynthesis, metabolism, and biological functions are intimately involved in many complex disease processes (1). Traditional clinical chemistry uses measurements of total cholesterol, triglycerides, and HDL as tools for determining health status and disease risk. The tests for these lipids are low cost, high throughput, and well established. The development of soft ionization techniques, particularly electrospray ionization has proven to be a watershed for lipidomics, allowing the detection and quantification of individual molecular species. Recently, the Lipid Maps Consortium described a detailed analysis of the plasma lipidome, reporting on the concentration of nearly 600 lipids in pooled human plasma from healthy individuals (1, 2). This analysis highlighted the complexity of the plasma lipidome and the potential of plasma lipid profiling for disease classification, risk assessment, and to uncover changes in lipid metabolism associated with disease states. To date, plasma lipid profiling has been used to identify lipidomic biomarkers associated with a variety of diseases and activities related to obesity (3), hypertension (4), smoking (5), cystic fibrosis (6), weight loss (7), and type 2 diabetes (8). These studies have, in general, have been conducted using relatively small cohorts (<100 participants) (3, 4, 6, 7) and/or limited coverage of the lipidome (<100 species) (4, 6, 8).
Large population-based studies with hundreds or thousands of samples, such as ours, necessitate the need for high throughput analytical methodology. A plethora of analytical strategies have been developed for performing lipidomic profiling (9). Here, we have combined a single phase extraction method with a targeted lipidomic approach using liquid chromatography electrospray ionization tandem mass spectrometry (LC ESI-MS/MS) to compare over 300 individual plasma lipids in a large population-based cohort, from the San Antonio Family Heart Study (SAFHS) (n = 1,076) (10). The large sample size provides us with statistical power to examine novel associations between circulating molecular lipid species and common anthropometric, physiological, and lifestyle measures (age, sex, obesity, and smoking) at a population level.
METHODS
Cohort
The SAFHS investigated the genetics and risk factors of cardiovascular disease (CVD) in Mexican Americans by profiling 1,431 individuals in 42 extended families at baseline (10). All procedures were approved by the institutional review board, and all subjects gave informed consent. Plasma cholesterol, HDL cholesterol, triglycerides, glucose, and insulin were measured (Table 1). Plasma samples were collected and stored at −75°C. Extensive genomic and gene expression profiling has been performed and genome wide association studies (GWAS) have identified many loci relating to type 2 diabetes, CVD, and other complex diseases (11–15).
TABLE 1.
Anthropometric and biochemical measurements of the participants of the SAFHSa
| 39.1% Male and 23.3% Smokers | Median | Interquartile range |
| Age (years) | 35.73 | 24.67–48.88 |
| BMI (kg/m2) | 28.47 | 24.58–32.95 |
| Systolic blood pressure (mm Hg) | 117 | 108–128 |
| Diastolic blood pressure (mm Hg) | 71 | 64–77 |
| Fasting blood glucose (mmol/l) | 4.9 | 4.5–5.3 |
| Two hour post load glucose (mmol/l) | 5.6 | 4.6–7.5 |
| Triglyceride (mmol/l) | 1.35 | 0.96–1.90 |
| Cholesterol (mmol/l) | 4.81 | 4.23–5.47 |
| HDL cholesterol (mmol/l) | 1.24 | 1.06–1.47 |
| LDL cholesterol (mmol/l) | 2.87 | 2.34–3.45 |
N = 1,076. Participants with incomplete data were excluded from the statistical analysis.
Extraction procedure
Plasma samples from the SAFHS for which we had complete data (n = 1,076) were randomized prior to lipid extraction. Samples were thawed and 1 μl of the anti-oxidant butylhydroxytoluene (BHT) (100 mM in ethanol) per 1,000 μl of plasma was added. To each plasma sample (10 μl) a mixture of internal standards in chloroform:methanol (1:1, 15 μl) was added. The internal standards comprised lipids which are either stable isotope labeled or nonphysiological, and so present in plasma at extremely low concentrations (Table 2). Lipids were extracted in a single phase chloroform:methanol (2:1) procedure as described previously (16).
TABLE 2.
Conditions for tandem mass spectrometry analysis of lipid species identified in human plasma
| Lipid Class or Subclass | No. of Species | Internal Standard | Pmola | Parent Ion | Experiment | Voltage Settings (V) | |||
| DP | EP | CollE | CXP | ||||||
| dhCer | 6 | dhCer 8:0 | 100 | [M+H]+ | PIS, m/z 284.3 | 90 | 30 | 28 | 10 |
| Cer | 6 | Cer 17:0 | 100 | [M+H]+ | PIS, m/z 264.3 | 50 | 10 | 35 | 12 |
| MHC | 6 | MHC 16:0 d3 | 50 | [M+H]+ | PIS, m/z 264.3 | 77 | 10 | 50 | 12 |
| DHC | 6 | DHC 16:0 d3 | 50 | [M+H]+ | PIS, m/z 264.3 | 100 | 10 | 65 | 12 |
| THC | 6 | THC 17:0 | 50 | [M+H]+ | PIS, m/z 264.3 | 130 | 10 | 73 | 12 |
| GM | 6 | THC 17:0 | 50 | [M+H]+ | PIS, m/z 264.3 | 155 | 10 | 105 | 16 |
| SM | 19 | SM 12:0 | 200 | [M+H]+ | PIS, m/z 184.1 | 65 | 10 | 35 | 12 |
| PC | 41 | PC 13:0/13:0 | 100 | [M+H]+ | PIS, m/z 184.1 | 100 | 10 | 45 | 11 |
| PC(O) | 18 | PC 13:0/13:0 | 100 | [M+H]+ | PIS, m/z 184.1 | 100 | 10 | 45 | 11 |
| PC(P) | 8 | PC 13:0/13:0 | 100 | [M+H]+ | PIS, m/z 184.1 | 100 | 10 | 45 | 11 |
| LPC | 21 | LPC 13:0 | 100 | [M+H]+ | PIS, m/z 184.1 | 90 | 10 | 38 | 12 |
| LPC(O) | 6 | LPC 13:0 | 100 | [M+H]+ | PIS, m/z 285.2 | 90 | 10 | 42 | 5 |
| PE | 18 | PE 17:0/17:0 | 100 | [M+H]+ | NL, 141 Da | 80 | 10 | 31 | 7 |
| PE(O) | 12 | PE 17:0/17:0 | 100 | [M+H]+ | NL, 141 Da | 80 | 10 | 31 | 7 |
| PE(P) | 9 | PE 17:0/17:0 | 100 | [M+H]+ | NL, 141 Da | 80 | 10 | 31 | 7 |
| LPE | 6 | PE 14:0/0:0 | 100 | [M+H]+ | NL, 141 Da | 80 | 10 | 31 | 7 |
| PI | 17 | PE 17:0/17:0 | 100 | [M+NH4]+ | PIS, m/z 184.1 | 51 | 10 | 43 | 14 |
| PS | 7 | PS 17:0/17:0 | 100 | [M+H]+ | NL, 185 Da | 86 | 10 | 29 | 16 |
| PG | 4 | PG 17:0/17:0 | 100 | [M+NH4]+ | NL, 189 Da | 60 | 10 | 25 | 12 |
| CE | 26 | CE 18:0 d6 | 1,000 | [M+NH4]+ | PIS, m/z 369.3 | 30 | 10 | 20 | 12 |
| COH | 1 | COH d7 | 1,000 | [M+NH4]+ | PIS, m/z 369.3 | 55 | 10 | 17 | 12 |
| DG | 21 | DG 15:0/15:0 | 200 | [M+NH4]+ | NL, fatty acid | 55 | 10 | 30 | 22 |
| TG | 43 | TG 17:0/17:0/17:0 | 100 | [M+NH4]+ | NL, fatty acid | 95 | 10 | 30 | 12 |
PIS, precursor ion scan; NL, neutral loss scan; DP, declustering potential; EP, entrance potential; CollE, collision energy; CXP, collision cell exit potential.
Amount of internal standard per sample.
