Abstract
Introduction: Endothelial progenitor colony forming unit-endothelial cells (CFU-EC) were first believed to be the progenitors of endothelial cells, named endothelial progenitor cells. Further studies revealed that they are monocytes regulating vasculogenesis. The main hindrance of these cells for therapeutic purposes is their low frequency and limited replicative potentials. This study was undertaken to determine telomerase activity and alternative splicing variants in CFU-EC as a potential cause of limited replicative capacity in these cells. Methods: CFU-EC were isolated from peripheral blood using a standard cell culture assay. Colonies were detached mechanically and alternative splicing variant mRNA were evaluated using real-time PCR. Telomerase enzyme activity was assessed using telomerase repeat amplification protocol. The same procedures were done on the cancer cell line Calu6 as the positive control. Results: The cultured peripheral blood mononuclear cells formed colonies with spindle-shaped monocytic cells sprouted from the clusters. These morphological characteristics fulfill the definition of CFU-EC. Telomere length amplification protocol assay revealed no telomerase activity and real-time PCR showed no expression of telomerase enzyme mRNA in CFU-EC. Both parameters were significantly higher in the cancer cell line Calu6 taken as the positive control. Conclusion: The absence of telomerase activity in the CFU-EC is a result of pre-transcriptional regulation of gene expression rather than other mechanisms for controlling telomerase activity such as post-transcriptional modifications. This finding can explain the limited proliferative activity of CFU-EC cells. We propose that absence of telomerase activity in CFU-EC can be attributable to their more mature monocytic nature that needs further investigations.
Key Words: Telomerase, Endothelial cells, Hemangioblast, Alternative splicing
INTRODUCTION
Angiogenesis, the model which was proposed that new vessels are produced by migration and proliferation of mature endothelial cells, was the dominant paradigm for new blood vessel formation in adults for a long time. In recent years, the discovery of endothelial progenitor cells (EPC) has shifted that paradigm to vasculogenesis, suggesting that new blood vessels are formed by EPC differentiation to endothelial cells [1].
As the first explanation of EPC, most citations refer to an article published by Asahara et al. [2] in 1997. They isolated, characterized and examined the in vivo function of putative EPC from human peripheral blood.
Hill et al. [3] further modified Asahara’s methodology and introduced a colony-forming assay in the field. The cells isolated by their method are now termed as colony-forming unit-endothelial cells (CFU-EC or CFU). They also showed that the number of CFU-EC in peripheral blood has a significant inverse correlation with cardiovascular risk score in human [3]. This finding triggered a lot of researches revealing the importance of peripheral blood CFU-EC frequency in many diseases.
Researchers have shown that the concentration of CFU-EC in culture is decreased in patients with type 1 diabetes, chronic obstructive pulmonary disease, congestive heart failure, acute stroke, rheumatoid arthritis, and in overweight and obese people compared to healthy non-obese adults [4]. Nevertheless, it has been shown currently that CFU-EC have mainly monocyte/macrophage characteristics while expressing some endothelial-lineage markers. CFU-EC do not differentiate into endothelial cells in vitro and cannot assemble into vascular networks [5]. Despite these findings, CFU-EC remain under the classification umbrella of EPC in the literature [4].
CFU-EC play a role in vascular remodeling and maintenance of the adult endothelium by releasing growth factors, that contribute to paracrine support and repair of the endothelial lining [5-10]. Furthermore, it has been shown that these cells can improve blood flow recovery and capillary density in animal models of hind-limb ischemia [9, 11].
The main hindrance of working with these cells for therapeutic purposes is their low frequency in peripheral blood and their limited replicative potentials. Limited replicative capacity is shown to be a characteristic of non-stem normal human cells in culture [12]. This happens due to a linear DNA replication defect known as "end replication problem", in which estimated 50-200 base pairs from the very end of chromosomes (telomeres) is lost each time the cell divides [13]. This shortening will continue until telomeres erode to a critical length [13, 14] enough to make the cells stop dividing.
A ribonucleoprotein complex enzyme called "telomerase" plays a critical role in telomere maintenance by adding new telomeric DNA repeats [15]. This complex is composed of an RNA subunit, human telomerase RNA and a protein component, human telomerase reverse transcriptase (hTERT).
