Summary
In response to environmental stress cells often generate pH signals that serve to protect vital cellular components and reprogram gene expression for survival. A major barrier to our understanding of this process has been the identification of signaling proteins that detect changes in intracellular pH. To identify candidate pH sensors we developed a computer algorithm that searches proteins for networks of proton-binding sidechains. This analysis indicates that Gα subunits, the principal transducers of G protein-coupled receptor signals, are pH sensors. Our structure-based calculations and biophysical investigations reveal that Gα subunits contain networks of pH-sensing sidechains buried between their Ras and helical domains. We show further that proton binding induces changes in conformation that promote Gα phosphorylation and suppress receptor-initiated signaling. Together, our computational, biophysical and cellular analyses reveal a new and unexpected function for G proteins as mediators of stress-response signaling.
Introduction
Coordinated changes in intracellular pH are triggered by extracellular chemical signals (Hellwig et al., 2004), nutrient limitation (Young et al., 2010), ischemia (Schroeder et al., 2010), and other cell stresses (Webb et al., 2011). This ability to adjust cytoplasmic pH is an essential feature of cellular physiology that enables a rapid response to adverse growth conditions. However, we have limited knowledge of the specific cellular components responsible for detecting and promoting changes in intracellular proton abundance. The identification and characterization of new pH-sensing proteins, particularly those in established signaling pathways, would lend support to the emerging view that protons can act as second messenger regulators of cell signaling, survival, and stress responses (Dechant et al., 2010; Rubenstein and Schmidt, 2010).
pH sensors are specialized proteins that couple their biological function to physiological changes in cellular pH (6 < pH < 8) (Srivastava et al., 2007). Perhaps the best-known proton sensor is hemoglobin, which exhibits reduced affinity for oxygen as a consequence of respiratory acidosis in the peripheral vasculature (the so-called Bohr effect) (Perutz, 1978). Hemoglobin and other pH sensors detect changes in cellular pH using sidechains (Asp, Glu, His, Lys, Arg) or sidechain modifications (phosphorylation) that bind protons. In these sensors, proton-binding groups are organized in structural arrangements that shift their pKa values into the physiological pH range (6 < pKa < 8). Prominent examples of these motifs include clusters of His residues and networks of surface-exposed basic and acidic sidechains. Lesser known, yet equally important, are examples of proton-binding motifs sequestered within protein cores or buried at the interface of protein-protein complexes (Tews et al., 2005). Although counterintuitive, pH-sensing by these core motifs exploits the effects of dehydration, which act to facilitate reversible proton binding to core sidechains by shifting their pKa values into the physiological pH range (Isom et al., 2008; Isom et al., 2011a; Isom et al., 2010a).
In an effort to predict and validate new pH sensors, we have developed a new computer algorithm (pHinder) that searches protein structures for buried proton-sensing motifs. The strategy of limiting our structure-based analysis to core networks is based on several compelling observations. Buried ionizable sidechains in proteins are relatively rare (Bush and Makhatadze, 2011; Gunner et al., 2000; Kajander et al., 2000; Kim et al., 2005; Rashin and Honig, 1984), often have pKa values within the physiological pH range (Isom et al., 2011a; Isom et al., 2010a), and are readily identifiable from protein structure. Furthermore, the large pKa shifts exhibited by buried sidechains correspond to sizeable increments of Gibbs free energy (1.36 kcal/mol per pKa unit) that can be harnessed by proteins to drive pH-dependent changes in structure, stability, and function. In contrast, surface-exposed ionizable residues are far more common, rarely exhibit highly perturbed pKa values, and rarely induce large changes in protein stability and structure.
