SUMMARY
The genetic interrogation and reprogramming of cells requires methods for robust and precise targeting of genes for expression or repression. The CRISPR-associated catalytically inactive dCas9 protein offers a general platform for RNA-guided DNA targeting. Here we show that fusion of dCas9 to effector domains with distinct regulatory functions enables stable and efficient transcriptional repression or activation in human and yeast cells with the site of delivey determined solely by a co-expressed short guide (sg)RNA. Coupling of dCas9 to a transcriptional repressor domain can robustly silence expression of multiple endogenous genes RNA-seq analysis indicates that CRISPR interference (CRISPRi)-mediated transcriptional repression is highly specific. Our results establish that the CRISPR system can be used as a modular and flexible DNA-binding platform for the recruitment of proteins to a target DNA sequence and reveal the potential of CRISPRi as a general tool for the precise regulation of gene expression in eukaryotic cells.
INTRODUCTION
Targeted gene regulation on a genome-wide scale is a powerful approach for interrogating gene function and re-wiring regulatory networks. Naturally occurring and engineered DNA-binding proteins, such as the tetracycline repressor, Gal4, zinc-fingers or the TALE proteins, have been fused to transcription activators and repressors to modulate gene expression (Cong et al., 2012; Deuschle et al., 1995; Gossen and Bujard, 1992; Hathaway et al., 2012; Maeder et al., 2013; Margolin et al., 1994; Perez-Pinera et al., 2013; Sadowski et al., 1988; Zhang et al., 2000). However, due to either fixed DNA sequence binding requirements or their repetitive composition and size, it remains time-consuming and expensive to develop large scale protein libraries for genome interrogation (Joung and Sander, 2013).
Recently, several groups have shown that a modified type II CRISPR (Clustered Regularly Interspaced Palindromic Repeats) system can be targeted to DNA using RNA, enabling genetic editing of any region of the genome in a variety of organisms (Cho et al., 2013; Cong et al., 2013; Dicarlo et al., 2013; Gratz et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2012, 2013; Mali et al., 2013; Wang et al., 2013). This single RNA – single protein CRISPR system is derived from a natural adaptive immune system in bacteria and archaea. Prokaryotes have evolved diverse RNA-mediated systems that use short CRISPR RNAs (crRNAs) and Cas (CRISPR-associated) proteins to detect and defend against invading DNA elements (Bhaya et al., 2011; Marraffini and Sontheimer, 2008, 2010; Wiedenheft et al., 2012). In the type II CRISPR/Cas system, a ribonucleoprotein complex formed from a single protein (Cas9), a crRNA, and a trans-acting crRNA (tracrRNA) can carry out efficient crRNA-directed recognition and site specific cleavage of foreign DNA (Deltcheva et al., 2011; Jinek et al., 2012). This system has been further simplified with the development of a chimeric single guide RNA (sgRNA) and a Cas9 protein from the Streptococcus pyogenes CRISPR that together are sufficient for targeted DNA binding and cleavage with the cleavage site dictated solely by complementarity to the sgRNA (Jinek et al., 2012). We have shown recently in bacterial and human cells that the endonuclease domains of the Cas9 protein can be mutated to create a programmable RNA-dependent DNA binding protein (Qi et al., 2013). Targeting of catalytically inactive Cas9 protein (dCas9) to the coding region of a gene can sterically block RNA polymerase binding or elongation, leading to dramatic suppression of transcription in bacteria. By contrast, only a modest block in transcription was seen in mammalian cells thus limiting the utility of the system as a tool for programmed knockdown of genes.
Transcriptional regulation in eukaryotes is complex. Most genes are controlled by the interplay of activating and repressive transcription factors acting at DNA regulatory elements which can be spread across large regions of the genome (Conaway, 2012). Further regulation occurs through epigenetic modification of histone acetylation and both histone and DNA methylation. Globally deciphering the mechanisms for establishing and maintaining these signals as well as the functional impact of such modifications has been hampered by a lack of tools for targeting transcription and epigenetic regulators to specific DNA sequences. Here, we show that dCas9 can be used as a modular RNA-guided platform to recruit different protein effectors to DNA in a highly specific manner in human cells and the budding yeast Saccharomyces cerevisiae. We show that both repressive and activating effectors can be fused to dCas9 to repress or activate reporter gene expression respectively. We also show CRISPRi can be used for multiplexed control of endogenous genes. Using a dCas9 fusion protein, we further show that the system can be used to stably repress genes with comparable gene silencing efficiency typically achieved by RNA interference (RNAi) while minimally impacting transcription of non-targeted genes.
