Abstract
Tuberculosis is a major cause of mortality and morbidity due to infectious disease. However, current clinical diagnostic methodologies such as PCR, sputum culture, or smear microscopy are not ideal. Antibody-based assays are a suitable alternative but require specific antibodies against a suitable biomarker. Mycolic acid, which has been found in patient sputum samples and comprises a large portion of the mycobacterial cell wall, is an ideal target. However, generating anti-lipid antibodies using traditional hybridoma methodologies is challenging and has limited the exploitation of this lipid as a diagnostic marker. We describe here the isolation and characterization of four anti-mycolic acid antibodies from a nonimmune antibody phage display library that can detect mycolic acids down to a limit of 4.5ng. All antibodies were specific for the methoxy subclass of mycolic acid with weak binding for α mycolic acid and did not show any binding to closely related lipids or other Mycobacterium tuberculosis (Mtb) derived lipids. We also determined the clinical utility of these antibodies based on their limit of detection for mycobacteria colony forming units (CFU). In combination with an optimized alkaline hydrolysis method for rapid lipid extraction, these antibodies can detect 105 CFU of Mycobacterium bovis BCG, a close relative of Mtb and therefore represent a novel approach for the development of diagnostic assays for lipid biomarkers.
Keywords: phage display, mycolic acid, biomarker, antibody-based assay
Tuberculosis (TB) is one of the world's most significant causes of mortality due to infectious disease, with an estimated 1.45 million deaths in 2010 (1). Rapid identification of new cases is key to the proper treatment of TB, the majority of which occur in resource-poor settings. However, current detection methods such as sputum smears, culture, PCR, and X-ray are unsuitable in such areas due to poor sensitivity, slow turnaround time, high operating costs, and infrastructure requirements, respectively (2–4). An alternative point-of-care diagnostic test is antibody-based assays, which rely on detecting Mycobacterium tuberculosis (Mtb)-derived antigens in patient specimen types such as serum, sputum, or urine. However, although a number of assays have been developed to detect a variety of protein and soluble glycolipid antigens such as ESAT-6 and lipoarabinomannan, meta-analyses of published work have shown that these assays do not improve on the sensitivity of current diagnostics or there are an insufficient number of independent studies to confirm their utility (5, 6).
Another potential biomarker for TB diagnostics is mycolic acid. It has been found using mass spectrometry, to be present in TB patient sputum samples and is a major component of the mycobacterial cell wall, comprising between 40 and 60% of the dry mass of the cell envelope (7, 8). It exists either as free lipid or is covalently bound to the peptidoglycan backbone and can be extracted either via organic solvents or alkaline hydrolysis, respectively (9, 10). To date, there is no method devised that allows for the detection of mycolic acids directly from culture or sputum samples. Instead, analysis of mycolic acids must be carried out on lipid extracted from such samples using high-performance liquid chromatography, electrospray ionization mass spectrometry, thin layer chromatography, or nuclear magnetic resonance (8, 11–14). However, these methods require highly sophisticated instruments and trained personnel and are not ideal for diagnostic assays in resource-poor settings.
Antibody-based detection has traditionally focused on water-soluble protein and carbohydrate antigens, as the antigens and antibodies are all soluble in the aqueous environment of the assay. As a result, insoluble molecules such as nonpolar lipids have remained neglected and represent a novel group of antigens whose potential as disease biomarkers has not been fully explored. A further complication is that traditional hybridoma methodologies typically do not generate high affinity antibodies against lipids due to their poor immunogenicity unless conjugated to a protein carrier that enables lipid-specific B-cells to obtain T-cell help (15). However, in vitro antibody selection techniques such as phage display have allowed the generation of antibodies against nonpolar lipids such as 15-ketocholestane, methyl palmitate, and 11-dehydro-thromboxane (16–18). These antibodies are able to detect free insoluble lipid deposited by evaporation of the carrier organic solvent, thus avoiding the need for the lipid to be emulsified in an aqueous environment. The development of such antibodies opens up the possibility of targeting such lipids as diagnostic biomarkers for point-of-care assays.
In this study, we screened a non-immune human Fab phage display library against Mtb-derived mycolic acid, and isolated four unique antibodies specific for the methoxy mycolic acid subclass. Concurrently, we have optimized a rapid lipid extraction protocol to enable these antibodies to detect mycolic acid present in mycobacterial culture via a simple immunoassay. We show that these antibodies are capable of distinguishing mycolic acid from common biological lipids present in most cells such as cholesterol and phospholipids as well as differentiating lipid extracts of mycobacterial and non-mycobacterial actinomycete species. These antibodies can also detect mycolic acids down to 4.5ng and between 105 and 106 colony-forming units (CFU) of Mycobacterium bovis Bacille Calmette-Guérin (BCG) strain. We therefore demonstrate that insoluble lipids previously inaccessible to antibody-based assays, such as mycolic acid, can be detected using novel recombinant anti-lipid antibodies in combination with rapid and simple lipid extraction techniques and thus such antibodies are useful for the detection of novel lipid biomarkers.
Material and methods
Reagents and bacterial strains
Mtb-derived mycolic acid extract, trehalose dimycolate, β-hydroxyl myristolic acid and n-hexane were purchased from Sigma-Aldrich, while all synthetic lipids (cholesterol, cholesterol derivatives, dimyristoyl phosphatidic acid, dimyristoyl phosphocholine (DMPC), sphingosine, α, keto, and methoxy mycolic acid) were purchased from Avanti Polar Lipids. phosphoinositol-capped lipoarabinomannan (PILAM) and mannose-capped lipoarabinomannan (ManLAM) were purchased from Invivogen and Nacalai Tesque, Japan, respectively. Mycobacterium smegmatis was a kind contribution from Dr Ayi Teck Choon, DSO National Laboratories and (BCG) was a kind contribution from Dr Sylvie Alonso, National University of Singapore. Nocardia cyriacigeorgica (ATCC BAA-1517) and Tsukamurella paurometabolum (ATCC 8368) were purchased from American Type Culture Collection.
