Abstract
The variant antigen, Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1), expressed on the surface of P. falciparum infected Red Blood Cells (iRBCs) is a critical virulence factor for malaria1. Each parasite encodes 60 antigenically distinct var genes encoding PfEMP1s, but during infection the clonal parasite population expresses only one gene at a time before switching to the expression of a new variant antigen as an immune evasion mechanism to avoid the host’s antibody responses2,3. The mechanism by which 59 of the 60 var genes are silenced remains largely unknown4–7. Here we show that knocking out the P. falciparum variant-silencing SET gene (PfSETvs), which encodes an ortholog of Drosophila melanogaster ASH1 and controls histone H3 lysine 36 trimethylation (H3K36me3) on var genes, results in the transcription of virtually all var genes in the single parasite nuclei and their expression as proteins on the surface of individual iRBCs. PfSETvs-dependent H3K36me3 is present along the entire gene body including the transcription start site (TSS) to silence var genes. With low occupancy of PfSETvs at both the TSS of var genes and the intronic promoter, expression of var genes coincides with transcription of their corresponding antisense long non-coding RNA (lncRNA). These results uncover a novel role of the PfSETvs-dependent H3K36me3 in silencing var genes in P. falciparum that might provide a general mechanism by which orthologs of PfSETvs repress gene expression in other eukaryotes. PfSETvs knockout parasites expressing all PfEMP1s may also be applied to the development of a malaria vaccine.
Besides histone deacetylases (HDACs)8,9, histone lysine methyltransferases (HKMTs) or histone lysine demethylases (HKDMs) may play critical roles in controlling gene expression in P. falciparum4–7,10,11. There are a total of 10 predicted PfHKMTs belonging to the SET domain superfamily, two PfHKDMs of the LSD1 family and three members of the Jumonji-related family10,12 (Supplementary Table 1). However, the major factor for var gene silencing remains unknown.
We therefore examined if PfHKMTs or PfHKDMs are key factors in controlling mutually exclusive expression of the var gene family by attempting to knock out all of the PfHKMT (PfSET) genes and three of the PfHKDM genes in 3D7 (Fig. 1a and Supplementary Fig. 1). Four of 9 PfSET genes and all 3 studied PfHKDM genes could be genetically disrupted (Fig. 1b and Supplementary Fig. 1), suggesting that the other 5 PfSET genes are essential for the parasite in the asexual blood stage. Gene expression microarray analyses showed that the knockout (Fig. 1c, d and Supplementary Fig. 1c) of a previously-called PfSET2 gene10 (PlasmoDB gene ID: PF3D7_1322100) led to expression of virtually all var genes in the ring stage (Fig. 1e and Supplementary Table 2). In contrast, knockout of any other PfSET or PfHKDM genes did not alter the transcription of the var gene family in 3D7 (Supplementary Fig. 1e–j and Supplementary Table 3–8). In addition, some members of other clonally variant gene families (rifin and stevor) plus the var gene family account for the majority of the genes upregulated in 3D7SET2Δ (Supplementary Fig. 2 and Supplementary Table 2). Therefore, we renamed this P. falciparum variant-silencing SET gene to PfSETvs. Activation of the majority of var genes by SETvsΔ was further corroborated by quantitative PCR (qPCR) at 18 h after invasion in both 3D7 (Fig. 1f) and Dd2 (Supplementary Fig. 3), indicating that PfSETvs is involved in broadly silencing var genes.
Figure 1. Knockout of PfSETvs leads to expression of all var genes.
a, Schematic diagram of the PfSETvs gene knockout strategy. E, EcoR V; 5′F, 5′ flanking fragment for crossover recombination; 3′F, 3′ flanking fragment for crossover recombination; hDHFR, human dihydrofolate reductase (hDHFR); TK, Thymidine Kinase; SET, SET domain; P, DNA probe for southern blot analysis. b, Summary of knockout studies for 9 PfHKMTs and 3 PfHKDMs (PfLSD1, PfJmjC1 and PfJmjC2) genes. KO, knockout; yes, succeeded in gene knockout; no, failed to knockout gene. c, d, Southern blot analysis using a DNA probe (P) from downstream of the knocked out SET domain of the PfSETvs gene (See also in a) for PfSETvsΔ in 3D7 (c) and Dd2 (d). The sizes of three different hybridization bands from the integrated (In) or wild-type (WT) genome and the episomal plasmid (EP) are indicated to the right. bp, base pairs. e, Comparative transcriptome analysis of wild-type 3D7 and 3D7SETvsΔ at 18 h after invasion. x axis (wild-type 3D7) and y axis (3D7SETvsΔ) are logarithmic and correspond to relative signal of hybridization to each gene shown as a dot (See also in Supplementary Table 2). All var genes with authentic hybridization signals are shown in red. The dominantly expressed var gene (PF3D7_1240600) in wild-type 3D7 is indicated by a red arrow. f, Quantitative PCR analyses of transcriptional upregulation (log2 ratio of PfSETvsΔ to wild-type parasites) of var genes in 3D7SETvsΔ at 18 h after invasion. Each type of var genes is shown on the top. The dominantly expressed var gene and a second gene expressed at a low frequency in the wild-type 3D7 population are indicated by red arrowheads. Experiments were repeated three times. Error bars represent s.e.m.