RESULTS
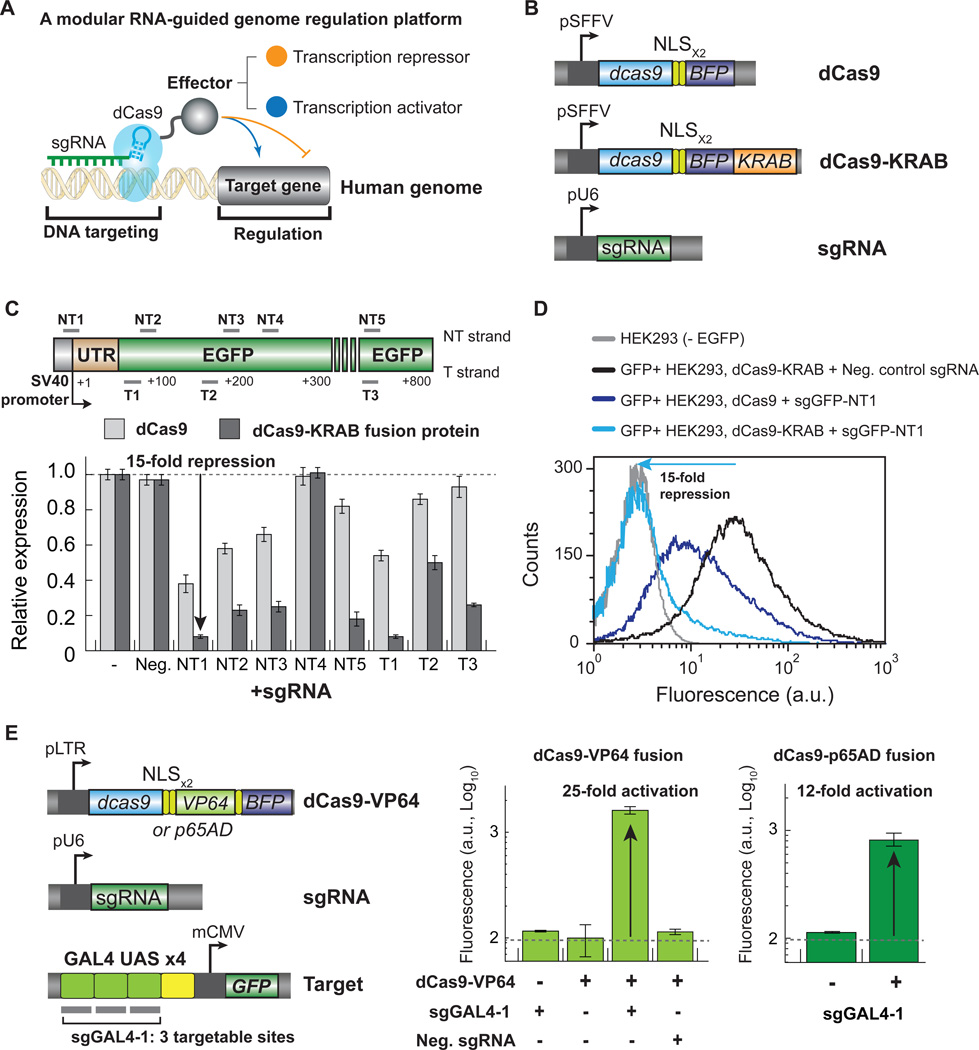
dCas9 fusion proteins can efficiently activate or silence transcription
We have shown recently that CRISPRi can decrease gene expression in human cells (Qi et al., 2013). In that initial study, the degree of repression achieved by CRISPRi was modest (~2-fold). To improve the efficacy of CRISPRi in human cells, we examined whether dCas9 could be fused to protein domains known to recruit repressive chromatin modifying complexes to improve transcriptional silencing (Figure 1A). We created a gene encoding a human codonoptimized dCas9 from S. pyogenes fused to two copies of a nuclear localization sequence (NLS), an HA tag, and blue fluorescent protein (BFP). We further fused this modified dCas9 gene with different repressive chromatin modifier domains, including the KRAB (Krüppel associated box) domain of Kox1 (Figure 1B), the CS (Chromo Shadow) domain of HP1α, or the WRPW domain of Hes1 (Fisher et al., 1996; Hathaway et al., 2012; Margolin et al., 1994). The sgRNAs were expressed from a murine RNA polymerase III U6 promoter (Figure 1B). To test whether dCas9 can recruit chromatin-modifying complexes to silence transcription, dCas9 or each dCas9-repressor fusion protein was co-transfected into GFP+ HEK293 reporter cells (in which an SV40 promoter driven GFP reporter gene is randomly genomically integrated) with an sgRNA targeting GFP. We found that cells expressing the dCas9-KRAB fusion protein show a 5-fold decrease in GFP signal while cells expressing dCas9 alone, dCas9-CS or dCas9-WRPW show a 2-fold decrease in GFP signal, suggesting that the dCas9-KRAB fusion protein might recruit chromatin modifying complexes to increase the potency of CRISPRi silencing.
Figure 1. A modular CRISPR fusion system for efficiently repressing and activating transcription in human cells.
(A) dCas9 fused to effector domains can serve as an RNA guided DNA binding protein to target any protein to any DNA sequence. (B) A minimal CRISPRi system in human cells contains an sgRNA expression plasmid and dCas9 or dCas9 fused to the repressive KRAB effector domain. Both dCas9 constructs are fused to two copies of a nuclear localization sequence and a blue fluorescent protein. (C) A dCas9-KRAB fusion protein efficiently silences GFP expression. 8 sgRNAs targeting GFP are transfected into GFP+HEK293 stably expressing either dCas9 (light grey) or dCas9-KRAB (dark grey). GFP fluorescence is quantified by flow cytometry 6 days following transfection and displayed as signal normalized to a vector control. The data is displayed as mean ± standard deviation for 3 independent experiments. See also Figure S1. (D) CRISPRi gene repression is stable over time. GFP+HEK293 cells were infected with lentivirus constructs expressing a negative control sgRNA or a sgRNA targeting GFP and either dCas9 or dCas9-KRAB. Cells were grown for 9 days and then analyzed for GFP expression. A histogram displays GFP fluorescence for each sample and a control population of HEK293 cells which do not express GFP. Data are representative of 3 independent experiments. (E) Two dCas9 fusion proteins were constructed with VP64 or p65AD. The sgRNA is expressed as before. A diagram of the Gal4 UAS-GFP reporter and data showing transient transfection of either dCas9-VP64 or dCas9-p65AD and sgGAL4–1 can activate gene expression in HEK293 cells. Cells were transfected with the indicated plasmids and 48 hours later analyzed by flow cytometry for GFP expression. The data are displayed as mean ± standard deviation for 2 independent experiments. See also Figure S2.