Here we report the use of our pHinder algorithm to identify a new pH sensor involved in cell signaling. This work reveals that Gα subunits, the principal transducers of G protein-coupled receptor signals (Neves et al., 2002; Rasmussen et al., 2011), use buried networks of acidic and basic sidechains to detect physiological changes in cellular pH. We show further that proton binding regulates Gα conformation and phosphorylation in a manner that suppresses GPCR signaling in cells. Taken together, our findings reveal a new and unexpected role for G proteins as integrators of hormone and metabolic stress-response pathways.
Results
Gα subunits contain pH-sensing structural motifs
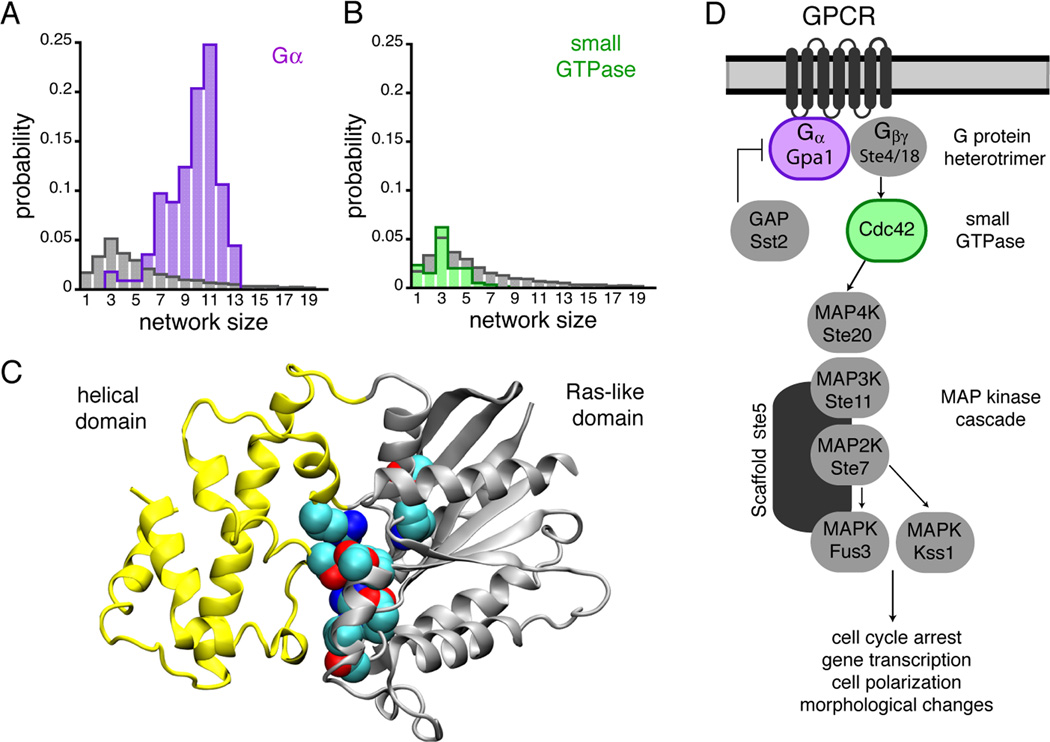
pHinder represents a structural proteomics calculation designed to infer pH-sensing capabilities from protein structure. This new algorithm predicts candidate pH sensors by identifying spatial networks of titratable sidechains using an automated, structure-based procedure (See Methods for details). Using pHinder to survey 11,890 non-redundant structures in the Protein Data Bank (PDB), we found that only 10% of proteins contained large (≥ 6 sidechains) proton-binding networks residing in their cores (Figures 1A and 1B). As expected, these calculations identified several documented pH sensors including phosphofructokinase (Trivedi and Danforth, 1966), gelsolin (Nag et al., 2009), and soluble-adenylyl cyclase (Tews et al., 2005). In addition, we made the unexpected observation that Gα subunits contained large core networks located at the interface of their Ras and helical domains (Figure 1C). When we expanded our pHinder analysis to all Gα structures (129 total structures) we found that 96% contained core networks (Figure 1A). In contrast, core networks were absent in small GTPases, which contain only the Ras-like domain (Figure 1B). These findings suggested that Gα subunits were pH sensors, and prompted us to quantify the effect of pH on Gα structure and function, using biochemical and biophysical approaches, and the effect of pH on Gα signal transmission, using a model GPCR pathway in yeast (Figure 1D).
Figure 1.
Gα subunits contain structural motifs capable of proton sensing. (A, B) pHinder results showing that core networks of ionizable sidechains identified in 129 Gα structures (purple bars in A) were much larger and more prevalent than core networks identified in a reference set of 11,890 non-redundant PDB chains (gray bars in A and B) and 595 small GTPases (green bars in B). (C) Gα subunits consist of a helical (yellow) and Ras-like (gray) domain, whereas small GTPases consist of only a Ras-like domain. In Gα proteins the core pH-sensing network identified by pHinder and the sidechains responsible for Gα function reside at the interface of the helical and Ras-like domains. Collectively, the core networks observed in structures of mammalian Gαi consistently included residues Lys46, Asp150, Asp200, Asp229, Arg242, Lys270, and Lys277. (D) The core GPCR signaling components of the yeast pheromone pathway.
Proton binding regulates Ga stability and conformation
Within GPCR signaling pathways, Gα subunits function as signaling switches that are turned on by binding to GTP and turned off by GTP hydrolysis. As expected for a pH sensor, the sidechains responsible for these Gα functions are found in close proximity to the pH-sensing network identified by pHinder. However, these catalytic sidechains also reside exclusively in the Ras-like domain, which itself was not predicted to be pH sensitive (Figure 1B). As anticipated, the effects of pH on nucleotide binding (Figure S1) and GTP hydrolysis (Figure 2A and Figure S1) were negligible in representative Gα subunits from yeast (Gpa1) and mammalian (Gαi) sources. Given that the pH-sensing network was positioned between the Ras-like and helical domains, we next considered whether there may be pH-driven changes in Gα stability, dynamics, and conformation. Indeed, the Ras and helical domains undergo a dramatic reorientation in response to receptor-mediated activation of the G protein (Chung et al., 2011; Rasmussen et al., 2011; Van Eps et al., 2011; Wall et al., 1995; Westfield et al., 2011).
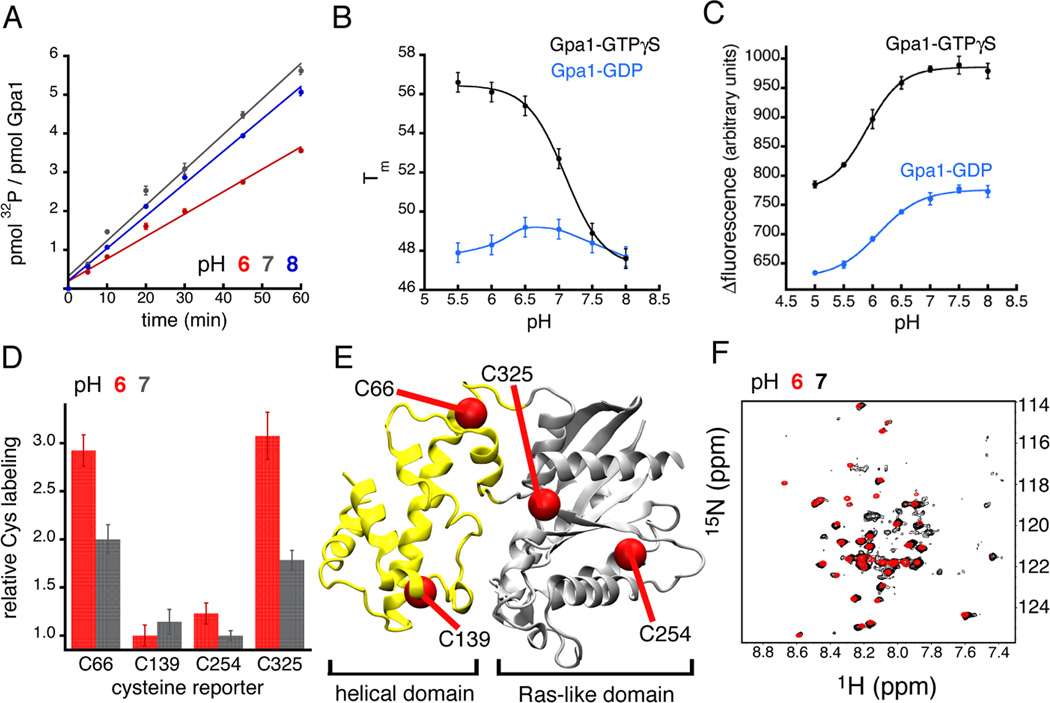
Figure 2.
