SUMMARY
DNA methylation at the 5-position of cytosine (5-mC) is a key epigenetic mark critical for various biological and pathological processes. 5-mC can be converted to 5-hydroxymethylcytosine (5-hmC) by the Ten-Eleven Translocation (TET) family of DNA hydroxylases. Here we report that “loss of 5-hmC” is an epigenetic hallmark of melanoma with diagnostic and prognostic implications. Genome-wide mapping of 5-hmC reveals loss of 5-hmC landscape in the melanoma epigenome. We show that down-regulation of Isocitrate Dehydrogenase 2 (IDH2) and TET family enzymes is likely one of the mechanisms underlying 5-hmC loss in melanoma. Rebuilding the 5-hmC landscape in melanoma cells by reintroducing active TET2 or IDH2 suppresses melanoma growth and increases tumor-free survival in animal models. Thus, our study reveals a critical function of 5-hmC in melanoma development and directly links the IDH and TET activity-dependent epigenetic pathway to 5-hmC-mediated suppression of melanoma progression, suggesting a new strategy for epigenetic cancer therapy.
INTRODUCTION
Melanoma is a unique, highly aggressive type of cancer, which occurs more frequently with increasing age and often with a significant contribution of environmental factors to its etiology (Jemal et al., 2001; Jemal et al., 2006; Marks, 2000). As one of the most virulent human cancers, melanoma is capable of distant and lethal metastases when the primary tumor volume is as little as 1 mm3. Studies of biomarkers predictive of clinical outcome are impeded by latent periods for detection of metastases that may range from several years to more than a decade, and thus clinically-annotated bio-specimen archives serve as valuable surrogates for the otherwise impractical prospective approaches. Such studies are further compounded by the difficulties inherent in the diagnosis of melanoma, since certain benign nevi and melanomas show significant histologic overlap. Presently, there is a dearth of molecular markers that facilitate detecting the differences between benign and malignant melanocytic lesions and assist in predicting their biological behaviors. Thus, there is a pressing need for novel biomarkers that define the malignant potential of primary lesions, predict clinical outcome, and forecast therapeutic responses.
Abnormal DNA methylation at the 5-position of cytosine (5-mC) is a well-known epigenetic feature of cancer. Melanoma exhibits global hypomethylation within the bulk genome and local hypermethylation at specific tumor suppressor genes (Hoon et al., 2004; Liu et al., 2008; Shen et al., 2007). Nonetheless, the degree of global hypomethylation in melanoma is not sufficient to distinguish benign nevus from melanoma (Paz et al., 2003). Gene-specific hypermethylation may be a better discriminator as recent studies indicate that multi-locus DNA-methylation signature genes may differentiate melanomas from nevi (Conway et al., 2011; Tellez et al., 2009). However, this requires sophisticated molecular biological tools that are not easily applicable in routine clinical practice, and the small biopsy size of melanocytic lesions presents another technical limitation. Thus, despite the increasing recognition that abnormal DNA methylation (and/or histone modification) is a crucial participant in melanoma progression; no characteristic epigenetic modifications have been discovered that can be readily used as molecular markers for diagnosis and evaluation of melanoma virulence.
