Abstract
Objectives:
To assess the association of established multiple sclerosis (MS) risk variants in 3,254 African Americans (1,162 cases and 2,092 controls).
Methods:
Human leukocyte antigen (HLA)-DRB1, HLA-DQB1, and HLA-A alleles were typed by molecular techniques. Single nucleotide polymorphism (SNP) genotyping was conducted for 76 MS-associated SNPs and 52 ancestry informative marker SNPs selected throughout the genome. Self-declared ancestry was refined by principal component analysis of the ancestry informative marker SNPs. An ancestry-adjusted multivariate model was applied to assess genetic associations.
Results:
The following major histocompatibility complex risk alleles were replicated: HLA-DRB1*15:01 (odds ratio [OR] = 2.02 [95% confidence interval: 1.54–2.63], p = 2.50e-07), HLA-DRB1*03:01 (OR = 1.58 [1.29–1.94], p = 1.11e-05), as well as HLA-DRB1*04:05 (OR = 2.35 [1.26–4.37], p = 0.007) and the African-specific risk allele of HLA-DRB1*15:03 (OR = 1.26 [1.05–1.51], p = 0.012). The protective association of HLA-A*02:01 was confirmed (OR = 0.72 [0.55–0.93], p = 0.013). None of the HLA-DQB1 alleles were associated with MS. Using a significance threshold of p < 0.01, outside the major histocompatibility complex region, 8 MS SNPs were also found to be associated with MS in African Americans.
Conclusion:
MS genetic risk in African Americans only partially overlaps with that of Europeans and could explain the difference of MS prevalence between populations.
Genome-wide association studies (GWAS) have contributed considerably to the understanding of multiple sclerosis (MS) susceptibility through the identification of genetic variants influencing risk and quantitation of their effects. To date, the results of 9 GWAS1–9 and 2 meta-analyses10,11 have been reported, generating a roster of nearly 60 candidate genes in support of a polygenic model of pathogenesis12,13 driven primarily by relatively common alleles. Most noteworthy is a recent multicenter GWAS8 that replicated nearly all of the previously GWAS-suggested associations together with the identification of 29 novel susceptibility loci. Immunologically relevant genes, including epistatic signals across the major histocompatibility complex (MHC) region, dominate the genomic signature of this disease. Invariably, all reported MS GWAS focused on high-prevalence datasets of European descent.14 The transferability of these results to other ancestral groups remains to be addressed.
African Americans have a lower disease risk compared with Europeans and white Americans, but appear to carry a greater risk of ambulatory disability.15,16 The differences in genome linkage disequilibrium (LD) patterns and clinical phenotype between African Americans and white Americans were successfully leveraged to probe and refine the genetics of MS using conventional association, admixture, and genotype-phenotype studies.15–21 Here, we extend these studies and test the updated MS genetic map in a well-characterized African American MS dataset. After controlling for admixture, we confirm the multiallelic human leukocyte antigen (HLA)-DRB1 association, but only 8 of 63 tested non-MHC MS loci replicated in this dataset.
METHODS
Subjects.
DNA samples from 1,162 African American patients with MS and 2,092 controls, and 577 white American cases and 461 controls were obtained through a recruitment effort initiated in 2000 using stringent inclusion and exclusion criteria as previously described.21 The African American control dataset included 1,178 de-identified samples from the National Marrow Donor Program. The African American dataset overlaps approximately 48% with our previous report.21
Standard protocol approvals, registrations, and patient consents.
The University of California at San Francisco Institutional Review Board and the National Marrow Donor Program Institutional Review Board approved this study. For all non-de-identified individuals participating in this study, written informed consent was obtained.
Single nucleotide polymorphism genotyping.
