Abstract
Periconceptional supplementation of folic acid to the diet of women is considered a great success for a public health intervention. Higher folate status, either by supplementation, or via the mandatory fortification of grain products in the United States, has lead to significant reduction in the incidence of neural tube defects. Besides birth defects, folate deficiency has been linked to a variety of morbidities, most notably to increased risk for cancer. However, recent evidence suggests that excess folate may be detrimental - for birth defect incidence or in the progression of cancer. How folate mediates beneficial or detrimental effects is not well understood. It is also unknown what molecular responses are elicited in women taking folate supplements, and thus experience a bolus of folate on top of the status achieved by fortification.
To characterize the response to a preconceptional regimen of supplementation with folinic acid, we performed gene expression profiling experiments on uterus tissue of pregnant mice with either wildtype alleles or targeted disruption at the folate receptor 4 locus. We observed that, depending on the genetic background, folinic acid supplementation affects expression of genes that contribute to lipid metabolism, protein synthesis, mitochondrial function, cell cycle, and cell activation. The extent of the response is strongly modulated by the genetic background. Finally, we provide evidence that folinic acid supplementation in the mutant paradigm affects histone methylation status, a potential mechanisms of gene regulation in this model.
Keywords: Epigenetics, Folate receptor, Folic acid, Gene expression, Genetic background, Microarray, Transcriptional response, Uterus
1. Background
Periconceptional folate supplementation has been a public health success story for the prevention of neural tube defects [1]. The seminal discovery by Smithells [2] was that the incidence of birth defects such as spina bifida was associated with low folate status. Subsequently, it was demonstrated that providing women who wish to become pregnant with folate supplements was correlated with a significantly lower incidence of neural tube defects [3], without any obvious detrimental effects [4]. In the last 15 years, many countries adopted the recommendation to encourage folate uptake in the periconceptual period as means of birth defect prevention. The use of folate supplements, and the introduction of mandatory fortification of grain products with folic acid in the United States in 1998, have resulted in significantly higher folate levels in serum and red blood cells [1]; these higher folate levels are credited with the concomitant reduction of birth defect incidence [5] in the last decade.
The mechanism by which folate prevents neural tube defects is still a mystery. Folate is a carrier for single-carbon moieties, with crucial roles for nucleotide synthesis, amino acid synthesis, as well as methylation reactions in the cell. Consequences of folate deficiency are best characterized in cancer research, where it is thought that low folate levels lead to insufficient nucleotide supply, increased incorporation of uracil into DNA, increased strand breaks, impaired DNA repair mechanisms, accumulation of mutations, and overall genome instability, a hallmark of neoplastic transformation [6, 7]. Furthermore, imbalances in DNA methylation were observed, which are also thought to contribute to genome instability. Therefore, low folate status is considered to be a risk for the development of neoplasms, implying that folate fortification may alleviate that risk and lead to beneficial effects for public health beyond prevention of birth defects.
Recently, concerns have arisen that high folate status may be associated with consequences that are not unequivocally advantageous [8]. Folate can act as a mitogen for cancer cells [9], and thereby stimulate the growth of pre-existing neoplasms. In patients with a history of colorectal adenoma, folic acid was associated with greater risk for multiple adenomas and non-colorectal cancer [10]. Tumors originating from ovarian and prostate cancer express high levels of folate receptors, indicating that those cells are exceptionally adept at scavenging folate to promote their proliferation. In fact, this feature has been exploited in the design of targeted chemotherapeutic agents [11], as well as in the invention of molecular tools to visualize folate receptor-expressing tumors during surgery [12]. High folate status clearly constitutes a concern with respect to the progression of such tumors. In the developing rodent embryo, evidence has recently been presented that very high folate levels in the dam can lead to an increase in birth defect frequency [13]. Furthermore, in some paradigms of genetic neural tube defect susceptibility, higher folate appeared to be detrimental and lead to higher neural tube defect incidence compared to lower folate status [14]. Together, these data suggest that high folate consumption may not be entirely beneficial.
On the molecular level, folate status has been associated with gene expression [15]. Initially, folate-dependent differences in gene expression levels were attributed to epigenetic events due to the presumed effects of folate on DNA methylation [16, 17]. Yet, methylation reactions shape other domains of the epigenome besides DNA methylation, and evidence has arisen that changes in gene expression occur in response to folate deficiency or supplementation without affecting DNA methylation [18, 19]. Many of these studies focused on cancer. However, it is reasonable to assume that the highest folate levels would be found in women who, because they want to become pregnant and prevent potential birth defects, take preconceptional folate supplements in addition to folate intake from fortified foods. The molecular consequences of such a bolus of folate, for instance on the female reproductive tract, are not well understood. The primary interface between mother and developing embryo is the endometrium of the uterus, where the nutritional interaction is established during the process of implantation, a crucial step for reproductive success. Whether folate modulates this interaction is currently unclear. Therefore, we decided to investigate the effects of preconceptional folate supplementation on the female reproductive tract, focusing on the uterus at the time of implantation. Using a mouse model system, we demonstrate a folate effect on gene expression in the uterus, and a broader response in mice lacking folate receptor 4, and suggest an effect on histone methylation.
2. Materials and Methods
2.1. Folate supplementation and tissue preparation
Purina 5001 (Lab Diet, Inc.) was used as diet for all mice. Female mice of the C57BL/6 strain (or from a strain carrying the Folr4 mutation on the C57BL/6 genetic background) were injected intraperitoneally with folinic acid at a dose of 12.5mg/kg body weight, or with saline as control. Dams received two injections, at 10 and 3 days before mating, respectively. Dams were mated to C57BL/6 males; noon of the day when presence of a vaginal plug could be detected was designated day 0.5 of gestation. At 3.5 days of gestation, mice were euthanized, uterus tissue was dissected, and either used to extract RNA, or processed for histology. RNA was extracted using Trizol (Invitrogen Life Technologies), quantified on a Nanodrop ND-1000 spectrophotometer, and stored at -80°C for further analysis.
2.2. Mice lacking Folr4
Mice with a mutation in the Folr4 gene were generated via homologous recombination in mouse embryonic stem cells. For this purpose, a construct was made where the first two coding exons of the gene were flanked by loxP recombination sites, followed by the gene for neomycin resistance and third loxP site. Electroporation of the construct, selection of ES cell clones with the correct constellation at the Folr4 gene, and generation of chimeric mice were done as described [20]. Homozygous mice carrying the floxed locus were viable. To generate a null mutation, homozygous Folr4FL mice were crossed to mice expressing Cre recombinase in the oocyte [21], resulting in heterozygous offspring with one allele of the Folr4 gene exhibiting a deletion of the first two coding exons. PCR analysis of the locus produced an amplicon of 413 bp indicative of the deletion, compared to an amplicon of 1365 bp from the wildtype allele. Mice with this Folr4 null allele were backcrossed to the C57BL/6 genetic background for more than 10 generations. Heterozygous mice were used to generate the homozygous Folr4 mutant female mice used in this study. All procedures involving animals were approved by the Institutional Animal Care and Use Committee.
