Abstract
In the intrinsic pathway of apoptosis, cell-damaging signals promote the release of cytochrome c from mitochondria, triggering activation of the Apaf-1 and caspase-9 apoptosome. The ubiquitin E3 ligase MDM2 decreases the stability of the proapoptotic factor p53. We show that it also coordinated apoptotic events in a p53-independent manner by ubiquitylating the apoptosome activator CAS and the ubiquitin E3 ligase HUWE1. HUWE1 ubiquitylates the antiapoptotic factor Mcl-1, and we found that HUWE1 also ubiquitylated PP5 (protein phosphatase 5), which indirectly inhibited apoptosome activation. Breast cancers that are positive for the tyrosine receptor kinase HER2 (human epidermal growth factor receptor 2) tend to be highly aggressive. In HER2-positive breast cancer cells treated with the HER2 tyrosine kinase inhibitor lapatinib, MDM2 was degraded and HUWE1 was stabilized. In contrast, in breast cancer cells that acquired resistance to lapatinib, the abundance of MDM2 was not decreased and HUWE1 was degraded, which inhibited apoptosis, regardless of p53 status. MDM2 inhibition overcame lapatinib resistance in cells with either wild-type or mutant p53 and in xenograft models. These findings demonstrate broader, p53-independent roles for MDM2 and HUWE1 in apoptosis and specifically suggest the potential for therapy directed against MDM2 to overcome lapatinib resistance.
INTRODUCTION
Mutation of the tumor suppressor p53 in ~50% of human tumors promotes both unrestrained cell proliferation and a failure of cells to die appropriately by apoptosis (1, 2). MDM2 is a key p53 inhibitor and prevents transcription of p53 target genes and induces p53 polyubiquitylation and degradation (3-7). In this regard, MDM2 has been viewed as an indirect, p53-dependent apoptotic regulator.
In a subset of cancers, tumorigenesis is driven by aberrantly activated tyrosine kinases that promote prosurvival and antiapoptotic signaling (8-10). In these tumors, targeted kinase inhibition triggers apoptosis and tumor regression. However, development of acquired therapeutic resistance has limited the clinical efficacy of this important class of targeted cancer drugs. Therapeutic resistance can result from mutations in the tyrosine kinase itself (for example, imatinib-resistance stemming from mutation of Bcr-Abl in chronic myeloid leukemias), but this is not always the case (11, 12). Under these circumstances, dissection of signal transduction pathways in resistant cells may reveal novel interactions among signaling molecules.
Overexpression or amplification of the HER2 (human epidermal growth factor receptor 2) receptor tyrosine kinase in 20 to 30% of breast cancers is associated with poor clinical outcomes (13). Several HER2-targeted therapeutics have been approved or are in clinical trials, including trastuzumab (trade name Herceptin), a monoclonal antibody directed against the extracellular domain of HER2, and lapatinib (trade name Tykerb), a small molecule that reversibly inhibits the kinase activities of HER2 and EGFR (epidermal growth factor receptor) (14, 15). Conventionally, a breast cancer patient undergoes lapatinib treatment after the development of therapeutic resistance to trastuzumab. However, the antitumor effects of lapatinib monotherapy are generally short-lived, with cancer cells inevitably developing resistance to this drug over time (16).
Because mutations in HER2 itself are not typically seen in acquired lapatinib resistance, alternative mechanisms underlying acquired lapatinib resistance have been evaluated (15, 17-22). Several studies have identified apoptotic inhibitors that show increased abundance in lapatinib-resistant cells, including X-linked inhibitor of apoptosis protein (XIAP) (18) and Mcl-1, an antiapoptotic Bcl-2 family member [seen in lapatinib-resistant colon cancer cells (19)]. It has been recently demonstrated that the simultaneous inhibition of the Bcl-2 family proteins Bcl-2, Bcl-xL, and Mcl-1 can synergize with lapatinib (23). Nevertheless, the molecular basis of acquired resistance remains to be fully elucidated.
In seeking to understand the molecular basis of lapatinib resistance, we undertook a systematic comparison of apoptotic signaling pathways in isotype-matched lapatinib-sensitive and lapatinib-resistant HER2+ breast cancer cells. We identified multiple antiapoptotic alterations both upstream and downstream of mitochondria in resistant cells. These studies revealed an unexpected signaling network in which the ubiquitin E3 ligase MDM2 ubiquitylated another E3 ligase, HUWE1, thereby increasing the abundance of its substrates, Mcl-1 and protein phosphatase 5 (PP5), an indirect inhibitor of the apoptosome. Thus, HUWE1 transmits a signal from MDM2 to control apoptotic events both upstream and downstream of mitochondria. These MDM2-dependent pathways were subverted in lapatinib-resistant cells, and regardless of cellular p53 status, inhibition of MDM2 rectified apoptotic defects, thereby overcoming drug resistance.
RESULTS
Mcl-1 stabilization in lapatinib-resistant breast cancer cells
To analyze the molecular mechanisms underlying lapatinib resistance, we derived four independent lapatinib-resistant HER2-positive breast cancer cell lines by continuous culture of BT474, SKBR3, SUM190, and AU565 cells in the presence of clinically relevant concentrations of lapatinib (20). Regardless of estrogen receptor, progesterone receptor, or p53 status (fig. S1 and table S1), all four parental sensitive cell lines died from apoptosis in response to lapatinib, whereas all four resistant lines (hereinafter referred to as rBT474, rSKBR3, rSUM190, and rAU565) did not (Fig. 1A and fig. S2). As previously reported (20), lapatinib inhibited the tyrosine autophosphorylation of HER2 even in resistant cells (Fig. 1B). Furthermore, phosphorylation of both Akt and extracellular signal-regulated kinase 1/2 (ERK1/2), two primary downstream effectors in HER2-positive breast cancer, was also attenuated in sensitive and resistant cells treated with lapatinib (Fig. 1, C and D). Note that we did not detect a consistent increase in the kinase activity of SRC (fig. S3), which has been implicated in acquired resistance to trastuzumab or lapatinib in HER2-positive breast cancer (21, 24), nor did the SRC inhibitor dasatinib affect MDM2 protein abundance or apoptotic resistance in lapatinib-resistant cells (fig. S4).
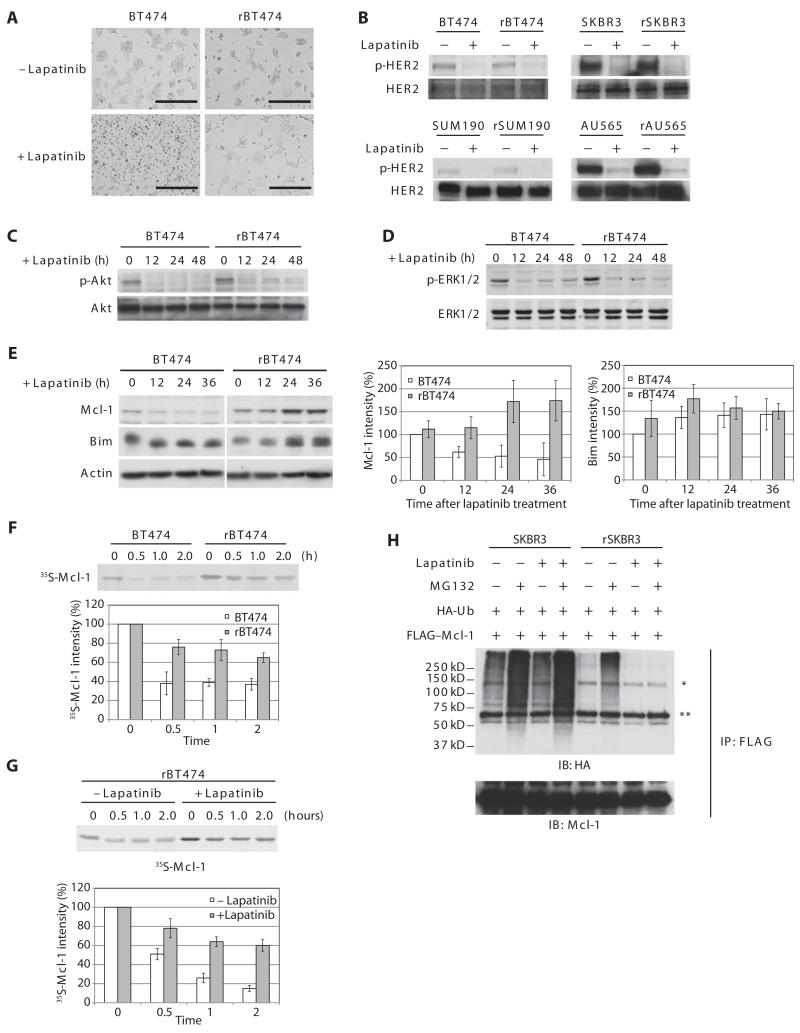
Fig. 1. Mcl-1 is stabilized in lapatinib-resistant cells.