Fifty-two ancestry informative marker (AIM) single nucleotide polymorphisms (SNPs) and 76 MS SNPs were genotyped using the TaqMan OpenArray genotyping technology (Life Technologies, Carlsbad, CA). AIM SNPs were selected from previously published studies22–27 to cover all chromosomes. MS SNPs were selected from the latest GWAS8 and meta-analysis.11 Samples were loaded into customized TaqMan OpenArray genotyping plates with the OpenArray Autoloader and amplified in a Dual Flat Block GeneAmp PCR System 9700, as recommended by the manufacturer. The OpenArray Genotyping Analysis Software was used for assigning genotypes.
Quality control.
SNPs had to meet several criteria to be included in the analysis: less than 10% missing genotypes, Hardy-Weinberg proportion test p > 0.001 in controls, and no significant SNP call rate differences between patients and controls in each population (p > 0.001), whereas samples needed to have missing genotype calls of less than 10%. This standard quality control removed 10 SNPs in total (7.8%): 2 AIM SNPs because of low SNP call rates, 7 AIM SNPs because of violation of Hardy-Weinberg proportion, and one MS SNP, rs2150702, located in MLANA, because of a significant difference in call rates between cases and controls in African Americans. The average sample call rate for 75 MS SNPs was 99.3% ± 1.4% in African Americans and 99.4% ± 1.2% in white Americans, whereas the call rate for 43 AIM SNPs was 99.0% ± 2.4% in both populations. Altogether, 118 SNPs, 1,144 African American MS cases, 1,986 African American controls, 569 white American MS cases, and 460 white American controls were included in the association analysis.
HLA allele typing.
Four-digit HLA-DRB1 allele data were available for 454 African American patients with MS and 1,124 controls. For an additional 487 patients and 470 controls, only HLA-DRB1*15:01, *15:03, *03:01, and *03:02 genotypes were available. Four-digit HLA-DQB1 allele data were available for 455 patients and 520 controls. Additionally, to explore the association of putative protective allele HLA-A*02:01,8,28 we generated HLA-A genotypes for 456 patients and 1,199 controls. In this dataset, 70.0% of HLA-DRB1 typing, 91.1% of HLA-DQB1 typing, and 95.3% of HLA-A*02:01 typing were obtained by the sequence-based typing technique, and the remaining data were typed by sequence-specific oligonucleotide hybridization or sequence-specific priming.
Molecular assessment of genetic ancestry.
The 43 AIM SNPs (including 5 in the extended MHC region) exceeding established quality-control thresholds were set in a principal component (PC) analysis to identify the degree of European ancestry admixture in each individual. Estimates were checked against our previously reported percentages of European ancestry19,20,29 using Pearson correlation test. To assess population stratification, the values of the major PCs between cases and controls in both of the populations were compared using Wilcoxon rank sum test.
Genetic association analysis.
An additive model of multivariate logistic regression was applied to assess the association of the genetic variants with MS. As covariates, the ancestral PC values and HLA class II alleles were used when relevant. For each MS variant, the Cochrane Heterogeneity Q Test was performed to test effect size differences between African Americans and Europeans. Finally, to evaluate the statistical power embedded in the African American dataset, 1,000 bootstrap simulations were performed in the white reference dataset. For each simulation, the number of MS SNPs validated by association analysis was returned and distribution of the numbers from those simulations was compared with the actual number of validated MS SNPs in the African American dataset. All of the analyses were conducted using R software (version 2.13.0; R Foundation for Statistical Computing, Vienna, Austria). Odds ratios (ORs) were used to reflect effect sizes of MS risk. Confidence intervals (CIs) are given at 95%, and all of the p values are provided uncorrected.
RESULTS
Clinical and demographic features of the enrolled participants.
In this study, African American patients with MS had a higher female to male ratio and younger age at onset compared with white Americans (table 1). The proportion of primary progressive or progressive relapsing MS was larger and the MS severity score was significantly higher in African American patients than in those of European origin, which can be partially explained by the lower proportion of progressive MS in this white American group compared with that of white Americans in the literature.8 Interestingly, family history of MS as reported by the proband was 17.3%, which is within the range of that reported for European datasets.30
Table 1.