Lipid analysis
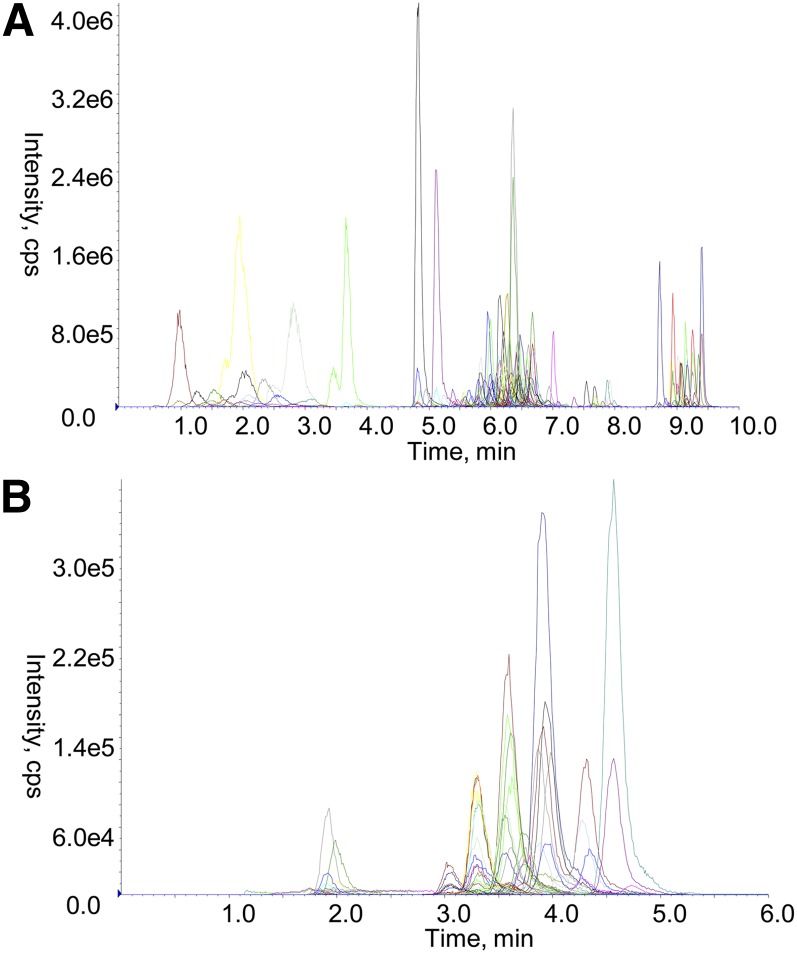
Lipid analysis was performed by LC ESI-MS/MS using an Agilent 1200 liquid chromatography system and Applied Biosystems API 4000 Q/TRAP mass spectrometer with a turbo-ionspray source (350°C) and Analyst 1.5 and Multiquant data systems. Liquid chromatography was performed on a Zorbax C18, 1.8 μm, 50 × 2.1 mm column (Agilent Technologies). Solvents A and B consisted of tetrahydrofuran:methanol:water in the ratio (30:20:50) and (75:20:5) respectively, both containing 10 mM ammonium formate. Columns were heated to 50°C and the auto-sampler regulated to 25°C. Diacylglycerol (DG) and triacylglycerol (TG) species (1 μl injection) were separated using an isocratic flow (100 μl/min) of 85% solvent B over 6 min. All other lipid species (5 μl injection) were separated under gradient conditions (300 μl/min) 0% solvent B to 100% solvent B over 8.0 min, 2.5 min at 100% solvent B, a return to 0% solvent B over 0.5 min then 10.5 min at 0% solvent B prior to the next injection. Representative chromatograms are shown in Fig. 1.
Fig. 1.
LC ESI-MS/MS of plasma lipids. Lipids were extracted from a QC plasma (10 μl) as described in Methods and analyzed by gradient LC ESI-MS/MS for all lipids except DG and TG species (A) or isocratic LC ESI-MS/MS for DG and TG species (B) as described in Methods. Each colored trace represents a different ion pair. cps, counts per second.
Method development and identification of plasma lipid species
Internal standards were available for most lipid classes and subclasses investigated. Using direct infusion experiments of these standards, declustering potential, collision energy, and exit potential were optimized to give maximum response. Using these values, precursor ion scans and neutral loss scans were performed on a lipid extract of pooled plasma obtained from healthy volunteers to identify the major lipid species of the following classes and subclasses: dihydroceramide (dhCer), ceramide (Cer), monohexosylceramide (MHC), dihexosylceramide (DHC), trihexosylceramide (THC), GM3 ganglioside (GM), sphingomyelin (SM), phosphatidylcholine (PC), alkylphosphatidylcholine [PC(O)], phosphatidylcholine plasmalogen [PC(P)], lysophosphatidylcholine (LPC), lysoalkylphosphatidylcholine [LPC(O)], phosphatidylethanolamine (PE), alkylphosphatidylethanolamine [PE(O)], phosphatidylethanolamine plasmalogen [PE(P)], lysophosphatidylethanolamine (LPE), phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylglycerol (PG), cholesterol ester (CE), free cholesterol (COH), DG, and TG (Table 2). Species that were chromatographically separated and gave a signal within the linear range of response (see Linearity of response below and Table 3) were subsequently incorporated into multiple reaction monitoring (MRM) experiments for comparative analysis. In the case of DG and TG species, the acyl chains were identified by chromatographically aligning the different neutral loss scans of each precursor mass. In cases where more than one combination of acyl chains was possible for one precursor mass, a unique fragment was chosen to distinguish between isomers that were not chromatographically separated. MRM experiments, established for each lipid species, were combined into two scheduled MRM experiments whereby data from each MRM was only collected during its retention time window (± 30 sec) (see Table 2 and supplementary Table I).
TABLE 3.
Assay performance of lipid classes and subclasses from plasma QC samples
| Lipid Class or Subclassa | Linear Range (μM)b | R2 | Recovery, %c | Intra-run Variation, %d | Assay Variation, %e |
| dhCer | 0.005–100 | 0.990 | 89.6 | 9.9 | 13.1 |
| Cer | 0.005–100 | 0.996 | 91.8 | 7.2 | 6.8 |
| Monohexosylceramidef | — | — | 88.4 | 8.5 | 13.6 |
| DHC | 0.01–100 | 0.999 | 91.3 | 9.2 | 9.6 |
| THC | 0.01–100 | 0.999 | 89.6 | 8.8 | 9.6 |
| GMf | — | — | — | 8.3 | 9.1 |
| SM | 0.005–500 | 0.988 | 90.4 | 5.4 | 6.4 |
| PC | 0.005–500 | 0.988 | 89.6 | 7.7 | 8.3 |
| PC(O)f | — | — | — | 6.9 | 8.9 |
| PC(P)f | — | — | — | 5.7 | 9.4 |
| LPC | 0.01–100 | 0.999 | 90.6 | 7.9 | 14.3 |
| LPC(O)f | — | — | — | 6.0 | 6.4 |
| PE | 0.005–100 | 0.994 | 88.2 | 5.5 | 6.3 |
| PE(O)f | — | — | — | 6.2 | 7.4 |
| PE(P) | 0.005–100 | 0.994 | — | 7.4 | 7.8 |
| LPE | 0.005–100 | 0.994 | 87.7 | 7.4 | 9.8 |
| PIf | — | — | — | 6.9 | 8.3 |
| PSf | — | — | 89.1 | 22.4 | 22.6 |
| PG | 0.01–100 | 0.998 | 89.1 | 13.2 | 13.8 |
| CE | 0.005–1000 | 0.998 | 86.3 | 12.4 | 13.2 |
| COH | 5–100 | 0.999 | 83.7 | 17.9 | 17.5 |
| DG | 0.05–100 | 0.996 | 92.3 | 20.1 | 17.5 |
| TG | 0.05–1000 | 0.997 | 93.3 | 23.7 | 20.4 |
Sum of the individual species within that class or subclass.
Calculated from one lipid species (spiked into QC plasma) and compared with the internal standard for that class or subclass.
Percent recovery compared with matrix spike.
Average intra-run CV from four separate analyses.
CV from 63 QC plasma samples across 1,076 samples in four analytical runs.
No suitable standards were available.
Acquisition of comparative lipidomic data
Comparative lipid abundances were calculated by relating the peak area of each species to the peak area of the corresponding internal standard. Peak integration was performed using AB Sciex MultiQuant software v1.2. Total measured lipids of each class were calculated by summing the abundance of individual lipid species. In a number of cases described below correction factors were applied.
DG and TG.