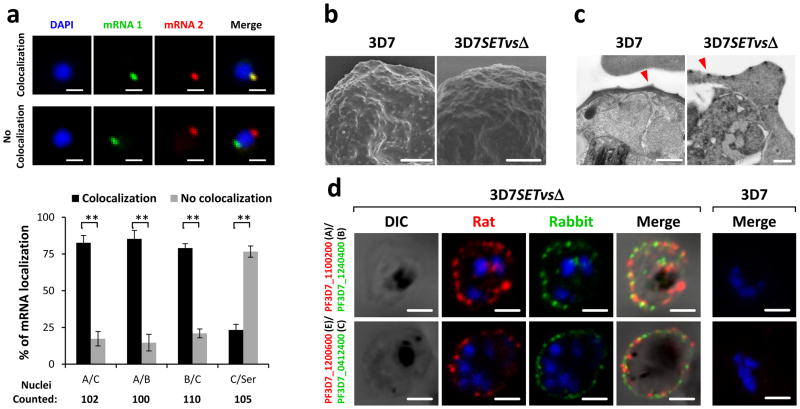
To determine if PfSETvsΔ activated multiple var genes in a single iRBC, we tested if different types of var genes could be transcribed in a single 3D7SETvsΔ iRBC by RNA fluorescence in situ hybridization (FISH). Each combined RNA FISH of two representative var transcripts indicated co-expression of all three types of var genes in individual 3D7SETvsΔ nucleus (Fig. 2a). The tested var transcripts colocalized with each other at a particular site of the nuclear periphery (Fig. 2a). Transcription of a control gene Seryl-tRNA synthetase (PF3D7_0717700) did not occur at this site (Fig. 2a), suggesting that var genes have a specific transcriptionally active site in agreement with previous findings6,13. Moreover, our results showed that multiple var transcripts also colocalized at the single peripheral site of 3D7SETvsΔ nuclei, even though the genomic loci of these var genes were diverse (Supplementary Fig. 4a–c). Taken together, our results demonstrate multiple var transcripts in one nucleus and suggest that a var-specific nuclear compartment exists for active transcription of multiple var genes.
Figure 2. Simultaneous expression of multiple var genes in single 3D7SETvsΔ iRBCs.
a, Two-color RNA FISH and statistical analyses of colocalization of each two types of var transcripts in 3D7SETvsΔ by using gene-specific probes (Supplementary Fig. 4a). Seryl-tRNA synthetase transcript served as a negative control. Average numbers of counted nuclei are listed under each tested group. n = 3. Error bars represent s.e.m; P values were obtained using a one-tailed Student’s t-test. **P < 0.01. b, c, Electron microscopy of gelatin selected 3D7 and 3D7SETvsΔ iRBCs. Typical knobs in scanning electron microscopy (SEM) (b) and transmission electron microscopy (TEM) (c) pictures are indicted by red arrowheads. d, Live-cell immunofluorescence assay (IFA) using rat and rabbit antisera to various PfEMP1s to detect co-expression of different PfEMP1s on the surface of 3D7SETvsΔ iRBCs. Wild-type 3D7 infected RBC is shown to the right. No staining is seen. DAPI (blue) is used to mark the parasite nucleus. Types of var genes are shown in parentheses. Scale bars: a, b, 1 μm; c, 0.5 μm; d, 1.5 μm.
To determine if parasites transcribing multiple var genes are able to translate and transport multiple PfEMP1s on the surface of iRBCs, live-cell immunofluorescence assay (IFA) was performed with rat and rabbit antibodies to different PfEMP1s. As expected, the gelatin-enriched parasite presented knobs on the surface of iRBCs in both 3D7 and 3D7SETvsΔ (Fig. 2b, c). Furthermore, surface expression of multiple PfEMP1s on a single 3D7SETvsΔ iRBC was observed by confocal microscopy (Fig. 2d and Supplementary Fig. 4d). It is important to note that in 3D7SETvsΔ iRBC double labeling of PfEMP1s was always observed (Fig. 2d). As reported previously14, no co-expression of different PfEMP1s in individual iRBCs by 3D7 clones was detected using different antibodies (Fig. 2d and Supplementary Fig. 4d). We were also unable to show surface labeling of the active PF3D7_1240600 in the wild-type 3D7 because we lacked an antibody to this PfEMP1.