To improve the utility of CRISPRi, we tested whether stable dCas9 or dCas9-KRAB expression could effectively silence gene expression. We cloned dCas9 and dCas9-KRAB into a minimal lentiviral construct under the control of the spleen focus forming virus promoter (SFFV) (Figure 1B). We generated lentivirus, infected GFP+ HEK293 cells, and isolated the cell sub-populations expressing dCas9 or dCas9-KRAB by flow cytometry sorting. After one week of growth, we transfected sgRNAs targeting GFP and measured the level of GFP expressed 3 and 6 days following transfection. We found that stable dCas9-KRAB expression is sufficient to silence GFP with substantial knockdown 3 days following transfection (Figure S1A–B) and strong silencing 6 days following transfection (Figure 1C). We observed that 6 out of 9 sgRNAs targeting GFP knocked down GFP expression by at least 75% with a ~15-fold repression for the best sgRNA (NT1) (Table S1). Our results also revealed a linear relationship between the level of expression from the sgRNA vector and the level of GFP remaining within a cell (Figure S2A). As a further technical refinement to CRISPRi, we also tested whether we could stably express the sgRNA with a lentivirus as this allows for stable long-term gene silencing. We transduced GFP+ HEK293 cells that stably express dCas9 or dCas9-KRAB proteins with a lentivirus to stably express an sgRNA targeting GFP or a negative control sgRNA. We then measured GFP expression 14 days following viral infection. We found that we can robustly silence GFP expression in HEK293 cells when both the RNA and protein components of CRISPRi are stably expressed in human cells (Figure 1D). We sequenced the GFP reporter locus in these cells to confirm that dCas9 is completely nucleolytically inactive. Here, GFP retained a wild type sequence with no detectable indels in GFP knockdown cells. In the ~5% of cells which still express some GFP, it is unclear why CRISPRi does not suppress transcription. These cells may express low levels of the sgGFP RNA or represent specific retroviral integration sites which are refractory to silencing.
We reasoned that the CRISPRi platform provides a modular protein effector recruitment system, which could also be used for gene activation when coupled with transcription activators. To test if we can activate gene expression in human cells with dCas9, we fused 4 copies of the well-characterized transcription activator VP16 or a single copy of p65 activation domain (AD) to dCas9. We co-transfected dCas9-VP64 or dCas9-p65AD and an sgRNA construct that targets the Gal4 UAS (upstream activation sequence) into a HEK293 reporter cell line expressing a Gal4 UAS-GFP reporter (Figure 1E, left panel). Two days following transfection we measured the levels of GFP expressed by flow cytometry. Our results revealed that both dCas9-VP64 and dCas9-p65AD can effectively activate reporter gene expression, suggesting that dCas9 can serve as a generic and modular platform for different types of transcriptional control including both repression and activation (Figure 1E). Interestingly, we observed varying amounts of activation with different fusion protein designs using VP64 (Figure S2B), arguing that the design of the fusion protein or fusion partner is an important parameter for future optimization efforts.
CRISPRi-mediated gene knockdown is highly specific to the target gene
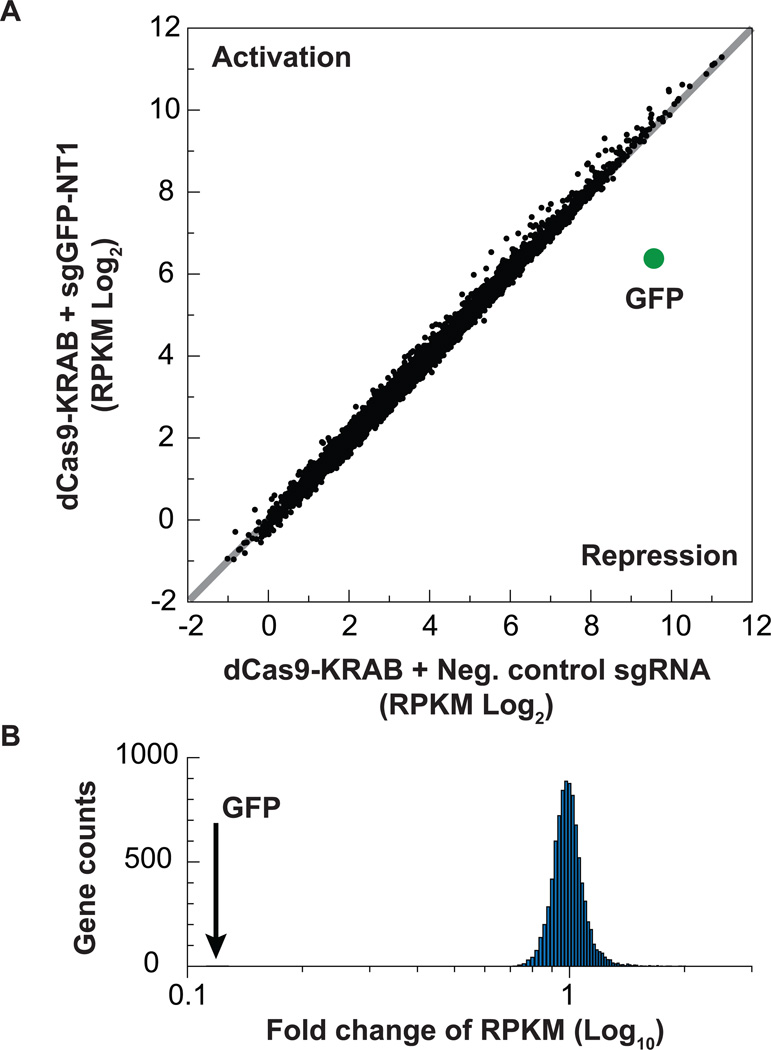
To test the specificity of CRISPRi in human cells, we used RNA-seq to quantify the transcriptome of GFP+ HEK293 cells expressing dCas9-KRAB and either a negative control sgRNA (sgGAL4–1) or an sgRNA targeting GFP (sgGFP-NT1). Our results show that CRISPRi is highly specific as GFP is the only gene that is significantly suppressed by the GFP-targeting sgRNA (Figure 2 and Figure S3A). If we average the data from two independent biological replicates, no gene other than GFP changes by more than 1.5 fold. For the few genes where we measured changes in expression, we find no evidence for the presence of a near match to the sgRNA seed sequence suggesting that the changes observed are due to a small level of noise in RNA-seq measurements. Furthermore, we observed few changes in gene expression in the negative control sample, suggesting that the negative control sgRNA triggers minimal off-target effects (Figure 2 and Figure S3A). We also found no evidence for significant on- or off-target effects due to expression of either sgGAL4 or sgGFP-NT1 alone (Figure S3B). Finally, expression of dCas9-KRAB resulted in minimal changes in gene expression (Figure S3C).
Figure 2. CRISPRi is highly specific in human cells.