Clinical and demographic features of the enrolled participants

PC analysis was used to assess ancestry and admixture, and to control for the pervasive effects of population stratification in follow-up analyses. The first component of the PC analysis explained 50.77% of the interindividual European ancestry variance as informed by the 43 AIM SNPs (figure e-1A on the Neurology® Web site at www.neurology.org). PC2 explained 2.30% of the variances, and the biplot showed that the main contributors for PC2 were SNPs located in the extended MHC region (figure e-1B). PC1 was correlated with the previously reported percentage of European ancestry in African Americans with MS18 (R = 0.84, p < 2.2e-16), and the mean percentage of European ancestry in African Americans was 20.4%. From the scree plot and biplot, both PC1 and PC2 were used to account for population stratification in the association analysis: PC1 as signals of genome-wide and PC2 as major MHC signals for ancestry. The values of PC1 differentiated the African American and white American groups (p < 2.2e-16), but were not different according to the affectation status in either population (figure e-1C). The variance of PC1 was smaller in cases than in controls within African Americans (p = 7.22e-06), suggesting higher heterogeneity in the African American control dataset. PC2 values were not statistically different based on the affectation status in African Americans, but a trend was observed in white Americans (p = 0.039) (figure e-1D).
Association analysis for HLA alleles.
HLA-DRB1*15:01 and HLA-DRB1*03:01 were significantly associated, as expected, with disease risk in African Americans (table 2). Adjustment of the association by the PC1 and (MHC-driven) PC2 retained the statistical significance. When fitting these 2 alleles in the multivariate model, HLA-DRB1*15:03 frequencies were elevated in African Americans with MS, as previously shown.31 HLA-DRB1*04:05 was confirmed as a risk allele, whereas 2 alleles, HLA-DRB1*11:01 and HLA-DRB1*04:01, were found to be associated with reduced MS risk. No association of HLA-DQB1 alleles was detected for African American MS (table 3). Additionally, association analysis for HLA-A*02:01 showed a protective effect for carriers, independent of the top 2 HLA-DRB1 risk alleles of HLA-DRB1*15:01 and HLA-DRB1*03:01 (OR = 0.72 [95% CI 0.55–0.93], p = 0.013).
Table 2.
Association of HLA-DRB1 alleles for MS in African Americansa

Table 3.
Association of HLA-DQB1 alleles for MS in African Americansa

In the association analysis for SNPs in the MHC region, rs3129889, tagging HLA-DRB1, was the most significantly associated with MS in both African Americans and white Americans, but with a different effect size between the 2 populations, i.e., a larger effect in whites (Q test p = 3.91e-07). No association was found in African Americans for the HLA-B SNP, rs2523393, tagging HLA-B*44 in Europeans (table e-1).10
Replication study of the MS-associated SNPs in non-MHC regions.
Among the 73 MS SNPs in non-MHC regions that passed quality control, 15 SNPs were replicated in African Americans with p values <0.05 (tables 4 and e-1). Using the p < 0.01 threshold, 8 MS-validated risk markers—rs7200786 (CLEC16A), rs1335532 (CD58), rs13333054 (IRF8), rs180515 (RPS6KB1), rs4613763 (PTGER4), rs1800693 (TNFRSF1A), rs7238078 (MALT1), and rs669607 (no gene)—were replicated in the African American dataset. Among those validated SNPs in non-MHC regions, no significant differences were detected for each effect size between African Americans and populations from European descent as reported in the literature (Q test p > 0.1). However, even though rs2300603 (BATF) was replicated (p = 0.017), the direction of the effect size was reversed in African Americans (OR = 0.81) compared with that reported in Europeans (Q test p = 0.002).
Table 4.
Replication study in African Americans for MS-associated SNPs in Europeansa

In the white American dataset, 4 non-MHC SNPs were replicated with p values <0.01. To investigate the role of sample size in our analysis, bootstrapping of the white American dataset was conducted up to the sample size of the African Americans. The mean number of the statistically associated non-MHC SNPs in each simulation in white Americans was higher (mean 23.8) than the number of the statistically associated SNPs in African Americans (n = 8), suggesting that the observed disparity in the number of validated SNPs between African Americans and white Americans is not entirely attributable to the size of the datasets.