Gα subunits are proton sensors in vitro. (A) Steady-state GTP hydrolysis by yeast and mammalian Gα (See Figure S1) was diminished at pH 6.0 relative to pH 7.0 and 8.0. (B) The thermostability of GTPγS-bound yeast and mammalian Gα (See Figure S1) was cooperatively dependent on pH (Hill coefficient of 2). (C) Intrinsic tryptophan fluorescence indicated that yeast and mammalian Gα subunits (See Figure S1) undergo a large-scale, pH-triggered conformational change. (D, E) Cysteine labeling and mass spectrometry of mammalian Gαi confirmed a pH-dependent conformational change. The cysteine sites that were exposed by the pH-triggered conformational change (Cys66 and Cys325) reside at the interface of the helical and Ras-like domains (E), the same region that contains the core network of pH-sensing residues (Figure 1D). (F) Overlay of heteronuclear single quantum coherence spectra of Gαi at pH 6.0 and 7.0 (See Figure S2). Error bars represent mean ± SEM.
We started by examining the effect of pH on Gpa1 and Gαi thermostability. Cooperative, pH-dependent changes in protein stability are an indicator of pH sensing. These changes reflect the thermodynamic coupling of global protein stability to the ionization free energy of proton-binding sidechains (Isom et al., 2008; Isom et al., 2011a; Isom et al., 2010a). Our thermostability measurements (Figure 2B and Figure S1), quantified by the midpoint of thermal unfolding (Tm) (Isom et al., 2011b; Isom et al., 2010b), indicated that both Gpa1 and Gαi were cooperatively sensitive (ΔTm of 8–10°C) to changes in pH when bound to GTPγS, a GTP analog that is resistant to hydrolysis. The resulting thermal stability profiles, which report an apparent pKa value of 7, indicate that the core networks within Gα-GTPγS contain ionizable (acidic and basic) sidechains having pKa values shifted into the physiological pH range. It is unlikely that this pH-dependence originates from the titration of the γ-phosphate of GTPγS, given the available evidence that the γ-phosphate is always charged above pH 6 due to a highly depressed pKa value (Schweins et al., 1995). As expected, mutating components of the core network dramatically altered the pH-dependent thermostability of GTPγS-bound Gpa1 and Gαi (Figure S3), or resulted in Gα variants that failed to express (e.g. Lys270 and Lys 277 of Gαi). Based on these findings, we conclude that Gpa1 and Gαi are equipped to function as proton sensors.
In contrast to ]GTP-bound Gα, there was a minimal effect of pH on the thermostability of the GDP-bound protein (Figure 2B and Figure S1). The difference between GDP- and GTPyS-bound Gα can be attributed to a number of underlying factors that, in some cases, limit the utility of our thermostability assay. First, the conformation of the GDP-bound state, which is distinctly different from the GTPγS-bound state, may have a smaller effective core network with fewer shifted pKa values. This idea is supported by our pHinder calculations (Figure 1 A), which revealed that GDP-bound Gα conformations tend to have smaller, or fragmented core networks. Second, multiple pKa shifts caused by the GDP-induced conformation may offset one another causing no net change in pH-dependent thermostability. Third, the presence of the negatively charged γ-phosphate on GTPγS, which is missing in GDP, may alter the pKa values of the one or more sidechains exclusively in the GTPγS-bound conformation. Fourth, the effects of pH sensing may be largely uncoupled from Gα thermal stability. Since our thermostability assay could not differentiate between these four scenarios, we investigated the relationship between pH, nucleotide occupancy, and Gα conformation using intrinsic tryptophan fluorescence, quantitative mass spectrometry, and NMR spectroscopy.
Intrinsic tryptophan fluorescence is often used to monitor the structural relationship between nucleotide binding and G protein activation (Higashijima et al., 1987). Here, we adapted the intrinsic fluorescence assay to interrogate the effect of pH on the conformation of GDP- and GTPγS-bound Gα subunits. The titration curves (Figure 2C and Figure S1) derived from the intrinsic fluorescence spectra of Gpa1 and Gαi (see Figure S1) revealed that both nucleotide-bound states exhibit cooperative changes in conformation as a function of pH. As expected, the pH titration of GTPγS-bound Gα corresponded to a higher level of intrinsic fluorescence than GDP-bound Gα (Figure 2C). This difference originates from a tryptophan residue (W211 in Gαi) within a regulatory motif (referred to as switch II) known to be sensitive to GTP binding. However, because Gα subunits contain multiple tryptophan residues, it is likely that the pH-dependent changes in intrinsic fluorescence we observe in the presence of both GDP and GTPγS reflect additional contributions from tryptophan residues elsewhere in the Ras-like and helical domains. Remarkably, these changes were similar in magnitude to that which accompanies GTPγS binding (Figure 2C), and G protein activation by receptors.
Next we corroborated our intrinsic fluorescence measurements by examining cysteine protection and backbone conformation as a function of pH. Using fast quantitative cysteine reactivity (fQCR) (Isom et al., 2011b; Isom et al., 2010b) and liquid chromatography-mass spectrometry (LC-MS) we identified specific cysteine sites that were exposed by pH-dependent conformational changes in mammalian Gαi (Figures 2D–E). Two protected cysteines (Cys66 and Cys325) at the interface of the Ras-like and helical domains became less protected as pH was reduced from 7.0 to 6.0. However, the reactivity of two cysteines buried within the Ras (Cys254) and helical domains (Cys139) was unaffected by pH. This pattern of cysteine labeling corroborated our intrinsic fluorescence data, and was consistent with a pH-driven conformational change that displaces the helical domain from the Ras-like domain. We were also able to observe this pH-dependent change in Gαi conformation by NMR spectroscopy (Figure 2F and S2). Our NMR spectra revealed that Gαi sampled multiple conformations at pH 7.0, but sampled single conformations at pH 6.0 and 8.0. Once again these behaviors mimic those resulting from receptor-dependent activation of the G protein (Chung et al., 2011; Rasmussen et al., 2011; Van Eps et al., 2011; Wall et al., 1995; Westfield et al., 2011).