The recent discovery of the Ten-Eleven Translocation (TET) family of 5-mC hydroxylases, including TET1, 2 and 3, which convert 5-mC to 5-hydroxymethylcytosine (5-hmC), also known as the “sixth base”, has added an additional layer of complexity to the epigenetic regulation of DNA methylation (Ito et al., 2010; Tahiliani et al., 2009; Zhang et al., 2010). 5-hmC exists at a high level in self-renewing and pluripotent stem cells (Szwagierczak et al., 2010; Tahiliani et al., 2009). However, 5-hmC levels are greatly reduced in most cultured, immortalized tumor cells (Haffner et al., 2011; Song et al., 2011; Yang et al., 2012). Frequent TET2 mutational inactivation has been reported to associate with decreased 5-hmC levels in various myeloid leukemias (Delhommeau et al., 2009; Langemeijer et al., 2009). In addition, the co-factor α-ketoglutarate (α-KG) is absolutely required and plays a positive and critical role in the conversion of 5-mC to 5-hmC (Xu et al., 2011a). Isocitrate dehydrogenases (IDHs) catalyze oxidative decarboxylation of isocitrate, producing α-KG and CO2 (Reitman et al., 2011; Xu et al., 2011a). There are two major IDH enzymes in mammalian cells, IDH1 in cytoplasm and its homologue, IDH2, in mitochondria, which catalyze the same reaction. It has been reported that gain-of-function mutations in IDH1 and IDH2 in cancer cells produce the oncometabolite 2-hydroxyglutarate (2-HG), an antagonist of α-KG (Chowdhury et al., 2011; Xu et al., 2011a), which inhibits the TET-mediated conversion of 5-mC to 5-hmC. Moreover, similar to the frequent mutation rate of IDH1 or IDH2 in glioma and myeloid leukemia (Dang et al., 2010; Krell et al., 2011), 10% of melanomas harbor a neomorphic mutation in IDH1 or IDH2 (Shibata et al., 2011). These studies suggest a role of 5-hmC, TET and IDH in malignancy. However, it remains elusive as to how 5-hmC is lost and what roles TET and IDH proteins play during tumor progression. In particular, it remains unknown as to how this epigenetic mark and these related enzymes partake in melanoma progression.
Using melanoma as a paradigm of aggressive cancer, here we report that “loss-of-5-hmC” is a new epigenetic hallmark of melanoma. We functionally characterize the significant impact of 5-hmC, IDH2 and TET2 in melanoma progression. Importantly, we show that the activity of IDH2 and TET2 enzymes required for the production of 5-hmC and the re-establishment of the 5-hmC landscape in melanoma cells is essential to regulation of melanoma virulence, contributing to our current understanding of cancer epigenetics.
RESULTS
5-hmC level is high in mature melanocytes and nevi and lost in human melanomas
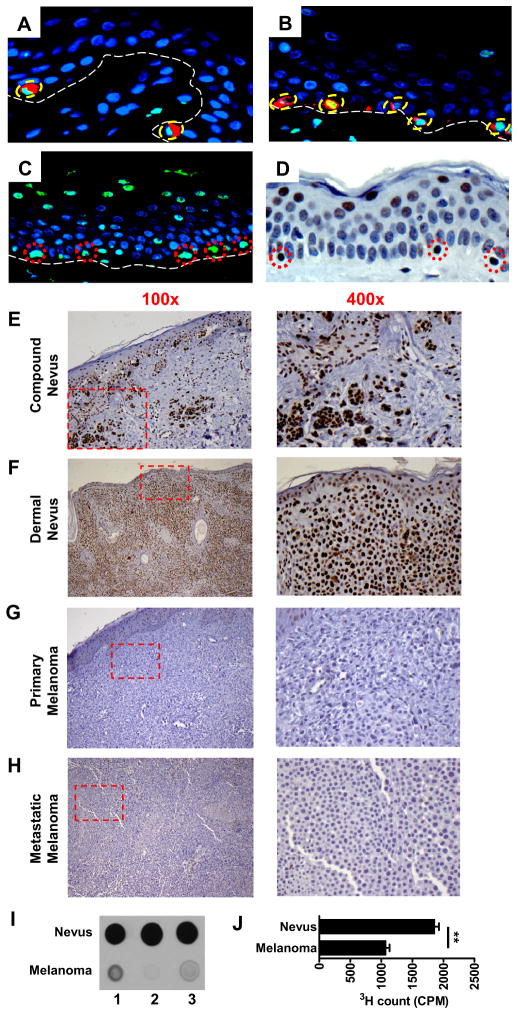
High levels of 5-hmC were detected by immunofluorescent (IF) staining in the nuclei of isolated melanocytes that co-expressed MART-1, a melanocyte specific marker, within the epidermal basal cell layer (Figures 1A and 1B). A more sensitive method for IF or immunohistochemical (IHC) staining of 5-hmC using formalin-fixed, paraffin-embedded tissue sections resulted in loss of the MART-1 epitope, but significantly improved the detection of 5-hmC as demonstrated by staining in other normal human tissues (Figure S1A). By this method, strong IF staining of 5-hmC was detected in melanocytes within the otherwise negative basal layer, as seen in Figures 1A and 1B, as well as variably within more differentiated suprabasal keratinocytes (Figure 1C). Occasional intradermal cells also showed 5-hmC reactivity (Figure 1C). The IF staining pattern was confirmed by IHC staining (Figure 1D), and this more sensitive method for 5-hmC detection was utilized for all subsequent studies of melanocytic nevi and melanomas.