(A) Photo micrograph of lapatinib-sensitive and resistant BT474 cells (BT474 and rBT474, respectively) treated with lapatinib. Scale bars, 400 μm. n = 3 independent experiments. (B) Cells treated with or without lapatinib in the presence of the caspase inhibitor z-VAD were immunoblotted with antibodies against phospho-HER2 (p-HER2) (Tyr877) and total HER2. n = 3 independent experiments. (C) BT474 and rBT474 cells treated with lapatinib and z-VAD for the indicated times were immunoblotted for phospho-Akt (p-Akt)(Thr308)andtotalAkt.n=2independentexperiments.(D)BT474 and rBT474 cellstreated with lapatinib and z-VAD for the indicated times were immunoblotted for phospho-ERK1/2 (p-ERK1/2) (Thr202 and Tyr204) and total ERK1/2. n = 3 independent experiments. (E) BT474 and rBT474 treated with lapatinib for the indicated times were immunoblotted for Mcl-1, Bim, and actin. A representative result is shown (left). Densitometric analysis of the abundance of Mcl-1 and Bim normalized to that of actin is shown (right). The protein abundance at the 0 time point in BT474 cells was set at 100%. n = 5 (Mcl-1) and n = 3 (Bim) independent experiments (means ± SEM). (F) 35S-labeled Mcl-1 protein was incubated in cell-free lysates prepared from BT474 and rBT474. Mcl-1 stability was monitored by detecting 35S-labeled Mcl-1 at the indicated time points. Representative result (top panel) and densitometric analysis of the abundance of Mcl-1 (bottom panel) are shown. The 35S-labeled Mcl-1 abundance at the 0 time point was set at 100%. n = 3 independent experiments (means ± SEM). (G) 35S-labeled Mcl-1 protein was incubated in lysates prepared from rBT474 cells treated with or without lapatinib for 7 days. Mcl-1 stability was monitored by detecting 35S-labeled Mcl-1 at the indicated time points. Representative result (top) and densitometric analysis of the abundance of Mcl-1 (bottom) are shown. The 35S-labeled Mcl-1 abundance at the 0 time point was set at 100%. n = 4 independent experiments (means ± SEM). (H) SKBR3 or rSKBR3 cells expressing FLAG-Mcl-1 and HA-ubiquitin (Ub) were treated with lapatinib in the presence of z-VAD and harvested after treatment with MG132 (Z-Leu-Leu-Leu-CHO). FLAG-Mcl-1 immunoprecipitates were immunoblotted for HA to analyze Mcl-1 ubiquitylation [*nonspecific band, immunoglobulin G (IgG) heavy chain]. IP, immunoprecipitation; IB, immunoblotting. n = 3 independent experiments.
Because lapatinib promotes tumor regression, at least in part, by inducing apoptosis, we speculated that apoptotic signaling might be altered in resistant cells. Indeed, lapatinib induced mitochondrial cytochrome c release in parental BT474 cells, but not rBT474 cells (fig. S5), suggesting that pro- and antiapoptotic Bcl-2 family proteins that govern the mitochondrial outer membrane permeability might be modulated in the lapatinib-treated resistant cells. Although there did not appear to be substantial differences in the abundance of the Bcl-2 family member Bim (Fig. 1E) between sensitive and resistant cells, as reported for colon cancer cells (19), the abundance of the antiapoptotic Bcl-2 member Mcl-1 was increased in the resistant breast cancer cells treated with lapatinib (Fig. 1E and fig. S6); in the sensitive cells, Mcl-1 abundance decreased upon treatment with lapatinib, whereas Mcl-1 abundance increased in the resistant cells maintained in the presence of lapatinib (Fig. 1E and fig. S6).
35S-labeled Mcl-1 was rapidly degraded in cell-free lysates prepared from sensitive cells, but was markedly delayed in those prepared from resistant cells, indicating that the observed differences in Mcl-1 abundance stemmed from differences in Mcl-1 stability (Fig. 1F and fig. S7). Although the Mcl-1 protein half-life was prolonged in resistant cell lysates, this was particularly prominent in lysates from cells treated with lapatinib (for example, lapatinib-treated rBT474 cells) (Fig. 1G). Addition of the deubiquitylase inhibitor ubiquitin aldehyde to the lysates did not prevent the failure to degrade 35S-labeled Mcl-1 in resistant cells (fig. S8), suggesting that Mcl-1 stabilization might be secondary to a defect in Mcl-1 ubiquitylation, rather than enhanced deubiquitylation. To examine Mcl-1 ubiquitylation directly, we co-transfected FLAG-tagged Mcl-1 together with hemagglutinin (HA)-tagged ubiquitin and examined the ubiquitylation of immunoprecipitated Mcl-1. In the absence of lapatinib, Mcl-1 protein was equally ubiquitylated in SKBR3 and rSKBR3 cells (Fig. 1H). Lapatinib treatment enhanced Mcl-1 ubiquitylation in SKBR3 cells but diminished Mcl-1 ubiquitylation in rSKBR3 cells (Fig. 1H), suggesting that the increased abundance of Mcl-1 protein in the lapatinib-resistant cells might result from a lapatinib-dependent defect in the Mcl-1 ubiquitylation machinery.
Incorrect apoptosome assembly in lapatinib-resistant cells
In analyzing the mechanism of lapatinib resistance, we noted that the resistant cells failed to activate caspases when purified cytochrome c was added directly to lysates, whereas cytochrome c induced robust caspase activation in parental cells, although the abundance of Apaf-1, caspase-9, and caspase-3 was similar in resistant and sensitive cells (Fig. 2A and fig. S9). As we observed for Mcl-1 protein stability, the failure of cytochrome c to induce caspase activation in lapatinib-resistant cells was lapatinib-dependent. Caspase activation did not occur in lysates from resistant cells cultured in the presence of lapatinib, a defect that was rescued by removing lapatinib from the culture medium (Fig. 2, A and B).
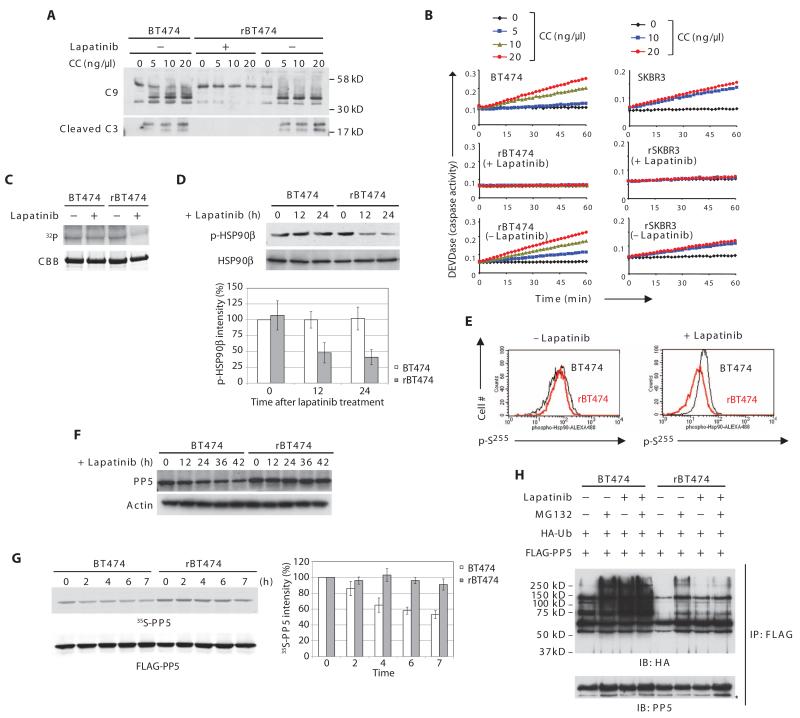
Fig. 2. PP5 stability and post-cytochrome c protection in lapatinib-resistant cells.
(A) rBT474 cells were cultured in the presence or absence of lapatinib for 1 week, whereas BT474 cells were maintained without lapatinib. Cytosolic lysates prepared from BT474 or rBT474 cells were incubated with 2′-deoxyadenosine 5′-triphosphate (dATP) and various amounts of cytochrome c (CC). Immunoblotting was performed for caspase-9 (cleaved and noncleaved) and cleaved caspase-3 (C9 and C3, respectively). n = 5 independent experiments. (B) Caspase-3 activity was assayed by measuring cleavage of DEVD-pNA after incubation of the cell lysates with dATP and various concentrations of cytochrome c. A representative result of five independent experiments is shown. (C) BT474 or rBT474 cells were cultured in the presence or absence of lapatinib for 6 hours and in the presence of z-VAD. Recombinant His-tagged HSP90β protein on nickel beads was incubated with the cell lysates in the presence of [γ-32P]ATP. HSP90β phosphorylation (32P) and total HSP90β protein [Coomassie brilliant blue (CBB)] are shown. n = 3 independent experiments. (D) BT474 or rBT474 cells treated with lapatinib for the indicated times were analyzed by immunoblotting for phospho-HSP90β (Ser226) and total HSP90β. Densitometric analysis of phospho-HSP90β (p-HSP90β) normalized to total HSP90β from four independent experiments is shown (means ± SEM). Phospho-HSP90β at the 0 time point in BT474 cells was set at 100%. (E) BT474 or rBT474 cells treated with lapatinib were fixed and incubated with phospho-Hsp90β (Ser255) antibody followed by an Alexa Fluor 488-conjugated secondary antibody. The phosphorylation status of Ser255 was analyzed by FACS (fluorescence-activated cell sorting; black: BT474; red: rBT474). n = 2 independent experiments. (F) BT474 and rBT474 treated with lapatinib for the indicated times were immunoblotted for PP5 and actin. n = 3 independent experiments. (G) BT474 or rBT474 cells expressing FLAG-PP5 were labeled by the addition of 35[S]Met and 35[S]Cys and cultured in the presence of lapatinib. 35S radioactivity (35S-labeled PP5) was measured in FLAG-PP5 immunoprecipitates collected at the indicated time points. Densitometric analysis of the abundance of 35S-labeled PP5 from five independent experiments is shown (means ± SEM). The 35S-labeled PP5 abundance at 0 time point was set at 100%. (H) BT474 or rBT474 cells expressing FLAG-PP5 and HA-Ub were cultured in the presence or absence of lapatinib and harvested after treatment with MG132. FLAG-PP5 immunoprecipitates were immunoblotted for HA to analyze PP5 ubiquitylation (*IgG heavy chain). n = 3 independent experiments.