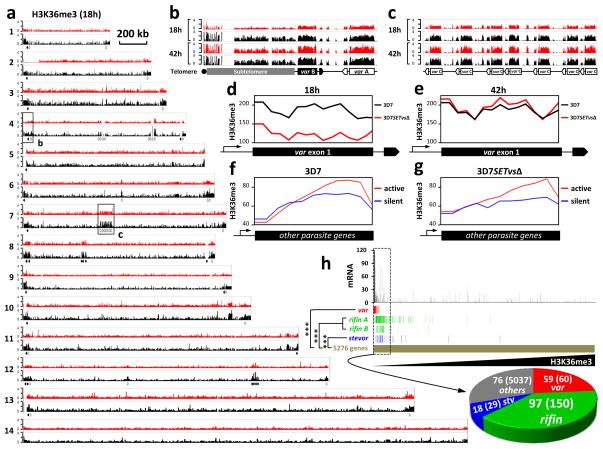
PfSETvs, an ortholog of Drosophila melanogaster ASH1, is the only representative of the SETD2-NSD-ASH1 clade in P. falciparum (Supplementary Fig. 5), which in addition to the SMYD clade are the two distinct occasions in the evolution of SET domains as H3K36-specific methyltransferases in eukaryotes12. To monitor changes of histone lysine methylations by PfSETvsΔ, antibodies that specifically recognized P. falciparum H3K36me3, H3K36me2 (Supplementary Fig. 6a, b), H3K4me3, H3K9me3 and H4K20me3 were used in ChIP-seq experiments. In the wild-type 3D7, a robust enrichment of H3K36me3 (Fig. 3a–c) but not H3K36me2 (Supplementary Fig. 7a) was observed only in the telomeric and subtelomeric heterochromatin regions of the 14 P. falciparum chromosomes plus several discrete genomic regions where all of the var genes are located at either 18 or 42 h after invasion. However, H3K36me3 other than other histone lysine methylations was greatly reduced in the entire gene body of var genes in 3D7SETvsΔ at 18 h after invasion (Fig. 3d and Supplementary Fig. 8 and 9), indicating a direct positive correlation of H3K36me3 with the PfSETvs activity. Considering the extremely low level of H3K36me2 at var loci in wild-type 3D7 (Supplementary Fig. 7a), H3K36me3 is functionally important for var gene regulation. PfSETvs may di- and tri- methylate H3K36 because these markers were also reduced at the TSS of activated var genes due to PfSETvsΔ (Supplementary Fig. 7c–g). Interestingly, similarly high levels of H3K36me3 were observed in both wild-type 3D7 and 3D7SETvsΔ at 42 h after invasion when var genes were silent (Fig. 3e), suggesting at least one other PfHKMT that catalyzes H3K36me3 in P. falciparum schizont iRBCs. In addition, our data showed that none of the var transcripts colocalized with H3K36me3 in the nuclei (Supplementary Fig. 10). Collectively, our data suggest that the PfSETvs-dependent H3K36me3 is specifically involved in var gene silencing.
Figure 3. PfSETvs-dependent H3K36me3 is specifically associated with var gene silencing.
a, Integrative genomic view of ChIP-seq analysis of H3K36me3 along 3D7 (black) and 3D7SETvsΔ (red) chromosomes at 18 h after invasion. Sixty var genes distributed along P. falciparum chromosomes 1–13 are indicated by solid (forward orientation) and open (reverse orientation) arrows. Each read was normalized by the total number of uniquely mapped ChIP-seq reads. Chromosomal numbers are shown to the left. Regions are boxed for a detailed view represented in (b) and (c). A scale bar representing 200kb is shown to the right of chromosome 1. b, c, At 18 h and 42 h after invasion, integrative genomic view of H3K36me3 distributed at the 5′ end of chromosome 4 representing a region that includes the telomere, subtelomere, type A and B var genes (b), and at the middle of chromosome 7 representing a type C var gene cluster (c) in 3D7 (black) and 3D7SETvsΔ (red). d, e, Distribution of H3K36me3 along exon 1 of 50 tested var genes in 3D7SETvsΔ (red) and wild-type 3D7 (black) at 18 h (d) or 42 h (e) after invasion. Exon1 of each vargene was equally divided into 14 bins. Total reads of each bin by ChIP-seq were normalized by total uniquely mapped reads. f, g, Distribution of H3K36me3 across the gene bodies of 400 ring-stage active genes (red) and 400 ring-stage silent genes (blue) (See the gene list in Supplementary Table 9) in wild-type 3D7 (f) and 3D7SETvsΔ (g). Each gene was equally divided into 20 bins. Total reads of each bin by ChIP-seq were normalized by total uniquely mapped reads. h, Statistical analysis of the correlation between reduction of H3K36me3 and upregulation of var, rifin and stevor gene families. 5276 parasite genes were sorted from low to high levels of H3K36me3 in 3D7SETvsΔ normalized by that in 3D7. Expression fold change of each gene by PfSETvsΔ was shown on the top panel (See also in Supplementary Table 10). Distribution of all of var (red), rifin including A- and B-type rifin genes (green) and stevor (blue) genes is shown along the parasite genes (gold). In the top 250 H3K36me3-reduced genes boxed by dash lines, numbers of var (red), rifin (green), stevor (blue) and other genes (grey) compared to their total numbers were shown in a pie chart at the bottom. Hypergeometric test was computed for the var (P = 3.4e-80), rifin (P = 9.7e-98) and stevor (P = 1.73e-17) gene families to gauge their significance of upregulation in the reduction of H3K36me3.
Strikingly, H3K36me3 was also observed for a high enrichment at the 3′ end of 400 ring stage-active genes (other than var, rifin and stevor genes) compared to 400 ring-stage silent genes (See gene lists in Supplementary Table 9) in both wild-type 3D7 and 3D7SETvsΔ (Fig. 3f, g), indicating that the PfSETvs-independent H3K36me3 may contribute to transcriptional elongation as reported in other eukaryotes15–17 and might compensate the global levels of H3K36me3 in 3D7SETvsΔ (Supplementary Fig. 6c, d). We next examined if reduction of H3K36me3 by PfSETvsΔ is specifically associated with activation of parasite clonally variant genes. Among 5276 P. falciparum genes, 59 out of 59 var genes, 97 out of 150 rifin genes (including 69 A-type and 28 B-type rifin genes) and 18 out of 29 stevor genes belonged to the top 250 genes with highest reduction of H3K36me3 by PfSETvsΔ (Fig. 3h). Furthermore, the same gene group is enriched for increased expression as determined by microarray experiments (Supplementary Table 10). Our data indicate that H3K36me3, controlled by PfSETvs, plays a repressive role in silencing parasite clonally variant gene families.