Fragmentation of the ammoniated adducts of DGs and TGs leads to the loss of ammonia and a fatty acid. However, DG can also lose water, which must also be considered to avoid erroneous assignment of the fatty acids in each DG species. For species containing different fatty acids, multiple product ions corresponding to the loss of each of the fatty acids will be formed and the signal divided between these competing pathways. While we would ideally monitor each of these losses with a separate MRM transition, the number of MRM transitions that would be required was too great to be compatible with the chromatographic timescale on which we were working. As a result, a single MRM transition was used to monitor each DG and TG. In this context it is important to recognize that for species which contain more than one of the same fatty acid, the loss of that fatty acid will result in an enhanced signal, as it is the end product from two competing pathways. Consequently, where we used an MRM transition that corresponded to the loss of a fatty acid that was present more than once, we divided by the number of times that fatty acid was present. While we recognize that the response factor for different species of TG varied substantially, the lack of suitable standards precluded the determination of suitable response factors for each TG species.
CE.
Response factors were determined with seven commercially available species and used to create a formula to extrapolate for all CE chain lengths and double bonds. Saturated species were characterized by the following relationship: y = 0.74x − 10.56, where y is the response factor relative to the CE 18:0 d6 internal standard and x is the carbon chain length. For monounsaturated species, the response factor was multiplied by 1.62 and for polyunsaturated species by 4.40.
Statistical analysis
Linear regression was used for analyzing and describing the linear relationship between lipids and selected characteristics in the SAFHS study. The β-coefficient describes the slope of the regression line and reflects the amount of variance of the dependent variable that is explained by variation of the independent variable. We applied linear regression to identify linear associations of each individual lipid species and each lipid class or subclass to age and body mass index (BMI) adjusting for appropriate covariates in each analysis. Logistic regression was used for analyzing and describing the relationship between lipids and dichotomous dependent variables in the SAFHS study. It relates the log odds of the probability of an event to a linear combination of the predictor variables. We applied logistic regression to identify associations of each individual lipid species and each lipid class or subclass to gender and smoking status adjusting for appropriate covariates in each analysis. The P values for linear and logistic regression were adjusted for multiple comparisons using the Benjamini-Hochberg approach (17).
Pearson correlation analysis was used to describe the linear relationship between each lipid species and all other lipid species.
RESULTS
Identification of major lipid species in human plasma
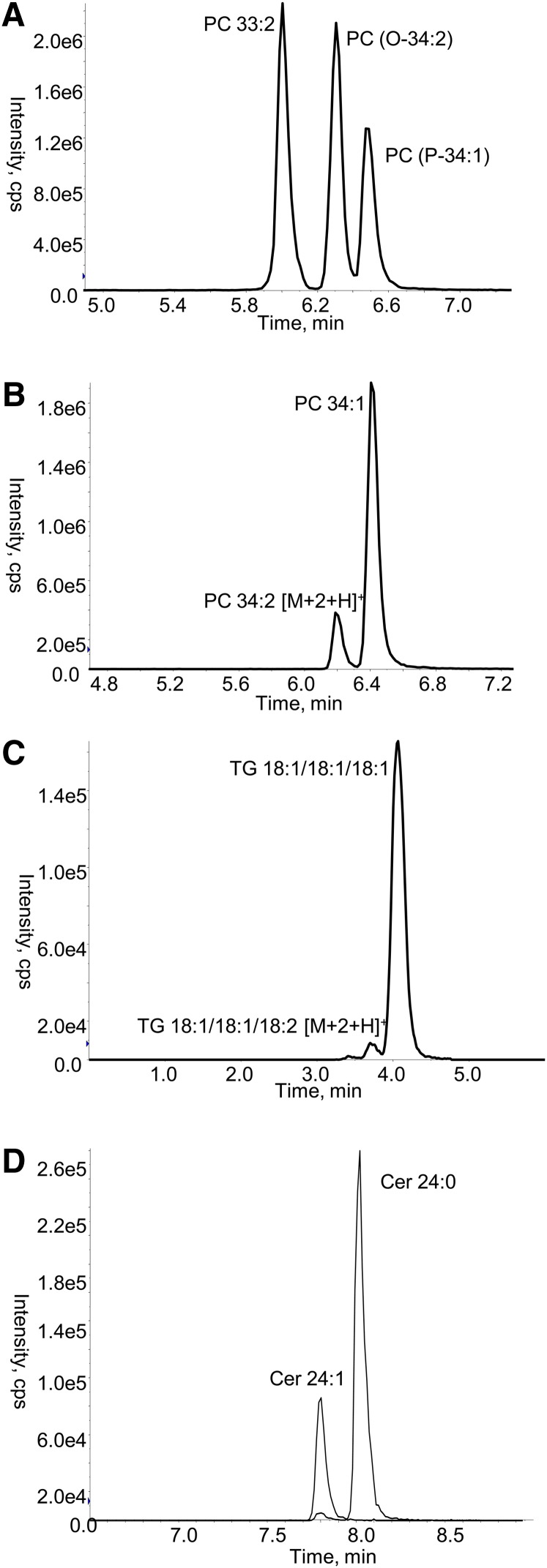
The lipids in each class and subclass examined were identified using pooled plasma samples from healthy volunteers by neutral loss and precursor ion scans (Table 2) and confirmed with product ion scans in positive and negative mode. Although the reverse phase chromatography resulted in the coelution of different lipid classes (Fig. 1), it did provide clear separation of isobaric species within each class such that isobaric species of PC(O), PC(P), and PC were well separated (Fig. 2A, supplementary Table I). Similarly, lipid species of the same class which differ only by a double bond are chromatographically separated. This is particularly important as the [M+2+H]+ ions of the more highly unsaturated species have the same nominal mass as the monoisotopic [M+H]+ ions of the less saturated species. As a result, the more highly unsaturated species give rise to a signal for the MRM transition used to monitor the less saturated species. This chromatographic resolution was possible for all lipid classes and subclasses (examples shown in Fig. 2B–D; full data in supplementary Table I).
Fig. 2.
LC ESI-MS/MS of isomeric and isobaric lipid species. Lipids were extracted from a QC plasma (10 μl) and analyzed by gradient LC ESI-MS/MS as described in Methods. A: Shows the chromatographic separation of three isomeric and isobaric species of PC, detected using the ion pair (744.6/184.1). B: Shows the separation of two isobaric species of PC detected by the ion pair (760.6/184.1). C: Shows the separation of two isobaric species of TG detected by the ion pair (902.9/603.6). D: Shows the separation of two species of ceramide detected by the ion pair (650.6/264.3). cps, counts per second.
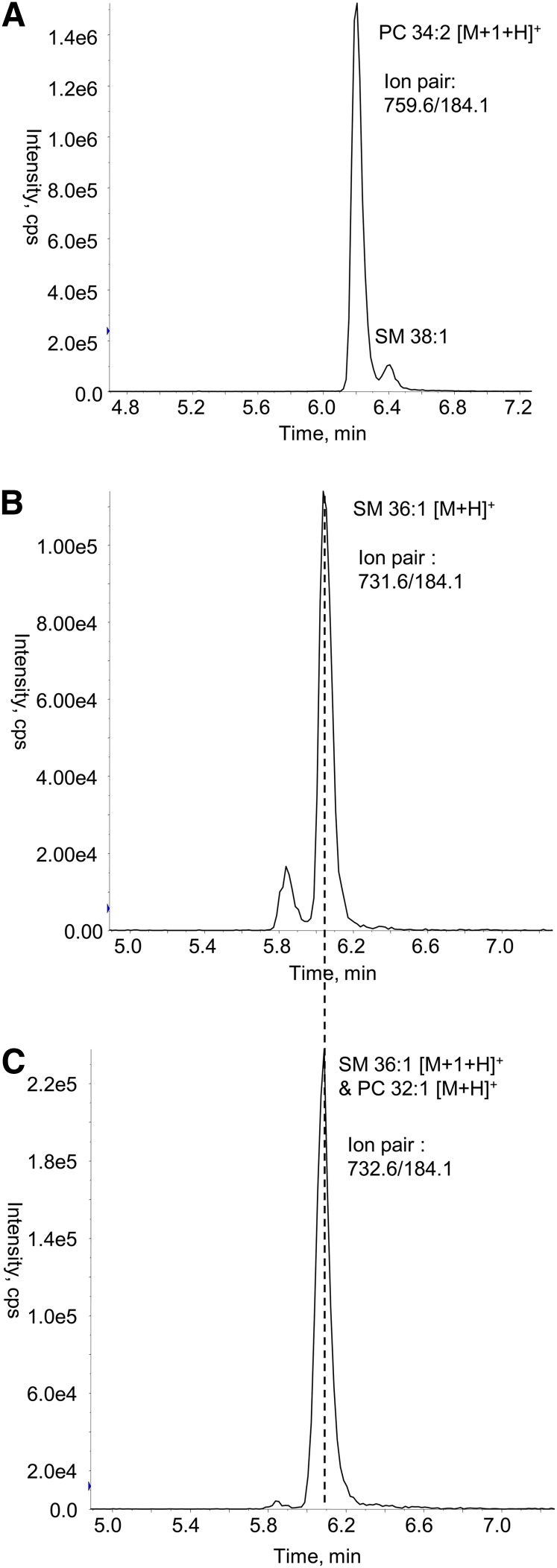
Further, as PC and SM undergo the same fragmentation to produce the phosphocholine head group with m/z 184, they can potentially be convoluted, if not separated chromatographically. The chromatography provided clear separation of the PC isotopologues from the isobaric SM species (Fig. 3A), but the SM isotopologues did coelute with the PC species (Fig. 3B, C). In many instances this did not make a significant difference to the PC species as the SM isotopologue was of relatively low abundance. In cases where the signal from the SM isotopologue represented more than 5% of the PC species, the PC species was excluded from the analysis (species excluded on this basis were PC 32:3, PC 32:2, PC 32:1, PC 35:1, and PC 36:1).