Figure 1. 5-hmC level is high in mature melanocytes and lost in melanomas.
(A–B) IF co-staining of 5-hmC and MART1 in normal human skin without HCl treatment. Green: 5-hmC; red: MART1; blue: DAPI counterstain of DNA. Among basal layer cells (dotted white line), 5-hmC-positive cells exclusively proved to be MART-1-positive melanocytes (dotted circles).
(C–D) Detecting 5-hmC in normal human skin by IF (C) and IHC staining (D) with HCl treatment. Both methods showed strong nuclear staining in isolated, solitary cells within the basal layer (dotted circles), in nuclei within the uppermost epidermal layers and occasional dermal cells.
(E–H) Representative histology of 5-hmC IHC staining in the individual cases of benign and malignant melanocytic lesions. Low power images (100×) on the left column with the dotted red area magnified at high power (400×) on the right column. All slides were counterstained with hematoxylin (light blue).
(I) Immunoblotting assay shows significantly higher 5-hmC levels in benign nevi than in melanomas. Three representative immunoblot images are shown here from the 10 cases of each group.
(J) 5-hmC glucosylation assay confirms that the 5-hmC level in the genomic DNA of nevi is significantly higher than that in melanomas. **P<0.01 by Student’s t-test. Data are shown as mean ± SD (n=3).
Over 50 individual cases of representative melanocytic lesions, including benign nevi, primary melanomas, and metastatic melanomas were initially evaluated. Benign nevus cases (n=30) showed strong nuclear 5-hmC staining, whereas virtually all tumor cells in primary (n=15) and metastatic (n=10) melanomas showed partial or complete loss of 5-hmC (Figures 1E-1H). Significant differences in other epimarks were not observed between benign nevi and melanomas (Figure S1B and Table S1), suggesting the unique discriminatory nature of 5-hmC staining as it relates to cells of melanocytic lineage. We next purified genomic DNA from nevi and melanomas and confirmed higher 5-hmC levels in nevi than in melanomas by two independent methods, the anti-5-hmC antibody-based dot-blot (Figure 1I) and T4 Phage β-glucosyltransferase-mediated 5-hmC glucosylation assay (Figure 1J). Taken together, these data demonstrate that while a high level of 5-hmC is a distinctive epigenetic signature for benign melanocytes and nevi, significantly diminished or complete loss of 5-hmC is a feature of melanomas.