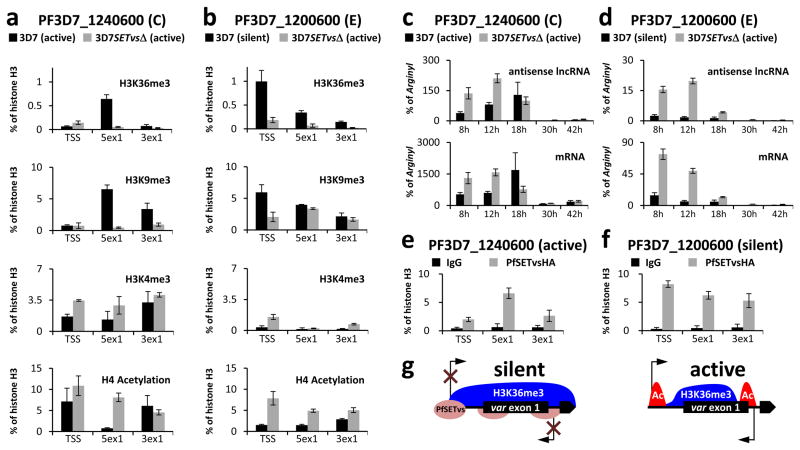
To further corroborate the role of H3K36me3 in var gene silencing, we examined the histone modification at the TSS of an active var gene (PF3D7_1240600) and a silent var gene (PF3D7_1200600) in the wild-type 3D7, both of which are active in 3D7SETvsΔ (Fig. 4a, b). Because of high sequence similarity in the 5′-UTR including the TSS and the intronic promoter of var genes, ChIP-qPCR but not ChIP-seq can be used in these regions (Fig. 3b, c). In wild-type 3D7, the TSS occupancy of H3K36me3 is considerably higher in the silent var gene compared to the active one (Fig. 4a, b). In contrast, two var genes studied exhibit low levels of H3K36me3 at the TSS in 3D7SETvsΔ, consistent with their active expression (Fig. 4a, b). H3K9me3, a transcriptional silent mark, showed similar profiles as H3K36me3 (Fig. 4a, b), whereas two active marks H3K4me3 and H4 acetylation were present at the TSS of active genes in both wild-type 3D7 and 3D7SETvsΔ (Fig. 4a, b). In addition, the similar results were observed in three other var genes representing type A (PF3D7_0400400), type B (PF3D7_0300100) and type C (PF3D7_0617400) (Supplementary Fig. 11). Altogether, our data support the idea that the high level of H3K36me3 at the TSS region is involved in transcriptional repression.
Figure 4. PfSETvs and H3K36me3 repress var gene expression at the TSS.
a, b, ChIP-qPCR of the active 3D7 var gene PF3D7_1240600 (a) and a silent 3D7 var gene PF3D7_1200600 (b) with antibodies to H3K36me3, H3K9me3, H3K4me3 and histone H4K5/K8/K12/K16 acetylation in both 3D7 and 3D7SETvsΔ at 18 h after invasion by using three different PCR primer sets schematized in Supplementary Fig. 7b. TSS, transcription start site; 5ex1, 5′ end of exon 1; 3ex1, 3′ end of exon 1. c, d, Expression profiles of mRNA and antisense lncRNA transcribed from PF3D7_1240600 (c) or PF3D7_1200600 (d) at five different time points after invasion as shown in the figures. Expression levels of var transcripts were normalized to expression of a housekeeping gene, arginyl-tRNA synthetase (PF3D7_0913900). The forward and reverse primers of the 3ex1 PCR primer set (Supplementary Fig. 7b) were used for antisense lncRNA and mRNA reverse transcription, respectively. Types and the transcription status of var genes are shown in parentheses, respectively. Experiments were repeated three times. Error bars represent s.e.m. e, f, ChIP-qPCR of the active 3D7 var gene PF3D7_1240600 (e) and a silent 3D7 var gene PF3D7_1200600 (f) with a mouse antibody to HA in 3D7SETvsHA at 18 h after invasion by using the same PCR primers in (a). g, Summary diagram showing that the PfSETvs-dependent H3K36me3 enriched along the entire gene body of silent var genes including the TSS of var genes and the respective intronic antisense promoter leads to silencing of both var mRNA and antisense lncRNA.
It is worth noting that each var gene harbors an intronic promoter driving the transcription of an antisense long non-coding RNA (lncRNA) with unknown function18. Our ChIP-seq data showed that two active var genes (PF3D7_1240600 and PF3D7_0900100) in wild-type 3D7 populations (Fig. 1f) had low levels of H3K36me3 at the 3′ end of the exon 1, whereas silent var genes had high levels of H3K36me3 at the same region (Supplementary Fig. 12), suggesting a positive correlation between PfSETvs-dependent H3K36me3 occupancy and var lncRNA silencing. To explore this concept further, histone modification profiles in the 3′ portion of var exon 1 as a proxy for lncRNA promoter was examined by ChIP-qPCR since the introns of var genes are highly conserved among the gene family. Our results showed similar trends of H3K36me3 between the TSS of var genes and their corresponding 3′ but not 5′ portion of the exon 1 (Fig. 4a, b and Supplementary Fig. 11), consistent with the observation by strand-specific qPCR that active transcription of var genes coincides with the expression of the corresponding antisense lncRNAs at 8–18 h after invasion (Fig. 4c, d and Supplementary Fig. 13). These results demonstrated a correlated upregulation of var genes and the corresponding lncRNAs, in association with low occupancy of the PfSETvs-dependent H3K36me3 at the TSS.