(A) RNA sequencing RPKM (reads per kilo base per million) are plotted for GFP+ HEK293 cells stably expressing dCas9-KRAB and either a negative control sgRNA targeting Gal4 or an sgRNA targeting GFP. Total RNA was collected 15 days following lentiviral transduction. The data are representative of 2 independent biological replicates. See also Figure S3. (B) A histogram showing the fold changes in gene expression from (A) plotting sgGFP-NT1 over the negative control. The GFP is indicated with an arrow. The data are representative of 2 independent biological replicates. See also Figure S3.
CRISPRi can silence endogenous human genes
As there can be important differences in how endogenous and reporter genes are transcribed, we tested whether CRISPRi can silence endogenous human genes. We designed sgRNAs to target genes encoding cell surface transmembrane proteins, enabling us to use flow cytometry with directly conjugated fluorescent antibodies to quantify gene expression at the single-cell level. We cloned 10 sgRNAs each for the transferrin receptor (CD71) and C-X-C chemokine receptor type 4 (CXCR4), which were designed to span a 500-bp region upstream or downstream of the transcription start site, targeting either template or non-template strand (Table S1). Each sgRNA was transfected into HeLa cells stably expressing dCas9 or dCas9-KRAB. Cells were dissociated and stained for CD71 and CXCR4 expression 3 days after transfection. In this experiment we found that both dCas9 and dCas9-KRAB proteins can knock down endogenous expression of CD71 and CXCR4 (Figure 3A–B), with 3 out of 10 sgRNAs for each gene showing significant repression (60–80% repression). Interestingly, a strong correlation exists between the degree of knockdown for dCas9 and dCas9-KRAB for endogenous genes, suggesting that sgRNA binding to DNA or local chromatin structure may be limiting factors which dictate gene knockdown (Figure S4A). Further work is required to systematically analyze why recruitment of a KRAB domain has an effect in some contexts but not others.
Figure 3. CRISPRi can stably suppress expression of endogenous eukaryotic genes.
(A) A diagram of sgRNA binding sites for CD71 and a graph displaying suppression of CD71 by dCas9 or dCas9-KRAB in HeLa cells. Cells stably expressing dCas9 or dCas9-KRAB were transfected with sgRNAs targeting CD71. After 72 hours, cells were harvested and analyzed by flow cytometry for CD71 protein expression. The data are displayed as mean ± standard deviation for 2 independent experiments. See also Figure S4A. (B) A diagram of sgRNA binding sites for CXCR4 and a graph displaying suppression of CXCR4 by dCas9 or dCas9-KRAB in HeLa cells. Cells stably expressing dCas9 or dCas9-KRAB were transfected with sgRNAs targeting CXCR4. After 72 hours, cells were harvested and analyzed by flow cytometry for CXCR4 protein expression. The data are displayed as mean ± standard deviation for 2 independent experiments. See also Figure S4A. (C) Stable suppression of CD71 by dCas9 or dCas9-KRAB in HeLa cells. Cells stably expressing dCas9 or dCas9-KRAB were transduced with sgRNAs targeting CD71. After 7 days, cells were harvested and analyzed by flow cytometry for CD71 protein expression. The data are displayed as mean ± standard deviation for 2 independent experiments. (D) Stable suppression of CXCR4 by dCas9 or dCas9-KRAB in HeLa cells. Cells stably expressing dCas9 or dCas9-KRAB were transduced with sgRNAs targeting CXCR4. After 8 days, cells were harvested and analyzed by flow cytometry for CXCR4 protein expression. The data are displayed as mean ± standard deviation for 2 independent experiments. (E) Double knockdown of both CD71 and CXCR4 using sgRNAs targeting CD71 and CXCR4 in HeLa cells that stably express dCas9-KRAB. Cells were dissociated and stained for CD71 and CXCR4 expression 5 days after transfection. The data are displayed as mean ± standard deviation for 2 independent experiments. See also Figure S4B. (F) Robust repression of endogenous genes in yeast. A bar graph shows fluorescence intensity of a strain expressing TEF1-GFP transformed with the indicated sgRNA and dCas9 or dCas9-Mxi1. The two dotted lines indicate fluorescent signals from untagged yeast cells that do not express GFP or TEF1-GFP yeast cells without dCas9. The data are displayed as mean ± standard deviation for 3 independent experiments. See also Figure S4C.
We also tested whether stable lentivirus-mediated expression of both the sgRNA and dCas9 or dCas9-KRAB could repress endogenous gene expression. We transduced HeLa cells expressing dCas9 or dCas9-KRAB with three sgRNAs targeting CD71 or CXCR4 and 7 or 8 days later measured the amount of CD71 or CXCR4 expressed. Our data show CD71 and CXCR4 can be efficiently and stably knocked down (Figure 3C–D), providing evidence that CRISPRi can be used like RNAi to suppress endogenous gene expression.
A valuable extension of CRISPRi is the capability for multiplexed gene regulation in human cells. To test whether we can suppress the expression of multiple genes, we transfected sgRNAs targeting CD71, CXCR4 or both into HeLa cells stably expressing dCas9-KRAB. Cells were dissociated and stained for CD71 and CXCR4 expression 5 days after transfection. In this experiment we found that we can simultaneously knock down endogenous expression of CD71 and CXCR4 (Figure 3E). This result was independently confirmed by qPCR (Figure S4B).
CRISPRi is a general method for silencing transcription of endogenous genes in eukaryotes
To test if CRISPRi can be used for transcription repression in other eukaryotes, we examined whether dCas9, alone or fused to a transcriptional repressor, will effectively silence gene expression in Saccharomyces cerevisiae. We cloned dCas9 with 2 NLS into a vector under control of the TDH3 promoter (Figure S4C). Using this same scheme, we also fused dCas9 to Mxi1, a mammalian transcriptional repressor domain reported to interact with the histone deacetylase Sin3 homolog in yeast (Figure S4C) (Harper et al., 1996; Kasten et al., 1996; Schreiber-Agus et al., 1995). Both dCas9 fusion proteins were transformed into a strain carrying a TEF1-GFP fusion expressed from the endogenous locus (Huh et al., 2003). We expressed an sgRNA that targets the endogenous TEF1 locus under control of the Pol III SNR52 promoter (Figure S4C) (Dicarlo et al., 2013). With dCas9 alone, we observed 18-fold repression, which is increased to 53-fold with addition of the Mxi1 domain (Figure 3F). As CRISPRi shows robust endogenous gene silencing in yeast, it could also prove a useful tool for studying organisms with limited genetic tools, such as the pathogenic yeast Candida albicans.