PP5 stabilization in lapatinib-resistant breast cancer cells
Recruitment of caspase-9 to the apoptosome is blocked by HSP90β binding to the CARD (caspase recruitment domain) domain of Apaf-1. In leukemia cells, decreased phosphorylation of HSP90β at residues Ser226 and Ser255 leads to a stronger interaction between Apaf-1 and HSP90β (25). Phosphorylation of HSP90β at both Ser226 and Ser255 was also markedly decreased in lapatinib-treated resistant cells compared to sensitive cells (Fig. 2, C, D, and E).
Because lapatinib did not prevent the phosphorylation of HSP90β in parental BT474 cells (Fig. 2C), lapatinib was unlikely to inhibit an HSP90β-directed kinase [such as casein kinase 2 (CK2) (26) (fig. S10)], suggesting that an HSP90β-directed phosphatase might be modulated in response to lapatinib. PP5 has been reported to control the phosphorylation status of HSP90 (27). Therefore, we hypothesized that the decreased phosphorylation of HSP90β in lapatinib-resistant cells might result from increased PP5 activity. Indeed, lapatinib treatment triggered a gradual decrease in PP5 abundance in sensitive parental cells, but not in resistant cells (Fig. 2F and fig. S11). Moreover, the protein half-life of PP5 was longer in resistant than in the parental cells (Fig. 2G). Accordingly, the ubiquitylation of PP5 was greater in sensitive than in resistant cells (Fig. 2H).
The role of the ubiquitin E3 ligase HUWE1 in the degradation of both Mcl-1 and PP5
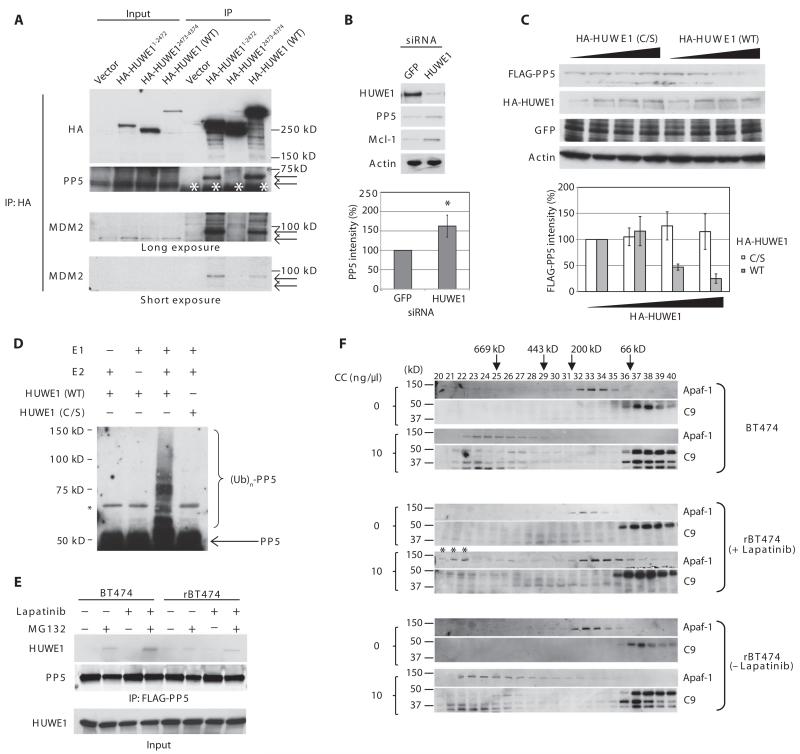
PP5 stabilization could result from a defect in an E3 ligase activity responsible for PP5 ubiquitylation in lapatinib-resistant cells. Hence, we sought to identify a PP5-directed E3 ligase through chromatographic purification. Cytosolic lysates prepared from SKBR3 cells were fractionated on a Q Sepharose column, and ubiquitylation of 35S-labeled PP5 by each fraction [supplemented with E1, E2, ATP (adenosine 5′-triphosphate), and ubiquitin] was analyzed by SDS-polyacrylamide gel electrophoresis. Most of the PP5-ubiquitylating activities were eluted with 500 mM NaCl (fig. S12). Because Mcl-1 stability changed in response to lapatinib, we speculated that an Mcl-1-directed E3 ubiquitin ligase might also regulate PP5 stability. Indeed, immunoblotting of Q Sepharose fractions revealed that HUWE1 (also known as Mule, ARF-BP1, and HectH9), a HECT domain E3 ligase that targets Mcl-1 for degradation (28), was also eluted with 500 mM NaCl, corresponding to the fraction with PP5-ubiquitylating activity (fig. S12).
In agreement with these observations, we found that HA-HUWE1 co-immunoprecipitated with endogenous PP5 (Fig. 3A). Moreover, knockdown of HUWE1 by small interfering RNA (siRNA) increased the abundance of both PP5 and Mcl-1 in BT474 cells (Fig. 3B). Conversely, overexpression of HA-HUWE1, but not catalytically inactive HA-HUWE1(C/S), reduced PP5 and Mcl-1 protein abundance (Fig. 3C and fig. S13). In the presence of both E1 and E2, wild-type recombinant HUWE1 protein, but not HUWE1(C/S), ubiquitylated recombinant PP5 in vitro (Fig. 3D). In BT474 cells, binding of PP5 to HUWE1 markedly increased after lapatinib treatment (Fig. 3E). In contrast, association of PP5 with HUWE1 was undetectable in rBT474 cells regardless of lapatinib treatment (Fig. 3E). Together, these results suggested that HUWE1 is a bona fide E3 ligase for PP5.
Fig. 3. HUWE1 is a PP5 ubiquitin ligase.
(A) BT474 cells were transfected with empty vector, HA-tagged HUWE1 encoding residues 1 to 2472 (HA-HUWE11-2472), 2473 to 4374 (HA-HUWE12473-4374), or the full-length protein [HA-HUWE1 WT (wild type)]. HA-HUWE1 immunoprecipitates were immunoblotted to detect the association of endogenous PP5 and MDM2 (*IgG heavy chain). After coimmunoprecipitation with HA-HUWE1, PP5 and MDM2 bands showed decreased mobility (arrows). n = 3 independent experiments. (B) BT474 cells transfected with GFP (green fluorescent protein)– or HUWE1-specific siRNA were immunoblotted with the indicated antibodies. A representative result is shown (top). Densitometric analysis of the abundance of PP5 normalized to that of actin is shown (bottom, n = 7 independent experiments). The band intensity in GFP siRNA-treated cells was set at 100%. *P < 0.05 by Student’s t test. (C) BT474 cells transfected with increasing amounts of HA-HUWE1 (WT) or its catalytically inactive mutant HA-HUWE1 (C/S) along with FLAG-PP5 and GFP (transfection control) were immunoblotted for FLAG, HA, GFP, and actin. A representative result is shown (top). Densitometric analysis of the abundance of FLAG-PP5 normalized to that of GFP is shown (bottom). The FLAG-PP5 abundance at the lowest amount of HA-HUWE1 transfected was set at 100%. n = 3 independent experiments. (D) Ubiquitylation of PP5 was reconstituted in vitro by incubating recombinant PP5 protein with E1, UbcH7 (E2), ubiquitin (Ub), and recombinant HUWE1 protein [WT or HUWE1 (C/S)] in reaction buffer. The reactions were immunoblotted for PP5. n = 3 independent experiments. (E) BT474 or rBT474 cells expressing FLAG-PP5 were treated with lapatinib and harvested after MG132 treatment. FLAG-PP5 immunoprecipitates were immunoblotted to detect the association of endogenous HUWE1 with PP5. n = 3 independent experiments. (F) Cell lysates prepared from BT474 or rBT474 cells were incubated with or without dATP and cytochrome c and subjected to gel filtration on a Superdex 200 column. Each fraction was immunoblotted for Apaf-1 and caspase-9. The asterisks show that a portion of Apaf-1 in the lapatinib-treated resistant cell lysate formed higher-order oligomers in response to cytochrome c. n = 3 independent experiments.