To further investigate the biological function of PfSETvs in var gene silencing, a triple HA tag was fused in frame to the C-terminus of PfSETvs in 3D7SETvsHA (Supplementary Fig. 14a–d). The resulting PfSETvsHA, like wild-type PfSETvs, still contributed to mutually exclusive expression of the var gene family (Supplementary Fig. 14e). Furthermore, IFA analysis showed that PfSETvsHA located at multiple nuclear sites, one of which colocalized with H3K36me3 in 3D7SETvsHA (Supplementary Fig. 14f), suggesting that the enzymatic activity of PfSETvs for H3K36me3 might require additional factors at the single perinuclear site. ChIP-qPCR results showed that, at 18 h after invasion, PfSETvsHA was not enriched at the TSS and in the intronic promoter region of the active var gene (Fig. 4e), instead tended to increase at these regions of silent var genes tested in 3D7SETvsHA (Fig. 4f and Supplementary Fig. 14g–i). No comparable enrichment of PfSETvsHA was observed in a var-unrelated silent gene (PF3D7_0424100) (Supplementary Fig. 14j). Taken together, our data indicate that PfSETvsHA specifically localizes to the TSS and intronic promoters for var gene silencing, in association with the PfSETvs-dependent H3K36me3 (Fig. 4g).
In this study, we showed that the H3K36 methylation system is differentiated into at least two distinct forms in P. falciparum, with the PfSETvs-dependent one functioning in a negative regulatory capacity (Fig. 4g), and the second independently of it alongside the elongating RNAPII (Supplementary Fig. 14k). Cognates of the PfSETvs-dependent mechanism for gene silencing might also exist in other eukaryotes in the cases of previously reported members of the ASH1-like subclade, such as Caenorhabditis elegans MES-419 and Drosophila melanogaster ASH120, and perhaps explain the association between H3K36me3 and silent genes in zebrafish sperm21 and the pericentromeric heterochromatin in mouse embryonic stem cells and fibroblasts22. In the RNAPII-related mechanism, H3K36me3 generated by the SETD2 subclade enzymes recruits HDACs15 and prevents incorporation of acetylated histones23 in transcribed gene bodies to prevent cryptic transcription initiation inside active genes. Given the role of lncRNAs as scaffolds recruiting Set2 histone methyltransferase and Set3 histone deacetylase complex to repress transcription initiation24,25, it would be interesting to investigate whether the antisense lncRNA might regulate var gene expression in a similar manner25.
The factor that activates individual var genes in the wild-type parasite still remains unknown. It may be a mechanism that randomly turns on var genes at a low rate. We previously found that only 1 in 200 parasites expresses the Rh4 gene in Dd226, controlled by H3K9me327 and a similar mechanism involving PfSETvs may exist for var genes. Recent work demonstrates that PfEMP1s are key targets of humoral immunity28. However, malaria immunity is acquired only slowly after years of repeated exposure that in part reflects the time required for an individual to experience a sufficient number of variant antigens. The SETvsΔ parasite could be used as an antimalarial vaccine via its ability to express all PfEMP1s to which the antibody would provide efficient protective immunity against malaria.
METHODS
Parasites Culture and Transfection
P. falciparum clones 3D7 initially isolated from Netherlands29 and Dd2 initially isolated from Vietnam30 were cultured in human O+ erythrocytes according to standard procedures 31. For gene deletion, PCR amplification was performed on P. falciparum strain 3D7 genomic DNA to obtain gene specific 5′ and 3′ flanking fragments, which were cloned into Spe I/Bgl II (5′) and EcoR I/Nco I (3′)-digested pHTK vector 32. Names of twelve targeted genes (Fig. 1d) and PCR primers are listed in Supplementary Table 11. Transfection and knockout selection were performed as described previously32. Briefly, 250 μl of packed iRBCs (5–10% ring parasites) were transfected by electroporation with 100 μg of the transfection pHTK plasmid. Positive (WR99210, 2 nM) and negative (Ganciclovir, 20 μM) drug selection were applied for selecting a population of parasites in which the plasmid-derived hDHFR gene (for WR99210 selection) has been integrated via double crossover homologous recombination into the endogenous targeted gene locus, and the episomal plasmid carrying the TK gene (for Ganciclovir self-killing selection). Selected knockout parasites were further confirmed by PCR screening (See also in Supplementary Fig. 1a) prior to being cloned by limiting dilution.
Antibody
A peptide (CNTKAFKSKKLKLRK) from the PfSETvs protein was synthesized, and rabbits were immunized to obtain the polyclonal antibody to PfSETvs by GenScript Inc. Various PfEMP1 domains (See also in Supplementary Fig. 3b) were recombinantly expressed in a baculovirus system and immunized to rats and rabbits for making polyclonal antibodies to different PfEMP1s as described previously13.
Southern blotting
Southern blot analyses on PfSETvsΔ or PfSETvsHA parasites were performed using the DIG High Prime DNA Labeling and Detection Starter Kit (Roche) according to the product manual. Briefly, genomic DNA was digested by EcoRV for 4 h at 37 °C and separated on a 0.8% agarose gel for Southern blotting onto the Hybond N+ nylon transfer membrane (Amersham). The target gDNA bands were hybrized by a Digoxigenin-labeled DNA probe complementary to the homologous 3′ flanking fragment (See P in Fig. 1a) and detected by anti-Digoxigenin-AP conjugated antibody. Primers for the amplification of the probe are listed in Supplementary Table 11.