Proton binding regulates Gα phosphorylation and signaling
Taken together, our in vitro experiments established that a network of core ionizable sidechains regulate Gα conformation in response to changes in pH. Significantly, these pH-sensing residues lie within the active site of the protein and become exposed following GCPR activation (Rasmussen et al., 2011; Westfield et al., 2011). To assess the biological relevance of Gα proton sensing, we monitored GPCR signaling as a function of cellular pH. We did these experiments in yeast because it was possible to manipulate cellular pH genetically (by gene knockdown of proton pumps), metabolically (via glucose limitation) (Orij et al., 2011), and biochemically (using the metabolite acetic acid) (Orij et al., 2011), while simultaneously monitoring the activation of pheromone pathway components. To quantify cellular pH we used the geneticallyencoded pH sensor, pHluorin (See Figure S4) (Miesenbock et al., 1998; Orij et al., 2011; Orij et al., 2009).
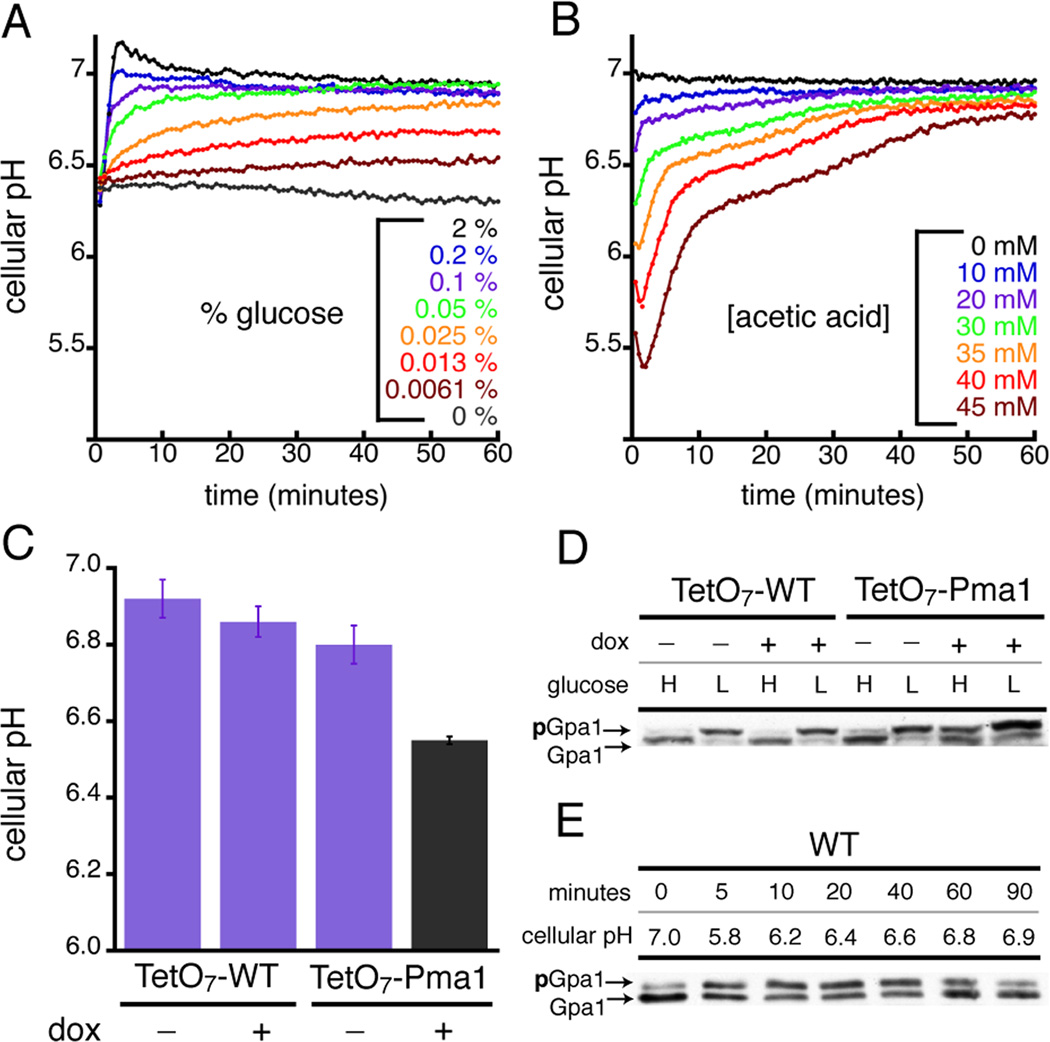
Using all three perturbations we found that intracellular proton concentrations increased by up to 10-fold before recovering to baseline (Figures 3A–B). Notably, the speed, duration, and magnitude of these changes resembled the behaviors of other wellknown second messengers, such as cAMP and calcium. Our experiments also revealed that yeast Gα was rapidly phosphorylated as cellular pH was lowered, and reversibly dephosphorylated as cellular pH recovered to resting levels (Figures 3D–E). Thus, the dynamics of Gα phosphorylation mirrored the changes in cellular pH. Moreover, we found that pH-dependent phosphorylation in vivo precisely matched the pH-dependent change in Gα conformation observed in vitro (Figures 4A–B). The kinase responsible for Gα phosphorylation (Elm1) (Hong et al., 2003; Torres et al., 2011) was itself not activated by pH, as another substrate for Elm1 (Snf1) was not phosphorylated under identical conditions (See Figure S3). Because Gpa1 phosphorylation was dependent on pH, and Elm1 activity was not, we conclude that proton binding within the Gα core network controls access to the kinase. Similar mechanisms of conformation-driven substrate phosphorylation have been reported for AMP-activated protein kinases and for G protein-coupled receptor kinases (Benovic et al., 1986; Rubenstein et al., 2008).