Among several identified factors that control hTERT transcription, transcriptional regulation seems to play the major role [16]. hTERT transcripts do undergo alternative splicing, which may be another regulatory mechanism for telomerase activity [17]. The most familiar alternatively spliced variants of hTERT are 2 deletions (α and β). The first is in the exon 6 (α site) leading to 36-bp deletion within the reverse transcriptase motif. It has been shown that α alternative splicing variant is an inhibitor of telomerase activity in immortal cell lines [18]. Premature translation termination up-stream of the essential reverse transcriptase motifs is the result of the other deletion in exons 7 and 8 (β site, 182 bp). The hTERT alternative splicing pattern among full-length, α-deletion, β-deletion, and α + β-deletion variants is shown to affect telomerase activity during embryonic development [17] and in some normal and neoplastic tissues [19-23]. It has been shown that each tissue has a specific alternative splicing pattern [24]. This study was designed to determine telomerase activity and alternative splicing variants in human CFU-EC as a potential cause of limited replicative potentials of these cells. We also compared results from CFU-EC with those of Calu6 cancer cell line as a positive control.
MATERIALS AND METHODS
Study population. The study population consisted of young (age<30 years old) healthy volunteers without cardiovascular risk factors. The exclusion criteria were: suffering from diabetes, malignancies, autoimmune disease, cardiovascular risk or disease (hypertension, chest pain induced by physical activities, vascular claudication and family history of premature cardiac events or severe abnormal lipid profiles.), current episode of infection, smoking and taking any prescribed medication for a long time.
Isolating peripheral blood cells. Under sterile condition, 10 ml of heparin anti-coagulated peripheral blood was drawn from antecubital vein. Samples were diluted 1:1 using PBS, transferred onto the layer of lymphosep (1.077 g/ml, Biosera, UK) and centrifuged at 300 g for 25 minutes without brake. Mononuclear cells (MNC) were isolated and the number of isolated cells was determined using a hemocytometer.
CFU-EC culture. Approximately 107 MNC were dissolved in 4 ml of EndoCult media (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco/Invitrogen, Carlsbad, CA, USA). Two milliliters of this suspension was plated per well in the 6-well fibronectin-coated plates (BD Biosciences, San Jose, CA, USA) and incubated at 37°C for two days in 5% CO2 with 95% humidity. After two days, the non-adherent cells were detached with forceful pipetting and 106 cells per well replated on 24-well fibronectin-coated plates (BD Biosciences, San Jose, CA, USA). After 5 days, the media was removed and plates were washed with PBS to remove non-adherent cells. Colonies were detached mechanically from the plates for further analysis.
Cancer cell line culture. The Calu6 cell line was purchased from National Cell Bank of Iran (NCBI, Pasture Institute of Iran, Tehran). The cells were cultured in RPMI-1640 (Biosera, UK) containing 10% fetal bovine serum (Gibco, USA) supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco/Invitrogen, Carlsbad, CA, USA) until they reached 80% confluency, which took 5-7 days. Then, the cells were harvested for the following experiments.
RNA extraction and real-time PCR. Total RNA was extracted from the CFU-EC and the Calu6 cells with Trizol reagent (Invitrogen, USA) based on the manufacturer’s instructions. Total RNA (5 µg) was reverse transcripted by Revert Aid First cDNA synthesis kit (Fermentas, Canada). This cDNA (2 µl) was used as the template for real-time PCR reactions. Primers were designed using Primer-BLAST online program with the sequences found in the genebank (NCBI). The location of primer adherence site and their sequences can be seen in Figure 1. Real-time PCR was performed using SYBR green PCR master mix (ABI, USA) in 40 cycles with an initial denaturation temperature of 95C for 10 minutes continued by cycling denaturation in 95C for 20 seconds, annealing at 60C for 1 minute and extension at 78C for 20 second. Finally, there was a melting curve analysis to confirm accuracy of PCR.
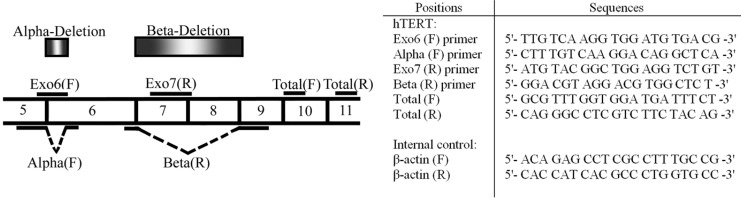
Fig. 1.