Fig. 3.
LC ESI-MS/MS of isobaric SM and PC species. Lipids were extracted from a QC plasma (10 μl) and analyzed by gradient LC ESI-MS/MS as described in Methods. A: Shows the chromatographic separation of two isobaric species, detected using the ion pair (759.6/184.1). B: Shows detection of SM 36:1 using the ion pair (731.6/184.1). C: Shows coelution of PC 32:1 and the isotope of SM 36:1 at 6.05 min, detected by the ion pair (732.6/184.1). cps, counts per second.
Recovery
Recovery of lipids was calculated by extracting plasma, spiked with internal standards, and comparing peak areas to those of plasma extracts that were spiked with the same standards after extraction. Samples were reconstituted according to the protocol and analyzed as described (Methods). The recoveries of all standards were >86% (mean 89.0%, median 89.6%, Table 3). We have made the assumption that recovery of the internal standard is representative of the recovery for associated lipid species in that class or subclass.
Linearity of response
The linearity and range of the lipid measurements was assessed using serial dilutions of lipid standards in a plasma matrix (Table 3). Under the assay conditions used, we observed detector saturation for the most abundant lipids; such saturation could lead to nonlinear responses. To circumvent this problem for those abundant lipids, in quadrapole 1 (Q1) we selected for the isotopologues which have a higher m/z (1 or 2 Da, designated as M+1 or M+2, respectively) but lower abundance than the monoisotopic parent ion. We incremented the corresponding Q3 m/z by the same amount as the Q1 m/z. These species were then normalized to the Q1 and Q3 ion pair of the internal standard corresponding to the equivalent isotopolgues. Using a combination of transitions corresponding to the monoisotopic ion and the Q1 and Q3 values increased by one m/z unit we observed linear responses up to 500 μM in plasma with R2 values between 0.984 and 0.999. Using the Q1 and Q3 values increased by two m/z units, this was extended to 1,000 μM for CEs (R2 = 0.998) and TGs (R2 = 0.997) to cover the full range of concentrations expected in plasma. As the difference between number of carbons in the internal standard and lipid of interest increases, so the relative intensities of the M+1 ions will also differ, potentially leading to greater difference in response factors and thereby compromising quantitative accuracy. While this is not an issue in comparative lipidomics, this differential should be adjusted for if accurate quantification is the goal.
Assay performance
We used 63 evenly spaced quality control (QC) plasma samples within the analysis of the 1,076 SAFHS samples. The analysis was performed over four LC-MS runs each of between 2 and 3 days duration. Two measures were used to assess the performance of the lipid measurements: i) the average intra-run %CV (coefficient of variation), where we calculate the average of the %CV for each of the four runs (each run contained between 13 and 17 QC samples); and ii) the %CVs across the entire analysis (63 QC samples), see Table 3. The median of the average intra-run %CV for the 23 lipid classes and subclasses (sum of the individual species) was 7.9% while the median %CV across the entire cohort was 9.6%. We also calculated %CVs of the 312 individual lipid species in the QC plasma extracts. The median average intra-run %CV was 10.6% with 90% of lipid species less than 27.6%, while the median %CV across the entire cohort was 13.8% with 90% lipid species less than 24.5%. Lipid species with %CV greater than 30% were typically of low abundance and/or had poor chromatography. Supplementary Fig. I shows histograms of the %CVs for the lipid species.
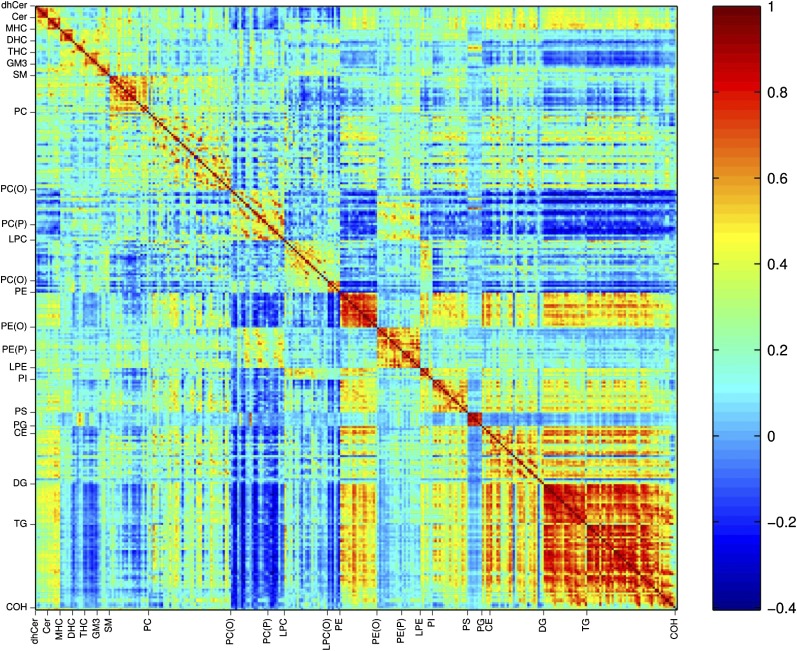
Correlation between lipid species
Pearson's correlation analysis identified positive correlations between lipid species within each lipid class/subclass, in particular PE, PS, DG, TG, and the sphingolipid classes were highly correlated (Fig. 4, supplementary Figs. II–IV). Between classes, PE displayed a strong correlation with PG, PI, DGs, and TGs as well as some species of PC and CEs. CEs, DG, and TG showed positive correlations for the majority of species. While the sphingolipid species were strongly positively correlated within classes, they displayed marginal positive or negative correlation across classes (Fig. 4, supplementary Fig. II). We also observed negative correlations between the PC(O) and plasmalogen with a number of lipid classes including PE, DG, and TG. PC species showed relatively weak correlation within the class and weak or negative correlations with the PC(O) and plasmalogen species (Fig. 4, supplementary Fig. III).
Fig. 4.
Pearson's linear correlation coefficients between all lipid species. Pearson's correlations were calculated between all pairs of lipid species. The correlation coefficients are plotted in a heat map. The color scale illustrates the magnitude and direction of correlation between lipid species. Correlation coefficients less than −0.063 or greater than 0.063 were significant (P < 0.05).