Figure 3.
Gα proton sensing leads to pH-dependent Gα phosphorylation. Changes in yeast cellular pH caused by diminished glucose (A), the addition of acetic acid (B), and genetic knockdown of the proton pump Pma1 (C) using a doxycycline(dox)/Tet-off(TetO7) repression system. In all three cases cellular pH was quantified using the genetically-encoded pH biosensor pHluorin (See Figure S4). Resting intracellular pH in yeast is ~0.2 pH units lower than in mammalian cells. (D) The amount of phosphorylated Gα (pGpa1) increased during a sustained reduction in cellular pH caused by lowering glucose concentration from 2% (H) to 0.05% (L), and by genetic knockdown of Pma1 (TetO7-Pma1 + dox). (E) Acetic acid treatment (45 mM) caused a transient change in cellular pH (refer to panel B) that triggered a reversible and pH-dependent change in Gα phosphorylation. Error bars represent mean ± SEM.
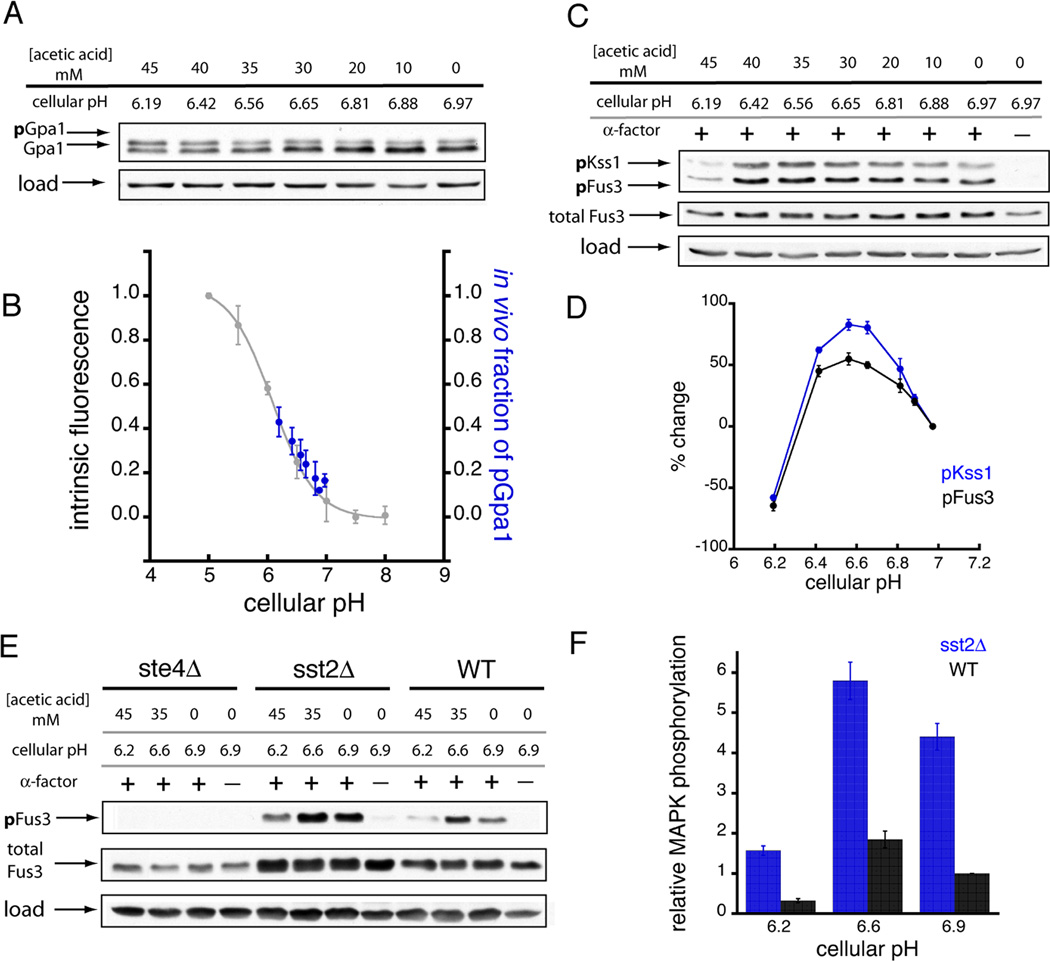
Figure 4.
pH-dependent changes in Gα conformation lead to concomitant changes in Gpa1 phosphorylation and MAPK activation. (A) Titration of yeast cellular pH using acetic acid resulted in the pH-dependent phosphorylation of Gpa1 (pGpa1). (B) The pattern of Gpa1 phosphorylation in vivo matched the pH-triggered Gpa1 conformational change observed by intrinsic tryptophan fluorescence in vitro. The in vitro data corresponds to the pH-dependent intrinsic fluorescence of the Gpa1-GDP state (normalized and inverted relative to Figure 2C). (C, D) Titration of yeast cellular pH using acetic acid revealed the pH-dependent activation of the pheromone-responsive MAP kinases (pFus3 and pKss1). (E, F) Comparison of pH-dependent MAP kinase activation in WT yeast with yeast strains lacking the Gβ subunit (ste4Δ), or the GTPase-activating protein Sst2 (sst2Δ). Data were normalized to MAPK activation in WT yeast at resting pH. Each sample (in panels A, C, and E) was treated with the indicated dose of acetic acid and 0.3 µM α-factor for 10 minutes. The pH reported for each sample corresponded to the observed cellular pH after 10 minutes of acetic acid treatment (See Figure 3B). Error bars represent mean ± SEM.