Western blotting
To determine knockout of PfSETvs at the protein level in 3D7SETvsΔ, total parasite proteins extracted at 18 h after invasion were separated on 4–12% NuPAGE denature gel (Life Technologies) for Western blot analysis using the rabbit antisera to the PfSETvs peptide and detected by an enhanced chemiluminescence (ECL) kit (Thermo Scientific). Total proteins from wild-type 3D7 were analysis as control. Anti-PfSETvs peptide diluted 1:300 and the secondary horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma) diluted 1:10,000 were incubated with Western blot PVDF membrane, respectively for ECL development. To determine reaction specificity of a commercial antibody to P. falciparum H3K36me3 (Cell Signaling), 1 μg of each four synthesized peptides with the P. falciparum specific histone H3K36 sequence (PfH3K36: biotin-GIKKPHRYRPG; PfH3K36me1: biotin-GIK(me)KPHRYRPG; PfH3K36me2: biotin-GIK(me2)KPHRYRPG; PfH3K36me3: biotin-GIK(me3)KPHRYRPG) was dotted on the PVDF membrane for Western blot analysis as described above. To detect effect of PfSETvsΔ on histone lysine methylations, total parasite proteins from wild-type 3D7 and 3D7SETvsΔ extracted at 18 h and 42 h after invasion were carried out for Western blot analysis using rabbit antibodies to H3K36me3 (Cell Signaling), H3K36me2 (Abcam), H3K4me3 (Abcam), H3K9me3 (Millipore), respectively. Antibody to histone H3 (Millipore) was used as a control. Rabbit anti-HA (Abcam) was used to detect PfSETvsHA in 3D7SETvsHA. The Western blot analysis was performed as mentioned above.
Microarray analyses
To analyze global gene expression profiles in the asexual stage, RNA from wild-type 3D7 and 3D7SETvsΔ were extracted from highly synchronized parasite cultures at 18 h (ring), 30 h (trophozoite) and 42 h (schizont) after invasion by using TRIzol (Life Technologies) according to the product manual and further digested with RNase free DNase (Ambion) to remove the DNA contamination. RNA hybridization was performed using the PFSANGER Affymetrix array at the microarray facility of the National Cancer Institute (Frederick, MD, USA). PFSANGER Affymetrix arrays are high-density 8-μm custom 25-mer oligonucleotide arrays, whose tiling-like design was based on the P. falciparum (3D7) genome. Briefly, 10 μg of total RNA was reverse-transcribed and biotin-labeled. Hybridizations were carried out at 45 °C for 16 h with constant rotation at 60 rpm. Gene arrays were then scanned at an emission wavelength of 570 nm at 1.56 μm pixel-resolution using a confocal scanner (Affymetrix GeneChip Scanner 3000 7G). After scanning, the hybridization intensity for each 25-mer feature was computed using Affymetrix GCOS version 1.3 software33. The raw data was then transferred to our in housing software for background adjustment, normalization, and summarization of the probe sets.
Quantitative PCR (qPCR)
For qPCR analysis, RNA was isolated and purified as described above. First strand cDNA was synthesized by either random primer mixes or gene specific primers using Superscript III Reverse Transcriptase (Life Technologies) according to product manual. PCR primers used for detecting mRNA expression of 3D7 var genes were as described previously34. Primers for detecting transcripts from each Dd2 var gene and for 3D7 var lncRNAs were designed in this study (Supplementary Table 11). qPCR was performed on a iQ5 Multi-color Real-time PCR Detection System (Bio-Rad) with a program of 1 cycle of 5 min at 95 °C; 40 cycles of 30 sec at 95 °C, 30 sec at 50 °C, 60 sec at 60 °C. A housekeeping gene, Arginyl-tRNA synthetase (PF3D7_0913900), was used to normalize transcriptional level of each var.
Live-cell infected RBC immunofluorescence assay (IFA)
Live-cell IFA for infected RBCs was performed as described previously with minor modifications13. Briefly, iRBCs were washed in 1% BSA in PBS (BSA/PBS) and the pellet was re-suspended in 200 μl BSA/PBS. Antibodies specific for various PfEMP1s listed in the Supplementary Fig. 3b were used at a 1:50 dilution and incubated at room temperature (RT) for 30 min. After washing three times in BSA/PBS, cells were fixed with 2.5% paraformaldehyde and 0.01% glutaraldehyde for 10 min at RT and washed with BSA/PBS. Subsequently, cells were incubated with Alexa488-conjugated goat anti-rabbit IgG (Life Technologies) and Alexa594-conjugated goat anti-rat IgG (Life Technologies) for 30 min at RT and washed with BSA/PBS containing 0.1% Triton X100 and mounted with prolong gold DAPI. Images were captured on a Leica SP2 confocal microscope and visualized using Bitplane Imaris software.