Associations of lipids with anthropometric and physiological measures
The SAFHS population cohort consisted of 1,076 individuals (39.1% male) from 15–91 years of age. Anthropometric and biochemical measurements are detailed in Table 1. Of the 23 lipid classes and subclasses analyzed, 15 showed a significant association with sex (P < 0.05, Benjamini and Hochberg corrected). In addition to the expected elevated levels of CE (6.9%) and TG (12.8%) in males relative to females, we observed significantly higher levels of Cer (6.9%), GM (9.1%), and LPC (14.1%) and significantly lower levels of SM (−7.1%), THC (−5.7%), and PS (−16.6%) after adjusting for age, BMI, systolic blood pressure (SBP), 2 h post load glucose, and smoking status (Table 4). Of the 312 individual species measured, 223 were significantly associated with sex, the most significant being SM 32:2, P = 8 × 10−36 (supplementary Table II). Sixteen lipid classes and subclasses were significantly associated with age (adjusted for sex, BMI, SBP, 2 h post load glucose, and smoking status) and 17 classes and subclasses were significantly associated with BMI (adjusted for sex, age, SBP, 2 h post load glucose, and smoking status) (Table 5). Of note were the strong associations of Cer with age (β-coefficient = 3.55, 95% confidence interval (CI) 2.50–4.60, P = 1.16 × 10−9) and dhCer with BMI (β-coefficient = 2.01, 95% CI 1.56–2.47, P = 2.22 × 10−16). Of the 312 individual lipid species, 148 were significantly associated with age and 200 were associated with BMI after adjusting for other covariates (supplementary Table III). Only two lipid subclasses showed a significant association with smoking status, these were PC(P) and PE(P) (Table 4), while 72 individual lipid species showed a significant association with smoking (supplementary Table II).
TABLE 4.
Logistic regression of lipid classes and subclasses with sex and smoking status
| Lipid Class or Subclassa | Sex | Smoking status | ||||
| IQR Odds Ratio (95% CI)b | c | Difference, %d | IQR Odds Ratio (95% CI)e | c | Difference, %f | |
| dhCer | 0.88 (0.74–1.05) | 1.87E-01 | −8.7 | 0.96 (0.80–1.17) | 7.93E-01 | −4.0 |
| Cer | 1.62 (1.35–1.95)g | 2.21E-06 | 6.9 | 0.96 (0.79–1.17) | 7.93E-01 | 0.5 |
| MHC | 1.05 (0.88–1.25) | 6.22E-01 | 2.7 | 1.03 (0.85–1.26) | 8.01E-01 | 1.1 |
| DHC | 0.80 (0.67–0.96) | 2.67E-02 | −1.9 | 1.01 (0.83–1.24) | 9.06E-01 | −0.4 |
| THC | 0.59 (0.49–0.70) | 1.34E-07 | −5.7 | 1.18 (0.97–1.42) | 2.14E-01 | 0.9 |
| GM | 1.57 (1.31–1.89) | 4.78E-06 | 9.1 | 1.11 (0.91–1.35) | 3.94E-01 | 3.5 |
| SM | 0.56 (0.45–0.68) | 1.34E-07 | −7.1 | 0.78 (0.63–0.97) | 8.79E-02 | −4.4 |
| PC | 0.92 (0.76–1.11) | 4.10E-01 | −1.1 | 1.18 (0.96–1.46) | 2.14E-01 | 0.9 |
| PC(O) | 1.16 (0.97–1.39) | 1.45E-01 | 1.7 | 0.76 (0.61–0.93) | 6.02E-02 | −2.9 |
| PC(P) | 1.05 (0.89–1.24) | 6.22E-01 | 1.1 | 0.75 (0.62–0.91) | 4.97E-02 | −3.9 |
| LPC | 1.78 (1.49–2.13) | 1.46E-08 | 14.1 | 1.09 (0.90–1.31) | 4.98E-01 | 4.9 |
| LPC(O) | 1.37 (1.15–1.64) | 1.28E-03 | 10.3 | 1.22 (1.00–1.48) | 1.54E-01 | 6.4 |
| PE | 0.76 (0.63–0.92) | 8.58E-03 | −8.8 | 1.25 (1.02–1.54) | 1.04E-01 | 1.8 |
| PE(O) | 0.86 (0.73–1.02) | 1.04E-01 | −4.4 | 0.86 (0.71–1.05) | 2.14E-01 | −4.5 |
| PE(P) | 1.36 (1.14–1.62) | 1.28E-03 | 4.6 | 0.75 (0.61–0.91) | 4.97E-02 | −5.3 |
| LPE | 1.35 (1.14–1.60) | 1.16E-03 | 9.4 | 1.23 (1.03–1.46) | 8.82E-02 | 6.4 |
| PI | 0.84 (0.70–1.02) | 9.56E-02 | −3.5 | 1.13 (0.92–1.39) | 3.54E-01 | 0.6 |
| PS | 0.74 (0.63–0.88) | 1.16E-03 | −16.6 | 0.86 (0.72–1.04) | 2.14E-01 | −11.9 |
| PG | 1.30 (1.10–1.54) | 4.58E-03 | 7.9 | 1.26 (1.06–1.50) | 7.57E-02 | 9.1 |
| CE | 1.57 (1.32–1.88) | 2.38E-06 | 6.9 | 1.16 (0.96–1.40) | 2.14E-01 | 4.0 |
| COH | 1.00 (0.84–1.19) | 9.30E-01 | −0.6 | 1.01 (0.83–1.23) | 9.06E-01 | −0.4 |
| DG | 1.52 (1.29–1.79) | 2.56E-06 | 14.6 | 1.12 (0.95–1.32) | 2.97E-01 | 7.0 |
| TG | 1.55 (1.31–1.83) | 1.97E-06 | 12.8 | 1.18 (0.99–1.40) | 1.87E-01 | 8.3 |
Sum of the individual species within each class or subclass.
Interquartile range (IQR) odds ratio, adjusted for age, BMI, SBP, 2 h post load glucose, and smoking status.
Benjamini and Hochberg corrected. Bold type indicates P < 0.05.
Male relative to female.
Interquartile range odds ratio, adjusted for sex, age, BMI, SBP and 2 hr post load glucose.
Current smokers relative to nonsmokers.
An individual with a 75th percentile level of Cer is 1.62 times more likely to be male than an individual with a 25th percentile level of Cer.
TABLE 5.
Linear regression of lipid classes and subclasses with age and BMI
| Lipid Class or Subclassa | Age (years) | BMI (kg/m2) | ||
| β-Coefficient (95% CI)b | Pc | β-Coefficient (95% CI)d | Pc | |
| dhCer | 0.72 (−0.30–1.74) | 2.01E-01 | 2.01 (1.56–2.47) | 2.22E-16 |
| Cer | 3.55 (2.50–4.60)e | 1.16E-09 | 0.14 (−0.38–0.65) | 6.66E-01 |
| MHC | 1.81 (0.76–2.85) | 2.31E-03 | −0.70 (−1.19–0.20) | 9.46E-03 |
| DHC | 1.05 (−0.03–2.12) | 7.73E-02 | −0.79 (−1.30–0.29) | 3.92E-03 |
| THC | 3.08 (2.08–4.08) | 2.73E-08 | −1.27 (−1.75–0.80) | 6.82E-07 |
| GM | 2.89 (1.83–3.95) | 5.87E-07 | −1.59 (−2.09–1.09) | 3.18E-09 |
| SM | 3.33 (2.22–4.44) | 4.55E-08 | 0.95 (0.41–1.49) | 1.30E-03 |
| PC | 1.53 (0.40–2.67) | 1.44E-02 | 0.93 (0.39–1.46) | 1.47E-03 |
| PC(O) | −1.46 (−2.55–0.36) | 1.65E-02 | −0.18 (−0.70–0.34) | 5.79E-01 |
| PC(P) | −0.36 (−1.38–0.65) | 5.05E-01 | −0.57 (−1.05–0.09) | 2.94E-02 |
| LPC | 0.73 (−0.30–1.77) | 2.01E-01 | −1.44 (−1.91–0.96) | 1.57E-08 |
| LPC(O) | 1.25 (0.17–2.33) | 3.54E-02 | −2.04 (−2.53–1.56) | 3.51E-15 |
| PE | 1.59 (0.48–2.70) | 1.18E-02 | 0.61 (0.08–1.14) | 3.40E-02 |
| PE(O) | 0.43 (−0.59–1.44) | 4.48E-01 | 0.07 (−0.41–0.55) | 7.90E-01 |
| PE(P) | 0.20 (−0.85–1.25) | 7.12E-01 | 0.45 (−0.05–0.94) | 9.54E-02 |
| LPE | 1.60 (0.61–2.60) | 4.74E-03 | −1.09 (−1.56–0.63) | 1.45E-05 |
| PI | 2.36 (1.26–3.46) | 1.09E-04 | 0.56 (0.04–1.09) | 4.96E-02 |
| PS | −0.39 (−1.26–0.49) | 4.48E-01 | −0.09 (−0.51–0.33) | 7.06E-01 |
| PG | 1.23 (0.23–2.23) | 2.70E-02 | 0.79 (0.32–1.26) | 1.95E-03 |
| CE | 1.63 (0.59–2.68) | 5.86E-03 | 1.53 (1.05–2.01) | 3.59E-09 |
| COH | 2.79 (1.76–3.82) | 5.87E-07 | 0.52 (0.02–1.01) | 5.38E-02 |
| DG | 1.04 (0.09–1.98) | 4.56E-02 | 0.99 (0.55–1.43) | 2.97E-05 |
| TG | 1.34 (0.35–2.33) | 1.44E-02 | 1.50 (1.04–1.95) | 1.30E-09 |
Sum of the individual species within that class or subclass.