Last we assessed the effect of cellular pH on GPCR signaling at the level of the effector MAPK (Figures 4C–F). These experiments revealed that GPCR signaling in yeast is pH-dependent, and maximal at pH 6.6 (Figures 4C–D). Moreover, although pheromone signaling could involve multiple proton sensors, there are several reasons to conclude that Gα is mostly responsible for the pH-dependent change in MAPK activity. First, pH-dependent activation of two MAPKs, Fus3 (which requires the scaffold Ste5) and Kss1 (which does not), was identical (Figures 4C–D), indicating that the proton sensor acts upstream of the scaffolded MAPK cascade. Second, neither MAPK was activated in yeast lacking the Gβ subunit (Ste4) (Figure 4E), establishing that protons were unlikely to regulate components downstream of the G protein heterotrimer. Third, deletion of the GTP aseactivating protein Sst2, which acts directly on Gα, led to increased MAPK activation (as expected), yet activation remained sensitive to changes in pH (Figures 4E–F). While several mammalian GPCRs are activated by binding to extracellular protons (Ludwig et al., 2003), this has not been observed in yeast. These genetic experiments indicate that pH sensing occurs at the level of the G protein. More generally, our computational, biophysical and cellular analysis reveals that the Gα protein functions as a pH sensor as well as a transducer of GPCR signaling.
Discussion
Although every protein binds to and releases protons, few cellular proteins are bona fide pH sensors. Prominent examples include proteins involved in oxygen transport (hemoglobin), metabolism (phosphofructokinase), and signal transduction (soluble adenylyl cyclase). Each of these proteins contains proton-binding sidechains with pKa values in the physiological range, and each is responsive to the transient increases in proton concentration caused by cellular stress conditions (6 < pH < 7.2). This situation is analogous to calcium sensors, such as calmodulin, that have binding affinities tuned to detect transient increases in calcium concentration elicited by extracellular chemical signals (0.1 to 1 µM) (Clapham, 2007).
Although it has long been appreciated that changes in cellular pH are essential to proper cell function, the molecular details that underlie pH signaling and regulation remain poorly understood. In this context, identifying candidate pH sensors and predicting the pKa values of their pH-sensing sidechains has been a major challenge (Alexov et al., 2011; Nielsen et al., 2011). Our pHinder software works to address this issue by identifying spatial arrangements of proton-binding sidechains that are likely to confer pH-sensing capabilities. Here we used the pHinder program to identify Gα subunits as pH sensors. We showed further that these proteins couple physiological changes in intracellular pH to large changes in cellular signaling.
Given that G proteins are conserved across different biological kingdoms, it is likely that pH sensing is a general feature of GPCR-mediated signaling in a variety of physiological contexts. For instance, in humans the ability to detect changes in cellular pH is important under conditions of ischemia and hypoxia (Bopassa, 2012), where diminished oxygen and glucose lead to the anaerobic metabolism of glycogen and intracellular acidosis. Similarly, in plants and fungi, acidic pH signals are critical for nutrient acquisition and sensing (Gjetting et al., 2012; Orij et al., 2011), responding to osmotic stress (Kader and Lindberg, 2010), and protecting vital cellular machinery (Peters et al., 2013). Within these contexts, proton signals facilitate signal transduction in a manner analogous to other known second messengers.
In summary, our computational, biophysical and cellular analyses reveal that the Gα protein functions as a pH sensor as well as a transducer of GPCR signaling. Furthermore, our findings indicate that protons can disrupt ]G protein-mediated signaling to limit hormone or neurotransmitter signaling during brief periods of nutrient or oxygen deprivation. In this way, it appears that evolution has adapted pre-existing signaling proteins to detect proton second messengers and reprogram cellular physiology in response to cell stress.
Experimental Procedures
pHinder calculation
The accession codes for the non-redundant PDB chain set used in this study (compiled on 12/26/2012) were downloaded from the National Center for Biotechnology Information (NCBI). The non-redundant database contained 12,486 unique PDB chain entries (out of a total of 81,418 chains). A subset of protein chains (596) that were much smaller (< 25) or larger (> 1000 residues) than typical proteins in the reference set was removed, bringing the total number of pHinder-analyzed protein chains to 11,890. The sets of 129 Gα proteins (PF00503) and 595 small GTPases (PF00071) were identified using the pFam database (Wellcome Trust Sanger Institute). A manuscript describing the pHinder algorithm, and web-based access to the pHinder software, are currently in preparation. Briefly, a Delaunay triangulation of the Cα atoms comprising the protein backbone was calculated using the 4D paraboloid method (O'Rourke, 1998). This triangulation was then used to calculate a surface (comprising triangular facets) that encapsulated the volume defined by the fold of protein backbone. Using a distance-to-plane calculation, the facets of the surface were used to classify the terminal atom of each sidechain as exposed (outside of the surface), marginal (intersecting or beneath the surface by < 3Å), or core (lying beneath the surface by ≥ 3Å). Based on this classification, core ionizable networks were identified using a three-step procedure. First, the terminal atom of each core ionizable sidechain was triangulated with the terminal atoms of the other ionizable sidechains in its immediate microenvironment (defined by a 10 Å distance cutoff). These surrounding sidechains could be classified as marginal or exposed, thus some networks could extend from the core to the protein surface. Second, using a node-to-node distance cutoff of 10 Å, the triangulation was searched for contiguous networks of ionizable sidechains. Third, any two networks containing shared components were consolidated into a single network. This consolidation step was repeated iteratively until all incident networks were matched and combined. The total number of interconnected nodes within each unique network comprised the pHinder score. Although a protein chain may have contained multiple core networks, here we reported only the largest network per chain (Figures 1A–B).
Preparation of stock solutions
Concentrations of GDP (Sigma-Aldrich, G7127), GTP (Sigma-Aldrich, G8877), and GTPγS (Roche Diagnostics, 11110349001) stock solutions were quantified spectroscopically using a molar extinction coefficient at 254 nm of 13,700 M−1 cm−1. A 500 mM stock solution of the fluorogenic cysteine probe 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole (ABD) (TCI America, A5597) was made in DMSO, aliquoted, and stored at −20°C. The concentration of all ABD solutions was quantified spectroscopically using a molar extinction coefficient at 313 nm of 4200 M−1 cm−1. Working stocks of ABD (26 mM) were made by combining 500 mM ABD stock in DMSO with 10 µL of PBS (pH 7.0) and 179.6 µL of water. It was necessary to store working ABD stocks at room temperature, instead of on ice, to prevent precipitation. All stock solutions of the reducing agent Tris(2-carboxyethyl)phosphine (TCEP) (TCI America, T1656) were made either fresh in water, or by adding TCEP crystals directly to premade buffer solutions.