Deletion sites of different variants of hTERT and position of gene specific primers and their sequences. α-deletion site is located on exon 6, while β-deletion site is located on exons7 and 8. Using specific primers, the expression of full-length hTERT and three alternatively spliced variants (α-, ß- and αß-) were analyzed. Sequence of primers used for real-time PCR evaluation of different variants of hTERT expression and β-actin as the house-keeping gene have been shown in the adjacent Table. Exo, exon
Telomere length amplification protocol. The telomere length amplification protocol assay was carried out using the TeloTAGGG Telomerase PCR ELISAPlus kit (Roche, Germany) for quantitative determination of telomerase activity regarding the manufacturer's instructions.
Statistical analysis. SPSS software version 19.0 for windows (IBM, USA) was used for statistical analysis. Mann-Whitney test was used for the analysis and P value < 0.05 was considered statistically significant.
RESULTS
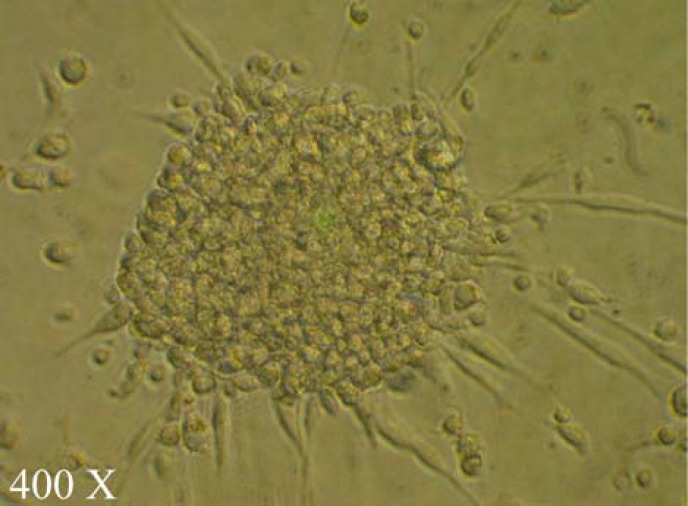
CFU culture. The cultured peripheral blood MNC in the Endocult media on the fibronectin coated culture plates, typically formed colonies with spindle-shaped monocytic cells, sprouted from the clusters. These morphological characteristics fulfill the definition of CFU-EC (Fig. 2).
Fig. 2.
A colony of colony-forming unit endothelial cells. The cultured peripheral blood mononuclear cells, typically formed colonies with spindle-shaped cells, sprouted from the clusters
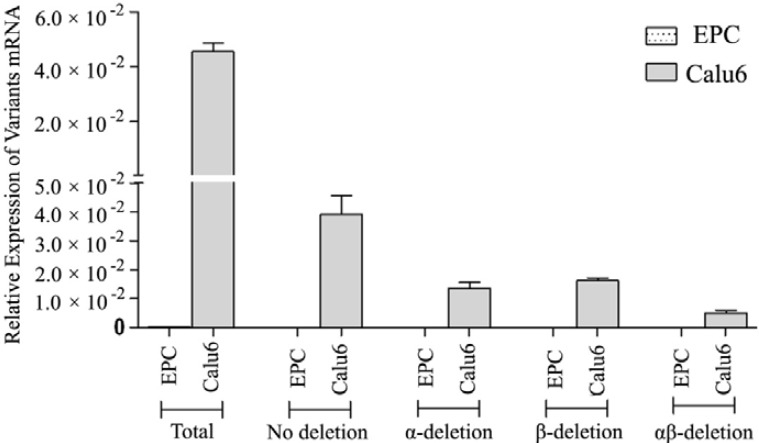
PCR results. Both CFU-EC and Calu6 cells showed expected levels of -actin as the internal control. CFU-EC did not show any alternative splicing variants of hTERT gene as well as the full-length mRNA (Fig. 3), while the Calu6 cells showed high levels of different variants and also the full-length mRNA. The difference among all levels of hTERT mRNA variants reached statistically significance (P<0.001, Fig. 4).