2.3. Gene expression profiling
Affymetrix Mouse 430 2.0 microarrays were used to perform gene expression profiling experiments as described previously [22]. We processed three RNA samples from wildtype C57BL/6 dams, four from wildtype C57BL/6 dams treated with folinic acid, three from Folr4 mutant dams, as well as four from Folr4 mutant dams supplemented with folinic acid for array hybridization. All samples were obtained from individual animals. Tissue dissection and RNA extraction was performed on the same day, and all further processing, including array hybridizations, were done in parallel to ensure technical consistency between samples. Arrays were normalized as described [22], and array data were processed to identify differential gene expression using CyberT software [23]. We used the implementation of the software available at http://cybert.ics.uci.edu/, employing a Bayes-regularized t-test after applying variance stabilized normalization [24]. Annotation for molecular function was performed using GO terms, with manual curation based on MGI (http://www.informatics.jax.org/) and GeneCards (http://www.genecards.org/). Results from probes for non-coding RNAs, or probes where gene identity was missing, were omitted. Data for one probe per gene were retained for the final list. When a gene was represented by more than one probe, duplicates were eliminated based on criteria of high P-value, low expression and low fold-change. In the very few cases where duplicates displayed opposite directions of change for the same gene, the gene was removed from consideration. Pathway analysis was performed using the DAVID (http://david.abcc.ncifcrf.gov/) [25] suite of tools; adjusted p-values given in the results section (padj) refer to p-values obtained after Benjamini-Hochberg correction. Visualization of expression differences was done in Microsoft Excel. Lists of folate- and genotype-responsive probes are presented in Supplementary Table 1. All microarray analyses reported here have been deposited to GEO (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE36974).
2.4. Expression measurements
Validation of expression measurements for selected genes was performed by real-time quantitative PCR on an Applied Biosystems 7900 system as described previously [26]. Expression measurements were generated using the gene for DNA polymerase subunit epsilon 4 as the reference gene for normalization [26]. Primer sequences are listed in Supplemental Table 2. Three replicate assays per sample were performed, and statistical evaluation of PCR data was done via two-tailed t-test based on at least n=4 samples; samples included those used for microarray as well as independently generated samples.
2.5. Histology
Paraffin-embedded tissue sections [27] or cryosections [28] were processed for in situ hybridization with digoxigenin-labeled antisense riboprobes as described previously. For immunohistochemistry with antibodies against histone H3 lysine K4 trimethylation (Millipore, Billerica, MA), frozen uterus tissue was sectioned at 16μm, fixed in 1% formaldehyde in PBS, washed, blocked in PBS containing 2% goat serum and 0.2% BSA, incubated with primary antibody over night at 4°C, washed in PBS, and developed with goat-anti rabbit-Alexa 594 secondary antibody (Invitrogen, Carlsbad, CA). Brightfield images were recorded on a Leica Z16 macroscope, whereas fluorescent images were captured on a Leica TCS SP5 confocal microscope, using the same instrument settings for all specimens, respectively.
3. Results
The focus of folate supplementation before and at onset of pregnancy has always been on pregnancy outcome and prevention of birth defects. Very little attention, however, has been paid to molecular responses to folate in the expecting mother, where the consequences of a boost in folate levels are not well understood. To gain insights into the molecular consequences of preconceptional folate supplementation on female reproductive tissues, we conducted an experiment where mice received two injections of folinic acid (a stable folate derivative) at 10 and 3 days before mating, respectively. Considering that implantation is a crucial step in reproductive success, we focused on uterus tissue at 3.5 days after mating, the time when the blastocyst reaches the uterus. With the main emphasis on the response to folinic acid, we processed uterus tissue for transcription profiling via microarrays, and analyzed (i) gene expression in the uterus of wildtype mice in response to folinic acid supplementation, (ii) gene expression in the uterus in response to folinic acid in the presence of a genetic liability in form of a mutation at the gene for Folate receptor 4, and (iii) explored to what extent supplementation with folinic acid may be able to reverse expression changes brought about by the Folr4 deficiency.
3.1. Identification of folate-responsive genes in peri-implantation mouse uterus
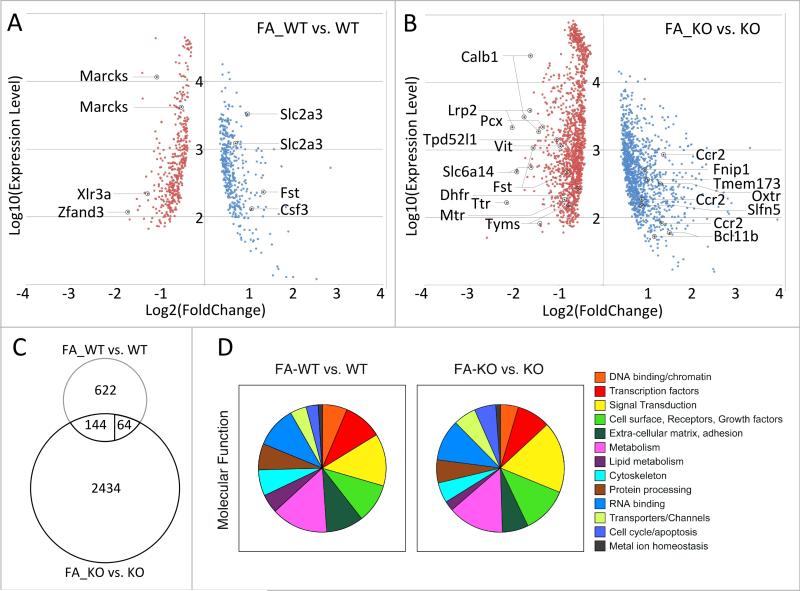
The response of mouse uterus tissue to folinic acid supplementation is shown in Figure 1A, based on statistical significance in a Bayes-regularized t-test (p<0.05). We found a moderate magnitude of the transcriptional response to folate supplementation before pregnancy, with 188 gene probes showing a folate-dependent 1.5-fold or higher increase in expression level, and 246 gene probes exhibiting a decrease of expression. Lowering the fold-change cutoff to 1.2-fold in either direction (using the same statistical test), we detected a total of 830 gene probes that change expression in response to folate, with 406 probes showing increase, and 424 probes showing a decrease in expression levels. This suggested that the response to folate in this paradigm is comprised by relatively small changes of expression levels for a moderately large number of genes. Genes of the folate cycle were unaffected, as were genes encoding methyltransferases. We chose five genes (2 up- and 3 down-regulated) that represented different levels of expression for validation of the array-based expression changes by qPCR. In all cases, we were able to confirm the direction of the expression change with statistical significance as determined by a t-test (Table 1); the assay for Zfand3 did not reach statistical significance.