Phage panning and IgG expression
A nonimmune human Fab phage display library (Humanyx Pte Ltd. was panned based on a previously developed protocol for generating anti-lipid antibodies (17). Briefly, a MaxiSorp immunotube (Nunc) was coated with 200 μg/ml of Mtb-derived mycolic acid dissolved in n-hexane, after which the hexane was allowed to evaporate overnight at 4°C. Screening of the library was performed on antigen coated immunotubes essentially as described previously for protein antigens and modified for lipid antigens (19). Briefly, Fab-phage were incubated with coated immunotubes for 2 h at room temperature followed by washing with PBS/0.5% Tween-20 to remove nonspecific bound phage; specifically bound phage were then eluted by digestion with trypsin solution for 30 min at 37°C. Stringency of washing was increased during subsequent pans. Following four rounds of panning, enrichment was established using ELISA with the polyclonal Fab-phage preparations from each pan and individual phage clones were analyzed from the eluted phage of the pan showing the highest level of enrichment by ELISA. To assess the uniqueness of positive clones, BstN1 restriction digest was performed following PCR amplification of the Fab coding region and resolved on 3% agarose gel. Clones with similar patterns were grouped and unique clones confirmed by Sanger sequencing. Unique clones with the highest ELISA signal was converted to IgG for subsequent characterization.
Unique high binding clones were converted to IgG1 by cloning the Fab containing fragment from pCES1 into pCMV based vectors harboring the human constant regions for either the lambda or kappa light chain and IgG1 heavy chain separated by an internal ribosome entry sequence (20). Constructs encoding the Fab-IgGs were transfected into human embryonic kidney (HEK293) cells by use of lipofectamine 2000 (Invitrogen). Culture media was collected 72 h posttransfection and fresh media was added to the cells and allowed to incubate for a further 72 h before collection. Secreted antibodies were purified using recombinant Protein-A Sepharose beads (ThermoScientific).
ELISA
Lipid antigens were dissolved first in a 9:1 mix of chloroform:methanol at 1 mg/ml and then further diluted in n-hexane to 25 μg/ml or indicated concentration. Lipid extracts from bacteria were processed as described below and diluted as indicated in n-hexane. Lipid/hexane mixtures were then evaporated onto 96-well MaxiSorp ELISA plates (Nunc) overnight at 4°C. ManLAM and PILAM were coated onto the same plates in aqueous solution (1× PBS) at 25 μg/ml concentration overnight at 4°C. Antigen-coated plates were washed twice with PBS, blocked with 360 μl/well 5% skim milk in PBS (MPBS) at room temperature for 2 h, then washed 2× with PBS. Monoclonal IgG at 1 μg/ml in 5% MPBS, polyclonal phage diluted 1:10 in 2% MPBS or monoclonal phage diluted 1:1 in 4% MPBS was then added. ELISA plates were washed four times in PBS (for polyclonal phage ELISA) or PBS/0.05% Tween-20 (for monoclonal IgG ELISA) and secondary antibody: αM13-HRP against phage major coat protein VIII (GE Healthcare) for phage ELISA or α-human IgG-Fc HRP (Thermo Scientific) for monoclonal IgG ELISA, added at 1:5000 dilution in 2% MPBS. Plates were washed as above and one additional time with PBS and then color developed with TMB (Thermo Scientific) and stopped with 2M sulphuric acid. All volumes are at 100 µl/well and incubations 1 h at room temperature unless specified.
Lipid extraction from bacterial cultures
M. smegmatis was cultured in Middlebrook 7H9 broth (BD Biosciences) while Nocardia cyriacigeorgica and Tsukamurella paurometabolum were cultured in Brain-Heart Infusion Broth (BD Biosciences) at 37°C in flasks in a shaking incubator. M. bovis (BCG) was cultured in 100 ml of Middlebrook 7H9 broth at 37°C with 5% CO2 in T175 cell culture flasks for 2 weeks. Cultures were grown until log-phase growth (optical density, OD600 ∼ 0.5) was reached. Cultures were then pelleted by centrifugation at 4, 000 g and lipids extracted by adapting a procedure from Minnikin et al (21). Briefly, pelleted bacteria was resuspended in 30 ml of n-hexane per gram of wet cell mass, vortexed and incubated at room temperature on rollers overnight for initial extraction based on wet cell mass. The next day, the mixtures were centrifuged at 16,000 g for 5 min and the organic layer containing lipids liberated from bacterial surfaces was recovered. Various methods of lipid extraction, as described below, were subsequently performed on M. smegmatis in order to derive at an optimal method for lipid extraction.
Optimization of lipid extraction
One milliliter of OD 0.1 M. smegmatis (equivalent to 5.8 × 107 cfu/ml) was centrifuged from pure culture grown as above and the pellet was resuspended in 1 ml of n-hexane. The mixture was vortexed for 15 min, incubated on a rotator at room temperature overnight, then centrifuged and the organic layer was recovered. To enhance the effectiveness of solvent extraction, a second methodology was tried where the sample was heated for 2 h at 70°C before vortexing. To lyse the bacteria to improve extraction, a third protocol was carried out where the same amount of mycobacteria was resuspended in 1ml of Milli-Q water and incubated at 99°C for 30 min using a Thermomixer heating block (Eppendorf). One milliliter of n-hexane was then added and processed as above without heating. A fourth protocol was also performed where the bacteria was pelleted after incubation in boiling water and then resuspended in 1 ml of n-hexane as above. Centrifugations were carried out at 16,000 g for 5 min.
For the fifth method of alkaline hydrolysis, which was subsequently found to be the most efficient, a simplified method adapted from Shui et al was used (10). Briefly, the pellet was resuspended in 500 μl of 1M potassium hydroxide (KOH) in methanol at 80°C for 1 h and then neutralized with concentrated 4M hydrochloric acid (HCl). One milliliter of n-hexane was added to each sample, which was then vortexed for 15 min, centrifuged, and the organic layer was recovered. One hundred microliters of extract was used for monoclonal IgG ELISA as described above. For determination of CFU limit of detection, various standard CFU amounts of mycobacteria (106 to 109 CFU) were treated by alkaline hydrolysis and the lipid extract used for ELISA in the same manner. t-tests with Bonferroni corrections for significant difference were carried out using Prism 5 software (GraphPad).