We found that both the Mxi1 and KRAB domain strongly suppress transcription when targeted to some regions of DNA but not to others. Additionally, some genes may require recruitment of repressive complexes other than Kap1/HP1 or Sin3. Ongoing work will define what regions of a gene are most effective for dCas9 fusion mediated gene silencing. However, the remarkable success in using RNA-guided Cas9 to enable DNA cutting for many genes in distinct organisms argues that Cas9 can bind to target DNA sites in a broad range of genes or regulatory regions (Cho et al., 2013; Cong et al., 2013; Dicarlo et al., 2013; Gratz et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2012, 2013; Mali et al., 2013; Wang et al., 2013). Taken together with the present study, these findings suggest that CRISPRi will be a general method for efficiently and specifically regulating transcription in many eukaryotes.
CRISPRi as a tool for mapping and perturbing regulatory elements
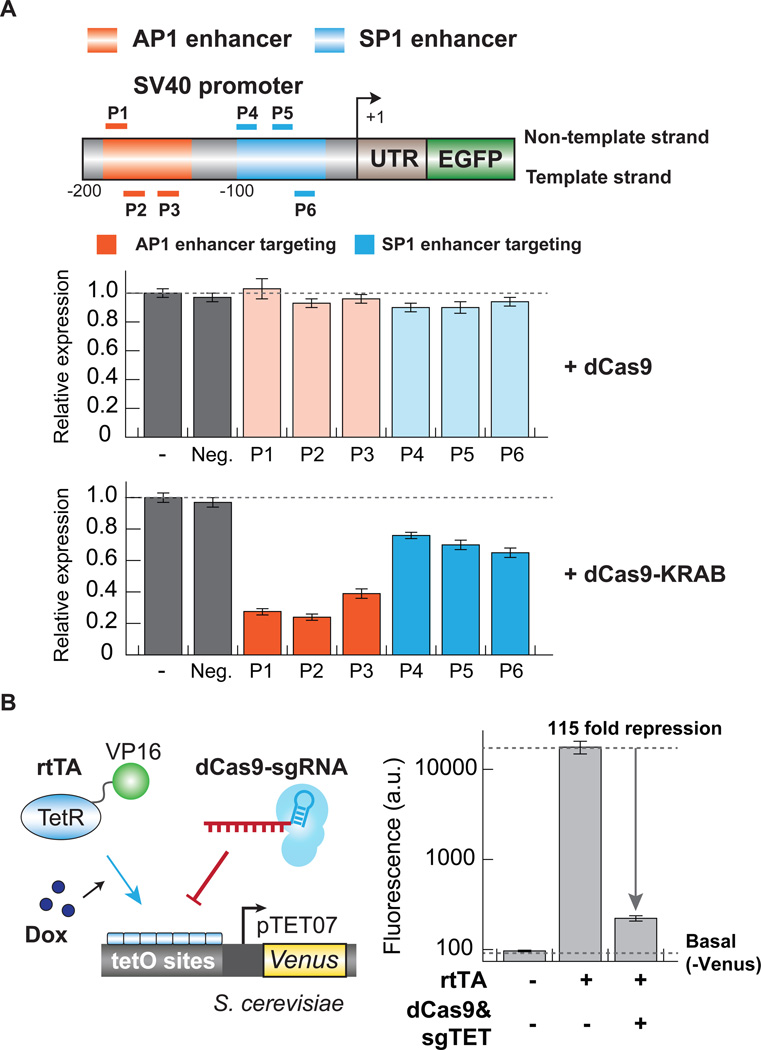
A method for functionally mapping DNA regulatory elements such as enhancers in a high-throughput manner does not currently exist. We reasoned that if CRISPRi was effective when targeted to promoters or introns we could map enhancer elements. We tested whether we could silence gene expression by targeting dCas9 or dCas9-KRAB fusion protein to cis-acting proximal elements within the SV40 early promoter of our GFP reporter (Table S1). We found that while dCas9 alone had no effect on transcription, we efficiently repressed GFP expression with dCas9-KRAB (Figure 4A). This suggests that while dCas9 may not be able to directly block the initiation of transcription, dCas9 fusion proteins can silence proximal regulatory elements within a promoter by recruiting chromatin modifiers. Previous studies have shown that the AP1 enhancer (for AP-1 transcription factor binding) is a more important DNA motif for SV40 promoter activity as compared to the SP1 enhancer (for SP-1 transcription factor binding), as SP1 enhancer deletion had little effect on transcription (Baty et al., 1984; Benoist and Chambon, 1981). Interestingly, we also observed that blocking the AP1 enhancer caused a stronger silencing effect than blocking the SP1 enhancer within the SV40 promoter (Figure 4A), implying that CRISPRi can be used as a tool for perturbing and mapping regulatory functions of DNA elements.
Figure 4. CRISPRi can suppress gene expression from the promoter of a gene.
(A) A dCas9-KRAB fusion protein efficiently silences GFP expression when targeted to the SV40 promoter of an SV40-GFP reporter. 6 sgRNAs targeting different regions of the SV40 promoter as indicated are transfected into GFP+HEK293 cells stably expressing either dCas9 (upper panel) or dCas9-KRAB (lower panel). GFP fluorescence is quantified by flow cytometry 6 days following transfection and displayed as signal normalized to a vector control. The data are displayed as mean ± standard deviation for 3 independent experiments. (B) The dCas9 construct and an sgRNA (sgTET) construct were co-transformed into a yeast strain expressing a TetON-Venus reporter and the rtTA protein. Doxycycline was added to cells expressing rtTA alone or rtTA, dCas9 and sgTET. The data are displayed as mean ± standard deviation for 3 independent experiments.