Figure 1. Transcriptional responses to folate supplementation in the uterus.
A. Comparison of gene expression profiles in normal (WT) and folinic acid (FA) supplemented normal (FA_WT) mice revealed a moderate response consisting of predominantly small changes: at a 1.2-fold or greater difference between average expression levels in each group, 830 probes were altered (406 with increased expression level, 424 with decreased expression level); at a 1.5-fold or greater difference, this number was reduced to 434 probes (188 increased, 246 decreased), whereas at 2-fold or greater change, only 89 gene probes were detected (47 increased, 42 decreased). Of the 830 gene probes, 4 passed a Benjamini-Hochberg correction (padj<0.1). All probes with a p-value below 0.05 in a Bayes-regularized t-test are shown, with their respective expression level on the array plotted against the observed fold change. Marked gene probes correspond to those genes validated by quantitative PCR (see Table 1). B. Comparison of gene expression profiles in Folr4-mutant (KO) and folinic acid (FA) supplemented mutant (FA_KO) mice demonstrated a more vigorous transcriptome response: at a 1.2-fold or greater difference between average expression levels, 2642 probes were altered (1177 expression level increased, 1465 expression level decreased); at a 1.5-fold or greater difference, this number was reduced to 1786 probes (885 increased, 901 decreased), whereas at 2-fold or greater change, only 451 gene probes were detected (299 increased, 152 decreased). Of the 2642 gene probes, 448 passed a Benjamini-Hochberg correction (padj<0.1). All probes with a p-value below 0.05 in a Bayes-regularized t-test are shown in the plot. Marked gene probes correspond to those genes validated by quantitative PCR (see Tables 1 and 3). C. Comparison of the response to folinic acid supplementation between wildtype and Folr4 mutant tissue. Only 208 gene probes are common between the two responses; out of those, 144 change expression in a fashion concordant between the two genotypes, whereas 64 show altered expression in a discordant manner. D. Annotation of differentially expressed genes by molecular function comprising the transcriptional response to folinic acid supplementation in either wildtype (FA_WT vs. WT) or Folr4 mutant (FA_KO vs. KO) tissue. Note the large segments for metabolism (including oxidative phosphorylation and mitochondrial function), protein processing (including localization), and RNA binding (including ribonucleoprotein complexes).
Table 1. Gene expression analysis by RT-PCR.
Representative genes at different expression levels were chosen to validate the microarray results. For the comparison of wildtype vs. folinic acid-supplemented wildtype, 5 genes are shown. We observed concordant expression changes in the array and the qPCR, although not all PCR assays reach statistical significance in a t-test. For the comparison of mutant vs. folinic acid-supplemented mutant, 6 genes are shown that represent differentially regulated genes at various expression levels according to the array data. Furthermore, three genes of the folate pathway were among the genes of the folinic acid response: Dhfr, Mtr, and Tyms all show decreased expression in response to folinic acid in the mutant.
| FA_WT vs. WT | ||||
|---|---|---|---|---|
| Microarray | qRT-PCR | |||
| Gene Symbol | Fold change | p-value | Fold change | p-value |
| Csf3 | 2.07 | 0.001 | 2.15 | 0.029 |
| Marcks | -2.11 | 0.000 | -1.34 | 0.027 |
| Scl2a3 | 1.95 | 0.002 | 2.02 | 0.032 |
| Xlr3a | -2.42 | 0.000 | -2.49 | 0.020 |
| Zfand3 | -3.29 | 0.000 | -1.17 | 0.051 |
| FA_KO vs. KO | ||||
|---|---|---|---|---|
| Microarray | qRT-PCR | |||
| Gene Symbol | Fold change | p-value | Fold change | p-value |
| Bcl11b | 2.80 | 0.002 | 5.00 | 0.009 |
| Calb1 | -3.41 | 0.000 | -5.05 | 0.024 |
| Lrp2 | -4.13 | 0.000 | -5.95 | 0.044 |
| Oxtr | 2.41 | 0.000 | 2.64 | 0.053 |
| Slc6a14 | -3.85 | 0.000 | -7.97 | 0.023 |
| Ttr | -4.49 | 0.000 | -8.37 | 0.001 |
| Dhfr | -1.48 | 0.029 | -1.53 | 0.049 |
| Mtr | -1.83 | 0.012 | -1.91 | 0.030 |
| Tyms | -2.12 | 0.000 | -2.49 | 0.005 |
In order to determine which biological pathways may be affected by folate supplementation, we used the genes derived with greater than 1.2-fold change as well as statistical significance. Annotation for molecular function of the products encoded by differentially expressed genes revealed (Figure 1D) that molecules involved in metabolism (11.4%), signal transduction (10.7%), RNA binding (8.5%), transcriptional regulation (8.3%), as well as receptors and growth factors (8.1%) comprised the largest groups. Using the DAVID suite of tools (http://david.abcc.ncifcrf.gov/) [25] for further functional annotation, we analyzed gene groups with increased or decreased expression separately; the results are summarized in Table 2, section A. Among the genes with increased expression, we found enrichment for the gene ontology term ‘lipid metabolic process’. In the group of genes with decreased expression, the gene ontology terms ‘ribonucleoprotein complex’ , ‘mRNA metabolic process’ and ‘structural constituent of ribosome’ were detected. Together, these results suggest that folinic acid supplementation decreases the expression of components of the protein synthesis mechanism in cells of the uterus, and may increase lipid metabolism.
Table 2. Summary of top-scoring categories in DAVID pathway analysis.
Pathway analysis was performed using the DAVID (http://david.abcc.ncifcrf.gov/) [25] suite of tools; adjusted p-values (padj) refer to p-values obtained after Benjamini-Hochberg correction. All analyses were performed with the default settings and classification stringency at “medium”.