Application of the frequently associated SNPs to the 1000 Genomes Project population.
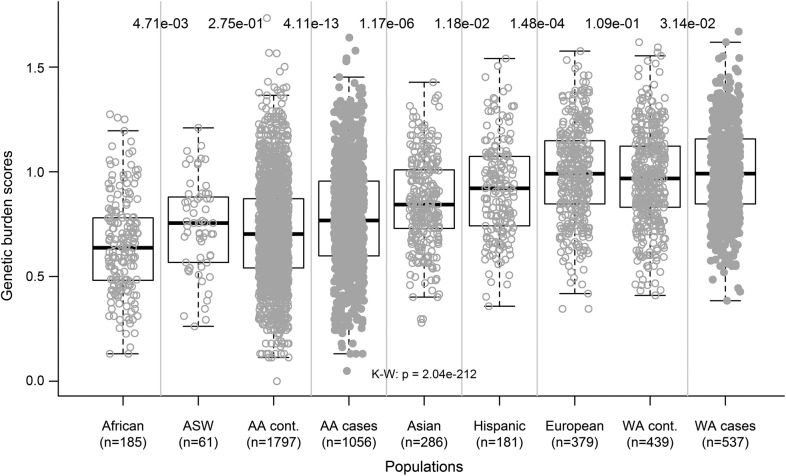
To address the hypothesis that the worldwide distribution of MS correlates with the frequency of the risk variants in each ancestral group, the cumulative genetic risk scores accounting for the minimal set of the 8 non-MHC SNPs with p values <0.01 in African Americans were computed for samples included in the 1000 Genomes Project dataset.32 The scores were significantly different according to the populations (figure 1). The African group had the lowest genetic risk scores and there was an increasing gradient from Africans to African Americans, Asians, Hispanics, and Europeans. This distribution mimics the worldwide disease prevalence. Additionally, the genetic burden scores of our MS case-control datasets fit within the observed ancestral gradient, and patients were more loaded than controls in both African Americans and white Americans (p = 4.11e-13 and p = 0.031, respectively).
Figure 1. MS genetic burden scores in the 1000 Genomes Project population.
Based on the 8 non-MHC MS SNPs for which the association with MS were replicated in African Americans (p < 0.01), aggregation of those SNPs was computed for individuals in the 1000 Genomes Project dataset, as previously described.40 Luhya in Webuye, Kenya, and Yoruba in Ibadan, Nigeria, are combined as “African”; Han Chinese in Beijing, China, Han Chinese South, and Japanese in Tokyo, Japan, are combined as “Asian”; Colombian in Medellin, Colombia, Mexican Ancestry in Los Angeles, CA, and Puerto Rican in Puerto Rico are combined as “Hispanic”; and Toscani in Italy, Iberian populations in Spain, Finnish from Finland, British from England and Scotland, and Northern and Western Europeans from Utah are combined as “European.” Additionally, genetic burden scores of our case-control dataset in AA and WA are included. A worldwide gradient of genetic burden scores was seen, matching MS prevalence in the world. Patients were more loaded than controls in both AA and WA. The p values at the top of the figure indicate the statistical significance of comparisons in groups next to each other (Wilcoxon rank sum test). The p value at the middle, bottom was calculated by Kruskal-Wallis test. Uncorrected p values are shown. AA = African Americans; ASW = African ancestry from southwest United States; K-W = Kruskal-Wallis test; MHC = major histocompatibility complex; MS = multiple sclerosis; SNP = single nucleotide polymorphism; WA = white Americans.