We next examined whether CRISPRi could block a transcription factor from binding to enhancer sites in Saccharomyces cerevisiae. Plasmids expressing the dCas9 protein and an sgRNA (sgTET) were co-transformed into a strain expressing the rtTA protein and a TetONVenus reporter in which the addition of doxycycline leads to expression of Venus. We added doxycycline to strains expressing rtTA alone or rtTA, dCas9 and sgTET (which targets dCas9 to the rTA binding site). Without dCas9, rtTA strongly activated the reporter, but this induction can be effectively blocked by dCas9 (115 fold repression, Figure 4B). This suggests that dCas9 can sterically compete with transcription factors which have tight binding affinity for DNA elements, further implying that CRISPRi can be used to perturb and map the regulatory roles of distal and proximal enhancers.
DISCUSSION
dCas9 provides a general platform for targeting proteins to DNA
Systematic characterization of gene function depends on the ability to manipulate genes through deletion, suppression or overexpression. Here, we have developed a modular method using a modified type II CRISPR system to target effector domains to specific genomic loci. We have shown that programmable dCas9 can repress or activate transcription in human cells. dCas9 fusion proteins can functionally mimic multiple native protein interactions, including the KRAB domain of Kox1, VP16 and p65AD (Groner et al., 2010). Our data demonstrate how dCas9 can be used to target a protein to a specific region of DNA. The modularity of the dCas9 system provides a new approach for studying transcription, epigenetic regulation and both DNA replication and repair.
The simple constitutive dCas9 system can be coupled to more complex inducible systems for precise temporal and spatial control, enabling refined analysis of chromatin remodeling processes during cell development and differentiation. For example, dCas9 could be used with existing optogenetic or chemical induced proximity systems to dynamically analyze recruitment of a broad range of chromatin modifiers to a specific DNA sequence (Hathaway et al., 2012; Levskaya et al., 2009). Targeting dCas9 fusions to distinct chromatin microenvironments could elucidate how distinct chromatin remodeling complexes propagate the spreading or insulation of repressive chromatin marks along a chromosome. Using multiple sgRNAs, one can titer how many chromatin modifying complexes are recruited to a gene to interrogate how the local concentration of any chromatin modifying complex modulates chromatin structure and transcription. dCas9 could also be used to recruit or tether a specific genomic locus to a sub-nuclear localization such as the nuclear pore, nucleolus or nuclear membrane. This system may enable spatial manipulation of DNA and chromatin, allowing the study of chromatin and transcription in qualitatively new ways, which to date have largely involved retrospective association studies or broad loss of function genetics.
At present all mammalian work on Cas9 has focused on the protein from a single species (S. pyogenes) and there is a huge amount of biological diversity that remains untapped (Chylinski et al., 2013; Zhang et al., 2013). It is likely that the current Cas9 is not optimal. Additionally, many of the different Cas9 species use distinct sequences to recognize the guide RNA. Thus two or more sgRNA variants can be expressed in the same cell and each will bind exclusively to its cognate dCas9 species. This will allow for orthogonal targeting of multiple independent sites within a genome, enabling the independent delivery of distinct effector domains and combinatorial manipulation of chromatin structure and epigenetic marks.
Mapping the function of enhancers and chromosome topology with dCas9
Our results provide evidence that dCas9 can target enhancers, introns, and other noncoding elements to map the regulatory functions of these elements on transcription. This could enable sophisticated high throughput mapping of enhancers at endogenous loci. In theory, using inducible dimerization systems, dCas9 could tether a distant enhancer close to the promoter of a gene to interrogate how chromosome conformation regulates transcription. Recent work has shown that during neural differentiation, CCCTC binding factor, cohesion and the mediator complex modulate chromosome topology to control development (Millau and Gaudreau, 2011; Phillips-Cremins et al., 2013). dCas9 could enable studying these processes in a much more dynamic and programmable manner by artificially creating different genomic conformations and measuring how these modulate transcription and development.
CRISPRi efficiently and specifically represses transcription in human cells
In our experiments, we observed robust gene knockdown of both reporter and endogenous genes (5~15-fold repression in human and 50-fold in yeast). Our method uses common lentiviral constructs to express both dCas9 and sgRNAs to achieve stable long term gene knockdown. This is comparable to the efficiency of existing gene knockdown techniques such as RNAi and TALE proteins. While we do not yet know what fraction of genes can be silenced by a dCas9 fusion protein, a significant portion of the genome should be targetable. In human cells, previous work has demonstrated that local chromatin context is a crucial determinant of KRAB mediated silencing. For example, recruitment of the KRAB domain to adjacent genomic regions can have no effect or can completely silence transcription (Groner et al., 2010). The biology underlying this observation remains to be characterized and CRISPRi could provide a tool to map chromatin structure and the function of chromatin remodeling complexes. Nonetheless, an exon trapping strategy in which Tet binding sites were randomly inserted in the genome allowing recruitment of a Tetracycline repressor-KRAB fusion protein revealed that ~80% of genes tested can be silenced by KRAB-mediated recruitment of Kap1 and HP1 proteins (Groner et al., 2010). Ongoing work will characterize additional fusion partners to understand which transcription repressor complexes are most useful for CRISPRi in different cell types.
Our RNA-seq results show that CRISPRi is highly specific with minimal off-target effects for two distinct sgRNA sequences. By contrast, genome editing by the catalytically active Cas9 nuclease can be accompanied by off-target genomic alterations which occur at loci where near cognate DNA seed matches exist (Fu et al., 2013). The specificity of CRISPRi remains to be determined across a broad range of sgRNA sequences. Nonetheless, it may be substantially easier to specifically modulate transcription as only a small fraction of the human genome is transcribed or required for controlling transcription thus binding of catalytically inactive dCas9, and it is derivatives, at near cognate sites will often not impact transcription.