| A: Folinic acid-supplemented wildtype vs. wildtype | |||||
|---|---|---|---|---|---|
| increase | decrease | ||||
| GO term | padj | genes | GO term | padj | genes |
| lipid metabolic process | 0.011 | 29 | ribonucleoprotein complex | 4.3×10-06 | 30 |
| structural constituent of ribosome | 2.5×10-05 | 11 | |||
| mRNA metabolic process | 6.4×10-04 | 20 | |||
| vasculature development | 0.016 | 15 | |||
| B: Folinic acid-supplemented Folr4 mutant vs. Folr4 mutant | |||||
|---|---|---|---|---|---|
| increase | decrease | ||||
| GO term | padj | genes | GO term | padj | genes |
| immune system process | 1.9×10-26 | 115 | structural constituent of ribosome | 2.5×10-59 | 83 |
| leukocyte activation | 1.5×10-09 | 38 | cell cycle | 1.5×10-10 | 83 |
| T cell activation | 1.3×10-06 | 23 | m-phase | 2.3×10-10 | 51 |
| regulation of immune response | 2.2×10-09 | 35 | mitochondrion | 2.6×10-10 | 141 |
| cytokine binding | 4.8×10-06 | 20 | oxidative phosphorylation | 0.064; 5.8×10-14 | 20 |
| mitochondrial lumen | 3.5×10-03 | 22 | |||
| C: Folr4 mutant vs. wildtype | |||||
|---|---|---|---|---|---|
| increase | decrease | ||||
| GO term | padj | genes | GO term | padj | genes |
| structural constituent of ribosome | 1.7×10-21 | 36 | defense response | 8.7×10-05 | 25 |
| mitochondrion | 0.1 | 33 | immune response | 1.8×10-07 | 31 |
| D: Folinic acid-supplemented Folr4 mutant vs. Folr4 mutant compared to Folr4 mutant vs. wildtype | |||||
|---|---|---|---|---|---|
| increase | decrease | ||||
| GO term | padj | genes | GO term | padj | genes |
| immune system process | 6.6×10-06 | 24 | structural constituent of ribosome | 2.4×10-29 | 36 |
| mitochondrion | 0.036 | 33 | |||
| glutathione transferase activity | 0.0054 | 6 | |||
3.2. Identification of folate-responsive genes in Folate receptor 4-deficient mice
Interestingly, we observed a much broader transcriptional response to folinic acid supplementation in uteri derived from mice with a deletion of the Folr4 gene (Figure 1B). Applying the same criteria as before for wildtype dams (more than 1.2-fold change in either direction, p<0.05), we detected 2642 gene probes in Folr4 mutants that responded to folate, with 1177 probes increasing, and 1465 probes decreasing in their expression levels. 451 probes showed more than 2-fold change; 299 of those showed increased expression in response to folate supplementation, whereas 152 responded with a decrease. The qPCR validation of the expression changes of nine genes representing different levels of expression on the array is shown in Table 1.
Among genes of the folate cycle, we find Dhfr, Mthfd1, Mthfd1l, Mthfd2, Mthfr, Mtr, Shmt1, Slc46a1, and Tyms mildly deregulated; only Tyms and Mthfd2 showed more than a 2-fold expression change. qPCR analysis of Dhfr, Mtr and Tyms confirmed direction and magnitude of the expression change initially observed in the microassay experiment. Notably, we also detected downregulation of Lrp2 (Megalin), which can bind soluble folate receptors; this change in expression was apparent on the array as well as in qPCR assays. We observed moderate deregulation of genes encoding methyltransferases, but no coordinate change in one direction: Ezh1, Mettl7a1, Mll5, Pcmtd1, Pcmtd2, Setd3, Suv420h1, Suv420h2, Tgs1 exhibited an average fold change of 1.51; and Alkbh8, Dnmt1, Lcmt1, Mtr, N6amt2, Nsd1, Prmt1, Setdb1, Trmt112 an average fold change of -1.51.
Similar to the situation observed in wildtype mice, the folate response In Folr4 mutant uterus was primarily composed of moderate magnitude changes for a large group of genes. Figure 1D shows that the distribution of encoded products follows a similar pattern as in the comparison of supplemented to unsupplemented wildtype females, although genes encoding ‘signal transduction’ molecules (15.2%) were enriched compared to wildtype. Groups comprising ‘metabolism’ (11.7%), 'receptors and growth factors’ (9.8%) and ‘RNA binding’ (8.7%) were similarly represented, but in absolute number, there were 3-4 fold more genes responding to folinic acid in the Folr4 mutants.
We obtained further annotation information using DAVID tools, analyzing the groups of up- and down-regulated genes separately (Table 2, section B). Among genes with increased expression in response to folinic acid, gene ontology terms related to immune response were highly enriched. In the group of genes that exhibit a decrease in expression as response to folinic acid supplementation in the mutant, the term ‘structural constituent of ribosome’ was strongly represented, paralleling the supplementation response in wildtype mice. However, considering individual gene identities, we noted relatively little concordance in the response to folinic acid between wildtype and Folr4 mutants (Figure 1C): only 208 gene probes were found to overlap between the two supplementation paradigms, and of these, only 144 showed a concordant direction of change.
Interestingly, strong associations to the GO terms ‘cell cycle’, ‘M-phase’, ‘mitochondrion’, ‘oxidative phosphorylation’ and ‘mitochondrial lumen’ were found among the genes with decreased expression. Together, this suggested that folinic acid supplementation in Folr4 mutant mice, in addition to eliciting a much broader transcriptomic response, may also lead to a dampening of cellular processes such as mitosis, possibly through decreased functionality of ribosomes which would affect protein synthesis, and to a reduction in mitochondrial functionality which could affect energy homeostasis of cells. The intriguing finding of extensive differences in the folinic acid response prompted us to investigate in more detail the differences between tissues from unsupplemented wildtype and mutant females.
3.3. Uterus gene expression modulated by a mutation in the gene for Folate receptor 4
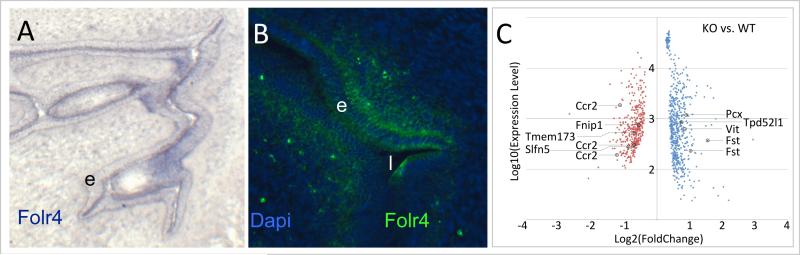
Folr4 is expressed in a restricted pattern in cells of the endometrium in uterus tissue obtained from a wildtype mouse (Figure 2 A and B). The Folr4 mutation deletes the first two coding exons of the Folr4 gene. Homozygous Folr4 mutant mice can be recovered from matings of heterozygote mice at Mendelian ratio. There was no evidence of birth defects, nor of overt morphological differences in the uterus at the time of isolation. Allowing a minimum fold-change of 1.2-fold in either direction at p<0.05 in a Bayes-regularized t-test, we detected 992 gene probes with altered expression when comparing Folr4 mutant to wildtype: 606 with increase, and 386 with decrease of expression (Figure 2C). At 2-fold change, 87 probes were detected, 40 with increase and 47 with decrease of expression. Such a moderate response is consistent with the fact that Folr4 is not expressed throughout the uterus, but limited to the endometrium and solitary cells in the uterine parenchyma. Other folate receptors and folate pathway genes were not changed in their expression. Using DAVID, we find that the gene ontology term ‘structural constituent of ribosome’ is significantly enriched in the group of genes with increased expression, whereas the terms ‘defense response’ and ‘immune response’ were found to be enriched in the group of genes with decreased expression in the Folr4 mutant. Results from the pathway analyses are summarized in Table 2 section C.