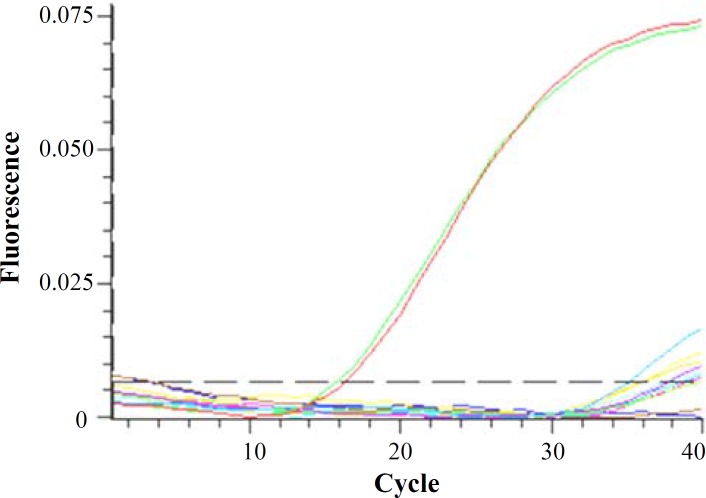
Fig. 3.
Real-time RT-PCR amplication graph. Low to absent levels of human telomerase reverse transcriptase alternative splicing variant was noted during real-time PCR cycles, despite the high levels of -actin defined as red and green lines
Fig. 4.
Real-time RT-PCR data. The analysis of RT-PCR data showed no expression of human telomerase reverse transcriptase alternative splicing variants in colony forming unit-endothelial cells. The α-actin mRNA, taken as the internal control, was significantly amplified
The results of telomere length amplification protocol assay . Telomere length amplification protocol assays showed no telomerase activity in CFU-EC (their OD were the same as blank wells), but high levels of telomerase activity (OD = 2.1) was detected in the Calu6 cells. The difference among OD from Calu6 cells and CFU-EC showed statistically significance (P<0.001).
DISCUSSION
Telomerase activity play an important role in cell surviving and one of the pitfalls in application of CFU-EC is short life time of these cells. Interestingly, our result showed that CFU-EC did not encompass any telomerase activity in adjunction with absence of any hTERT variant; despite -actin amplification, as well as detection of all telomerase variants in calu6 cell line.
Telomerase activity is regulated by a complex process with several steps including: up-regulation and down-regulation of gene expression and post-transcriptional modifications such as alternative splicing and protein to protein interactions at the site of telomere [16]. Among these steps, post-transcriptional modifications play a role in determining the telomerase activity in the cells. It is shown that the expression of alpha variant of hTERT mRNA roles an inhibitory effect on the activity of telomerase [18]. In our study, no expression of any variants of hTERT mRNA could be noticed, which logically correlated with the absence of any telomerase enzyme activity. This issue shows that regulation of telomerase activity in CFU-EC is done on the level of pre-transcriptional gene expression and is not attributable to other mechanisms such as alternative splicing.
High level expression of hTERT mRNA is mostly seen in cancerous cells, but this finding can be noticed in some stem cells. At the year 2004, Ingram et al. [25] reported the isolation of true EPC, which are now called “endothelial colony-forming cells”. They showed a high level of proliferative activity in adjunction with high levels of telomerase activity for these cells. In addition, hematopoietic stem cells (HSC) show low levels of telomerase activity, but their descendent progenies such as monocytes do not exhibit such characteristics [26]. The combination of the cell surface markers CD34 and CD133 have long been used to isolate HSC as they are CD34+CD133+ [27]. CFU-EC also do express low levels of CD34 and high levels of CD133 [5]. This action may make a misinterpretation that CFU-EC are a subtype of HSC and should express low levels of telomerase activity. However, this presumption can be disproved by the following findings. First of all it is worthy to mention that highly pure isolated CD34+ cells cannot produce CFU-EC in vitro [28]. van Beem et al. [29] showed that CD34 depleted MNC can produce CFU-EC properly in regular culturing conditions. These finding shows that CFU-EC are distinct from myeloid progenitors. On the other hand, it has been demonstrated that depletion of CD14+ cells from MNC will totally eradicate the presence of CFU-EC in culturing systems the same as CD2+ cells [30]. The dominant idea about the formation of CFU-EC considers that the interaction of activated CD4+ T cells with CD14+ cells is necessary to form these colonies [29]. The spindle shaped cells radiating away from the core are monocytes expressing many endothelial markers and characteristics.
In this study, we have clearly showed that CFU-EC cells lack telomerase alternative splicing variant mRNA and activity. This finding, at least in part, can propose an explanation for limited replicative potentials of CFU-EC. The absence of telomerase activity in the CFU-EC is a result of pre-transcriptional regulation of gene expression rather than other mechanisms for controlling telomerase activity such as post-transcriptional modifications. We propose that absence of telomerase activity in CFU-EC can be attributable to their more mature monocytic nature and the presence of CD34 and CD133 markers do not necessarily mean that they are progenitor or stem cells. Therefore, this suggestion needs further investigations.