Adjusted for sex, BMI, SBP, 2 h post load glucose, and smoking status.
Benjamini and Hochberg corrected. Bold type indicates P < 0.05.
Adjusted for sex, age, SBP, 2 h post load glucose, and smoking status.
An interquartile range increase in Cer levels is associated with a mean increase in age of 3.55 years.
DISCUSSION
We have presented a LC ESI-MS/MS lipid profiling technique for analyzing more than 300 molecular lipid species across 23 lipid classes and subclasses in 20 min. The methodology is also applicable to tissue or cell extracts with as little as 25 μg of cellular protein (18–22).
The Folch extraction method (23) has been used extensively to extract lipids from biological matrices. It involves the partitioning of the lipids in a biphasic solution of chloroform:methanol (2:1) and water. We identified two major drawbacks in this method: a) it does not lend itself to high sample throughput; and b) polar lipids are differentially partitioned between the aqueous and organic phases. A modification of this method was developed utilizing a single phase extraction and small plasma volume (10 μl). Reconstitution in water saturated BuOH and 10 mM ammonium formate in methanol provided high recoveries and a suitable injection solvent for subsequent chromatography. Using the method described, our laboratory can routinely extract 400 samples per day. While robotics has obvious advantages (24, 25), we have demonstrated that our method, even with manual handling and extraction of samples, is robust and applicable to cohorts of 1,000 or more samples.
Chromatographic separation prior to ESI-MS/MS allows for analysis of lipid mixtures and importantly, detection of low abundance lipids (26, 27). However, HPLC conditions that allow the separation of a wide range of lipids in large sample cohorts and are compatible with downstream ESI-MS/MS have not been widely reported. We have used a gradient method that provides good separation of most lipid classes and subclasses and importantly resolves most isomeric and isobaric species in 14 min. However, we observed high background signals for some DG and TG species on the gradient method, resulting from solvent impurities, and so developed a short isocratic LC method that is able to minimize this background.
To ensure accurate peak fitting and integration when using LC, it is necessary to maintain a sampling rate (in this experiment, 1 s) that provides a sufficient number of points across a chromatographic peak. Because MRM is a serial technique, as the number of MRM transitions increases, the time spent on each MRM transition is reduced with a deleterious effect on sensitivity. In order to circumvent this limitation, the MRMs may be scheduled so that an MRM pair is only monitored around its expected retention time, thus reducing the number of transitions monitored at any one point in time. Coupled with the robust RP-HPLC used here, this has allowed us to monitor 261 lipids from a single injection on the gradient method and 66 lipids on our isocratic analysis.
Full structural elucidation of lipid species by mass spectrometry is rarely achieved and practically impossible in a single MS experiment. For example, negative mode fragmentation of PCs provides information on the chain length and number of double bonds present in each acyl chain, but does not provide empirical evidence on either the position of those acyl chains on the glycerol backbone or the position of the double bonds within the acyl chain. While mass spectrometry-based experiments have been developed to obtain further structural information on both these points (28), in the current context it is impractical to attempt to fully elucidate the composition of isomeric lipids. Due to the necessity for high throughput in the context of population profiling, only limited structural elucidation is achieved, as reflected in the nomenclature. For example, PCs are reported with the sum of the number of carbons in the two esterified fatty acids and the sum number of double bonds, e.g., PC 36:3.
The gold standard for quantification in mass spectrometry is to employ a stable isotope internal standard for each analyte of interest. For lipidomic studies analyzing large numbers of lipid species, this approach is impractical due to the high cost and limited availability of suitable standards. While it has been widely recognized that lipid class is the primary determinant of ionization efficiency (29, 30), carbon chain length, number of double bonds, and changes in solvent composition (due to different elution times) will also influence ionization efficiency and signal response. While these factors will adversely influence quantitative measurements, they have minimal effect on analytical precision. For quantitative lipidomics by LC-MS the use of multiple internal standards can been used, often combined with algorithms to interpolate response factors for those species not covered by the internal standards. Alternatively, calibration curves can be used to determine relative response factors of species for which no internal standard is available (2, 31, 32). While such approaches can provide accurate quantitative data, the time and cost required may preclude such quantitative approaches in some situations.
For large population studies such as described here, where the objective is to identify associations and relationships between lipids and anthropometric and physiological measures, an alternate approach is to perform comparative lipidomics. In this analysis the emphasis is on precision of the assays to provide maximum power to identify significant associations and correlations within the resulting dataset. Thus, we have not, with the exception of DGs, TGs, and CEs, utilized correction factors to account for variation of signal response between lipids and their corresponding internal standard. However, prior to regression analysis, we standardize each lipid measurement to the interquartile range. The calculated odds ratios then reflect the change in likelihood of belonging to the outcome group as you increase the relative level of the lipid from the 25th to the 75th percentile of the population. Similarly the β-coefficients reflect the change in outcome measure as you increase the lipid level from the 25th to the 75th percentile. This approach provides for the rigorous statistical analysis of the biological interactions of lipids without the need for accurate quantification and is particularly suited for the analysis of large population cohorts.
Despite the limitation in accuracy, when we compare values from our study (expressed as μM) with published quantitative measures of plasma lipids [LIPID MAPS Consortium (2)], we observe a close alignment, with most lipid classes giving comparable values (less than 50% difference). For example, CEs, DGs, and TGs were −16, −8, and −47% respectively in our study relative to the LIPID MAPS study, while PC and SM were −41 and 17% respectively. In contrast, PI was 3-fold higher and PE was 7-fold lower in our measurements. The major factor leading to these large differences is most likely the response factors of individual species relative to the internal standards used.
Our lipidomic analysis is sufficiently rapid to have allowed us to analyze a large population cohort of over 1,000 samples. This provided statistical power to identify significant associations between circulating plasma lipids that reflect the overlapping, intersecting, or competing metabolic pathways involved in the synthesis and catabolism of these lipid species and the lipoproteins they comprise. In addition to this, we were able to identify significant independent associations of individual lipid species and lipid classes and subclasses with key anthropometric, physiological, and lifestyle factors (age, sex, BMI, and smoking).
Given the clear difference in risk of CVD between men and women and the well-established association with plasma lipids, this study provides the opportunity to examine whether differences in circulating lipids may contribute to this effect. Our results show that the traditional risk factors of cholesterol and triglycerides are higher in men than women. Furthermore, we, and others, have previously reported that plasma Cer is positively associated with CVD (33–35) and further with unstable CVD (16). Here, we observed that men have significantly higher plasma levels of Cer than women (+6.9%, P = 2.21 × 10−6, Table 4). Analysis of the Cer subspecies shows that the increase is primarily driven by the long chain species of Cer, 22:0, 24:0, and 24:1, which are highly correlated (supplementary Table II, supplementary Fig. II). This implies that the difference may be related to differential expression of ceramide synthase 2, which preferentially uses long chain (C20–C26) fatty acyl CoA species to produce Cer (36), and is the most abundant species expressed in the liver (37). We also observed that Cer is strongly associated with age (Table 5); however in this instance, all Cer species contribute to the association (supplementary Table III), suggesting regulation of Cer synthesis at the rate-limiting step of the de novo biosynthetic pathway, the condensation of the serine and palmitate by serine palmitoyl transferase. While Cer level is not associated with BMI, we did observe a strong and specific association between Cer 18:0 and BMI (P = 1.29 × 10−12), suggesting yet another regulatory mechanism involving ceramide synthase 1 which has specificity for C18 acyl chains and is expressed primarily in skeletal muscle (37). In addition to this finding, we observed significant associations of all species of dhCer with BMI. dhCer is the metabolic precursor to Cer indicating an upregulation of the de novo biosynthetic pathway without increasing Cer per se, but rather downstream metabolites. The glycoplipid classes (MHC, DHC, and THC) are all significantly negatively associated with BMI, but in contrast, SM, which has been reported to be proatherogenic (38), shows a positive association and so appears to be the end-product of this upregulation.