Figure 2. Expression of Folr4 in the uterus and altered gene expression in Folr4 mutant uterus.
A. Expression of Folr4 mRNA in the uterus at 3.5 days of gestation as visualized by in situ hybridization. Folr4 mRNA was detected in cells of the endometrium. B. Expression of Folr4 protein visualized by immunohistochemistry. Similar to the in situ hybridization, Folr4 protein expression (green) is limited to the endometrium. In addition a few solitary cells in the uterine parenchyma were detected, suggesting that the antibody is more sensitive than the DIG-labeled riboprobe under these conditions; e, endometrium; l, lumen of the uterus. C. Transcriptome analysis comparing gene expression in uterus tissue from Folr4 mutant mice (KO) to uterus tissue of wildtype mice (WT). At a 1.2-fold or greater difference between average expression levels in each group, 992 probes were altered (606 with increased expression level in the mutant, 386 with decreased expression level); at a 1.5-fold or greater difference, this number was reduced to 567 probes (305 increased, 262 decreased), whereas at 2-fold or greater change, only 87 gene probes were detected (40 increased, 47 decreased). Of the 992 gene probes, 20 passed a Benjamini-Hochberg correction (padj<0.1). All probes with a p-value below 0.05 in a Bayes-regularized t-test are shown, with their respective expression level on the array plotted against the observed fold change. Marked gene probes correspond to those genes examined by quantitative PCR.
This suggested that the lack of Folr4 may have a rather moderate effect on the development of uterus tissue, consistent with the absence of overt morphological differences in uterus tissue of mutant dams compared to wildtype. Furthermore, the decreased expression of genes associated with the term ‘immune response’ and ‘defense response’ in the mutant may be related to the expression of Folr4 on regulatory T-cells [29].
3.4. Reversal of genotype-dependent changes by folate supplementation
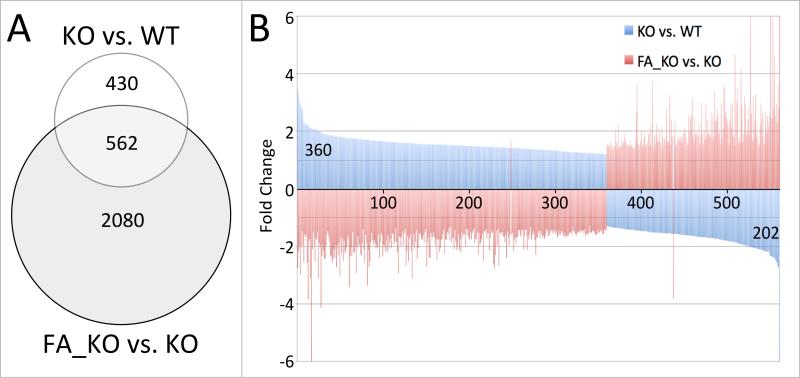
When analyzing genotype-mediated differential gene expression in Folr4 mutant dams, we noted that 562 gene probes (representing 451 genes) were also responsive to folinic acid supplementation (Figure 3A), changing their expression levels between normal mutant and supplemented mutant. Intriguingly, except for 2 genes, all those gene probes show a statistically significant discordant response in the two comparisons: 359 of 360 probes (representing 279 genes) with increased expression in the unsupplemented mutant compared to wildtype displayed a decrease of expression in the folinic acid-supplemented mutant compared to the unsupplemented mutant (Figure 3B). Likewise, 201 of 202 probes (representing 172 genes) with decreased expression in the mutant when compared to wildtype exhibited an increase of expression in the folinic acid supplemented mutant uterus. The two genes that do not conform are Cxcl17 and Pdgfd. In total, approximately 57% of the gene probes showing changes in expression attributable to genotype also respond to folinic acid supplementation in the mutant; notably, the magnitude of changes imposed by the mutation is comparable to the magnitude of changes induced by folinic acid in the mutant: absolute fold-changes for each gene are highly similar, yet, the direction of change is back towards levels comparable to unsupplemented wildtype.
Figure 3. Folinic acid can correct changes in expression caused by the Folr4 mutation.
A. Comparison of statistically significant responses (p<0.05 in a Bayes-regularized t-test) in Folr4-/- and normal dams (KO vs. WT, 992 probes), and in FA-supplemented Folr4-/- and non-supplemented Folr4-/- dams (FA_KO vs. KO, 2642 probes), respectively, revealed 562 probes that responded under both conditions. B. These 562 probes showed discordant directions for the changes in expression levels when compared between the two conditions: 360 probes show increased expression in the mutant vs. the wildtype, whereas 202 show decreased expression. These probes exhibit the opposite expression change when comparing the supplemented mutant to the mutant (except for 2 probes), indicating that FA can, in part, rectify the effect of the Folr4 mutation. Y-axis: fold-change between average expression levels for any given gene in each comparison.
Interestingly, gene ontology terms ‘structural constituent of ribosome’ and ‘mitochondrion’, which were associated with increased gene expression levels due to mutant genotype, are now found in the group of genes with decreased expression in response to folinic acid. Conversely, the term ‘immune system process’ attached to genes with decreased expression level due to mutant genotype, but these are now found reversed to normal by folinic acid supplementation (Table 2, section D).
We sought to confirm this finding of inverse direction of change for selected genes by measuring expression levels by quantitative real-time PCR. Eight genes were chosen for this analysis: Fst, Pcx, Tpd52l1, and Vit are four genes that were up-regulated in the mutant compared to wildtype, whereas Ccr2, Fnip1, Slfn5, and Tmem173 represent four genes with decreased expression in the mutant (Figure 2C). In the microarray experiment, all eight genes showed a reversal of the initial expression levels in the mutant by folinic acid supplementation. In the validation study (Table 3), we were able to confirm the initial observations: Fst, Pcx, Tpd52l1, and Vit expression levels were increased in the mutant compared to the wildtype, and Ccr2, Slfn5, Fnip1, and Tmem173 levels were decreased, although the comparison between groups did not reach statistical significance for the latter two genes. As in the microarrays, opposing direction of changes in expression of these genes were found when comparing folinic acid-supplemented mutant samples to non-supplemented mutant samples: supplementation decreased Fst, Pcx, Tpd52l1, and Vit, and increased Ccr2, Fnip1, Slfn5, and Tmem173. Together, the results demonstrate the reversal of mutation-dependent molecular alterations by folinic acid supplementation.
Table 3. Folinic acid-mediated reversal of gene expression changes in Folr4 mutant mice.