Protein production
The proteins used in this study were overexpressed to high density using the auto-induction introduced by F.W. Studier (Studier, 2005). All purification steps were done in a PBS-GMT buffer (25 mM KPO4, 100 mM KCl, 50 µM GDP, 50 µM MgCl2, 1 mM TCEP, pH 7.0). The remaining details of our purification protocol can be found in Supplemental Experimental Procedures.
pH dependence of nucleotide hydrolysis
Steady-state GTP hydrolysis by yeast Gpa1 and mammalian Gαi was initiated by combining 800 µL of 2X HOT-PBS-GMT (10 µM GTP, 2 mM MgCl2, 2 mM TCEP, 1 µL GTPγ32P, pH 7.0) with 800 µL of protein sample (1 µM protein in PBS) at 37°C. After each time interval, 90 µL aliquots (in duplicate) were quenched in 750 µL of icecold mixture of activated charcoal (5% w/v charcoal (Sigma-Aldrich, 260010), 50 mM phosphoric acid), vortex-mixed for 2 seconds, pulse-centrifuged to sequester protein and nucleotides in the pelleted charcoal fraction, and 200 µL of supernatant (containing hydrolyzed 32Pi) was transferred to a clean PCR tube. The amount of 32Pi in each tube was quantified in a scintillation counter via Cerenkov counting. The data in Figure 2A and Figure S1 correspond to the mean (values) and standard deviation (error bars) of two independent experiments done at each of the indicated pH values.
pH dependence of Gα thermostability
Thermal unfolding of yeast Gpa1 and mammalian Gαi was quantified by fast quantitative cysteine reactivity (fQCR). The analysis of fQCR unfolding curves has been described previously (Isom et al., 2011b; Isom et al., 2010b). The details of the pH-dependent fQCR experiment can be found in the Supplemental Experimental Procedures.
pH dependence of Gα conformation
pH-dependent intrinsic fluorescence was measured using Gα protein stock (in PBS-GMT) diluted to 0.5 µM in PBS (pH 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, or 8.0) with 5 µM GDP in a 1.5 mL tube. The samples were then vortex-mixed for 2 seconds, and equilibrated at ambient room temperature for 15 minutes. Because changes in chloride concentration affect the fluorescence measurement, the panel of PBS buffers used in this experiment was made as a self-consistent set by combining volumes of mono- and dibasic phosphate stock solutions composed of 25 mM potassium phosphate and KCl. Starting with pH 5.0, fluorescence spectra were collected in a quartz cuvette using an excitation wavelength of 280 nm, and matching excitation and emission bandwidths of 7.5 nm for Gαi, and 6.5 nm for Gpa1. The cuvette was then emptied and filled with the next protein sample (i.e. pH 5.5) without washing. Titration curves were assembled from the individual spectra using the fluorescence value at the emission wavelength of 345 nm. The data in Figure 2C and Figure S1 represent the mean (values) and standard deviation (error bars) of four experiments. In the case of Gαi, the pH-dependent fluorescence of the GTPγS-bound state could not be measured because the amount of GTPγS required for half-saturation (~50 µM) absorbed most of the excitation light. The detailed procedures for our pH-dependent mass spectrometry and NMR experiments can be found in the Supplemental Experimental Procedures.
In vivo quantification of intracellular pH and GPCR signaling
The details regarding our yeast strains and media, pHluorin calibration, pH manipulation procedures, and Western blotting can be found in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Cellular pH is altered by nutritional and metabolic stress
A novel structure-based algorithm predicts pH sensing by Gα proteins
Proton binding alters Gα conformation, phosphorylation and signal transmission
G proteins serve as integrators of receptor- and stress-mediated cell signaling
Acknowledgments
H.G.D., D.G.I., V.S., R.B., and S.T.C. are supported by the National Institutes of Health (NIH) (grant GM059167 and GM073180). D.G.I. acknowledges the support of the UNC University Research Council and the James Moeser Award for Distinguished Research. V.S. acknowledges support from the National Science Foundation REU Site Award 1156840 to UNC-Chapel Hill. D.M.S acknowledges funding from the NIH (grant CA016086) for support of the UNC Proteomics Center. We thank Sharon Campbell for assistance with NMR, Ashutosh Tripathy and Gary Johnson for access to instrumentation, and UNC Research Computing for access to computing clusters.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexov E, Mehler EL, Baker N, Baptista AM, Huang Y, Milletti F, Nielsen JE, Farrell D, Carstensen T, Olsson MH, et al. Progress in the prediction of pKa values in proteins. Proteins. 2011;79:3260–3275. doi: 10.1002/prot.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovic JL, Strasser RH, Caron MG, Lefkowitz RJ. Beta-adrenergic receptor kinase: identification of a novel protein kinase that phosphorylates the agonist-occupied form of the receptor. Proc Natl Acad Sci U S A. 1986;83:2797–2801. doi: 10.1073/pnas.83.9.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopassa JC. Protection of the ischemic myocardium during the reperfusion: between hope and reality. Am J Cardiovasc Dis. 2012;2:223–236. [PMC free article] [PubMed] [Google Scholar]
- Bush J, Makhatadze GI. Statistical analysis of protein structures suggests that buried ionizable residues in proteins are hydrogen bonded or form salt bridges. Proteins. 2011;79:2027–2032. doi: 10.1002/prot.23067. [DOI] [PubMed] [Google Scholar]
- Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. Embo J. 2010;29:2515–2526. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjetting KS, Ytting CK, Schulz A, Fuglsang AT. Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J Exp Bot. 2012;63:3207–3218. doi: 10.1093/jxb/ers040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunner MR, Saleh MA, Cross E, ud-Doula A, Wise M. Backbone dipoles generate positive potentials in all proteins: origins and implications of the effect. Biophys J. 2000;78:1126–1144. doi: 10.1016/S0006-3495(00)76671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig N, Plant TD, Janson W, Schafer M, Schultz G, Schaefer M. TRPV1 acts as proton channel to induce acidification in nociceptive neurons. J Biol Chem. 2004;279:34553–34561. doi: 10.1074/jbc.M402966200. [DOI] [PubMed] [Google Scholar]