DISCUSSION
This is the largest genetic study of an MS dataset of non-European ancestry. After adjusting for admixture, we replicated established HLA-DRB1 allelic associations as well as 8 of the 73 validated non-MHC MS SNPs discovered in European populations. African American patients with MS carry the 2 most prominent susceptibility HLA-DRB1 alleles found in Europeans, HLA-DRB1*15:01 and HLA-DRB1*03:01, as well as the African allele HLA-DRB1*15:03.8,20,31 The role of HLA-DRB1*04:05 observed in this study was also reported in other populations, including Japanese and Sardinian.33,34 We also confirmed the protective association of HLA-A*02:01 in African Americans, independent of the HLA-DRB1 risk alleles. Also consistent with previous reports,35 a lower frequency for HLA-DRB1*11 was observed. Although the protective association of HLA-DRB1*09:01 lost statistical significance after adjustment, the direction of the effect matched previous results in Asians.33,36 No association of HLA-DQB1 alleles was detected for African American patients with MS, consistent with our previous study highlighting the primary role of HLA-DRB1 vs HLA-DQB1.31 Interestingly, all of these MS-associated alleles are found in various HLA haplotypes in different populations,13,37 consistent with a primary role for HLA-DRB1 and HLA-A alleles rather than other HLA genes. When the effect size of HLA-DRB1*15 was compared in our African American and white American datasets, it was different (Q test p = 1.65e-06). Whether this difference reflects a true biological effect of the MHC in these 2 populations remains to be addressed.
Outside the MHC, the number of statistically associated MS SNPs in African Americans was far lower than the mean number of MS-associated SNPs in the bootstrapped white American dataset. There are 3 possible explanations for this partial replication. First, differences in minor allele frequencies (MAF), especially for those SNPs with lower MAF in African Americans, could have affected the power to detect the associations. Indeed, among the MS SNPs with a MAF less than 15% in African Americans, only the SNP rs3129889 in the HLA-DRB1 region was replicated. Second, the smaller size of African haplotype blocks32,38 may have caused some of the MS SNPs originally identified in Europeans to fail tagging putative causative variants in African Americans. For example, our previous study in African Americans showed associations for 2 SNPs in EVI5 (rs10735781 and rs6680578).21 Although the SNP genotyped in this study (rs11810217) locates between rs10735781 and rs6680578 and appears to be within the same haplotype block in Europeans (measures of LD, R2, and d′ are strong among the 3 SNPs), the R2 between rs11810217 and the other variants, rs10735781 and rs6680578, is extremely low in African Americans. Similarly, we can attribute the lack of replication for CD6 and TYK2, despite suggestive association signals in our previous report,21 to SNP selection and LD haplotype differences between African Americans and Europeans. The observed opposite direction of the association of the SNP rs2300603 in BATF can be interpreted as a possible difference in the LD pattern between this SNP and the putative causative variants in African Americans, considering the observed smaller haplotype blocks at the locus in African Americans than in Europeans.39 It is important to note that for the 57 non-MHC SNPs reported by Sawcer et al.,8 the number of possible genes corresponding to one SNP signal ranges between 1 and 33. Finally, genetic heterogeneity across populations is an additional conceivable explanation for absence of replication, at least for some of the loci (table e-2). It is of interest that the individual accumulation of genetic risk alleles across populations for this minimal set of common risk alleles matches the disease prevalence in each ancestral group, suggesting that the global frequency of the disease is influenced, at least in part, by the distribution of risk alleles and that these validated SNPs tag causative variants across different populations.
Building on the success of the GWAS approach, meta-analyses and dense genotyping in large datasets will most likely result in the rapid identification of additional risk alleles. Studies comparing the genomes of different ancestral groups may prove to be highly informative by leveraging the differences in genome LD patterns and clinical phenotypes.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the MS patients and healthy controls who participated in this study. The authors acknowledge the contributions of H. Mousavi and R. Guerrero (UCSF) for sample processing and management, R. O’Shea and J. More (UCSF) for the management of the clinical data, and L. Maddireddy (UCSF) for support in writing R commands.