We observed 562 gene probes that appear to reverse the expression change elicited by the Folr4 mutation when supplemented with folinic acid. Eight genes were chosen for validation by qPCR: four genes that were initially up-regulated in the mutant vs. the wildtype, yet showed a decrease in expression in folinic acid-supplemented mutants, and four genes with the opposite pattern of gene expression changes: reduced expression in mutant compared to wildtype, and increased expression in folinic acid supplemented mutant compared to unsupplemented mutant. In all cases, the direction of the expression change could be confirmed by qPCR, although not all qPCR assays reached statistical significance in a t-test.
| Microarray | qRT_PCR | |||||||
|---|---|---|---|---|---|---|---|---|
| KO vs WT | FA_KO vs KO | KO vs WT | FA_KO vs KO | |||||
| Gene Symbol | Fold change | p-value | Fold change | p-value | Fold change | p-value | Fold change | p-value |
| Fst | 2.92 | 0.042 | -2.98 | 0.048 | 3.31 | 0.018 | -3.28 | 0.130 |
| Pcx | 1.85 | 0.009 | -2.56 | 0.035 | 2.13 | 0.042 | -5.03 | 0.026 |
| Tpd52l1 | 1.67 | 0.008 | -2.07 | 0.003 | 1.62 | 0.023 | -1.49 | 0.296 |
| Vit | 1.82 | 0.041 | -1.96 | 0.037 | 1.68 | 0.032 | -2.04 | 0.003 |
| Ccr2 | -2.19 | 0.017 | 2.55 | 0.012 | -1.98 | 0.002 | 2.27 | 0.028 |
| Fnip1 | -1.47 | 0.046 | 1.91 | 0.024 | -1.40 | 0.118 | 1.34 | 0.036 |
| Slfn5 | -1.84 | 0.042 | 1.81 | 0.047 | -1.53 | 0.021 | 1.24 | 0.361 |
| Tmem173 | -1.65 | 0.009 | 2.49 | 0.025 | -1.30 | 0.156 | 1.87 | 0.042 |
3.5. Histone methylation patterns in Folr4 mutant uterus
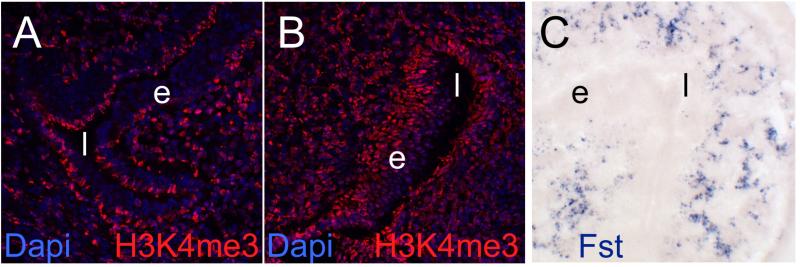
Finally, to explore potential mechanisms for folate-dependent gene regulation, we investigated whether the status of histone H3 lysine K4 trimethylation (H3K4me3), a histone modification typically associated with promoters of actively transcribed genes, would be responsive to folinic acid supplementation. To this end, we stained cryosections of uterus tissue with an antibody specific for trimethylation at lysine 4 of Histone 3 (H3K4me3); due to the stronger transcriptomic response to supplementation, we chose uterus tissue from mutant dams in this study and compared supplemented mutant to non-supplemented mutant. We observed a signal for H3K4me3 in uterus tissue from a non-supplemented Folr4 mutant mouse (Figure 4A) in many, but not all, cell nuclei on a representative section of uterus tissue. In parallel, staining for H3K4me3 in uterus tissue from a folinic acid-supplemented mutant revealed a brighter signal in nearly all cell nuclei of a section (Figure 4B). This suggests that supplementation with folinic acid leads to a stimulation of H3K4 trimethylation levels in uterus tissue, with strong representation in the endometrium. As the H3K4me3 staining pattern, which included the domain of Folr4 expression, also contained a majority of Folr4 non-expressing cells in the uterus, the altered histone methylation status may contribute to the altered gene expression levels in response to folinic acid.
Figure 4. Histone methylation responses to folinic acid supplementation.
A. Cryosection of a Folr4-/- uterus stained with an antibody against trimethylated histone H3 lysine 4 (H3K4me3, red). B. Folr4-/- uterus after supplementation with folinic acid (FA), stained, processed and imaged in parallel under identical conditions as in A. Stronger and more widely distributed staining indicates that folinic acid stimulates histone methylation in the uterus, in a pattern that appears broader than the expression pattern of Folr4. Sections in A, and B were counterstained with Dapi to reveal nuclei. C. Expression pattern of Follistatin (Fst) in uterus tissue at 3.5 days of gestation occurs in a pattern different from the expression of Folr4. Fst was never detected in the endometrium, but appears to be restricted to uterine glands.
4. Discussion
4.1. Epigenetic effects of folate supplementation in the mouse uterus
In this study, we demonstrate that preconceptional supplementation with folinic acid alters the transcriptome of uterus tissue in a mouse model. The overall responses were moderate in magnitude of expression changes, which may be related to the fact that we measured transcriptome responses seven days after the last injection of folinic acid. We find that components of RNA processing and protein synthesis pathways respond with decreased gene expression. Genes in these categories were also downregulated by folinic acid in mice carrying the Folr4 mutation, suggesting that under our experimental conditions, folinic acid may have a dampening effect on fundamental cellular processes. Such an action would be contrary to what one might expect in light of the notion that folate can act as a mitogen in cancer cells [9, 30][31]. Our histological observations provided no indication for the notion that folate acts as a mitogen in the uterus: cell density (as detected by nuclear staining with DAPI) did not differ between folinic acid-supplemented and unsupplemented specimen (data not shown). Yet, given that we investigated one time point only, this aspect warrants further investigation.
4.2. Identification of mitochondrial pathways as targets of folate supplementation
In Folr4 mutants, the uterine response to folinic acid supplementation involved a much larger number of genes, and this broader response contained, in top scoring categories, genes associated with immune function, cell cycle regulation, and mitochondrial function. Pathways related to immune function were increased by folinic acid in the mutant, whereas genes associated with cell cycle and mitochondrial function were decreased by supplementation. The link to mitochondrial function is in line with a recent report on gene expression profiles in response to dietary folate status in mice that were heterozygous for a mutation in Lrp6, a gene encoding a Wnt co-receptor [30]. In that study, genes for oxidative phosphorylation and mitochondrial function were found to respond to folate, both in embryonic crania at 9.5 days of gestation, as well as in adult liver. Our experiments, in Folr4 mutant mice, now provide further support for the idea that mitochondrial biology may be affected by folate. Folates themselves have been considered to be antioxidants [31], and as such could potentially influence levels of oxidative stress, a factor that has been implicated in birth defect etiology. Notable from our results is that in mutant tissue, folinic acid supplementation downregulated the expression of genes encoding components of the respiratory chain, suggesting that high doses of folinic acid may negatively affect mitochondrial energy metabolism, and possibly mitochondrial function in general.