ACKNOWLEDGEMENTS
This work was supported by the grant number 88-4759 of Shiraz University of Medical Sciences (Iran). The authors wish to thank Dr. Parsanezhad M.E. (Chief of Obstetrics and Gynecology Department, School of Medicine, Shiraz University of Medical Sciences) and Dr. Alireza Attar for improving and preparation of the manuscript. This project has been performed in Students’ Research Lab, Pathology Department, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran.
References
- 1.Shi Q, Rafii S, Wu MH, Wijelath ES, Yu C, Ishida A, et al. Evidence for circulating bone marrow-derived endothelial cells. Blood. 1998 Jul;92(2):362–7. [PubMed] [Google Scholar]
- 2.Asahara T, Murohara T, Sullivan A, Silver M, Zee RVD, Li T, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997 Feb;275:964–7. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 3.Hill JM, Zalos G, Halcox JP, Schenke WH, Waclawiw MA, Quyyumi AA, et al. Circulating endothelial progenitor cells, vascular function, and cardiovascular risk. N Engl J Med. 2003 Feb;348(7):593–600. doi: 10.1056/NEJMoa022287. [DOI] [PubMed] [Google Scholar]
- 4.Lin C, Rajakumar A, Plymire DA, Verma V, Markovic N, Hubel CA. Maternal Endothelial Progenitor Colony-Forming Units With Macrophage Characteristics Are Reduced in Preeclampsia. Am J Hypertens. 2009 Sep;22(9):1014–9. doi: 10.1038/ajh.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirschi KK, Ingram DA, Yoder MC. Assessing identity, phenotype, and fate of endothelial progenitor cells. Arterioscler Thromb Vasc Biol. 2008 Sep;28(9):1584–95. doi: 10.1161/ATVBAHA.107.155960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbich C, Heeschen C, Aicher A, Dernbach E, Zeiher AM, Dimmeler S. Relevance of monocytic features for neovascularization capacity of circulating endothelial progenitor cells. Circulation. 2003 Nov;108(20):2511–6. doi: 10.1161/01.CIR.0000096483.29777.50. [DOI] [PubMed] [Google Scholar]
- 7.Urbich C, Aicher A, Heeschen C, Dernbach E, Hofmann WK, Zeiher AM, et al. Soluble factors released by endothelial progenitor cells promote migration of endothelial cells and cardiac resident progenitor cells. J Mol Cell Cardiol. 2005 Nov;39(5):733–42. doi: 10.1016/j.yjmcc.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Hur J, Yoon CH, Kim HS, Choi JH, Kang HJ, Hwang KK, et al. Characterization of two types of endothelial progenitor cells and their different contributions to neovasculogenesis. Arterioscler Thromb Vasc Biol. 2004 Feb;24(2):288–93. doi: 10.1161/01.ATV.0000114236.77009.06. [DOI] [PubMed] [Google Scholar]
- 9.Barber CL, Iruela-Arispe ML. The ever-elusive endothelial progenitor cell: identities, functions and clinical implications. Pediatr Res. 2006 Apr;59(4 Pt 2):26R–32R. doi: 10.1203/01.pdr.0000203553.46471.18. [DOI] [PubMed] [Google Scholar]
- 10.Schatteman GC, Dunnwald M, Jiao C. Biology of bone marrow-derived endothelial cell precursors. Am J Physiol Heart Circ Physiol. 2007;292:H1–18. doi: 10.1152/ajpheart.00662.2006. [DOI] [PubMed] [Google Scholar]
- 11.Yoon CH, Hur J, Park KW, Kim JH, Lee CS, Oh IY, et al. Synergistic neovascularization by mixed trans-plantation of early endothelial progenitor cells and late outgrowth endothelial cells: the role of angiogenic cytokines and matrix metalloproteinases. Circulation. 2005 Sep;112(11):1618–27. doi: 10.1161/CIRCULATIONAHA.104.503433. [DOI] [PubMed] [Google Scholar]