5-hmC is a putative molecular marker of melanoma progression
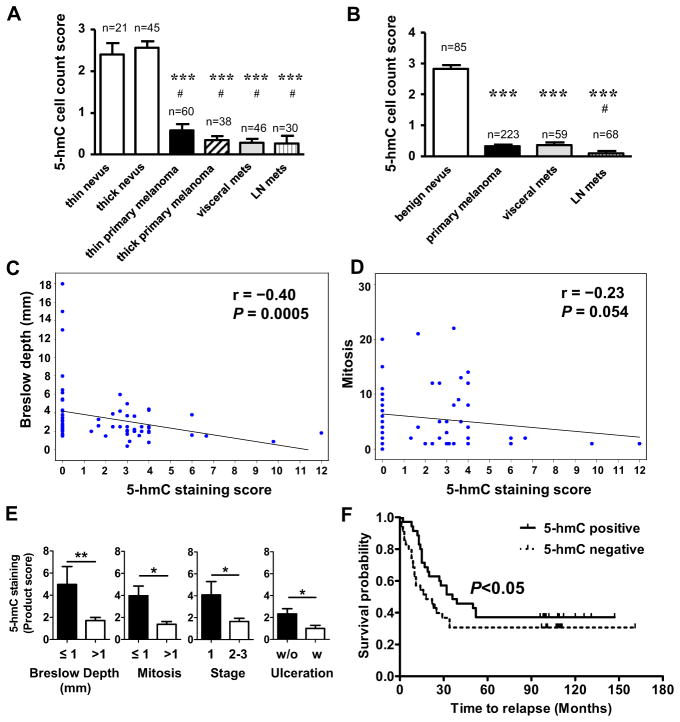
We next examined 5-hmC levels by IHC using a melanoma progression tissue microarray (TMA) representing four major diagnostic tumor types: benign melanocytic nevus, primary cutaneous melanoma, melanoma metastases to lymph nodes and metastases to viscera (Kabbarah et al., 2010; Schatton et al., 2008). Consistent with the individual cases examined above (Figure 1), the TMA confirmed significant 5-hmC loss in primary melanomas and metastatic melanomas compared with nevi (P<0.001; Figures 2A and S2, Tables S2 and S3). In two additional commercially available melanoma TMAs, there was significant loss of 5-hmC in melanomas compared to benign nevi (P=1.1×10−7) and loss in nodal compared to visceral metastases (P=0.016) (Figure 2B). Taken together, these data further support “loss of 5-hmC” as a distinctive epigenetic event in melanoma, and suggest 5-hmC may represent a new epigenetic mark for melanoma recognition and progression.
Figure 2. Loss of 5-hmC correlated with melanoma progression.
(A) Analysis of 5-hmC levels in the SPORE TMA represented by positive cell count score. Each column represents a category of melanocytic lesion (n=number of cases, each case has duplicated tissue cores). Data are shown as mean ± SEM. ***P<0.001 compared to benign thin nevi; #P<0.001 compared to benign thick nevi.
(B) Combined cell count scores of 5-hmC staining of three tissue microarrays. Each column represents a category of melanocytic lesion (n=number of cases). Data are shown as mean ± SEM. ***P<0.001 compared to benign nevus, #P<0.05 compared to visceral metastases.
(C–D) The Spearman correlation between Breslow depth and 5-hmC staining product score (C) or between mitosis and 5-hmC staining product score (D).
(E) 5-hmC staining product scores are correlated with critical melanoma staging parameters. Data are shown as mean ± SEM. *P<0.05, **P<0.01 by Student’s t-test.
(F) Kaplan-Meier survival curves of melanoma patients with positive 5-hmC staining (solid line) and negative 5-hmC staining (dashed line). P < 0.05 by Gehan-Breslow-Wilcoxon Test.
See also Figure S2 and Tables S2–S6.
We next correlated the 5-hmC level with critical melanoma staging parameters (tumor depth and mitotic rate) using the melanoma specimens from a clinically-annotated cohort including 70 superficial spreading and nodular melanomas (Table S4). There was a negative correlation between 5-hmC staining score and primary melanoma Breslow depth, a standard predictor of prognosis (Figure 2C, r=−0.4, P=0.0005), as well as between 5-hmC level and mitotic rate (Figure 2D, r=−0.23, P=0.054). Furthermore, 5-hmC levels were significantly reduced in melanomas with Breslow values of >1 mm compared to those with Breslow values of ≤1 mm (P<0.01), and melanomas with the presence of >1 mitosis (a current predictor of nodal metastasis) had less staining than those with ≤1 mitosis (P<0.05) (Figure 2E and Tables S5 and S6). Similarly, 5-hmC levels in pathological stage 1 melanomas were significantly higher than in stage 2–3 melanomas (P<0.05), and were significantly lower in melanoma patients with ulceration (an important staging parameter) than those without ulceration (P<0.05) (Figure 2E and Tables S5 and S6). We further analyzed the association between 5-hmC levels and the survival probability based on data for all 70 patients (Table S4). Importantly, Kaplan-Meier curves revealed that patients with 5-hmC-positive melanomas (staining score≥1) had significantly higher survival probabilities than patients with 5-hmC-negative melanomas (staining score=0) at diagnosis (Figure 2F). Thus, loss of 5-hmC in melanoma has both diagnostic and prognostic value in these biospecimen cohorts.