4.3. Maternal genotype modulates the response to folate supplementation
Transcriptional responses to external stimuli, such as presence or absence of a micronutrient, are highly dependent on the status of the cell in question: pre-existing conditions, such as transcription factor repertoire, or epigenetic states, shape the capability of the cell to respond. The experiment conducted with Folr4 mutant mice underscores this notion. Under identical conditions of folinic acid concentration and schedule of supplementation, the Folr4-deficient mutants exhibited a much broader transcriptional response to folate compared to wildtype mice (see Figure 1C). Yet, when we compared the group of genes affected by folinic acid in the wildtype to the genes affected by folinic acid in the mutant, we found that the overlap between the responses was small, indicating that the mutant genotype fundamentally alters the transcriptome response to supplementation. This result in the mutant paradigm indicates that a significant interaction occurred between folate supplementation and genotype in the mutant female tissue. Thus, our work extends the cases of folate-dependent gene-environment interactions to uterus.
Interestingly, the much broader transcriptional response in Folr4 mutants occurs even though the majority of cells in the wildtype uterus do not express Folr4 (see Figure 2), indicating that other transporters are involved. Uterus tissue also expresses folate receptors Folr1 and Folr2, as well as the folate transporters Slc46a1 (proton-coupled folate transporter) and Slc19a1 (reduced folate carrier). Slc19a1, specifically, is ubiquitously expressed, which should permit folate entry into nearly every cell, particularly so under folate supplementation conditions. The wide distribution of the H3K4me3 signal in the supplemented mutant uterus underscores this interpretation, as it indicated that almost the entire tissue is capable of responding to folinic acid.
Under the theoretical assumption that the targeted disruption of the Folr4 gene has cell-autonomous effects, the parsimonious prediction would be that in the mutant, molecular responses to the genetic manipulation would be restricted to cells in the uterus that normally express only Folr4. One would further expect that cells negative for Folr4 respond similarly to folinic acid supplementation, regardless whether they are in a wildtype or mutant animal; considering that the majority of cells in uterus are Folr4 negative, one might even expect the overall tissue responses to be quite similar. However, our results clearly demonstrate that this is not the case: quantity (number of genes) and quality (types of genes and associated pathways) of the response is altered by Folr4 deletion. This prompts our conclusion that the Folr4 mutation has non-cell-autonomous effects on neighboring cells, and that these effects also alter the response to folinic acid supplementation. Intriguingly, more than 50% of the genes affected by Folr4 deficiency in the mutant (Figure 3A) respond to folinic acid supplementation, providing evidence that the Folr4 mutation and folinic acid supplementation converge on some pathways common to both paradigms. The most prominent pathways in our analysis are related to immune function and mitochondrial biology (Table 2 section D).
Additionally, it is conceivable that the Folr4 disruption, through biological action at earlier stages in development of females, may affect the uterine tissue cell-non-autonomously. Support for this possibility comes from genes like Follistatin: Fst responds to genotype and supplementation in opposing fashion, yet it is expressed in cells outside of the Folr4 expression domain (Figure 4C), constituting an example for non-cell-autonomous regulation of expression by genotype. Taken together, our results strongly suggest that the initial lack of Folr4 predisposes cells outside of the Folr4 expression domain to mount a broader transcriptomic response than in the wildtype. Thus, genotype has a strong modifying effect on the response to folate supplementation. While the underlying mechanism for cell-non-autonomous actions of Folr4 remain to be investigated, we conclude that preexisting conditions, such as e.g. genetic liabilities, or susceptibilities introduced by environmental exposure, can exert a significant influence on size and repertoire of the response to folate supplementation. Thus, responses to folate can be modulated, and may depend on pre-existing conditions such as genetic, nutritional, or metabolic susceptibilities.
Furthermore, the notion that responses to folate are strongly modulated by genetic factors is underscored by the recent finding that folate status in mouse mutants of neural tube defect genes is not unequivocally beneficial: higher folate status relieved the neural tube defect burden in some of these mutant mouse strains, but aggravated it in others [14], demonstrating that the response to folate was highly dependent on the genetic background. In contrast to the mouse strains utilized in that study, homozygous Folr4 mutant embryos of both sexes develop normally and do not display neural tube defects; thus, they are clearly unsymptomatic with respect to neural tube defects. Yet, the significant modulation of the response to folinic acid by the Folr4 genotype raises the question how many other ‘phenotypically unsymptomatic’ genetic variations or mutations could have the potential to modulate the response to folate. The answer to this question has considerable implications for folate supplementation and fortification in humans. While functional consequences of human DNA variation are often milder than what is observed in inbred mouse models with targeted gene disruptions, it still stands to reason that folate also interacts with the genetic background in the human population, and may thus elicit distinct molecular responses in different individuals. If this were the case, health benefits from folate may vary by individual genetic constitution as well. Therefore, the concept that folate is unequivocally beneficial may deserve critical re-evaluation.
Supplementary Material
Supplementary Table 2 lists sequences of primers used in the validation studies by quantitative real-time PCR.
We provide lists (Supplementary Table 1) of Folate responsive genes in uterus of pregnant wildtype FVB females, of Folate responsive genes in uterus of pregnant FolR4 knockout females, and of Folate responsive genes that are differentially expressed between wiltype and Folr4 mutant uterus tissues. Genes are listed that fulfilled criteria of statistically significant difference between folinic acid supplemented and non-supplemented control tissues at a P-value smaller than 0.05 and fold-change of 1.2 or greater.
Highlights.
Folinic acid supplementation alters gene expression profiles in mouse uterus
Folate receptor 4 mutation changes the repertoire of folinic acid-responsive genes
Partial rescue of genotype-dependent changes by folinic acid in Folr4 mutants
Folinic acid supplementation increases histone methylation in mouse uterus
Identification of epigenetic and mitochondrial pathways as possible folate targets
Acknowledgments
This work was supported by grant NIH-R21-HD48516 to JMS. Support by COBRE NIH-P20GM103528 and NORC NIH-P30DK072476 to the PBRC Cell Biology and Bioimaging Core and the PBRC Genomics Core is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ Contributions.
J. Michael Salbaum and Claudia Kappen designed and supervised the study, Claudia Kruger contributed to the microarray analyses, J. Michael Salbaum performed the histological experiments; all three authors participated in analysis and interpretation of the data, and J. Michael Salbaum composed the figures and wrote the manuscript, which Claudia Kappen edited, with J. Michael Salbaum bearing primary responsibility for final content. All authors read and approved the submitted manuscript.
The authors declare that they have no conflicts of interest.