Decreased abundance of CAS in lapatinib-resistant cells
Decreased phosphorylation of HSP90β downstream of PP5 stabilization might explain apoptosome inhibition in the lapatinib-treated resistant cells, but when we resolved cell lysates by gel filtration, we saw not only a failure to recruit caspase-9 to Apaf-1 (which could be explained if HSP90β blocked caspase-9 recruitment) but also a failure of the majority of Apaf-1 to oligomerize (Fig. 3F). Moreover, upon addition of cytochrome c to resistant cell lysates, we detected a small fraction of Apaf-1 and caspase-9 in large (1000 to 1400 kD) fractions (Fig. 3F, shown by asterisks), which has been previously described as incorrectly assembled inactive complexes (29).
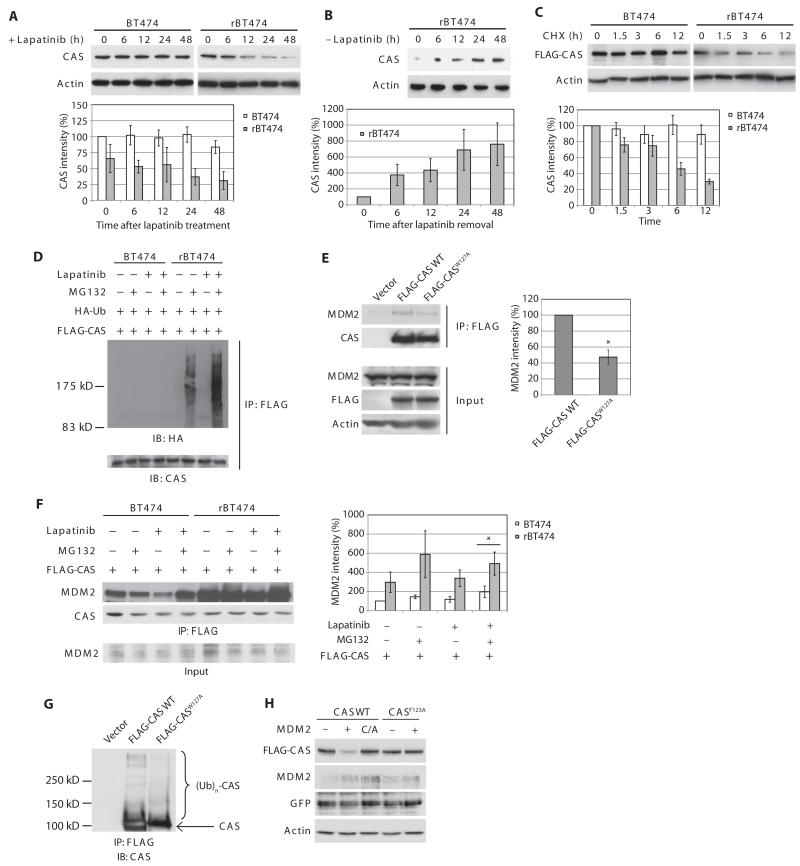
Without nucleotide exchange, cytochrome c induces Apaf-1 to form non-functional aggregates (29). Accordingly, we examined the abundance of PHAP1, HSP70, and CAS (also known as CSE1L or Exportin-2), essential components of the nucleotide exchange factor required for loading Apaf-1 with dATP (29). We found that CAS protein abundance was decreased in resistant cells compared with parental cells, most notably in the presence of lapatinib (Fig. 4A and fig. S14). CAS protein abundance declined over time after the addition of lapatinib to resistant cells, but lapatinib removal restored the abundance of CAS to levels comparable to the parental cells (Fig. 4, A and B). Treatment of parental cells with lapatinib did not affect CAS protein abundance (Fig. 4A). CAS protein was stabilized in sensitive cells, but destabilized in lapatinib-treated resistant cells (Fig. 4C). Accordingly, ubiquitylation of CAS was nearly undetectable in sensitive cells even in the presence of MG132, whereas ubiquitylated species of CAS were readily detected in resistant cells and more prominent in the presence of lapatinib (Fig. 4D and fig. S15). These data suggest that decreased CAS abundance in lapatinib-resistant cells results from augmented CAS protein ubiquitylation.
Fig. 4. CAS degradation in lapatinib-resistant cells.
(A) BT474 and rBT474 (left) cultured in the absence of lapatinibfor1weekwere treated with lapatinib for the indicated times and immunoblotted for CAS and actin (top). Densitometric analysis of the abundance of CAS normalized to that of actin from four independent experiments (means ± SEM) is shown (bottom). CAS abundance at time 0 in BT474 cells was set at 100%. (B) rBT474 cells maintained in the presence of lapatinib were harvested at various time points after removal of lapatinib and immunoblotted for CAS and actin (top). Densitometric analysis of the abundance of CAS normalized to that of actin from four independent experiments (means ± SEM) is shown (bottom). CAS abundance at time 0 was set at 100%. (C) BT474 and rBT474 cells expressing FLAG-tagged CAS were treatedwith cycloheximide(CHX)andlapatinibfortheindicated times and immunoblotted for FLAG and actin (top). Densitometric analysis of the abundance of FLAG-CAS normalized to that of actin from four independent experiments (means ± SEM) is shown (bottom). FLAG-CAS abundance at time 0 in each cell line was set at 100%. (D) BT474 and rBT474 expressing FLAG-CAS and HA-Ub were treated with lapatinib and z-VAD and harvested after MG132 treatment. FLAG-CAS immunoprecipitates were immunoblotted for HA to analyze ubiquitylation of CAS. n = 2 independent experiments. (E) H1299 cells were transfected with empty vector, FLAG-CAS WT, or FLAG-CASW127A. Immunoprecipitates of the FLAG-tagged proteins were immunoblotted for MDM2. Densitometric analysis of the abundance of immunoprecipitated MDM2 normalized to that of FLAG-CAS WT or FLAG-CASW127A from four independent experiments is shown (means ± SEM). The amount of MDM2 bound to FLAG-CAS WT was set at 100% (*P < 0.05 by paired, two-tailed t test). (F) BT474 and rBT474 cells expressing FLAG-CAS were treated with lapatinib and z-VAD and harvested after MG132 treatment. FLAG-CAS immunoprecipitates were immunoblotted to detect association of endogenous MDM2. Densitometric analysis of the abundance of immunoprecipitated MDM2 normalized to that of immunoprecipitated FLAG-CAS is shown (means ± SEM; n = 4 independent experiments). The immunoprecipitated MDM2 in BT474 cells without any treatments was set at 100% (*P < 0.05 by paired, two-tailed t test). (G) H1299 cells expressing empty vector, FLAG-CASW127A, or FLAG-CAS WT were treated with MG132. Ubiquitylation of CAS was analyzed after immunoprecipitation. n = 2 independent experiments. (H) H1299 cells expressing MDM2 (WT, C/A mutant, or empty vector), FLAG-CAS (WT or F123A mutant), and GFP (transfection control) were immunoblotted for FLAG, MDM2, GFP, and actin. n = 3 independent experiments.
To determine the mode of CAS degradation, we analyzed the CAS protein sequence and identified a potential binding motif for the RING finger ubiquitin ligase MDM2 (30, 31) (fig. S16). CAS interacts with p53 (32); however, MDM2 coimmunoprecipitated with CAS in BT474 and p53-null H1299 cells (Fig. 4, E and F). When Trp127, a conserved residue in the MDM2 binding motif, was replaced with Ala, binding of CAS to MDM2 was diminished (Fig. 4E). Moreover, when expressed in H1299 cells lacking p53, ubiquitylation of CASW127Awas suppressed compared with that of wild-type CAS (Fig. 4G). Furthermore, transient overexpression of MDM2 decreased the abundance of wild-type CAS, but not that of the F123A mutant in which the conserved Phe123 in the MDM2 binding motif was replaced with Ala (Fig. 4H). Together, these results indicate that CAS is an MDM2 target. In BT474 cells, the CAS-MDM2 interaction decreased in the presence of lapatinib, whereas binding was enhanced in rBT474 cells under the same conditions (Fig. 4F), consistent with the idea that augmented ubiquitylation and subsequent degradation of CAS protein in lapatinib-resistant cells are mediated by MDM2.
Identification of MDM2 as a HUWE1-directed E3 ligase
High HUWE1 and low MDM2 activities in sensitive cells resulted in reduced abundance of Mcl-1 and PP5 and sustained abundance of CAS, respectively, rendering the sensitive cells prone to apoptosis. In parental cells, HUWE1 protein abundance remained stable in the presence of lapatinib, whereas MDM2 rapidly decreased (Fig. 5A and fig. S17). Conversely, when resistant cells were treated with lapatinib, HUWE1 abundance decreased, whereas MDM2 abundance was maintained (Fig. 5A and fig. S17). Reduction of HUWE1 in resistant cells appeared to be due to lapatinib-induced protein degradation. In the absence of lapatinib, HUWE1 was stable in both BT474 and rBT474 cells. However, in lapatinib-treated rBT474 cells, HUWE1 protein half-life became considerably shorter, compared to that in BT474 cells (Fig. 5B). Although both HUWE1 and MDM2 are p53-targeting E3 ligases (5-7, 33) (and MDM2 abundance can also be modulated in a feedback loop by p53-induced transcription), mutant p53 is present in three of the four cell lines (BT474, SKBR3, and SUM190) (table S1 and fig. S1), suggesting that the observed changes in HUWE1 and MDM2 protein abundance are independent of the transcriptional activity of p53.