Mass spectrometric profiling and quantification of mycolic acids
Mycolic acid profiles were determined by means of high-resolution tandem mass spectrometry. Briefly, lipid extract from M. smegmatis was diluted to 2:1 (v/v) with isopropanol:methanol:chloroform 4:2:1 containing 2 mM ammonium acetate. Ten microliter samples were infused into a LTQ Orbitrap XL mass spectrometer (Thermo Scientific) using nanomate interface (Advion). The nanospray voltage was −1.35kV (negative ionization) and mass spectra were acquired in duplicate at a resolution of 30,000 and 100,000. α-alkyl chains were identified by product ion scans, using collision energies between 35 and 45eV. Once the mycolic acid profile of the samples was determined, individual mycolic acid molecular species were quantified by multiple reaction monitoring (MRM) analysis. A comprehensive set of MRM transitions was set up, including all mycolic acid species and α-alkyl chains identified by high-resolution mass spectrometry (HR-MS) and MS/MS. A total of 90 mycolic acid species were analyzed. Five microliters of lipid extract were diluted to 50 μl in chloroform:methanol 1:1 containing two internal standards (keto- and methoxy- mycolic acids, m/z 1266 and 1254, final concentration: 0.5 μg/ml each; Avanti Polar Lipids) and injected into a 4000QTrap mass spectrometer (ABSciex). These two mycolic acids were not detected in the samples during HR-MS analysis and could then be used safely as internal standards. The carrier solvent was chloroform:methanol 1:1 containing 10 mM piperidine, flowing at 250 μl/min. Each sample was injected in duplicate (injection volume: 20 μl). The MS parameters were as follows: negative ESI, capillary voltage -4.5kV, source temperature 250°C, DP -120, EP -10, CE -90, CXP -5. For quantification, the signal intensity of each MRM transition value was normalized to the internal standard transition intensity.
Determination of CFU limit of detection
To standardize lipid extraction by CFUs, bacteria were pelleted by centrifugation, weighed, and resuspended in PBS containing 50% (v/v) glycerol to OD600 of 1.0, then serially diluted 1:10 six times and plated onto Middlebrook 7H9 and 7H11 (BD Biosciences) agar plates and incubated at 37°C for 3 days and 37°C with 5% CO2 for 21 days for M. smegmatis and BCG, respectively. The procedure was performed in duplicate and the average CFU per unit OD calculated to determine the appropriate dilutions for subsequent experiments. Subsequently, bacteria were diluted to the appropriate CFU/ml concentrations, pelleted by centrifugation at 16,000 g for 5 min and extracted by alkaline hydrolysis as described above. Extraction was carried out in triplicate and then tested by ELISA as described above to determine the limit of detection.
Results
Generation and identification of mycolic acid-specific antibodies
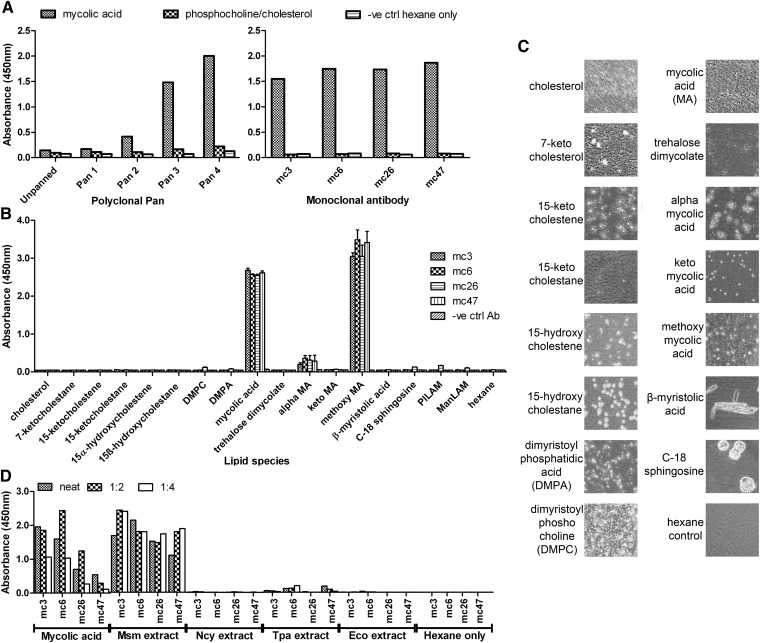
Purified Mtb-derived mycolic acid was coated onto solid polystyrene surface by evaporation from hexane, allowing the Humanyx Fab phage library to be panned against it. This was done for four successive rounds and the subsequent polyclonal ELISA indicated successful enrichment for binders to mycolic acid as shown by the increase in specific absorbance (Fig. 1A, left panel). To verify that enrichment for high affinity Fab-phage clones occurs and to identify suitable high affinity monoclonal antibodies, 376 individual clones from the fourth pan (the pan with the highest signal) were screened for binding activity by monoclonal ELISA. The ELISA signals were classified by signal-to-noise ratio (SNR) as follows: negative binders (SNR<2), weak binders (SNR 2-5), moderate binders (SNR 5-10), and strong binders (SNR >10). The distribution of the various types of binders is given in Table 1. The strong binders formed the largest group with 51% of all sampled clones while the negative binders formed only 4% of all tested clones indicated clear preference for strong binders. The top 48 binders were sequenced to identify unique clones and retested by monoclonal phage ELISA. The four best unique binders mc3, mc6, mc26, and mc47 were converted to IgG and retested for mycolic acid binding by IgG monoclonal ELISA. All four antibodies retained binding activity as IgG (Fig. 1A right panel). The complementarity determining regions (CDR) sequences and original germline genes of the antibodies are given in supplementary Table I.
Fig. 1.