GLOSSARY
- AIM
ancestry informative marker
- CI
confidence interval
- GWAS
genome-wide association studies
- HLA
human leukocyte antigen
- LD
linkage disequilibrium
- MAF
minor allele frequencies
- MHC
major histocompatibility complex
- MS
multiple sclerosis
- OR
odds ratio
- PC
principal component
- SNP
single nucleotide polymorphism
Footnotes
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
Dr. Isobe participated in study design, performed experiments and statistical analysis, and drafted and edited the manuscript. Dr. Gourraud participated in study design, supervised and performed the statistical analyses, and critically revised the manuscript. Dr. Harbo conducted experiments and critically revised the manuscript. Ms. Caillier was responsible for sample preparation and management. Mr. Santaniello was responsible for data management. Mr. Khankhanian supported the statistical analyses. Mr. Maiers, Mr. Spellman, Dr. Cereb, and Dr. Yang contributed samples and data and critically revised the manuscript. Dr. Pando contributed data and critically revised the manuscript. Dr. Piccio, Dr. Cross, and Dr. De Jager contributed samples and critically revised the manuscript. Dr. Cree and Dr. Hauser contributed to the phenotypic characterization of the study participants and critically revised the manuscript. Dr. Oksenberg designed and supervised the study and edited the manuscript.
STUDY FUNDING
This study was supported by NIH grants RO1NS076492 and RO1NS046297. Recruitment of study participants and sample acquisition were supported by NMSS RG2899-D11 and RC2 GM093080. The National Marrow Donor Program is supported in part by funding from the Office of Naval Research N00014-11-1-0339 and the Health Resources and Services Administration (HRSA/DHHS contract HHSH234200637020C).
DISCLOSURE
N. Isobe received fellowship from the Uehara Memorial Foundation and has received funding from a postdoctoral fellowship for research abroad from the Japan Society for the Promotion of Science. P.-A. Gourraud is a recipient of the Nancy Davis Foundation young investigator award. H. Harbo is supported by the Research Council of Norway (grants 189639/196776). S. Caillier, A. Santaniello, P. Khankhanian, M. Maiers, and S. Spellman report no disclosures. N. Cereb is a CEO and cofounder of Histogenetics LLC, which performed HLA typing for this manuscript. S. Yang is a chairman and founder of Histogenetics LLC, which performed HLA typing for this manuscript. M. Pando reports no disclosures. L. Piccio is a Harry Weaver Neuroscience Scholar of the National MS Society (JF2144A2/1) and received support from the Fondazione Italiana Sclerosi Multipla (FISM) (2009/R/33). A. Cross reports no disclosures. P. De Jager is a Harry Weaver Neuroscience Scholar of the National MS Society (JF2138A1). B. Cree, S. Hauser, and J. Oksenberg report no disclosures. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafler DA, Compston A, Sawcer S, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007;357:851–862 [DOI] [PubMed] [Google Scholar]
- 3.Comabella M, Craig DW, Camina-Tato M, et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS One 2008;3:e3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranzini SE, Wang J, Gibson RA, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet 2009;18:767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet 2009;41:824–828 [DOI] [PubMed] [Google Scholar]
- 6.Jakkula E, Leppa V, Sulonen AM, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet 2010;86:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanna S, Pitzalis M, Zoledziewska M, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet 2010;42:495–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matesanz F, Gonzalez-Perez A, Lucas M, et al. Genome-wide association study of multiple sclerosis confirms a novel locus at 5p13.1. PLoS One 2012;7:e36140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 2009;41:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patsopoulos NA, Esposito F, Reischl J, et al. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol 2011;70:897–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bush WS, Sawcer SJ, De Jager PL, et al. Evidence for polygenic susceptibility to multiple sclerosis: the shape of things to come. Am J Hum Genet 2010;86:621–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gourraud PA, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: an up-to-date review. Immunol Rev 2012;248:87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugliatti M, Sotgiu S, Rosati G. The worldwide prevalence of multiple sclerosis. Clin Neurol Neurosurg 2002;104:182–191 [DOI] [PubMed] [Google Scholar]