Fig. 5. MDM2-mediated ubiquitylation and degradation of HUWE1 in lapatinib-resistant cells.
(A) Cells treated with lapatinib for various times were immunoblotted for HUWE1, MDM2, and actin. See fig. S17 for the densitometric analysis. n = 3 independent experiments except MDM2 in SUM190/rSUM190 cells (n = 5) and HUWE1 in AU565/rAU565 cells (n = 4). (B) BT474 and rBT474 cells expressing HA-tagged HUWE1 were treated with cycloheximide and lapatinib for various times and immunoblotted for HA and actin. Densitometric analysis of the abundance of HA-HUWE1 normalized to that of actin is shown (means ± SEM; n = 4 independent experiments). HA-HUWE1 abundance at time 0 was set at 100%. (C) H1299 cells were transfected with empty vector, HA-HUWE1W1202A, or HA-HUWE1WT.Immunoprecipitates of HA-tagged proteins were immunoblotted to detect the association of MDM2 (left). Densitometric analysis of the abundance of immunoprecipitated MDM2 normalized to that of immunoprecipitated HA-HUWE1 is shown (means ± SEM; n=4 independentexperiments). The amount of MDM2 that precipitated with HA-HUWE1 WT was set at 100% (*P < 0.05 by paired, two-tailed t test). (D) Fluorescence polarization binding assays were performed with recombinant human MDM2 protein and the indicated peptides. IC50 ± SD, Ki ± SD, and a representative binding curve from threeindependentexperiments are shown. (E) BT474 cells expressing increasing amounts of MDM2 were immunoblotted for HUWE1 and actin. n = 2 independent experiments. (F) SUM190 cells transfected with scrambled or MDM2-specific siRNA were immunoblotted for HUWE1, MDM2, and actin. n = 3 independent experiments. (G) Ubiquitylation of HUWE1 was reconstituted in vitro by incubating recombinant HUWE11-2472 protein with E1, UbcH5 (E2), ubiquitin (Ub), and recombinant MDM2 protein in reaction buffer. The reactions were immunoblotted for HUWE1. n = 4 independent experiments. (H) rBT474 cells were initially transfected with scrambled or MDM2-specific siRNA, then with HA-tagged HUWE1 (WT). Cells were treated with lapatinib. After 24 hours, cycloheximide was added to the culture medium for various times, and cells were immunoblotted for HA and actin. Densitometric analysis of the abundance of HA-HUWE1 normalized to that of actin at each time point is shown (means ± SEM; n = 3 independent experiments). HA-HUWE1 abundance at time 0 was set at 100%.
Through protein sequence analysis, we determined that HUWE1 contained a putative MDM2 binding motif at residues 1198 to 1205 (fig. S18); the interaction of these two E3 ligases was confirmed by immunoprecipitation of endogenous proteins (fig. S19). The N-terminal portion of the HUWE1 protein (amino acids 1 to 2474) as well as the full-length protein interacted with MDM2 (Fig. 3A). Moreover, replacement of the conserved Trp1202 with Ala (HUWE1W1202A) reduced binding between the two proteins (Fig. 5C). The HUWE1-MDM2 interaction was evident not only in BT474 cells but also in H1299 cells, again indicating that their binding is neither specific for breast cancer cells nor mediated by p53 (Figs. 3A and 5C and fig. S19). To examine the binding affinity between HUWE1 and MDM2, we performed fluorescence polarization-based competitive binding assays using recombinant MDM2 protein and modified p53 peptide labeled with FAM as a fluorescent probe (34). The HUWE1 peptide competitively inhibited binding of the fluorescent probe to MDM2 with a Ki (inhibition constant) value of 1.5 μM and was a threefold more potent inhibitor of probe binding than the p53 peptide (amino acids 13 to 29, which constitutes the MDM2 binding motif) (Fig. 5D).
To determine whether MDM2 is a HUWE1-directed E3 ligase, we overexpressed MDM2 in H1299 cells. MDM2 expression reduced HUWE1 protein abundance in a dose-dependent manner (Fig. 5E). In contrast, siRNA knockdown of MDM2 increased HUWE1 protein abundance (Fig. 5F). Using a truncated mutant of HUWE1 (HUWE11–2474) to allow visualization of an electrophoretic mobility shift, we confirmed that, in the presence of both E1 and E2, MDM2 could ubiquitylate HUWE1 in vitro (Fig. 5G). Last, we demonstrated that lapatinib treatment triggered degradation of HUWE1 in resistant cells transfected with control siRNA, but when these cells were transfected with MDM2 siRNA, HUWE1 stability was markedly increased, even in the presence of lapatinib (Fig. 5H). Together, these results are consistent with MDM2 acting as a HUWE1-directed ligase.
Reversal of lapatinib resistance by inhibition of MDM2
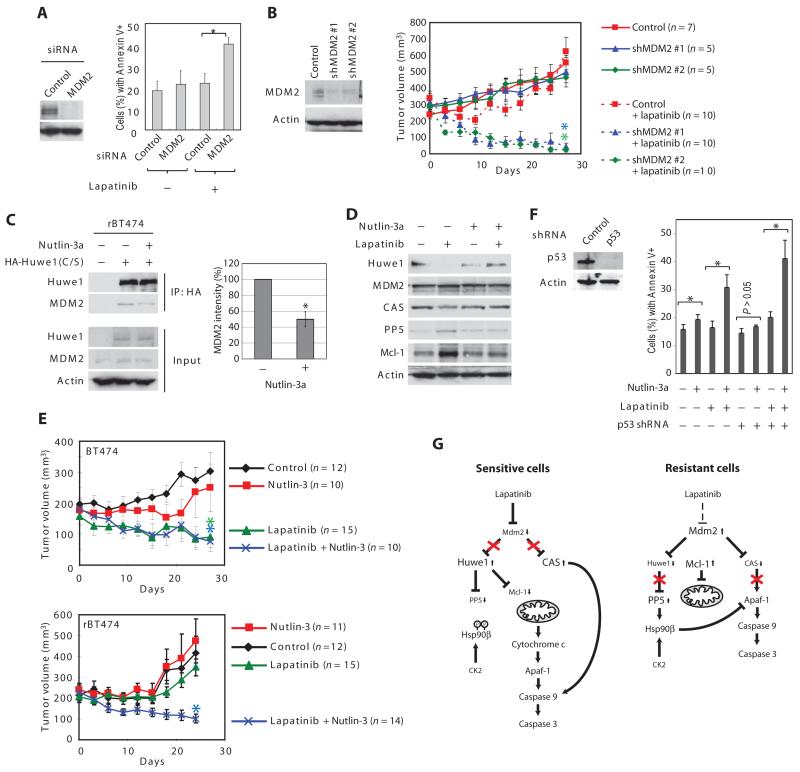
Our findings raised the interesting possibility that the changes in Mcl-1, PP5, and CAS in lapatinib-resistant cells could all be traced to high MDM2 activity in the resistant cells. Accordingly, when resistant cells were co-treated with lapatinib, MDM2 knockdown induced significant apoptotic cell death (Fig. 6A). To evaluate this further, we implanted xenografts of rBT474 cells stably expressing control or MDM2 short hairpin RNAs (shRNAs) into the mammary fat pads of female nude mice (Fig. 6B). Without lapatinib treatment, tumors developed similarly for all tumors. Oral lapatinib administration induced regression of xenografted tumors expressing MDM2 shRNAs, whereas control tumors continued to grow (Fig. 6B). Moreover, by the end of the treatment period, 6 of 10 mice in the MDM2 knockdown group had tumors smaller than 50 mm3.
Fig. 6. MDM2 inhibition can reverse lapatinib resistance.