A: Polyclonal ELISA (left panel) and monoclonal ELISA (right panel) showing enrichment of successive phage pans and identification of four monoclonal IgG specific for Mtb-derived mycolic acid respectively. Each ELISA plate was coated with either mycolic acid or phosphocholine/cholesterol in a 2:1 w/w ratio as a negative. Hexane was added as a negative vehicle control. B: Monoclonal ELISA showing specificity of isolated recombinant monoclonal IgGs mc3,6,26 and 47 for Mtb-derived mycolic acid and methoxy mycolic acid in contrast to other synthetic cholesterol derivatives, membrane lipids (DMPC and DPMA), β-hydroxyl lipids (myristolic acid and sphingosine) and mycobacterial lipids (mycolic acids, mycolate and LAMs). All antibodies including the control antibody (anti-dengue E protein antibody 14C10) were at 1 µg/ml concentration and the lipids were coated at 25 µg/ml. Results are the average of three independent experiments and the error bars show the SEM. C: Phase contrast light microscopy images of ELISA well bottoms indicating adherence of evaporated lipid onto the ELISA plate. Images are at 100× magnification. D: Monoclonal ELISA showing specificity of IgGs for M. smegmatis lipid extract (Msm) compared with lipid extract from other actinomycete genera (N. cyriacigeorgica-Ncy, T. paurometabolum-Tpa). E. coli (Eco) and hexane were added as negative controls and Mtb-derived mycolic acid (6.25 µg/ml) as a positive control.
TABLE 1.
Proportion of negative, weak, moderate and strong binding clones isolated from the 4th Pan as determined by monoclonal phage ELISA
| Signal-to-noise ratio (A450/background) | No. of clones | Percentage of total (%) |
| Negative (SNR < 2) | 15 | 3.99 |
| Weak (SNR 2-5) | 36 | 9.57 |
| Moderate (SNR 5-10) | 132 | 35.11 |
| Strong (SNR > 10) | 193 | 51.33 |
All four antibodies were subsequently tested against a panel of synthetic and purified lipids. Earlier studies had indicated that mycolic acid might have a similar 3D structure to cholesterol and evidence of cross-reactivity to cholesterol was observed in anti-mycolic acid antibodies previously isolated from a chicken phage display library (22, 23). We therefore tested all four antibodies against purified cholesterol and oxidized cholesterol derivatives. We also tested for cross-reactivity to common membrane lipids such as DMPC and dimyristoyl phosphatidic acid, as well as the structurally related β-hydroxyl lipids sphingosine and β-hydroxyl myristolic acid. Finally, to determine the exact specificity of our antibodies, we tested for binding against synthesized subclasses of mycolic acid in Mtb, namely α, keto, and methoxy mycolic acid, which contain the identical polar head group but differ in the position of functional groups in the acyl chain (supplementary Fig II). In addition, binding to purified extracts of trehalose dimycolate and ManLAM, which are present in Mtb, and the PILAM found in M. smegmatis was also performed. No binding was observed to any other cholesterol lipid, membrane lipid, β-hydroxyl lipid, or mycobacterial lipid, even trehalose dimycolate, which is comprised of two mycolic acid molecules attached to a trehalose sugar (Fig. 1B). All antibodies bound the original purified Mtb-derived mycolic acid as well as the synthesized methoxy mycolic acid strongly, but bound α mycolic acid weakly and not to keto mycolic acid at all (Fig. 1B). To ensure that negative signals were not due to lack of effective antigen coating, the ELISA wells were imaged under phase contrast light microscopy to visualize the adsorbed insoluble lipids prior to the blocking step. All wells except the hexane control clearly contained evaporated lipid droplets (Fig. 1C). ManLAM and PILAM could not be visualized as they are coated in aqueous solution and are not evaporated onto the well surface as droplets. Adherence of these glycolipids to the ELISA plates used had been previously verified using a rabbit anti-LAM polyclonal (data not shown).
The specificity of all four antibodies was also tested against lipid extracts of various species from various actinomycete genera that are known to contain mycolic acids, along with the mycobacterial species M. smegmatis (9). All four antibodies bound M. smegmatis lipid extract strongly but did not bind lipid extracts from related Gram positive actinomycete species Nocardia cyriacigeorgica or Tsukamurella paurometabolum or the Gram negative bacterial species Escherichia coli with the exception of mc6 and mc47, which bound weakly to T. paurometabolum lipid extract only (Fig. 1D). Binding of mc26 and mc47 to mycolic acid and mycobacterial lipid extract was noticeable weaker compared with mc3 and mc6.
Determination of limit of detection and optimization of mycolic acid extraction protocol
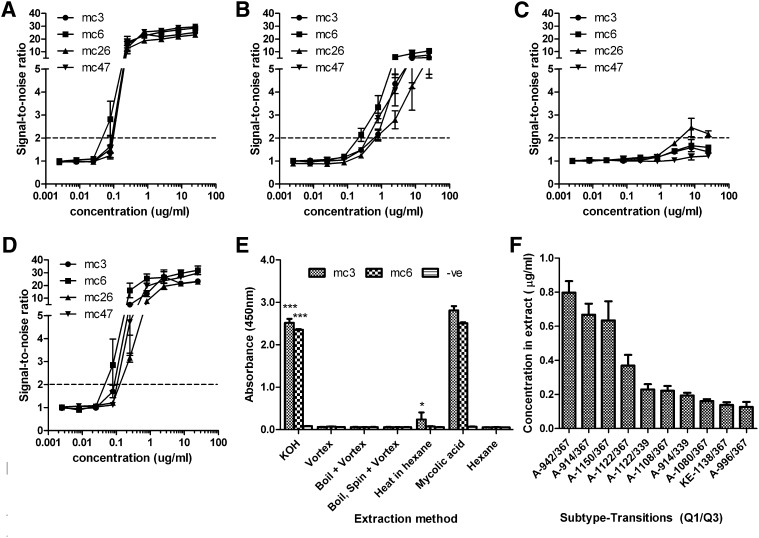
To determine the sensitivity of the antibodies, all four antibodies were tested against serially diluted concentrations of purified Mtb-derived mycolic acid and synthetic mycolic acid subclasses. All four antibodies had similar binding curves although mc3 and mc6 had the highest affinity as defined by SNR. The limit of detection (defined by a SNR of 2) for mycolic acid extract and methoxy mycolic acid was similar at about 45ng/ml for mc6 while the rest had a slightly poorer sensitivity of about 80 to 100 ng/ml (Fig. 2A, 2D). The limit of detection range for α mycolic acid was 200 ng/ml for the mc6 to 800 ng/ml for mc26 and mc47 (Fig. 2B). Keto mycolic acid could only be detected weakly by mc26 but not the other antibodies at concentrations above 4 µg/ml (Fig. 2C).