Interestingly, SM showed a strong positive association with age but in contrast to Cer was significantly lower in males than females. In a study of 1,102 recruited participants with coronary artery disease (CAD), Schlitt et al. (39) found no association of SM with age or BMI and no difference in SM levels between male and female participants. However, a positive association of plasma SM with CAD was reported. Based on the finding that SM was strongly associated with triglyceride levels, they hypothesized that, rather than an inflammatory marker, SM is related to atherogenic lipoprotein aggregation, a process involved in the progression of atherosclerosis. The absence of an association with age in the study by Schlitt et al. (39) may relate to the older and narrower range of ages in their study compared with SAFHS (average 61.1 years, standard deviation 10.1 years vs. average 38.3 years, standard deviation 16.1 years). Elevated levels of LPC and LPC(O), the precursor to platelet activating factor, in men relative to women (Table 4, supplementary Table II), suggests an increase in phospholipase activity (specifically lipoprotein-associated phospholipase A2 (Lp-PLA2), also known as platelet activating factor acetylhydrolase). This enzyme is highly expressed in atherosclerotic plaque and associated with pathogenesis of atherosclerosis (40, 41). This is further supported by several reports that Lp-PLA2 activity is higher in men than women and is linked to a higher risk of CAD, reviewed by Gregson et al. (42). We also observed a strong negative association of LPC with BMI. Because BMI is not reported to be associated with Lp-PLA2 activity, we must look for an alternate regulation of LPC levels. Lecithin:cholesterol acyltransferase (LCAT) activity is a major source of LPC in circulation and studies have shown LCAT activity is decreased in obesity (43, 44) potentially leading to a decrease in circulating LPC.
Our data showed relatively few lipid species associated with smoking status (Table 4, supplementary Table II). At a class and subclass level, only PC(P) and PE(P) were significantly decreased in individuals who smoked (Table 4). Plasmalogens are particularly susceptible to oxidation (45–47) and the negative association may be explained by an increase of oxidative stress associated with smoking (48, 49). Analysis of a subset of the KORA (Cooperative Health Research in the Region of Augsburg) study (5) quantified 198 metabolites, including 44 sphingolipids and 89 glycerophospholipids in 238 participants, and examined differences between smokers and nonsmokers. Twenty-three lipids, including three plasmalogens were identified as nicotine-dependant biomarkers. This study suggested smoking was associated with plasmalogen deficiency disorders specifically related to the reduced activity of the enzyme alkylglycerone phosphate synthase.
Our study highlights the power of performing lipidomic analysis at a population level where we have identified significant associations of many lipid classes, subclasses, and individual species with common anthropometric and lifestyle traits. In the context of population profiling, this highlights the power available in large cohort studies to identify independent associations between lipid species and anthropometric and lifestyle traits. Our study also suggests that lipid metabolism could play a role in the pathogenesis of various diseases, and our lipidomic methodology provides a means to further investigate these hypotheses.
Supplementary Material
Acknowledgments
The authors are grateful to the participants of the SAFHS.
Footnotes
Abbreviations:
- BMI
- body mass index
- CAD
- coronary artery disease
- CE
- cholesterol ester
- Cer
- ceramide
- CI
- confidence interval
- COH
- free cholesterol
- CV
- coefficient of variation
- CVD
- cardiovascular disease
- DG
- diacylglycerol
- DHC
- dihexosylceramide
- dhCer
- dihydroceramide
- GM
- GM3 ganglioside
- LC ESI-MS/MS
- liquid chromatography electrospray ionization tandem mass spectrometry
- LPC
- lysophosphatidylcholine
- LPC(O)
- lysoalkylphosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- Lp-PLA2
- lipoprotein-associated phospholipase A2
- MHC
- monohexosylceramide, MRM, multiple reaction monitoring
- PC
- phosphatidylcholine
- PC(O)
- alkylphosphatidylcholine
- PC(P)
- phosphatidylcholine plasmalogen
- PE
- phosphatidylethanolamine
- PE(O)
- alkylphosphatidylethanolamine
- PE(P)
- phosphatidylethanolamine plasmalogen
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- QC
- quality control
- SAFHS
- San Antonio Family Heart Study
- TG
- triacylglycerol
- THC
- trihexosylceramide
This work was supported by funding from the National Health and Medical Research Council (NHMRC) of Australia, the Operational Infrastructure (OIS) Program of the Victorian Government, Australia, and by award number 1R01DK-088972-01 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the aforementioned funding bodies. P.J.M. and J.S. were supported by NHMRC senior research fellowships and J.B.M.J. was supported by a NHMRC career development award. Data collection for the San Antonio Family Heart Study was supported by grant P01 HL-045522 from the National Institutes of Health (to J.B.). Additional work was supported by National Institutes of Health Grants R01 DK-082610 and R01 DK-079169 (to J.E.C.), and R01 HL-091035 (to J.B.). Parts of this investigation were conducted in facilities constructed with support from Research Facilities Improvement Program Grants C06 RR-013556 and C06 RR-017515 from the National Center for Research Resources of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures and three tables.
REFERENCES
- 1.Quehenberger O., Dennis E. A. 2011. The human plasma lipidome. N. Engl. J. Med. 365: 1812–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietiläinen K. H., Sysi-Aho M., Rissanen A., Seppänen-Laakso T., Yki-Järvinen H., Kaprio J., Oresic M. 2007. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects–a monozygotic twin study. PLoS ONE. 2: e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graessler J., Schwudke D., Schwarz P. E., Herzog R., Shevchenko A., Bornstein S. R. 2009. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS ONE. 4: e6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang-Sattler R., Yu Y., Mittelstrass K., Lattka E., Altmaier E., Gieger C., Ladwig K. H., Dahmen N., Weinberger K. M., Hao P., et al. 2008. Metabolic profiling reveals distinct variations linked to nicotine consumption in humans–first results from the KORA study. PLoS ONE. 3: e3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ollero M., Astarita G., Guerrera I. C., Sermet-Gaudelus I., Trudel S., Piomelli D., Edelman A. 2011. Plasma lipidomics reveals potential prognostic signatures within a cohort of cystic fibrosis patients. J. Lipid Res. 52: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwab U., Seppänen-Laakso T., Yetukuri L., Agren J., Kolehmainen M., Laaksonen D. E., Ruskeepää A-L., Gylling H., Uusitupa M., Oresic M. ; GENOBIN Study Group 2008. Triacylglycerol fatty acid composition in diet-induced weight loss in subjects with abnormal glucose metabolism–the GENOBIN study. PLoS ONE. 3: e2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee E. P., Cheng S., Larson M. G., Walford G. A., Lewis G. D., McCabe E., Yang E., Farrell L., Fox C. S., O'Donnell C. J., et al. 2011. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 121: 1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X., Yang K., Gross R. W. 2012. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 31: 134–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell B. D., Kammerer C. M., Blangero J., Mahaney M. C., Rainwater D. L., Dyke B., Hixson J. E., Henkel R. D., Sharp R. M., Comuzzie A. G., et al. 1996. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 94: 2159–2170 [DOI] [PubMed] [Google Scholar]
- 11.Arar N. H., Voruganti V. S., Nath S. D., Thameem F., Bauer R., Cole S. A., Blangero J., MacCluer J. W., Comuzzie A. G., Abboud H. E. 2008. A genome-wide search for linkage to chronic kidney disease in a community-based sample: the SAFHS. Nephrol. Dial. Transplant. 23: 3184–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hixson J. E., Blangero J. 2000. Genomic searches for genes that influence atherosclerosis and its risk factors. Ann. N. Y. Acad. Sci. 902: 1–7 [DOI] [PubMed] [Google Scholar]