(A) rBT474 cells transfected with scrambled or MDM2-specific siRNA were treated with lapatinib and subjected to annexin V staining. The percentage of annexin V-positive cells was analyzed by FACS. Results are mean percentages ± SEM and analyzed by paired, two-tailed t test (*P < 0.05, n = 4 independent experiments). (B) rBT474 cells stably expressing control or MDM2-specific shRNA (#1 or #2) were injected into the mam-mary fat pad of female nude mice. The oral administration of lapatinib began when the average tumor volume surpassed 300mm3.Resultsaremean percentages ± SEM. The statistical difference in tumor volume was analyzed by one-way analysis of variance (ANOVA) (P < 0.001) followed by pairwise comparisons using the Bonferroni correction for multiple comparisons. *P < 0.05 between control and shMDM2 #1, and between control and shMDM2 #2 when the mice were treated with lapatinib. n values in the figure indicate the number of mice. (C) H1299 cells transfected with HA-HUWE1 (C/S) were treated with Nutlin-3a. HA-HUWE1 (C/S) immunoprecipitates were immunoblotted for MDM2 (left). Densitometric analysis of the abundance of immunoprecipitated MDM2 normalized to that of immunoprecipitated HA-HUWE1 (C/S) is shown (means ± SEM; n = 4 independent experiments). The amount of MDM2 that precipitated with HUWE1 in the absence of Nutlin-3a was set at 100% (*P < 0.05 by paired, two-tailed t test). (D) rSKBR3 cells were cultured in the presence or absence of lapatinib for a week, and then treated with Nutlin-3a and z-VAD for the indicated times. Cells were immunoblotted for HUWE1, CAS, MDM2, PP5, Mcl-1, and actin. See fig. S17 for the densitometric analysis. n = 6 independent experiments. (E) BT474 and rBT474 cells were injected into the mammary fad pad of each mouse. When tumors developed to a size of 200 mm3, the mice were randomly assigned to receive vehicle, lapatinib, Nutlin-3, or lapatinib and Nutlin-3. Results are mean percentages ± SEM. The statistical difference in tumor volume was analyzed by one-way ANOVA (P = 0.003 and P < 0.001 for BT474 and rBT474 xenografts. respectively) followed by pairwise comparisons using the Bonferroni correction for multiple comparisons. (Top) *P < 0.05 between control and lapatinib and between control and lapatinib + Nutlin-3, whereas the difference was not significant between control and Nutlin-3 in BT474 xenografts. (Bottom) *P < 0.05 between control and lapatinib + Nutlin-3, whereas there was no significant difference between control and Nutlin-3, and between control and lapatinib in rBT474 xenografts. n values in the figure indicate the number of mice. (F) rAU565 cells stably expressing control or p53-specific shRNA were treated with DMSO (dimethyl sulfoxide) or Nutlin-3a in the presence or absence of lapatinib. The percentage of apoptotic cell death was measured by FACS analysis with annexin V. Results are mean percentages ± SEM of annexin V-positive cells (*P < 0.05 by paired, two-tailed t test). n = 4 independent experiments. (G) Model for coordinate control of multiple apoptotic regulators by the MDM2 and HUWE1. In sensitive cells, lapatinib promotes degradation of MDM2, which leads to decreases in the degradation of CAS. Loss of MDM2 also results in increased abundance of HUWE1, which results in more PP5 and Mcl-1 degradation. In lapatinib-resistant cells, MDM2 promotes decreased CAS abundance, decreased HUWE1 abundance, and, consequently, increased abundance of PP5 and Mcl-1. In aggregate, these changes render the cells resistant to lapatinib-induced apoptosis.
A hydrophobic pocket in the N terminus of MDM2 binds to the trans-activation domain of p53 (30). If MDM2 bound HUWE1 in the same fashion as suggested by the fluorescence polarization-based assays (Fig. 5D), the MDM2 antagonist Nutlin-3a, which disrupts the p53-MDM2 complex, might also be expected to interfere with HUWE1-MDM2 binding (35). To exclude the possibility that any effects of Nutlin-3a might be mediated by the p53-MDM2 interaction, we used p53-null H1299 cells. Catalytically inactive HUWE1 [HUWE1(C/S)] bound to endogenous MDM2, and this interaction was reduced by Nutlin-3a (Fig. 6C). Moreover, Nutlin-3a also prevented lapatinib-induced degradation of HUWE1 (Fig. 6D and fig. S20), suggesting that Nutlin-3a stabilizes HUWE1 by disrupting HUWE1-MDM2 binding. Consequently, Nutlin-3a also suppressed lapatinib-induced up-regulation of Mcl-1 and PP5 (Fig. 6D and fig. S20). CAS degradation was also inhibited by Nutlin-3a (Fig. 6D and fig. S20).
To examine the effect of Nutlin-3a in vivo, mice bearing BT474 or rBT474 xenografts were randomly assigned to vehicle, lapatinib, Nutlin-3 (a racemic mixture of the enantiomers Nutlin-3a and Nutlin-3b), or lapatinib and Nutlin-3. Lapatinib alone or a combination of lapatinib and Nutlin-3 significantly suppressed growth of BT474-derived tumors, whereas Nutlin-3 alone only modestly inhibited tumor growth (Fig. 6E). Neither lapatinib nor Nutlin-3 inhibited rBT474-derived tumor growth as a single agent (Fig. 6E). However, combination treatment of lapatinib and Nutlin-3 significantly suppressed tumor growth, resulting in ~50% reduction in tumor volume (Fig. 6E). These experiments were repeated using p53-positive AU565 and rAU565 cells. Lapatinib substantially inhibited the growth of tumors derived from AU565 cells, but not those derived from rAU565 cells (fig. S21). Nutlin-3 treatment alone moderately inhibited the growth of tumors derived from these p53-positive cells, but combined lapatinib and Nutlin-3 treatment reduced tumor volume by ~50% by the end of the evaluation period (fig. S21). Tissue culture experiments conducted in parallel with all four sensitive and resistant cell line pairs using the MDM2 antagonist MI-219 (34) suggest that similar effects are likely to be obtained with other MDM2 inhibitors (fig. S2). Moreover, the ability of the MDM2 inhibitors to resensitize resistant cells to lapatinib was not compromised by RNA interference (RNAi)-mediated knockdown of p53 in rAU565 cells (Fig. 6F). Together, these results demonstrate that regardless of p53 status, an MDM2 inhibitor such as Nutlin-3a or MI-219 can reverse lapatinib resistance, potentially by stabilizing CAS and HUWE1, thereby promoting loss of PP5 and Mcl-1 (Fig. 6G).
DISCUSSION
Here, we have demonstrated that multiple changes in the abundance of apoptotic regulators, both upstream and downstream of mitochondrial cytochrome c release, cause resistance to lapatinib. Furthermore, these changes could be attributed to a previously unsuspected network of MDM2 (and HUWE1) substrates that does not include p53. Failure to degrade MDM2 in response to lapatinib in all four independently derived lapatinib-resistant cell lines was the key factor in increasing the abundance of components of the antiapoptotic machinery in these cells (Fig. 6G).
We identified changes in mitochondrial cytochrome c release after lapatinib treatment, but we have also found that the apoptosome is refractory to cytochrome c–induced activation in the resistant cells. There is some debate as to the importance of apoptosome inhibition in cancers that already have a block to mitochondrial cytochrome c release. However, we postulate that the block is more profound when several loci in the apoptotic pathway are affected: Even if small amounts of cytochrome c were released in response to lapatinib in the resistant cells, efficient caspase activation would be inhibited at the level of the apoptosome. Because the abundance of both PP5 and Mcl-1 is controlled by HUWE1, apoptotic inhibition both upstream and downstream of the mitochondria could be controlled coordinately.
We also determined that the abundance of CAS was decreased through MDM2-mediated ubiquitylation. Knockdown of CAS renders breast cancer cells resistant to apoptosis (36). CAS also binds to a subset of p53 target genes to regulate p53-mediated gene expression (32). In this regard, our results raise the interesting possibility that, in addition to ubiquitylation of p53, MDM2 might also modulate the p53 signaling pathway by targeting CAS for degradation.
Notably, high MDM2 activity promotes not only degradation of CAS protein but also a decrease in HUWE1 abundance (and thus increased stability of its substrates). Although several proteins have been identified as substrates of HUWE1, such as Cdc6, p53, and N-Myc/c-Myc, little is known about the regulation of this E3 ligase (28, 33, 37-39). It should be noted that MDM2 substrates in addition to HUWE1 and CAS may also contribute to lapatinib resistance. For example, MDM2 enhances IRES (internal ribosome entry site)-mediated translation of XIAP (40). In this regard, it is noteworthy that increased IRES-mediated XIAP translation has been implicated in inflammatory breast cancer cells with acquired resistance to lapatinib (18).
The basis for the difference in MDM2 abundance in resistant and sensitive cells is not clear. This could be secondary to differences in protein stability or in transcription between resistant and sensitive cells. It is possible that a mutation, modification, or unidentified binding partner renders MDM2 unable to auto-ubiquitylate, but still able to ubiquitylate substrates in trans. Alternatively, failure to degrade MDM2 in the resistant cells could result from a mutation in MDM2 itself, rendering it insensitive to degradative signals triggered by lapatinib in sensitive cells. Answering these questions will provide a stepping stone from MDM2 to the relevant signaling factor(s) primarily responsible for the lapatinib resistance while revealing additional nodes in the MDM2 and HUWE1 regulatory network.
MATERIALS AND METHODS
Cell culture
BT474, SKBR3, and AU565 cells were obtained from the American Type Culture Collection (ATCC). SUM190 cells were obtained from Asterand Inc. All cell lines were cultured in RPMI medium containing 10% fetal bovine serum (FBS). Lapatinib-resistant cells (rBT474, rSKBR3, rAU565, and rSUM190) were established as previously described (20). Lapatinib-resistant cells were maintained in the presence of 1 μM lapatinib unless otherwise stated. However, the resistant cells were cultured in lapatinib-free medium for 7 days before the treatment with lapatinib. H1299 cells (from ATCC) were maintained in Dulbecco’s modified Eagle’s medium with 10% FBS.