Fig. 2.
Limit of detection curves for Mtb- derived mycolic acid (A), and synthetic α (B), keto (C) and methoxy (D) mycolic acids. Antibodies were used at a concentration of 1 µg/ml and lipids were coated in hexane at the indicated concentrations, both with a volume of 100 µl/well. Results are given as SNR and the limit of detection of a SNR of 2 is shown by the dashed line E: Optimization of mycolic acid extraction method using the two most sensitive IgGs, mc3 and 6, and 1 ml of OD0.1 (5.8 × 107 CFU) M. smegmatis as a test sample. Mycolic acid was extracted using either overnight incubation at room temperature with hexane after vigorous vortexing only (Vortex) or with additional prior heating (Heat in hexane). Two more methods using a lysis step with boiling water where hexane was added directly to the water after heating (Boil + Hexane) or sequentially following an intermediate centrifugation to pellet the bacteria (Boil, spin + Hexane) were also carried out. A fifth protocol involving rapid alkaline hydrolysis (KOH) to release covalently bound mycolic acid was also done. *** P < 0.001, alkaline hydrolysisversus all other methods; *P < 0.05 for heat in hexane versus all other methods. F: Detection of individual species of mycolic acid in M. smegmatis KOH extract by mass spectrometry in MRM mode; the concentrations of the top ten mycolic acids, identified by Q1/Q3 transitions, are indicated along with the subtype (A or KE for Alpha and Keto/Epoxy, respectively). No methoxy mycolic acid was detected. All results are the average of three independent experiments and the error bars show SEM.
We then optimized the sensitivity and efficiency of the assay by introducing various heating and alkaline hydrolysis steps to the basic lipid extraction protocol of vortexing in hexane. To ensure that enhancement of signal could be observed, we carried out this extraction at a much lower concentration of M. smegmatis bacteria (1 ml of OD600 0.1, 5.8 × 107 CFU/ml) as compared with the earlier extraction where vortexing alone gave a high signal. We tested incubation with or without prior heating in hexane to improve lipid extraction or boiling in water to lyse the bacteria. An additional alkaline hydrolysis protocol was also tested as the majority of mycolic acid is known to be covalently bound to the peptidoglycan cell, which can be liberated by treatment with alkali. Lipid extraction via these various methods was carried out independently in triplicate and evaporated onto ELISA plates and tested with mc3 and mc6, the two antibodies with the best limit of detection.
At this low concentration of bacteria, simple vortexing did not give a significant signal over background (hexane only). However, lipid extracted by alkaline hydrolysis gave a strong signal compared with all other methods by a large and statistically significant margin. Extraction by heating in hexane also gave significantly higher signal for mc3 only over vortexing and background although the increase was slight (Fig. 2E). Other methods showed no significant increase in signal over vortexing in hexane or background. Given that alkaline hydrolysis only requires several hours to complete as compared with the other methods, which require overnight incubation in hexane, this is the method we selected to determine the limit of detection on standardized CFU amounts of mycobacteria.
To confirm that the alkaline hydrolysis procedure released mycolic acid from the bacteria and that our antibodies were indeed detecting mycolic acid, triplicate alkaline hydrolysis extracts were first analyzed by HR-MS/MS. Identification was based on accurate mass measurement and product ion scans (supplementary Fig. IA, IB). Mycolic acids were ionized as [M-H]- ions and observed m/z were within 2 ppm of expected values. Product ion analysis allowed for the identification of α-alkyl chains (the most abundant being C24:0, supplementary Fig. IB). Identified mycolic acids were then quantified in the same triplicate extracts by MRM analysis using Q1/Q3 transitions previously identified in mycolic extracts of mycobacterial species and adjusted based on an internal synthetic mycolic acid standard (8). Identified unique mycolic acid transitions were ranked by concentration and analysis of the top ten mycolic acids identified indicated that the majority of these were α mycolic acids and their total concentration was approximately 3.4 µg/ml (Fig. 2F). The levels of methoxy mycolic acids were insignificant compared with α mycolic acid. Nonetheless, the concentration of α mycolic acid alone is well within the detection limit of our antibodies indicated earlier and can account for the high signal detected.
Determination of CFU limit of detection
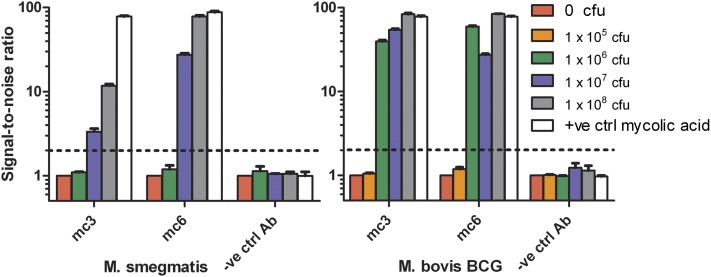
Various standard amounts of bacteria from 109 to 106 CFU/ml were treated using the alkaline hydrolysis method to extract lipids into 1 ml of n-hexane and 100 µl of lipid extract was evaporated onto ELISA plates and tested with mc3 and mc6. We tested both the nonpathogenic mycobacterium M. smegmatis as well as a less pathogenic (BSL2) close relative of Mtb- M. bovis BCG. Both antibodies were able to bind the lipid extract of both mycobacterial species (Fig. 3). As before, mc6 had superior sensitivity compared with mc3, giving higher signal at the same bacterial concentration. Based on the limit of detection of SNR of 2, the range of detection for both antibodies was roughly similar at between 106 to 107 CFU of M. smegmatis and one order of magnitude better for BCG at 105 to 106 CFU. Given that the volume of lipid extract evaporated was 100 µl, this translates into a lower concentration limit of 107 to 108 CFU/ml for M. smegmatis and 106 to 107 CFU/ml for BCG.
Fig. 3.