Scanning and Transmission Electron Microscopy
Scanning and transmission electron microscopy were performed as described previously with modifications35,36. For scanning electron microscopy (SEM), iRBCs were gently allowed to settle on silicon chips for 20 min at RT in an 8-well chamber slide (Labtek). Freshly prepared fixative (2.5% glutaraldehyde, 3% paraformaldehyde, 0.05 M phosphate buffer, and 4% sucrose) was added to the cells and incubated at room temperature for 1 hr. All subsequent processing was carried out in a Pelco Biowave laboratory microwave system (Ted Pella, Inc.) at 250W and 20 in. Hg vacuum. The chips were postfixed with1% osmium tetroxide–0.8% potassium ferricyanide in 0.1M sodium cacodylate, followed by rinsing with water and dehydration in a graded ethanol series. The specimen was critical point dried in a Bal-Tec CPD 030 drier (Bal-Tec AG) and coated with 80 Å of iridium using an IBS ion beam sputter (South Bay Technology, Inc.). SEM samples were imaged using a Hitachi SU8000 SEM (Hitachi High Technologies). For transmission electron microscopy (TEM), parasites were fixed with 2.5% glutaraldehyde, 3% paraformaldehyde, 0.05 M phosphate buffer, and 4% sucrose at room temperature for 2 h. The cells were postfixed in a microwave with 1% osmium tetroxide–0.8% potassium ferricyanide in 0.1 M sodium cacodylate, followed by 1% tannic acid in distilled water, and stained en bloc with 1% aqueous uranyl acetate. They were then rinsed with distilled water and dehydrated in a graded ethanol series. The pellets were then infiltrated and embedded in Spurr’s resin which was polymerized overnight in a 68°C oven. Thin sections (90 nm) were cut using a UC6 ultramicrotome (Leica Microsystems) and stained with 4% aqueous uranyl acetate and Reynold’s lead citrate prior to viewing on a 120 kV Tecnai Biotwin Spirit TEM (FEI, Hillsboro, OR). Digital images were acquired with a Hammamatsu XR-100 digital camera system.
FISH
Synchronized ring-stage parasites were released from infected red blood cells by 0.15% saponin treatment followed by fixation with 4% paraformaldehyde in 1xPBS overnight at 4°C. The fixed parasites were washed twice with 1xPBS, then deposited on a microscope slide (Fisher Scientific) as a monolayer and subjected to RNA FISH in the conditions as described previously6. For combined immuno-RNA FISH, parasites were deposited on slides and treated with 0.1% Trition-X100 in 1xPBS for 5 min prior to hybridization of RNA FISH. After incubation of parasites with FISH probe at 42°C for 16hr, the slides were washed three times with 2xSSC, and fixed again in 4% paraformaldeyde for 15 min, then subjected to Immunofluorescence assay (IFA) for detection of H3K36me3 by using the antibody to H3K36Me3 (Cell Signaling) with 1:100 dilution. For the individual var gene-specific RNA FISH probes, DNA templates were amplified by PCR from 3D7 genomic DNA with primers shown in Supplementary Table 11. For the template of the exon 2 probe for var gene family, the exon 2 regions were amplified with types A, B, and C primer sets as described previously6. The products were pooled for labeling. The PCR products were purified by Gel Extraction kit (Qiagen) and used in probe preparation with a Biotin- or a Fluorescein-High Prime kit (Roche). Images were captured by using a Nikon Eclipse 80i microscope with a CoolSnap HQ2 camera (Photometrics). Primers used in amplification of individual var probes were described in Supplementary Table 11.
ChIP-seq and ChIP-qPCR
Highly synchronous cultures of ring-, trophozoite-, and schizont-stage parasites were used for the ChIP study. Cross-linked chromatin was prepared by adding 1% formaldehyde to the culture for 5 min followed by addition of glycine to 0.125 M final concentration. After saponin lysis, nuclei were isolated by homogenization in 10 mM Tris at pH 8.0, 3.0 mM MgCl2, 0.2 % Nonidet P-40, collected on a 0.25M sucrose-buffer cushion and suspended in SDS buffer (1 % SDS, 50 mM Tris, pH 8.0, 10 mM EDTA, protease inhibitors). Chromatin was sheared by sonication in a Bioruptor UCD-200 (Diagenode) for 10 min at 30-s intervals, power setting high, to a size of 300–800 bp. Chromatin samples were frozen and stored at −80°C. ChIP was performed as described previously17. Briefly, commercially available antibodies to H3K36me3 (Cell Signaling), H3K4me3 (Abcam), H3K9me3 (Millipore), H3K20me3 (Abcam) and histone H4K5/K8/K12/K16 acetylation (Abcam) were added to cross-linked samples of wild-type 3D7 and 3D7SETvsΔ, or a mouse anti-HA (Abcam) to 3D7SETvsHA samples, and incubated at 4°C, followed by the addition of 10 μl A/G beads and further incubation for 2 h. After washing with buffers containing 100, 150 and 250 mM NaCl, immuno-precipitated DNA was eluted and purified using PCR purification columns (Qiagen). The resulting dsDNA was then end repaired, followed by adding an ‘A’ base at the ends. Illumina paired-end index adaptor was ligated and size selected. A 16-cycle PCR was then carried out with Phusion Hot Start High-Fidelity DNA Polymerase (Finnzymes) to generate the final ChIP-seq library. We use Illumina HiSeq 2000 to perform the single-end sequencing (50 cycles). Quality sequencing reads were mapped against the Plasmodium falciparum genome assembly (PlasmoDB v8.2) with Burrows-Wheeler Alignment tool (BWA) using default parameters. ChIP-qPCR was performed for different gene regions (TSS, 3′ end of exon1) as well as antisense transcription level in iQ5 Multi-color Real-time PCR Dection System (Bio-Rad) using primer sets described in Supplementary Table 11.