References
- 1.Obican SG, Finnell RH, Mills JL, Shaw GM, Scialli AR. Folic acid in early pregnancy: a public health success story. FASEB J. 2010;24:4167–4174. doi: 10.1096/fj.10-165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smithells RW, Sheppard S, Schorah CJ. Vitamin dificiencies and neural tube defects. Arch Dis Child. 1976;51:944–950. doi: 10.1136/adc.51.12.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smithells RW, Sheppard S, Schorah CJ, Seller MJ, Nevin NC, Harris R, Read AP, Fielding DW. Apparent prevention of neural tube defects by periconceptional vitamin supplementation. Arch Dis Child. 1981;56:911–918. doi: 10.1136/adc.56.12.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milunsky A, Jick H, Jick SS, Bruell CL, MacLaughlin DS, Rothman KJ, Willett W. Multivitamin/folic acid supplementation in early pregnancy reduces the prevalence of neural tube defects. JAMA : the journal of the American Medical Association. 1989;262:2847–2852. doi: 10.1001/jama.262.20.2847. [DOI] [PubMed] [Google Scholar]
- 5.Locksmith GJ, Duff P. Preventing neural tube defects: the importance of periconceptional folic acid supplements. Obstetrics and gynecology. 1998;91:1027–1034. doi: 10.1016/s0029-7844(98)00060-x. [DOI] [PubMed] [Google Scholar]
- 6.Fenech M. The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutation research. 2001;475:57–67. doi: 10.1016/s0027-5107(01)00079-3. [DOI] [PubMed] [Google Scholar]
- 7.Sieber O, Heinimann K, Tomlinson I. Genomic stability and tumorigenesis. Seminars in cancer biology. 2005;15:61–66. doi: 10.1016/j.semcancer.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Ulrich CM, Potter JD. Folate supplementation: too much of a good thing? Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15:189–193. doi: 10.1158/1055-9965.EPI-152CO. [DOI] [PubMed] [Google Scholar]
- 9.Kim YI. Role of folate in colon cancer development and progression. The Journal of nutrition. 2003;133:3731S–3739S. doi: 10.1093/jn/133.11.3731S. [DOI] [PubMed] [Google Scholar]
- 10.Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, McKeown-Eyssen G, Summers RW, Rothstein RI, Burke CA, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA : the journal of the American Medical Association. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 11.He W, Kularatne SA, Kalli KR, Prendergast FG, Amato RJ, Klee GG, Hartmann LC, Low PS. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. International journal of cancer Journal international du cancer. 2008;123:1968–1973. doi: 10.1002/ijc.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kularatne SA, Zhou Z, Yang J, Post CB, Low PS. Design, synthesis, and preclinical evaluation of prostate-specific membrane antigen targeted (99m)Tc-radioimaging agents. Molecular pharmaceutics. 2009;6:790–800. doi: 10.1021/mp9000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickell L, Brown K, Li D, Wang XL, Deng L, Wu Q, Selhub J, Luo L, Jerome-Majewska L, Rozen R. High intake of folic acid disrupts embryonic development in mice. Birth Defects Res A Clin Mol Teratol. 2011;91:8–19. doi: 10.1002/bdra.20754. [DOI] [PubMed] [Google Scholar]
- 14.Marean A, Graf A, Zhang Y, Niswander L. Folic acid supplementation can adversely affect murine neural tube closure and embryonic survival. Human molecular genetics. 2011;20:3678–3683. doi: 10.1093/hmg/ddr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jhaveri MS, Wagner C, Trepel JB. Impact of extracellular folate levels on global gene expression. Mol Pharmacol. 2001;60:1288–1295. doi: 10.1124/mol.60.6.1288. [DOI] [PubMed] [Google Scholar]
- 16.Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;193:1184–1190. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- 17.Sohn KJ, Stempak JM, Reid S, Shirwadkar S, Mason JB, Kim YI. The effect of dietary folate on genomic and p53-specific DNA methylation in rat colon. Carcinogenesis. 2003;24:81–90. doi: 10.1093/carcin/24.1.81. [DOI] [PubMed] [Google Scholar]
- 18.Crott JW, Liu Z, Keyes MK, Choi SW, Jang H, Moyer MP, Mason JB. Moderate folate depletion modulates the expression of selected genes involved in cell cycle, intracellular signaling and folate uptake in human colonic epithelial cell lines. J Nutr Biochem. 2008;19:328–335. doi: 10.1016/j.jnutbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Protiva P, Mason JB, Liu Z, Hopkins ME, Nelson C, Marshall JR, Lambrecht RW, Pendyala S, Kopelovich L, Kim M, et al. Altered folate availability modifies the molecular environment of the human colorectum: implications for colorectal carcinogenesis. Cancer Prev Res (Phila) 2011;4:530–543. doi: 10.1158/1940-6207.CAPR-10-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kappen C. Disruption of the homeobox gene Hoxb-6 results in increased numbers of early erythrocyte progenitors. Am J Hematol. 2000;65:111–118. doi: 10.1002/1096-8652(200010)65:2<111::aid-ajh4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic research. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 22.Pavlinkova G, Salbaum JM, Kappen C. Maternal Diabetes alters Transcriptional Programs in the Developing Embryo. BMC Genomics. 2009;10:274. doi: 10.1186/1471-2164-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 24.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic acids research. 2012;40:W553–559. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Kruger C, Kappen C. Expression of cartilage developmental genes in Hoxc8- and Hoxd4-transgenic mice. PLoS One. 2010;5:e8978. doi: 10.1371/journal.pone.0008978. e8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salbaum JM, Kruger C, Zhang X, Delahaye NA, Pavlinkova G, Burk DH, Kappen C. Altered gene expression and spongiotrophoblast differentiation in placenta from a mouse model of diabetes in pregnancy. Diabetologia. 201154:1909–1920. doi: 10.1007/s00125-011-2132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salbaum JM. Punc, a novel mouse gene of the immunoglobulin superfamily, is expressed predominantly in the developing nervous system. Mechanisms of Development. 1998;71:201–204. doi: 10.1016/s0925-4773(98)00005-7. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi T, Hirota K, Nagahama K, Ohkawa K, Takahashi T, Nomura T, Sakaguchi S. Control of immune responses by antigen-specific regulatory T cells expressing the folate receptor. Immunity. 2007;27:145–159. doi: 10.1016/j.immuni.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Gray JD, Nakouzi G, Slowinska-Castaldo B, Dazard JE, Rao JS, Nadeau JH, Ross ME. Functional interactions between the LRP6 WNT co-receptor and folate supplementation. Human molecular genetics. 2010;19:4560–4572. doi: 10.1093/hmg/ddq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanger O, Wonisch W. Enzymatic and Non-enzymatic Antioxidative Effects of Folic Acid and Its Reduced Derivates. Sub-cellular biochemistry. 2012;56:131–161. doi: 10.1007/978-94-007-2199-9_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 2 lists sequences of primers used in the validation studies by quantitative real-time PCR.
We provide lists (Supplementary Table 1) of Folate responsive genes in uterus of pregnant wildtype FVB females, of Folate responsive genes in uterus of pregnant FolR4 knockout females, and of Folate responsive genes that are differentially expressed between wiltype and Folr4 mutant uterus tissues. Genes are listed that fulfilled criteria of statistically significant difference between folinic acid supplemented and non-supplemented control tissues at a P-value smaller than 0.05 and fold-change of 1.2 or greater.