- 15.Cree BA, Khan O, Bourdette D, et al. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology 2004;63:2039–2045 [DOI] [PubMed] [Google Scholar]
- 16.Cree BA, Reich DE, Khan O, et al. Modification of multiple sclerosis phenotypes by African ancestry at HLA. Arch Neurol 2009;66:226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barcellos LF, Begovich AB, Reynolds RL, et al. Linkage and association with the NOS2A locus on chromosome 17q11 in multiple sclerosis. Ann Neurol 2004;55:793–800 [DOI] [PubMed] [Google Scholar]
- 18.Reich D, Patterson N, De Jager PL, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet 2005;37:1113–1118 [DOI] [PubMed] [Google Scholar]
- 19.Caillier SJ, Briggs F, Cree BA, et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J Immunol 2008;181:5473–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McElroy JP, Cree BA, Caillier SJ, et al. Refining the association of MHC with multiple sclerosis in African Americans. Hum Mol Genet 2010;19:3080–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson BA, Wang J, Taylor EM, et al. Multiple sclerosis susceptibility alleles in African Americans. Genes Immun 2010;11:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MW, Patterson N, Lautenberger JA, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 2004;74:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seldin MF, Price AL. Application of ancestry informative markers to association studies in European Americans. PLoS Genet 2008;4:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz-Narvaez EA, Rosenberg L, Wise LA, Reich D, Palmer JR. Validation of a small set of ancestral informative markers for control of population admixture in African Americans. Am J Epidemiol 2011;173:587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lao O, Vallone PM, Coble MD, et al. Evaluating self-declared ancestry of U.S. Americans with autosomal, Y-chromosomal and mitochondrial DNA. Hum Mutat 2010;31:E1875–E1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kidd JR, Friedlaender FR, Speed WC, Pakstis AJ, De La Vega FM, Kidd KK. Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig Genet 2011;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang CW, Gajdos ZK, Korn JM, et al. Rapid assessment of genetic ancestry in populations of unknown origin by genome-wide genotyping of pooled samples. PLoS Genet 2010;6:e1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brynedal B, Duvefelt K, Jonasdottir G, et al. HLA-A confers an HLA-DRB1 independent influence on the risk of multiple sclerosis. PLoS One 2007;2:e664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson N, Hattangadi N, Lane B, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet 2004;74:979–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahana E. Epidemiologic studies of multiple sclerosis: a review. Biomed Pharmacother 2000;54:100–102 [DOI] [PubMed] [Google Scholar]
- 31.Oksenberg JR, Barcellos LF, Cree BA, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet 2004;74:160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, Altshuler D, Auton A, et al. ; 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka T, Matsushita T, Osoegawa M, et al. Association of the HLA-DRB1 alleles with characteristic MRI features of Asian multiple sclerosis. Mult Scler 2008;14:1181–1190 [DOI] [PubMed] [Google Scholar]
- 34.Cocco E, Sardu C, Pieroni E, et al. HLA-DRB1-DQB1 haplotypes confer susceptibility and resistance to multiple sclerosis in Sardinia. PLoS One 2012;7:e33972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramagopalan SV, Morris AP, Dyment DA, et al. The inheritance of resistance alleles in multiple sclerosis. PLoS Genet 2007;3:1607–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu W, James I, Carroll WM, Mastaglia FL, Kermode AG. HLA-DR allele polymorphism and multiple sclerosis in Chinese populations: a meta-analysis. Mult Scler 2011;17:382–388 [DOI] [PubMed] [Google Scholar]
- 37.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 2007;68:779–788 [DOI] [PubMed] [Google Scholar]
- 38.Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet 2004;36:S21–S27 [DOI] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 40.Gourraud PA, McElroy JP, Caillier SJ, et al. Aggregation of multiple sclerosis genetic risk variants in multiple and single case families. Ann Neurol 2011;69:65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.