Future optimization for CRISPRi technology
The observation that the sgRNA is a limiting factor for CRISPRi function suggests that improvements to RNA stability or loading into dCas9 could improve both CRISPRi and Cas9 mediated genome editing in human cells (Jinek et al., 2013). While we are able to target dCas9 to both endogenous and reporter genes, not all sgRNA targeting constructs work efficiently. Specific sgRNA sequence composition may improve sgRNA stability, loading into dCas9, or binding to DNA. Alternately, dCas9 binding may be strongly dictated by the local chromatin structure of the target sequence. The recently published genome-wide maps of chromatin structure can inform where one targets dCas9 for both transcription repression and activation (Thurman et al., 2012). High-throughput analysis of sgRNAs that are functional will discriminate between sequence requirements and local chromatin states to determine rules for dCas9 targeting and function. While the experiments described here have examined a set of fusion partners and fusion design strategies, future work will be required to refine and improve CRISPRi fusion proteins with different effectors for optimal performance. Finally, as additional Cas9 proteins are characterized, the use of parallel orthogonal dCas9 proteins with cognate sgRNAs will enable controlling multiple genes differentially (Chylinski et al., 2013; Zhang et al., 2013).
CRISPRi provides an alternative strategy to RNAi for gene regulation
Our data here describes a dramatic improvement from our previously published work on CRISPRi in human cells and argues that CRISPRi represents an alternate strategy to RNAi for repressing gene expression in mammalian cells. RNAi is a transformative technique for studying mammalian biology and more than a decade of effort has led to sophisticated vectors, algorithms and libraries for knocking down genes in human and mouse cells (Chang et al., 2006; Fellmann et al., 2011). Genome wide shRNA libraries enable high throughput interrogation of gene function and genetic networks in human cells (Bassik et al., 2013). It should be possible to develop pooled sgRNA libraries that will allow CRISPRi to be used in an analogous manner to pooled genome-wide shRNA libraries.
We believe that in many applications RNAi and CRISPRi could be complementary methods with CRISPRi providing a number of potential advantages. As an exogenous system, CRISPRi does not compete with endogenous machinery such as microRNA expression or function. Furthermore, because CRISPRi acts at the DNA level, one can target transcripts such as non-coding RNAs, microRNAs, antisense transcripts, nuclear localized RNAs, and polymerase III transcripts. Finally, CRISPRi possesses a much larger targetable sequence space, as we show that promoters and in theory introns can also be targeted.
METHODS
RNA sequencing
Total RNA was purified from the cells using Trizol (Invitrogen). 150 µg of total RNA was mixed with oligo (dT)25 Dynabeads and mRNA was purified according to the manufacturers protocol (Invitrogen). 1 µg mRNA was resuspended in 20 µL of 10 mM Tris pH 7 mixed with an equal volume of 2× alkaline fragmentation solution (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3, pH 9.3) and incubated for 30 min at 95 °C to generate fragments ranging from 30–100 nt. The fragmentation reaction was stopped by adding 0.56 mL of ice-cold precipitation solution (300 mM NaOAc pH 5.5 plus GlycoBlue (Ambion)), and the RNA was purified by a standard isopropanol precipitation. The fragmented mRNA was then dephosphorylated in a 50 µL reaction with 25 U T4 PNK (NEB) in 1× PNK buffer (without ATP) plus 0.5 U SUPERase•In (Ambion), and precipitated with GlycoBlue via standard isopropanol precipitation methods. A linker was ligated to the fragmented RNA using truncated T4 RNA Ligase 2 (NEB) in 25% PEG-8000 in 50 mM Tris-HCl 10 mM MgCl2 1 mM DTT pH 7.5 at 25°C for 3 hours. The RNA was then reverse transcribed to DNA using SuperScript III (Invitrogen) and circularized using Circligase (Epicentre) following the manufacturers recommended protocol. Barcodes were added by PCR with Phusion polymerase (NEB). The DNA library was sequenced on an Illumina HiSeq 2000. Reads were processed using the HTSeq Python package and other custom software written in Python.
Plasmid design and construction for CRISPRi in human cells
The sequence encoding mammalian codon optimized Streptococcus pyogenes dCas9 (DNA 2.0) was fused with two C-terminal SV40 nuclear localization sequences (NLS) and tagBFP alone or with the KRAB, CSD, WRPW, VP64, or p65AD domains. Using Gibson cloning we cloned these fusion proteins into MSCV-Puro (Clontech) or pHR (Addgene). sgRNAs were expressed using a lentiviral U6 based expression vector derived from pSico which co-expresses mCherry-T2A–Puro from a CMV promoter. The sgRNA expression plasmids were cloned by inserting annealed oligos into the lentiviral U6 based expression vector that was digested by BstXI and XhoI. To detect indel formation in GFP in human cells, we purified genomic DNA from cells expressing dCas9-KRAB and either a negative control sgRNA or sgGFP-NT1 that had been grown in culture for 3 weeks. We PCR amplified the SV40 promoter and GFP, and sequenced the PCR products.
Cell culture, DNA transfections, viral production and fluorescence measurements for CRISPRi in human cells
HEK293 and HeLa cells were maintained in Dulbecco’s modified eagle medium (DMEM) in 10 % FBS, 2 mM glutamine, 100 units/mL streptomycin and 100 µg/mL penicillin. Lentivirus and retrovirus were produced by transfecting HEK293 or amphotropic Phoenix packaging cell lines with standard packaging vectors. HEK293 or HeLa cell lines were generated by transducing cells with a MSCV retrovirus expressing GFP from the SV40 promoter or lentivirus expressing dCas9, dCas9-KRAB from an SFFV promoter, or the sgRNA from a U6 promoter or a Gal4-GFP reporter. Pure populations of each stable cell line were sorted by flow cytometry using a BD FACS Aria2 for stable GFP, BFP or mCherry expression. For transient transfection experiments, reporter HEK293 cell lines were transfected using TransIT-LT1 transfection reagent (Mirus) with the manufacturers recommended protocol in 24 well plates. Cells were transfected with 0.5 µg of the dCas9 expression plasmid and 0.5 µg of the RNA expression plasmid, or if dCas9 is stably expressed with 1 µg of the RNA expression plasmid. Two sgRNA plasmids (0.75 µg each) were co-transfected for double knockdown experiments. For 6–7 day experiments, the cells are split at day 3. At 3 or 6 days following transfection or 7–14 days following transduction, cells were trypsinized to a single cell suspension, gated on mCherry positive population (top 30– 60% for transient transfection experiments) and GFP expression was analyzed. To analyze CXCR4 or CD71 expression cells were dissociated in 10 mM EDTA PBS and then stained in PBS/10% FBS for 1 hour at room temperature. Isotype control, CXCR4, and CD71antibodies for flow cytometry were purchased from MBL or eBioscience and used at 0.05 mg/mL. All flow cytometry analysis was performed using a LSR II flow cytometer (BD Biosciences).