Reagents and antibodies
Lapatinib and dasatinib were purchased from LC Laboratories. MG132 and z-VAD were purchased from Enzo Life Sciences. E1, E2, ubiquitin, and ubiquitin aldehyde were purchased from Boston Biochem. Nutlin-3a and racemate Nutlin-3 were synthesized and purified at the Duke Small Molecule Synthesis Facility. Nutlin-3a and Nutlin-3 were also purchased from Cayman Chemical. Cycloheximide and purified cytochrome c were purchased from Sigma.
The following antibodies were used: antibodies against HER2, phospho-HER2 (Tyr877), Akt, phospho-Akt (Thr308), caspase-9, cleaved caspase-3, Bcl2, Bcl-xL, ERK1/2, phospho-ERK1/2 (Thr202/Tyr204), phospho-SRC (Tyr416), SRC, FLAG, cytochrome c oxidase IV (Cell Signaling Technology), Apaf-1 (2E12; Enzo Life Sciences), HSP90β (5E12), MDM2 (2A10; Calbiochem), HSP90β (Millipore), phospho-HSP90β (Ser226), phospho-HSP90β (Ser255), α-tubulin (Abcam), actin, MDM2 (SMP14), HA (F-7) (Santa Cruz Biotechnology), FLAG M2 (Sigma), Bim, cleaved caspase-3, Mcl-1,PP5,CAS,HSP70(BDTransduction Laboratories),Mcl-1(BioLegend), HUWE1 (Bethyl Laboratories), and PHAP1 (ProSci).
Plasmids
HA-HUWE1 constructs in pCMV (porcine cytomegalovirus; wild type, C4341S mutant, and amino acids 2473 to 4374 mutant) were gifts from K. Helin (University of Copenhagen). HUWE1 in pENTR was a gift from J. G. Cook (University of North Carolina, Chapel Hill, NC). HUWE1 in pENTR was recombined with the destination vector pDEST10 (Invitrogen) for production of His-tagged protein. FLAG-PP5 in pcDNA3 and FLAG-CAS in p3XFLAG-CMV10 were gifts from X.-F. Wang (Duke University) and C. Prives (Columbia University), respectively. CAS was also cloned into pENTR and subsequently into pDEST10 for baculoviral protein expression. PP5 was cloned into pGEX-KG for production of glutathione S-transferase (GST) fusion protein. Mcl-1 with the N-terminal FLAG tag was generated from human Mcl-1 (a gift from J. Rathmell, Duke University) and cloned into pcDNA3. The plasmids encoding GST-MDM2, GST-MDM2C464A, and HA-tagged ubiquitin were obtained from Addgene (Addgene plasmids 11492, 11493, and 17608) (41, 42). MDM2 was also cloned into pcDNA3 with an N-terminal FLAG tag. All point mutations (HA-HUWE11-2474, HA-HUWE1W1202A, His-HUWE1C4341S, FLAG-CASW127A, FLAG-CASF123A) were generated with the QuikChange mutagenesis kit (Stratagene).
RNA interference
Transient siRNA transfections were performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. MDM2-specific siRNA (sense: 5′-CACCUCACAGAUUCCAGCUUCGGAA-3′) and its scrambled siRNA control (sense: 5′-CACACACUUAGGACCCUUCGU-CGAA-3′) were designed and synthesized by Invitrogen. siRNA oligos targeting HUWE1 or GFP were previously described (37).
For stable knockdown of MDM2, an shRNA construct in the lentiviral vector pLKO.1-puro (shMDM2 #1) was purchased from Open Biosystems (sense: 5′-GATTCCAGAGAGTCATGTGTT-3′). Additional MDM2 shRNA construct (shMDM2 #2; sense: 5′-TTGAAGTTATTAAAGTCTGTT-3′) and control shRNA construct (sense: 5′-CTGTGCTGTAGGTGAAACTGT-3′) were also created in the vector pLKO.1-puro according to Addgene’s pLKO.1 protocol (http://www.addgene.org/plko). Stable knockdown of p53 was performed with the vector shp53-pLKO.1-puro (Addgene plasmid 19119) (43).
Mouse xenograft experiments
Six-week-old female nude mice were purchased from the National Cancer Institute (NCI) Frederick National Laboratory. Xenograft tumors were produced by injection into the mammary fat pad with cells (10 × 106). Tumor volumes were calculated every 3 to 4 days by caliper measurements of the short (a) and long (b) tumor diameters (volume = a × 2 × b/2). Lapatinib and Nutlin-3 were formulated in vehicle (water with 0.5% hydroxypropyl methylcellulose and 0.1% Tween). Mice were dosed orally with vehicle alone, lapatinib (100 mg/kg), Nutlin-3 (100 mg/kg), or combination of both twice daily by gavage.
Immunoprecipitation
Cells were transiently transfected with the indicated plasmid by FuGene 6 (Roche) according to the manufacturer’s instructions. Cells were harvested and lysed with coimmunoprecipitation buffer [10 mM Hepes (pH 7.4), 150 mM KCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, leupeptin (5 μg/ml), and aprotinin (5 μg/ml)]. Cell lysate was incubated with anti-FLAG M2 agarose (Sigma) or anti-HA affinity matrix (Roche) at 4°C for 2 to 3 hours. The bead pellet was washed three times with coimmunoprecipitation buffer and then incubated with SDS sample buffer.
Apoptosis assays
Cell lysates for apoptotic assays were prepared as described previously with buffer A [20 mM Hepes (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, leupeptin (5 μg/ml), and aprotinin (5 μg/ml)] (25). Cell lysate (10 μg/μl) was incubated in the presence or absence of 1 mM dATP and various concentrations of cytochrome c at 37°C for 30 min and subjected to gel filtration, caspase assays, or Western blotting.
Gel filtration was performed as previously described (25). After incubation with cytochrome c, the cell lysate (in a volume of 250 μl) was loaded onto a Superdex 200 column at a flow rate of 0.3 ml/min.
Colorimetric caspase assays were performed by incubating cell lysate (3 μl) in 90 μl of DEVDase buffer [50 mM Hepes (pH 7.5), 100 mM NaCl, 0.1% CHAPS, 10 mM DTT, 1 mM EDTA, 10% glycerol] containing the peptide substrate, Ac-DEVD-pNA (200 mM final concentration; BIOMOL Research Laboratories). Reactions were incubated at 37°C for 30 min. Absorbance of the colorimetric product was measured at 405 nm with a Bio-Rad microplate reader.
In vitro ubiquitylation assays
Recombinant human PP5 and MDM2 proteins were bacterially expressed with a GST tag. After purification by glutathione Sepharose 4B (GE Healthcare), the GST tag was cleaved off by thrombin. Recombinant human wild-type HUWE1, the C4341S mutant (HUWE1 C/S), and HUWE11-2474 were expressed in the baculovirus system and purified from Sf9 cells.
For in vitro ubiquitylation assays, 100 ng of purified recombinant PP5 or HUWE11-2474 was incubated with 10 ng of E1, 100 ng of E2 (UbcH5b or UbcH7), 100 μg of ubiquitin, and 0.5 μg of a purified E3 enzyme (HUWE1 wild type, HUWE1 C/S, or MDM2) in 40 μl of reaction buffer [50 mM tris (pH 7.5), 5 mM MgCl2, 2 mM ATP, 2 mM DTT]. After incubation at 30°C for 3 hours, the reaction was terminated by the addition of SDS sample buffer.
Fluorescence polarization binding assays
The binding affinities of HUWE1 and p53 peptides to MDM2 protein were determined by a quantitative fluorescence polarization-based competitive binding assay, with a recombinant human His-tagged MDM2 protein (residues 1 to 118) and a FAM-tagged modified p53 peptide as the fluorescent probe (34). The following peptide sequences were tested: p53, Ac-LSQETFSDLWKLLPENNVL-NH2; HUWE1, Ac-DGTGEFLDAWLMLVEKMVN-NH2.
Statistics
The statistical analysis was carried out with GraphPad Prism software version 4.0c and R software version 2.15.2. For the densitometric analyses of the immunoblots, significance was computed with a paired, two-tailed t test on Western blot intensities after normalizing to the relevant control (CAS, HUWE1, or actin) and after log transforming to correct for skewness in the distribution.
Supplementary Material
Fig. S1. p53 status of the cell lines used in this study.
Fig. S2. MI-219 can reverse lapatinib resistance.
Fig. S3. SRC abundance in the cell lines used in this study.
Fig. S4. The SRC inhibitor dasatinib does not affect MDM2 protein abundance.
Fig. S5. BT474 cells with acquired lapatinib resistance prevent mitochondrial cytochrome c release in response to lapatinib treatment.
Fig. S6. Mcl-1 abundance increases in SKBR3 cells with acquired lapatinib resistance following lapatinib treatment.
Fig. S7. Mcl-1 is stabilized in lapatinib-resistant SKBR3 cells.
Fig. S8. Ubiquitin aldehyde does not prevent the failure to degrade Mcl-1 in the resistant cells.
Fig. S9. Unaltered abundance of Apaf-1.
Fig. S10. CK2-mediated phosphorylation of HSP90β at Ser226 and Ser255.
Fig. S11. PP5 is stabilized in lapatinib-resistant AU565 cells.
Fig. S12. Biochemical purification of the PP5-targeted E3 ligase.