Limit of detection of the two most sensitive IgGs mc3 and mc6 using the rapid alkaline hydrolysis extraction method for various CFU amounts of M. smegmatis(left panel) and M. bovis BCG (right panel) based on a SNR of 2 (indicated by dashed line). Both antibodies mc3 and mc6 can detect between 107 to 108 CFU/ml of M. smegmatis and 106 to 107 CFU/ml of M. bovis BCG. All results are the average of three independent experiments and the error bars show SEM.
Discussion
Earlier studies had already indicated the feasibility of isolating antibodies from phage display libraries against lipids (16–18). Using this methodology, we panned the Humanyx nonimmune Fab phage library against mycolic acid purified from Mtb evaporated onto a solid surface. Enrichment of the library through repeated panning was successfully achieved as demonstrated by the increase in signal for mycolic acid in the polyclonal ELISA (Fig. 1A, left panel). This was further confirmed by the dominance of high affinity clones in the population of monoclonal antibodies tested from the fourth pan (Table 1), indicating that selection of fast replicating but poor expressing, nonspecific clones is not occurring and that this methodology has utility for the selection of high affinity anti-lipid antibodies.
Interestingly, our antibodies had specificity primarily for the methoxy subclass of mycolic acid with weak binding to α mycolic acid and no binding to any other lipid tested (Fig. 1B). The difference in these mycolic acid subclasses lies in long fatty acid tail, referred to as the meromycolate branch. This contains various functional groups such as keto and methoxy for which these subclasses are named and which are lacking in the α subclass (supplementary Fig. II) (9, 14). Conformational studies have shown that keto and methoxy mycolic acids tended to fold in a W-shaped conformation that exposed their polar functional groups, which could be hydrated by water molecules (supplementary Fig. II) (24). This could allow for interaction with antibodies via electrostatic and hydrogen bonds in an aqueous environment. It is therefore surprising that the isolated antibodies bound methoxy mycolic acid only. This is even more unexpected given that keto mycolic acid appears to be the subclass with the greatest tendency to fold in this manner (25). However, serological studies have shown that anti-trehalose dimycolate antibodies produced in the course of TB infection in humans are generally methoxy mycolic acid specific (26). It may be that the human antibody repertoire is predisposed toward recognition of this particular subclass and may explain the tendency of our nonimmune library to produce methoxy mycolic acid antibodies. The ability of our antibodies to distinguish between similar lipids with only small changes in molecular structure has been showed previously for antibodies raised in the same manner against cholesterol derivatives and indicates the ability of antibodies to provide fine specificity for lipid detection (17).
As some related bacterial genera such as Nocardia and Tsukamurella contain mycolic acid in their cell walls as well, we wanted to determine whether our antibodies could specifically recognize mycobacterial mycolic acid so as to reduce the potential for false negatives. We showed that our antibodies strongly bound M. smegmatis lipid extract only and not extract from other nonmycobacterial species (Fig. 1D). Interestingly, there was also some weak binding in two of the antibodies, mc6 and mc47, to T. paurometabolum lipid extract. This could be because its mycolic acids in terms of fine structure such as total number of carbon atoms and fatty acid chain length more closely resemble mycobacterial mycolic acids as compared with some other genera such as Nocardia and Corynebacteria, which have fewer carbon atoms and shorter chain length (9). A previous study indicated that a phage display-derived antibody against methyl palmitate, a nonpolar lipid, could distinguish between this lipid and others with similar structure but longer carbon aliphatic tail (18). Therefore, while it is generally assumed that antibodies principally target and recognize polar epitopes even on lipids, it is possible that our antibodies can differentiate apolar portions of the mycolic acid molecule as well, such as the fatty acid tail. This ability to discriminate mycobacterial mycolic acids is important for diagnostic purposes as the abovementioned species of actinomycetes have been identified in sputum samples from TB patients (27).
Our most sensitive antibody, mc6, was able to detect 4.5 ng of total mycolic acid or methoxy mycolic acid by ELISA (Fig. 2A, 2D). While this is the first time the sensitivity of an anti-mycolic acid antibody has been determined, this is not too dissimilar to other antibody-based TB diagnostic assays such as the urinary lipoarabinomannan test, which also detect in the low- to sub-nanogram level at similar SNRs (28, 29). To determine whether such sensitivity is sufficient for clinical use, we sought to establish a limit of detection based on bacterial count (CFU). However, to ensure that our assay was as sensitive as possible while remaining suitable for use as a point-of-care test in a resource-poor setting, we first had to optimize the protocol for maximum efficiency of lipid extraction from the sample while minimizing time, reagents, and equipment required. While the initial method of vortexing in hexane was the simplest and did not require heating or neutralization and was ideal for a point-of-care assay, it was unlikely to give the best lipid yield. Instead, alkaline hydrolysis proved to be the method of choice for optimal lipid extraction. It enables release of covalently bound mycolic acids in addition to being the most rapid method (Fig. 2E). A variant of this method, which utilized a more complex protocol requiring overnight incubation with organic solvent to inactivate and delipidate cells, had also been applied previously in combination with mass spectrometry to give a limit of detection of 104 CFU Mtb in spiked sputum and was also used to extract detectable levels of mycolic acid from patient samples, thus indicating its suitability for clinical use (8). We therefore chose the alkaline hydrolysis method to determine the CFU limit of detection for the two most sensitive antibodies mc3 and mc6 and verified its ability to extract mycolic acids through HR-MS/MS and MRM analyses of the lipid extract which confirmed the presence of mycolic acid.
To determine the equivalent CFU detection limit, we used both M. smegmatis and M. bovis BCG mycobacterial species. The choice of M. bovis BCG was due to fact that while these two mycobacterial species along with Mtb have α mycolic acids, both M. bovis BCG and Mtb but not M. smegmatis produce methoxy mycolic acids (9). Thus, they have a greater similarity in the subclasses of mycolic acids detected by our antibodies although the levels of methoxy mycolic acid in BCG have reported to be low (9, 14). Our results indicate that mc3 and mc6 recognize M. bovis BCG at a limit of between 105 to 106 CFU (Fig. 3) and are less sensitive to M. smegmatis by an order of magnitude, which is not surprising given the partial dissimilarity in mycolic acid composition and the higher sensitivity of our antibodies to methoxy mycolic acid over α mycolic acid. This higher sensitivity would be an advantage if the antibodies are to be used for diagnostic purposes as there are only a very limited number of mycobacterial species other than the Mtb complex group of species (Mtb, M. bovis, M. africanum, M. microti) that contain methoxy mycolic acids (9).