Quantitative RT-PCR
Cells were harvested using trypsin (Invitrogen), and total RNA was isolated using the RNeasy Mini Kit (Qiagen), according to manufacturer’s instructions. RNA was converted to cDNA using AMV reverse transcriptase under standard conditions with oligo dT primers and RNasin (Promega). Quantitative PCR reactions were prepared with Go-Taq (Promega) and SYBR Green (Invitrogen), according to the manufacturer’s instructions. Reactions were run on a LightCycler thermal cycler (Roche). Primer sequences for CXCR4, CD71, and GAPDH are listed below.
CXCR4: 5’ GAAGCTGTTGGCTGAAAAGG 3’ CTCACTGACGTTGGCAAAGA
CD71: 5’ AAAATCCGGTGTAGGCACAG 3’ GCACTCCAACTGGCAAAGAT
GAPDH: 5’ ACAGTCAGCCGCATCTTCTT 3’ ACGACCAAATCCGTTGACTC
Strain construction and fluorescence measurements for CRISPRi in yeast
The mammalian codon optimized dCas9 fused with two C-terminal SV40 nuclear localization signal sequences was PCR amplified and inserted into pJED103 CEN/ARS plasmid (BglII / BamHI digested) using In-Fusion HD cloning (Clontech). The Mxi1 repressive domain was synthesized using IDT gBlocks gene fragments and cloned into the vector using Gibson assembly (Gibson et al., 2009). A contiguous IDT gene block containing the SNR52 promoter, sgRNA, and SUP4 terminator 3’ flanking sequence was amplified using Phusion HF PCR, and digested using EcoRI and PstI. The digested product was inserted into the AH057 CEN/ARS plasmid using standard ligation/insertion protocols. Inverse PCR was used to generate sgRNA cassettes with new 20 bp complementary regions. Standard lithium acetate transformations were carried out to transform 1 µg of plasmid per transformation into TEF1-GFP tagged strain or a strain with a genomic integrated TetON-Venus reporter and an rtTA gene. After transformation, cells were grown on selective media (SC-uracil and -leucine) for 2 days. Strains were grown overnight at 30°C and the levels of fluorescence protein were determined using a LSRII flow cytometer (BD Biosciences). For Doxycycline induction experiments, the overnight culture without Doxycycline was diluted to OD 0.05 in the same selective media supplemented with 100 µg/mL Doxycycline. The levels of fluorescence protein were assayed by flow cytometer after 5 hours of growth. Triplicate cultures were measured for each experiment and their standard deviation was indicated as the error bar.
Supplementary Material
Highlights.
-
-
CRISPRi enables robust gene repression and activation in human cells
-
-
CRISPRi knockdown is specific with minimal off-target effects in human cells
-
-
CRISPRi can effectively repress endogenous genes in human and yeast
-
-
dCas9 enables modular and programmable RNA guided genome regulation in eukaryotes
ACKNOWLEDGMENTS
The authors thank Martin Jinek for discussion and distribution of the dCas9 gene. The authors also thank Jason Park, Martin Kampmann, Mike Bassik, and Xin Xiong for lentiviral vectors, technical advice and helpful discussion. L.S.Q. acknowledges support from the UCSF Center for Systems and Synthetic Biology. This work was supported by NIH P50 GM102706 (J.S.W., J.A.D), NIH P50 GM081879 (L.S.Q., W.A.L., Z.L., E.H.W.), NIH U01 CA168370 (J.S.W), Howard Hughes Medical Institute (L.A.G., G.A.B., O.B., J.A.D., J.S.W., W.A.L.), NSF SynBERC EEC-0540879 (W.A.L.), NIGMS IMSD (S.E.T.), the Human Frontiers Scientific Program (N.S., L.M.), and a Ruth L. Kirschstein National Research Service Award (M.H.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baty D, Barrera-Saldana HA, Everett RD, Vigneron M, Chambon P. Mutational dissection of the 21 bp repeat region of the SV40 early promoter reveals that it contains overlapping elements of the early-early and late-early promoters. Nucleic Acids Res. 1984;12:915–932. doi: 10.1093/nar/12.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C, Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981;290:304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteriaarchaea versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat. Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA Cas9 families of type II CRISPR-Cas immunity systems. Rna Biol. 2013;10 doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway JW. Introduction to theme “Chromatin, epigenetics, and transcription.”. Annu. Rev. Biochem. 2012;81:61–64. doi: 10.1146/annurev-biochem-090711-093103. [DOI] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo Y-C, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U, Meyer WK, Thiesen HJ. Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loophelix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. Highfrequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013 doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracyclineresponsive promoters. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. Plos Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SE, Qiu Y, Sharp PA. Sin3 corepressor function in Myc-induced transcription and transformation. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8536–8540. doi: 10.1073/pnas.93.16.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten MM, Ayer DE, Stillman DJ. SIN3-dependent transcriptional repression by interaction with the Mad1 DNA-binding protein. Mol. Cell. Biol. 1996;16:4215–4221. doi: 10.1128/mcb.16.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Reyon D, Angstman JF, Fu Y, Sander JD, Joung JK. Robust, synergistic regulation of human gene expression using TALE activators. Nat. Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ., 3rd Krüppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millau J-F, Gaudreau L. CTCF, cohesin, and histone variants: connecting the genome. Biochem. Cell Biol. Biochim. Biol. Cell. 2011;89:505–513. doi: 10.1139/o11-052. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Zhang L, Spratt SK, Liu Q, Johnstone B, Qi H, Raschke EE, Jamieson AC, Rebar EJ, Wolffe AP, Case CC. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J. Biol. Chem. 2000;275:33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol. Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.