- Higashijima T, Ferguson KM, Sternweis PC, Ross EM, Smigel MD, Gilman AG. The effect of activating ligands on the intrinsic fluorescence of guanine nucleotide-binding regulatory proteins. J Biol Chem. 1987;262:752–756. [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci U S A. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Cannon BR, Castaneda CA, Robinson A, Garcia-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc Natl Acad Sci U S A. 2008;105:17784–17788. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci U S A. 2011a;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Castaneda CA, Cannon BR, Velu PD, Garcia-Moreno EB. Charges in the hydrophobic interior of proteins. Proc Natl Acad Sci U S A. 2010a;107:16096–16100. doi: 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Marguet PR, Oas TG, Hellinga HW. A miniaturized technique for assessing protein thermodynamics and function using fast determination of quantitative cysteine reactivity. Proteins. 2011b;79:1034–1047. doi: 10.1002/prot.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Vardy E, Oas TG, Hellinga HW. Picomole-scale characterization of protein stability and function by quantitative cysteine reactivity. Proc Natl Acad Sci U S A. 2010b;107:4908–4913. doi: 10.1073/pnas.0910421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader MA, Lindberg S. Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal Behav. 2010;5:233–238. doi: 10.4161/psb.5.3.10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander T, Kahn PC, Passila SH, Cohen DC, Lehtio L, Adolfsen W, Warwicker J, Schell U, Goldman A. Buried charged surface in proteins. Structure. 2000;8:1203–1214. doi: 10.1016/s0969-2126(00)00520-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Mao J, Gunner MR. Are acidic and basic groups in buried proteins predicted to be ionized? J Mol Biol. 2005;348:1283–1298. doi: 10.1016/j.jmb.2005.03.051. [DOI] [PubMed] [Google Scholar]
- Ludwig MG, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, Hofstetter H, Wolf RM, Seuwen K. Proton-sensing G-protein-coupled receptors. Nature. 2003;425:93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Nag S, Ma Q, Wang H, Chumnarnsilpa S, Lee WL, Larsson M, Kannan B, Hernandez-Valladares M, Burtnick LD, Robinson RC. Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc Natl Acad Sci U S A. 2009;106:13713–13718. doi: 10.1073/pnas.0812374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- Nielsen JE, Gunner MR, Garcia-Moreno BE. The pKa Cooperative: a collaborative effort to advance structure-based calculations of pKa values and electrostatic effects in proteins. Proteins. 2011;79:3249–3259. doi: 10.1002/prot.23194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke J. Computational Geometry in C. New York: Cambridge University Press; 1998. pp. 182–189. [Google Scholar]
- Orij R, Brul S, Smits GJ. Intracellular pH is a tightly controlled signal in yeast. Biochim Biophys Acta. 2011;1810:933–944. doi: 10.1016/j.bbagen.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Orij R, Postmus J, Ter Beek A, Brul S, Smits GJ. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology. 2009;155:268–278. doi: 10.1099/mic.0.022038-0. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Electrostatic effects in proteins. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- Peters LZ, Hazan R, Breker M, Schuldiner M, Ben-Aroya S. Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J Cell Biol. 2013;201:663–671. doi: 10.1083/jcb.201211146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashin AA, Honig B. On the environment of ionizable groups in globular proteins. J Mol Biol. 1984;173:515–521. doi: 10.1016/0022-2836(84)90394-2. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein EM, McCartney RR, Zhang C, Shokat KM, Shirra MK, Arndt KM, Schmidt MC. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J Biol Chem. 2008;283:222–230. doi: 10.1074/jbc.M707957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein EM, Schmidt MC. The glucose signal and metabolic p[H+]lux. Embo J. 2010;29:2473–2474. doi: 10.1038/emboj.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MA, Swietach P, Atherton HJ, Gallagher FA, Lee P, Radda GK, Clarke K, Tyler DJ. Measuring intracellular pH in the heart using hyperpolarized carbon dioxide and bicarbonate: a 13C and 31P magnetic resonance spectroscopy study. Cardiovasc Res. 2010;86:82–91. doi: 10.1093/cvr/cvp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweins T, Geyer M, Scheffzek K, Warshel A, Kalbitzer HR, Wittinghofer A. Substrate-assisted catalysis as a mechanism for GTP hydrolysis of p21ras and other GTP-binding proteins. Nat Struct Biol. 1995;2:36–44. doi: 10.1038/nsb0195-36. [DOI] [PubMed] [Google Scholar]
- Srivastava J, Barber DL, Jacobson MP. Intracellular pH sensors: design principles and functional significance. Physiology (Bethesda) 2007;22:30–39. doi: 10.1152/physiol.00035.2006. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Tews I, Findeisen F, Sinning I, Schultz A, Schultz JE, Linder JU. The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science. 2005;308:1020–1023. doi: 10.1126/science.1107642. [DOI] [PubMed] [Google Scholar]
- Torres MP, Clement ST, Cappell SD, Dohlman HG. Cell cycle-dependent phosphorylation and ubiquitination of a G protein alpha subunit. J Biol Chem. 2011;286:20208–20216. doi: 10.1074/jbc.M111.239343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi B, Danforth WH. Effect of pH on the kinetics of frog muscle phosphofructokinase. J Biol Chem. 1966;241:4110–4112. [PubMed] [Google Scholar]
- Van Eps N, Preininger AM, Alexander N, Kaya AI, Meier S, Meiler J, Hamm HE, Hubbell WL. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A. 2011;108:9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.