- 12.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961 Dec;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 13.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990 May;345(6274):458–60. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 14.Chang E, Harley CB. Telomere length and replicative aging in human vascular tissues. Proc Natl Acad Sci USA. 1995 Nov;92(24):11190–4. doi: 10.1073/pnas.92.24.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985 Dec;43(2 Pt 1):405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 16.Pendino F, Flexor M, Delhommeau F, Buet D, Lanotte M, Segal-Bendirdjian E. Retinoids down-regulate telomerase and telomere length in a pathway distinct from leukemia cell differentiation. Proc Natl Acad Sci USA. 2001 Jun;98(12):6662–7. doi: 10.1073/pnas.111464998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998 Sep;58(18):4168–72. [PubMed] [Google Scholar]
- 18.Colgin LM, Wilkinson C, Englezou A, Kilian A, Robinson MO, Reddel RR. The hTERT alpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia. 2000 Sep-Oct;2(5):426–32. doi: 10.1038/sj.neo.7900112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahjoub S, Krams M. Comparison of telomerase activity in prostate cancer, prostatic intraepithelial neoplasia and benign prostatic hyperplasia. J Res Med Sci. 2006;11(4):229–33. [Google Scholar]
- 20.Yokoyama Y, Wan X, Takahashi Y, Shinohara A, Tamaya T. Alternatively spliced variant deleting exons 7 and 8 of the human telomerase reverse transcriptase gene is dominantly expressed in the uterus. Mol Hum Reprod. 2001 Sep;7(9):853–7. doi: 10.1093/molehr/7.9.853. [DOI] [PubMed] [Google Scholar]
- 21.Zaffaroni N, Porta CD, Villa R, Botti C, Buglioni S, Mottolese M, Daidone MG. Transcription and alternative splicing of telomerase reverse transcriptase in benign and malignant breast tumours and in adjacent mammary glandular tissues: implications for telomerase activity. J Pathol. 2002 Sep;198(1):37–46. doi: 10.1002/path.1178. [DOI] [PubMed] [Google Scholar]
- 22.Krams M, Claviez A, Heidorn K, Krupp G, Parwaresch R, Harms D, et al. Regulation of telomerase activity by alternate splicing of human telomerase reverse transcriptase mRNA in a subset of neuroblastomas. Am J Pathol. 2001 Nov;159(5):1925–32. doi: 10.1016/S0002-9440(10)63039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villa R, Porta CD, Folini M, Daidone MG, Zaffaroni N. Possible regulation of telomerase activity by transcription and alternative splicing of telomerase reverse transcriptase in human melanoma. J Invest Dermatol. 2001 Jun;116(6):867–73. doi: 10.1046/j.1523-1747.2001.01343.x. [DOI] [PubMed] [Google Scholar]
- 24.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer. 2001 Mar;91(5):644–9. [PubMed] [Google Scholar]
- 25.Ingram DA, Mead LE, Tanaka H, Meade V, Fenoglio A, Mortell K, et al. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood. 2004 Nov;104(9):2752–60. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann S, Martens UM. Telomeres, senescence, and hematopoietic stem cells. Cell Tissue Res. 2008 Jan;331(1):79–90. doi: 10.1007/s00441-007-0469-4. [DOI] [PubMed] [Google Scholar]
- 27.Giebel B, Punzel M. Lineage development of hematopoietic stem and progenitor cells. Biol Chem. 2008 Jul;389(7):813–24. doi: 10.1515/BC.2008.092. [DOI] [PubMed] [Google Scholar]
- 28.Case J, Mead LE, Bessler WK, Prater D, White HA, Saadatzadeh MR, et al. Human CD34+AC133+VEGFR-2+ cells are not endothelial progenitor cells but distinct, primitive hematopoietic progenitors. Exp Hematol. 2007 Jul;35(7):1109–18. doi: 10.1016/j.exphem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 29.van Beem RT, Noort WA, Voermans C, Kleijer M, ten Brinke A, van Ham SM, et al. The presence of activated CD4(+) T cells is essential for the formation of colony-forming unit-endothelial cells by CD14(+) cells. J Immunol. 2008 Apr;180(7):5141–8. doi: 10.4049/jimmunol.180.7.5141. [DOI] [PubMed] [Google Scholar]
- 30.Rohde E, Bartmann C, Schallmoser K, Reinisch A, Lanzer G, Linkesch W, et al. Immune cells mimic the morphology of endothelial progenitor colonies in vitro. Stem Cells. 2007 Jul;25(7):1746–52. doi: 10.1634/stemcells.2006-0833. [DOI] [PubMed] [Google Scholar]