Tree construction and topology testing
Sequences of the SETD2-NSD-ASH1 clade to span a comprehensive phyletic range across eukaryotes were collected using the PSI-BLAST program. The SET domains and the associated AWS domains were aligned using the MUSCLE program. The tree was constructed using two methods: 1) A preliminary tree was obtained using the approximately-maximum-likelihood method implemented in the FastTree 2.1 program under default parameters. This gave an idea of the positions of key members. 2) A complete tree was constructed using the MEGA 5.1 program with the following parameters: 4 distinct gamma distributed rate categories and one invariant were used for modeling among site variation, the WAG matrix with frequencies, was used as the substitution model; the ML searched used the close neighbor exchange method. The tree was bootstrapped using 10000 RELL-BP resamplings with the Molphy package. The tests for alternative topology were carried out using the CONSEL program for the SH test and these overwhelmingly rejected the grouping of the apicomplexan clade with either the NSD subclade (p<10−4) or the SETD2 subclade (p<10−7).
Supplementary Material
Acknowledgments
We thank S.K. Pierce in the National Institute of Allergy and Infectious Diseases, National Institutes of Health for critical suggestions on the manuscript, A.F. Cowman for providing the pHTK transfection plasmid. We also thank V. Nair and E. Fisher in the NIH Research Technology Branch for assistance with electron microscopy. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Disease and the National Heart, Lung and Blood Institute, National Institutes of Health, and also supported by National Natural Science Foundation of China (81271863) and the Key Research Program of the Chinese Academy of Sciences (KSZD-EW-Z-003-1-2). A.S. was supported by an ERC grant PlasmoEscape (250320). All authors have reviewed and agreed with the content of the manuscript.
Footnotes
Author Contributions
L.J. and L.H.M. conceived and designed experiments. L.J. and J.M. performed the majority of the experiments. L.J., Q.Z. and A.S. performed FISH assays and analyzed the data. L.J. and P.S. performed IFA and EM assays. L.J., J.M., T.N., W.Y., W.P., Y. W., Y. L., J.Z. and K.Z. performed ChIP-seq assays and analyzed the data. L.J., J.M. and J.M.C.R. performed microarray analysis. K.R., L.T., T.L., T.G.T., A.B., G.W. and J.Q. generated reagents. L. A. performed phylogenetic analyses. L.J. J.M. and L.H.M. analyzed all the data and wrote the manuscript. All authors discussed and edited the manuscript.
The authors declare no competing financial interests.
Microarray raw data have been deposited in MIAME format into the GEO database under accession GSE47349 and ChIP-seq raw data have been deposited into the SRA database under accession SRP022761 (http://www.ncbi.nlm.nih.gov/sra/?term=SRP022761).
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–9. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–70. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 3.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chookajorn T, et al. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci U S A. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Rubio JJ, et al. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–90. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Salcedo-Amaya AM, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci U S A. 2009;106:9655–60. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonkin CJ, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duraisingh MT, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Cui L, Fan Q, Miao J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int J Parasitol. 2008;38:1083–97. doi: 10.1016/j.ijpara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volz JC, et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe. 2012;11:7–18. doi: 10.1016/j.chom.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L, Abhiman S, Iyer LM. Natural history of the eukaryotic chromatin protein methylation system. Prog Mol Biol Transl Sci. 2011;101:105–76. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 13.Joergensen L, et al. Surface co-expression of two different PfEMP1 antigens on single plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathog. 2010;6:e1001083. doi: 10.1371/journal.ppat.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newbold CI, Pinches R, Roberts DJ, Marsh K. Plasmodium falciparum: the human agglutinating antibody response to the infected red cell surface is predominantly variant specific. Exp Parasitol. 1992;75:281–92. doi: 10.1016/0014-4894(92)90213-t. [DOI] [PubMed] [Google Scholar]
- 15.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–16. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–27. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, et al. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PLoS One. 2011;6:e28171. doi: 10.1371/journal.pone.0028171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–89. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chantalat S, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21:1426–37. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesh S, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–5. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 24.Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–69. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Werven FJ, et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–81. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soubes SC, Wellems TE, Miller LH. Plasmodium falciparum: a high proportion of parasites from a population of the Dd2 strain are able to invade erythrocytes by an alternative pathway. Exp Parasitol. 1997;86:79–83. doi: 10.1006/expr.1997.4153. [DOI] [PubMed] [Google Scholar]
- 27.Jiang L, et al. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc Natl Acad Sci U S A. 2010;107:2224–9. doi: 10.1073/pnas.0913396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan JA, et al. Targets of antibodies against Plasmodium falciparum-infected erythrocytes in malaria immunity. J Clin Invest. 2012;122:3227–38. doi: 10.1172/JCI62182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walliker D, et al. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–6. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 30.Wellems TEOA, Fenton B, Desjardins R, Panton LJ, do Rosario VE. Chromosome size variation occurs in cloned Plasmodium falciparum on in vitro cultivation. REVISTA BRASILEIRA DE GENETICA. 1988;11:813–825. [Google Scholar]
- 31.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 32.Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol. 2002;32:81–9. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salanti A, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–91. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasan P, et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci U S A. 2011;108:13275–80. doi: 10.1073/pnas.1110303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahlan K, et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:1797–809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.