Fig. S13. Overexpression of HUWE1 leads to reduced abundance of PP5 and Mcl-1.
Fig. S14. CAS degradation in lapatinib-resistant AU565 cells.
Fig. S15. Enhanced CAS polyubiquitylation in lapatinib-resistant SKBR3 cells.
Fig. S16. Schematic diagram of the CAS protein sequence.
Fig. S17. Densitometric analysis of HUWE1 and MDM2 abundance after lapatinib treatment.
Fig. S18. Schematic representation of HUWE1 protein and the amino acid sequence surrounding the MDM2 binding motif.
Fig. S19. HUWE1 and MDM2 interact in cells.
Fig. S20. Densitometric analysis of the abundance of HUWE1, MDM2, CAS, PP5, and Mcl-1 upon lapatinib-Nutlin-3a cotreatment.
Fig. S21. Nutlin-3 can reverse lapatinib resistance in rAU565 xenografts.
Table S1. Biomarker status of breast cancer cell lines used in this study.
Acknowledgments
We would like to thank C. Kent Osborne for the valuable discussion. We would also like to thank J. G. Cook (University of North Carolina, Chapel Hill, NC), K. Helin (University of Copenhagen), A. Lasorella (Columbia University), and J. Opferman (St. Jude Children’s Research Hospital) for HUWE1 constructs; C. Prives (Columbia University) for CAS cDNA (complementary DNA); X.-F. Wang (Duke University) for PP5 constructs; J. Rathmell (Duke University) for Mcl-1 constructs; X. Chen and E. Toone (Duke Small Molecule Synthesis Facility) for Nutlin-3 synthesis; B. Harvat and M. Cook for FACS analyses; and D. Ribar and K. Fernald for excellent technical assistance.
Funding:
This work was supported by NIH RO1 CA102707 (to S.K.), NCI Career Development Award K99 CA140948 (to M.K.), the Balderacchi Gift and Susan G. Komen Scholars Research Program (to N.L.S.), and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012496768-0000475) (to W.-J.K.).
Footnotes
Author contributions:
M.K., N.L.S., and S.K. designed the research. X.R. made the peptides for the fluorescence polarization binding assays. M.K., J.K., K.M., L.L., and T.J.R. performed experiments. M.K., J.G., L.L., R.H., W.-J.K., J.E.L., N.L.S., and S.K. analyzed the data. W.X., M.W.D., S.W., and N.L.S. provided reagents and analytic tools. M.K. and S.K. wrote the manuscript.
Competing interests:
M.K., M.W.D., N.L.S., and S.K. have filed a patent application based on this work. S.W. owns stocks at Ascenta and is a consultant for Ascenta and is a co-inventor of MI-219 and related MDM2 inhibitors. Ascenta has licensed MI-219 and related MDM2 inhibitors from the University of Michigan for clinical development and has sublicensed these MDM2 inhibitors to Sanofi.
REFERENCES
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Feki A, Irminger-Finger I. Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit. Rev. Oncol. Hematol. 2004;52:103–116. doi: 10.1016/j.critrevonc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 4.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 5.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 6.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 7.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 8.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 9.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 10.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 11.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 12.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Jänne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 13.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 14.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J. Clin. Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 15.Spector N, Xia W, El-Hariry I, Yarden Y, Bacus S, Small molecule HER-2 tyrosine kinase inhibitors HER2 therapy. Breast Cancer Res. 2007;9:205. [Google Scholar]
- 16.Johnston S, Trudeau M, Kaufman B, Boussen H, Blackwell K, LoRusso P, Lombardi DP, Ben Ahmed S, Citrin DL, DeSilvio ML, Harris J, Westlund RE, Salazar V, Zaks TZ, Spector NL. Phase II study of predictive biomarker profiles for response targeting human epidermal growth factor receptor 2 (HER-2) in advanced inflammatory breast cancer with lapatinib monotherapy. J. Clin. Oncol. 2008;26:1066–1072. doi: 10.1200/JCO.2007.13.9949. [DOI] [PubMed] [Google Scholar]
- 17.Rexer BN, Engelman JA, Arteaga CL. Overcoming resistance to tyrosine kinase inhibitors: Lessons learned from cancer cells treated with EGFR antagonists. Cell Cycle. 2009;8:18–22. doi: 10.4161/cc.8.1.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aird KM, Ghanayem RB, Peplinski S, Lyerly HK, Devi GR. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol. Cancer Ther. 2010;9:1432–1442. doi: 10.1158/1535-7163.MCT-10-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin AP, Miller A, Emad L, Rahmani M, Walker T, Mitchell C, Hagan MP, Park MA, Yacoub A, Fisher PB, Grant S, Dent P. Lapatinib resistance in HCT116 cells is mediated by elevated MCL-1 expression and decreased BAK activation and not by ERBB receptor kinase mutation. Mol. Pharmacol. 2008;74:807–822. doi: 10.1124/mol.108.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia W, Bacus S, Hegde P, Husain I, Strum J, Liu L, Paulazzo G, Lyass L, Trusk P, Hill J, Harris J, Spector NL. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7795–7800. doi: 10.1073/pnas.0602468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rexer BN, Ham AJ, Rinehart C, Hill S, deMatosGranja-Ingram N, González-Angulo AM, Mills GB, Dave B, Chang JC, Liebler DC, Arteaga CL. Phosphoproteomic mass spectrometry profiling links Src family kinases to escape from HER2 tyrosine kinase inhibition. Oncogene. 2011;30:4163–4174. doi: 10.1038/onc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sánchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, Manning HC, Chang J, Arteaga CL. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang JY, Hsu JL, Meric-Bernstam F, Chang CJ, Wang Q, Bao Y, Yamaguchi H, Xie X, Woodward WA, Yu D, Hortobagyi GN, Hung MC. BikDD eliminates breast cancer initiating cells and synergizes with lapatinib for breast cancer treatment. Cancer Cell. 2011;20:341–356. doi: 10.1016/j.ccr.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, Sahin AA, Esteva FJ, Hortobagyi GN, Yu D. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat. Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa M, Zhao C, Reya T, Kornbluth S. Inhibition of apoptosome formation by suppression of Hsp90β phosphorylation in tyrosine kinase-induced leukemias. Mol. Cell. Biol. 2008;28:5494–5506. doi: 10.1128/MCB.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees-Miller SP, Anderson CW. Two human 90-kDa heat shock proteins are phosphorylated in vivo at conserved serines that are phosphorylated in vitro by casein kinase II. J. Biol. Chem. 1989;264:2431–2437. [PubMed] [Google Scholar]
- 27.Wandinger SK, Suhre MH, Wegele H, Buchner J. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 2006;25:367–376. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Kim HE, Jiang X, Du F, Wang X. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol. Cell. 2008;30:239–247. doi: 10.1016/j.molcel.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 31.Uesugi M, Verdine GL. The α-helical FXXΦΦ motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14801–14806. doi: 10.1073/pnas.96.26.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 34.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yang D, Wang S. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 36.Brinkmann U, Brinkmann E, Gallo M, Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell. 2007;18:3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Zhao X, Heng JI-T, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–375. doi: 10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 42.Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: Implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, McAllister SS, Jacks T, Weinberg RA. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. p53 status of the cell lines used in this study.
Fig. S2. MI-219 can reverse lapatinib resistance.
Fig. S3. SRC abundance in the cell lines used in this study.
Fig. S4. The SRC inhibitor dasatinib does not affect MDM2 protein abundance.
Fig. S5. BT474 cells with acquired lapatinib resistance prevent mitochondrial cytochrome c release in response to lapatinib treatment.
Fig. S6. Mcl-1 abundance increases in SKBR3 cells with acquired lapatinib resistance following lapatinib treatment.
Fig. S7. Mcl-1 is stabilized in lapatinib-resistant SKBR3 cells.
Fig. S8. Ubiquitin aldehyde does not prevent the failure to degrade Mcl-1 in the resistant cells.
Fig. S9. Unaltered abundance of Apaf-1.
Fig. S10. CK2-mediated phosphorylation of HSP90β at Ser226 and Ser255.
Fig. S11. PP5 is stabilized in lapatinib-resistant AU565 cells.
Fig. S12. Biochemical purification of the PP5-targeted E3 ligase.
Fig. S13. Overexpression of HUWE1 leads to reduced abundance of PP5 and Mcl-1.
Fig. S14. CAS degradation in lapatinib-resistant AU565 cells.
Fig. S15. Enhanced CAS polyubiquitylation in lapatinib-resistant SKBR3 cells.
Fig. S16. Schematic diagram of the CAS protein sequence.
Fig. S17. Densitometric analysis of HUWE1 and MDM2 abundance after lapatinib treatment.
Fig. S18. Schematic representation of HUWE1 protein and the amino acid sequence surrounding the MDM2 binding motif.
Fig. S19. HUWE1 and MDM2 interact in cells.
Fig. S20. Densitometric analysis of the abundance of HUWE1, MDM2, CAS, PP5, and Mcl-1 upon lapatinib-Nutlin-3a cotreatment.
Fig. S21. Nutlin-3 can reverse lapatinib resistance in rAU565 xenografts.
Table S1. Biomarker status of breast cancer cell lines used in this study.