- 13.Burke J. P., Duggirala R., Hale D. E., Blangero J., Stern M. P. 2000. Genetic basis of acanthosis nigricans in Mexican Americans and its association with phenotypes related to type 2 diabetes. Hum. Genet. 106: 467–472 [DOI] [PubMed] [Google Scholar]
- 14.Atwood L. D., Samollow P. B., Hixson J. E., Stern M. P., MacCluer J. W. 2001. Genome-wide linkage analysis of pulse pressure in Mexican Americans. Hypertension. 37: 425–428 [DOI] [PubMed] [Google Scholar]
- 15.Cai G., Cole S. A., Freeland-Graves J. H., MacCluer J. W., Blangero J., Comuzzie A. G. 2004. Genome-wide scans reveal quantitative trait Loci on 8p and 13q related to insulin action and glucose metabolism: the San Antonio Family Heart Study. Diabetes. 53: 1369–1374 [DOI] [PubMed] [Google Scholar]
- 16.Meikle P. J., Wong G., Tsorotes D., Barlow C. K., Weir J. M., Christopher M. J., MacIntosh G. L., Goudey B., Stern L., Kowalczyk A., et al. 2011. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 31: 2723–2732 [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57: 289–300 [Google Scholar]
- 18.Henstridge D. C., Bruce C. R., Pang C. P., Lancaster G. I., Allen T. L., Estevez E., Gardner T., Weir J. M., Meikle P. J., Lam K. S., et al. 2012. Skeletal muscle-specific overproduction of constitutively activated c-Jun N-terminal kinase (JNK) induces insulin resistance in mice. Diabetologia. 55: 2769–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam Y. Y., Hatzinikolas G., Weir J. M., Janovska A., McAinch A. J., Game P., Meikle P. J., Wittert G. A. 2011. Insulin-stimulated glucose uptake and pathways regulating energy metabolism in skeletal muscle cells: the effects of subcutaneous and visceral fat, and long-chain saturated, n-3 and n-6 polyunsaturated fatty acids. Biochim. Biophys. Acta. 1811: 468–475 [DOI] [PubMed] [Google Scholar]
- 20.Kamili A., Wat E., Chung R. W., Tandy S., Weir J. M., Meikle P. J., Cohn J. S. 2010. Hepatic accumulation of intestinal cholesterol is decreased and fecal cholesterol excretion is increased in mice fed a high-fat diet supplemented with milk phospholipids. Nutr. Metab. (Lond). 7: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth I., Hacking D. F., Hilton A. A., Mukhamedova N., Meikle P. J., Ellis S., Satterley K., Collinge J. E., de Graaf C. A., Bahlo M., et al. 2008. A mouse model of harlequin ichthyosis delineates a key role for Abca12 in lipid homeostasis. PLoS Genet. 4: e1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sozo F., Vela M., Stokes V., Kenna K., Meikle P. J., De Matteo R., Walker D., Brien J., Bocking A., Harding R. 2011. Effects of prenatal ethanol exposure on the lungs of postnatal lambs. Am. J. Physiol. Lung Cell. Mol. Physiol. 300: L139–L147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509 [PubMed] [Google Scholar]
- 24.Jung H. R., Sylvanne T., Koistinen K. M., Tarasov K., Kauhanen D., Ekroos K. 2011. High throughput quantitative molecular lipidomics. Biochim. Biophys. Acta. 1811: 925–934 [DOI] [PubMed] [Google Scholar]
- 25.Löfgren L., Ståhlman M., Forsberg G. B., Saarinen S., Nilsson R., Hansson G. I. 2012. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 53: 1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu C., van der Heijden R., Wang M., van der Greef J., Hankemeier T., Xu G. 2009. Analytical strategies in lipidomics and applications in disease biomarker discovery. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2836–2846 [DOI] [PubMed] [Google Scholar]
- 27.Sommer U., Herscovitz H., Welty F. K., Costello C. E. 2006. LC-MS-based method for the qualitative and quantitative analysis of complex lipid mixtures. J. Lipid Res. 47: 804–814 [DOI] [PubMed] [Google Scholar]
- 28.Mitchell T. W., Pham H., Thomas M. C., Blanksby S. J. 2009. Identification of double bond position in lipids: from GC to OzID. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2722–2735 [DOI] [PubMed] [Google Scholar]
- 29.Han X., Gross R. W. 1994. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA. 91: 10635–10639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han X., Yang K., Yang J., Fikes K. N., Cheng H., Gross R. W. 2006. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J. Am. Soc. Mass Spectrom. 17: 264–274 [DOI] [PubMed] [Google Scholar]
- 31.Hutchins P. M., Barkley R. M., Murphy R. C. 2008. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 49: 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Mello V. D., Lankinen M., Schwab U., Kolehmainen M., Lehto S., Seppanen-Laakso T., Oresic M., Pulkkinen L., Uusitupa M., Erkkila A. T. 2009. Link between plasma ceramides, inflammation and insulin resistance: association with serum IL-6 concentration in patients with coronary heart disease. Diabetologia. 52: 2612–2615 [DOI] [PubMed] [Google Scholar]
- 34.Ichi I., Nakahara K., Miyashita Y., Hidaka A., Kutsukake S., Inoue K., Maruyama T., Miwa Y., Harada-Shiba M., Tsushima M., et al. 2006. Association of ceramides in human plasma with risk factors of atherosclerosis. Lipids. 41: 859–863 [DOI] [PubMed] [Google Scholar]
- 35.Holland W. L., Summers S. A. 2008. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr. Rev. 29: 381–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy M., Futerman A. H. 2010. Mammalian ceramide synthases. IUBMB Life. 62: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr, Futerman A. H. 2003. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278: 43452–43459 [DOI] [PubMed] [Google Scholar]
- 38.Nelson J. C., Jiang X. C., Tabas I., Tall A., Shea S. 2006. Plasma sphingomyelin and subclinical atherosclerosis: findings from the multi-ethnic study of atherosclerosis. Am. J. Epidemiol. 163: 903–912 [DOI] [PubMed] [Google Scholar]
- 39.Schlitt A., Blankenberg S., Yan D., von Gizycki H., Buerke M., Werdan K., Bickel C., Lackner K. J., Meyer J., Rupprecht H. J., et al. 2006. Further evaluation of plasma sphingomyelin levels as a risk factor for coronary artery disease. Nutr. Metab. (Lond). 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolodgie F. D., Burke A. P., Skorija K. S., Ladich E., Kutys R., Makuria A. T., Virmani R. 2006. Lipoprotein-associated phospholipase A2 protein expression in the natural progression of human coronary atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 26: 2523–2529 [DOI] [PubMed] [Google Scholar]
- 41.Schmitz G., Ruebsaamen K. 2010. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 208: 10–18 [DOI] [PubMed] [Google Scholar]
- 42.Gregson J., Stirnadel-Farrant H. A., Doobaree I. U., Koro C. 2012. Variation of lipoprotein associated phospholipase A2 across demographic characteristics and cardiovascular risk factors: a systematic review of the literature. Atherosclerosis. 225: 11–21 [DOI] [PubMed] [Google Scholar]
- 43.Bajnok L., Seres I., Varga Z., Jeges S., Peti A., Karanyi Z., Juhasz A., Csongradi E., Mezosi E., Nagy E. V., et al. 2007. Relationship of endogenous hyperleptinemia to serum paraoxonase 1, cholesteryl ester transfer protein, and lecithin cholesterol acyltransferase in obese individuals. Metabolism. 56: 1542–1549 [DOI] [PubMed] [Google Scholar]
- 44.Angell R., Mitsuhashi Y., Bigley K., Bauer J. E. 2009. Plasma LCAT activity and lipid subfraction composition in obese beagles undergoing weight loss. Lipids. 44: 415–424 [DOI] [PubMed] [Google Scholar]
- 45.Felde R., Spiteller G. 1995. Plasmalogen oxidation in human serum lipoproteins. Chem. Phys. Lipids. 76: 259–267 [DOI] [PubMed] [Google Scholar]
- 46.Engelmann B. 2004. Plasmalogens: targets for oxidants and major lipophilic antioxidants. Biochem. Soc. Trans. 32: 147–150 [DOI] [PubMed] [Google Scholar]
- 47.Ford D. A. 2010. Lipid oxidation by hypochlorous acid: chlorinated lipids in atherosclerosis and myocardial ischemia. Clin. Lipidol. 5: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J. F., Yan X. F., Guo F. Z., Sun N. Y., Qian Z. J., Ding D. Y. 2000. Effects of cigarette smoking and smoking cessation on plasma constituents and enzyme activities related to oxidative stress. Biomed. Environ. Sci. 13: 44–55 [PubMed] [Google Scholar]
- 49.Rahman I., Morrison D., Donaldson K., MacNee W. 1996. Systemic oxidative stress in asthma, COPD, and smokers. Am. J. Respir. Crit. Care Med. 154: 1055–1060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.