While our assay is currently less sensitive than mass spectrometry, which detects 104 CFU, this is not surprising given that mass spectrometry is generally far more sensitive than immunological assays and can detect as low as 1pg of pure lipid (8). Nevertheless, based on CFU alone, this result is still less sensitive as compared with current diagnostic methods such as sputum culture, which can detect levels as low as 102 CFU/ml of mycobacteria and even sputum smear, which requires 104 CFU/ml for reliable detection (30). However, both these methods require intact bacteria and, in the case of culture, live bacteria for successful detection. As our technique relies on antigen detection instead. It can also detect mycolic acids in sputum released by dead bacterial fragments and thus may be more sensitive than indicated based on CFU counts alone. Furthermore, the higher affinity of our antibodies for methoxy mycolic acid over other subclasses (45 ng/ml limit versus 200 ng/ml) probably results in an artificially poor CFU limit of detection as our test species M. smegmatis and M. bovis BCG have no or low levels of methoxy mycolic acid, respectively, as compared with α mycolic acids as indicated by our own mass spectrometry data and previous publications (9). On the other hand, NMR analysis has indicated that the proportion of methoxy to α mycolic acid is 50% or above in various Mtb strains (14). Mass spectrometry data has also indicated that the most common species of methoxy and α mycolic acids are present in TB patient sputum at an average concentration of over 100 ng/ml (8). This is well within the detection limits of mc3 and mc6 and samples can be further concentrated by evaporation of organic carrier solvent, thus allowing for enhanced sensitivity when mycolic acid concentrations are low.
We have described the novel application of antibody phage display and recombinant antibody technology for the detection of nonpolar lipid biomarkers, in this particular case, mycolic acid, for TB diagnostics. Further testing on actual clinical sputum samples will be required to validate the clinical utility of these antibodies. While the avidity of the antibodies or efficiency of extraction can be improved to make this assay more clinically useful, in principle this method can be applied to a whole range of insoluble lipids that previously may have been inaccessible to antibody-based detection. We have demonstrated that such lipids can be extracted efficiently and rapidly and be detected in an assay format, which could be conducted rapidly if required without the need for extensive training or expensive equipment. These methods could therefore prove useful in the future for the detection of a wide variety of novel lipid biomarkers in a point-of-care format.
Supplementary Material
Footnotes
Abbreviations:
- BCG
- Bacille Calmette-Guérin
- CFU
- colony forming units
- DMPC
- dimyristoyl phosphocholine
- HR-MS
- high-resolution -mass spectrometry
- ManLAM
- mannose-capped lipoarabinomannan
- Mtb
- Mycobacterium tuberculosis
- MPBS
- skim milk in PBS
- OD
- optical density
- MRM
- multiple reaction monitoring
- Mtb
- PILAM, phosphoinositol-capped lipoarabinomannan
- SNR
- signal-to-noise ratio
- TB
- tuberculosis
This research was supported by the National Research Foundation, Singapore (Grant No. 182-000-218-281), Biomedical Research Council-Science and Engineering Research Council, Singapore (Joint Grant No. R182-000-206-30) and National University of Singapore–Life Sciences Institute (SLING) (Grant No. R-711-000-021-133).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table and two figures.
REFERENCES
- 1.WHO Global Tuberculosis Control. 2011, Geneva: World Health Organisation. [Google Scholar]
- 2.WHO Diagnostics for Tuberculosis: Global demand and market potential. 2006, Geneva: World Health Organisation [Google Scholar]
- 3.Steingart K. R., Ng V., Henry M., Hopewell P. C., Ramsay A., Cunningham J., Urbanczik R., Perkins M. D., Aziz M. A., Pai M. 2006. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 6: 664–674 [DOI] [PubMed] [Google Scholar]
- 4.Kambashi B., Mbulo G., McNerney R., Tembwe R., Kambashi A., Tihon V., Godfrey-Faussett P. 2001. Utility of nucleic acid amplification techniques for the diagnosis of pulmonary tuberculosis in sub-Saharan Africa. Int. J. Tuberc. Lung Dis. 5: 364–369 [PubMed] [Google Scholar]
- 5.Minion J., Leung E., Talbot E., Dheda K., Pai M., Menzies D. 2011. Diagnosing tuberculosis with urine lipoarabinomannan: systematic review and meta-analysis. Eur. Respir. J. 38: 1398–1405 [DOI] [PubMed] [Google Scholar]
- 6.Flores L. L., Steingart K. R., Dendukuri N., Schiller I., Minion J., Pai M., Ramsay A., Henry M., Laal S. 2011. Systematic review and meta-analysis of antigen detection tests for the diagnosis of tuberculosis. Clin. Vaccine Immunol. 18: 1616–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rastogi N., Legrand E., Sola C. 2001. The mycobacteria: an introduction to nomenclature and pathogenesis. Rev. Sci. Tech. 20: 21–54 [DOI] [PubMed] [Google Scholar]
- 8.Shui G., Bendt A. K., Jappar I. A., Lim H. M., Laneelle M., Herve M., Via L. E., Chua G. H., Bratschi M. W., Zainul Rahim S. Z., et al. 2012. Mycolic acids as diagnostic markers for tuberculosis case detection in humans and drug efficacy in mice. EMBO Mol Med. 4: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry III C. E., Lee R. E., Mdluli K., Sampson A. E., Schroeder B. G., Slayden R. A., Yuan Y. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37: 143–179 [DOI] [PubMed] [Google Scholar]
- 10.Shui G., Bendt A. K., Pethe K., Dick T., Wenk M. R. 2007. Sensitive profiling of chemically diverse bioactive lipids. J. Lipid Res. 48: 1976–1984 [DOI] [PubMed] [Google Scholar]
- 11.Viader-Salvadó J. M., Molina-Torres C. A., Guerrero-Olazaran M. 2007. Detection and identification of mycobacteria by mycolic acid analysis of sputum specimens and young cultures. J. Microbiol. Methods. 70: 479–483 [DOI] [PubMed] [Google Scholar]
- 12.Song S. H., Park K. U., Lee J. H., Kim E. C., Kim J. Q., Song J. 2009. Electrospray ionization-tandem mass spectrometry analysis of the mycolic acid profiles for the identification of common clinical isolates of mycobacterial species. J. Microbiol. Methods. 77: 165–177 [DOI] [PubMed] [Google Scholar]
- 13.Leite C. Q., de Souza C. W., Leite S. R. 1998. Identification of mycobacteria by thin layer chromatographic analysis of mycolic acids and conventional biochemical method: four years of experience. Mem. Inst. Oswaldo Cruz. 93: 801–805 [DOI] [PubMed] [Google Scholar]
- 14.Watanabe M., Aoyagi Y., Ridell M., Minnikin D. E. 2001. Separation and characterization of individual mycolic acids in representative mycobacteria. Microbiology. 147: 1825–1837 [DOI] [PubMed] [Google Scholar]
- 15.Singh K. V., Kaur J., Varshney G. C., Raje M., Suri C. R. 2004. Synthesis and characterization of hapten-protein conjugates for antibody production against small molecules. Bioconjug. Chem. 15: 168–173 [DOI] [PubMed] [Google Scholar]
- 16.Tsuruta L. R., Tomioka Y., Hishinuma T., Kato Y., Itoh K., Suzuki T., Oguri H., Hirama M., Goto J., Mizugaki M. 2003. Characterization of 11-dehydro-thromboxane B2 recombinant antibody obtained by phage display technology. Prostaglandins Leukot. Essent. Fatty Acids. 68: 273–284 [DOI] [PubMed] [Google Scholar]
- 17.Islam M. O., Lim Y. T., Chan C. E., Cazenave-Gassiot A., Croxford J. L., Wenk M. R., Macary P. A., Hanson B. J. 2012. Generation and characterization of a novel recombinant antibody against 15-ketocholestane isolated by phage-display. Int. J. Mol. Sci. 13: 4937–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gargir A., Ofek I., Meron-Sudai S., Tanamy M. G., Kabouridis P. S., Nissim A. 2002. 2002. Single chain antibodies specific for fatty acids derived from a semi-synthetic phage display library. Biochim. Biophys. Acta. 1569: 167–173 [DOI] [PubMed] [Google Scholar]
- 19.Chan C. E., Chan A. H., Lim A. P., Hanson B. J. 2011. Comparison of the efficiency of antibody selection from semi-synthetic scFv and non-immune Fab phage display libraries against protein targets for rapid development of diagnostic immunoassays. J. Immunol. Methods. 373: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim A. P., Chan C. E., Wong S. K., Chan A. H., Ooi E. E., Hanson B. J. 2008. Neutralizing human monoclonal antibody against H5N1 influenza HA selected from a Fab-phage display library. Virol. J. 5: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minnikin D. E. 1988, Isolation and purification of mycobacterial wall lipids. In Bacterial Cell Surface Techniques, I. Hancock and I. Poxton, editors. John Wiley & Sons, New Jersey. p. 125–135. [Google Scholar]
- 22.Benadie Y., Deysel M., Siko D. G., Roberts V. V., Van Wyngaardt S., Thanyani S. T., Sekanka G., Ten Bokum A. M., Collett L. A., Grooten J., et al. 2008. Cholesteroid nature of free mycolic acids from M. tuberculosis. Chem. Phys. Lipids. 152: 95–103 [DOI] [PubMed] [Google Scholar]
- 23.Beukes M., Lemmer Y., Deysel M., Al Dulayymi J. R., Baird M. S., Koza G., Iglesias M. M., Rowles R. R., Theunissen C., Grooten J., et al. 2010. Structure-function relationships of the antigenicity of mycolic acids in tuberculosis patients. Chem. Phys. Lipids. 163: 800–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villeneuve M., Kawai M., Watanabe M., Aoyagi Y., Hitotsuyanagi Y., Takeya K., Gouda H., Hirono S., Minnikin D. E., Nakahara H. 2010. Differential conformational behaviors of alpha-mycolic acids in Langmuir monolayers and computer simulations. Chem. Phys. Lipids. 163: 569–579 [DOI] [PubMed] [Google Scholar]
- 25.Villeneuve M., Kawai M., Kanashima H., Watanabe M., Minnikin D. E., Nakahara H. 2005. Temperature dependence of the Langmuir monolayer packing of mycolic acids from Mycobacterium tuberculosis. Biochim. Biophys. Acta. 1715: 71–80 [DOI] [PubMed] [Google Scholar]
- 26.Pan J., Fujiwara N., Oka S., Maekura R., Ogura T., Yano I. 1999. Anti-cord factor (trehalose 6,6’dimycolate) IgG antibody in tuberculosis patients recognizes mycolic acid subclasses. Microbiol. Immunol. 43: 863–869 [DOI] [PubMed] [Google Scholar]
- 27.Dheda K., Davids V., Lenders L., Roberts T., Meldau R., Ling D., Brunet L., van Zyl Smit R., Peter J., Green C., et al. 2010. Clinical utility of a commercial LAM-ELISA assay for TB diagnosis in HIV-infected patients using urine and sputum samples. PLoS ONE. 5: e9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehme C., Molokova E., Minja F., Geis S., Loscher T., Maboko L., Koulchin V., Hoelscher M. 2005. Detection of mycobacterial lipoarabinomannan with an antigen-capture ELISA in unprocessed urine of Tanzanian patients with suspected tuberculosis. Trans. R. Soc. Trop. Med. Hyg. 99: 893–900 [DOI] [PubMed] [Google Scholar]
- 29.Tessema T. A., Hamasur B., Bjun G., Svenson S., Bjorvatn B. 2001. Diagnostic evaluation of urinary lipoarabinomannan at an Ethiopian tuberculosis centre. Scand. J. Infect. Dis. 33: 279–284 [DOI] [PubMed] [Google Scholar]
- 30.Tiruviluamala P., Reichman L. B. 2002. Tuberculosis. Annu. Rev. Public Health. 23: 403–426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.