Genome-wide mapping of 5-hmC in nevi and melanomas reveals a demolished 5-hmC landscape in the epigenome of melanomas
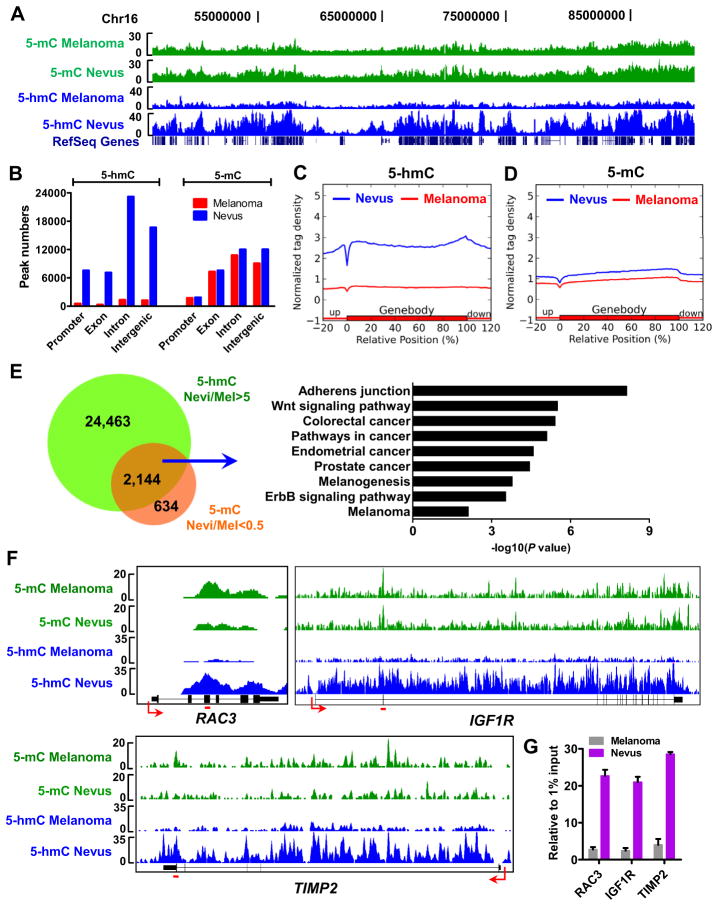
We next asked if 5-hmC loss in melanoma is genome-wide or loci-specific. We first investigated the 5-hmC level changes at specific genomic loci by mapping the genome-wide 5-hmC distribution in nevus and melanoma tissues using a barcoded hydroxymethylated DNA immunoprecipitation (hMeDIP) approach coupled with deep sequencing (hMeDIP-seq) (Xu et al., 2011b) that permits quantitative comparisons of genome-wide changes of 5-hmC between nevi and melanomas (Figure S3A). Similar to our previous finding in mouse ES cells (Xu et al., 2011b), we found that 5-hmC is associated with gene-rich regions in the nevus genome (Figure 3A),. Using MACS software (Zhang et al., 2008), we identified a total 54,454 5-hmC peaks in nevi (P<10−5, FDR<0.01, fold enrichment >10) among which more than half are located either in exons (13%) or introns (42.6%) and 13.9% are located at promoters (Figures 3B and S3B). These 5-hmC peaks are associated with 17,468 Refseq genes, in which 15,750 and 10,065 genes are modified by 5-hmC in gene bodies and promoters (−2k to +2k of transcriptional start sites (TSSs)), respectively. There are 8,347 Refseq genes that are modified by 5-hmC both at promoters and in gene bodies. However, we only identified 3,362 5-hmC peaks in melanomas using the MACS software with the same cut-off values as for nevi (Figures 3B and S3B). These peaks are only associated with 3,219 Refseq genes. Importantly, we observed a significantly decreased 5-hmC level within the averaged gene bodies and 20% of their up- and down-stream regions in melanomas in comparison to nevi (Figure 3C). Indeed, we identified 41,886 out of 54,454 (77%) total 5-hmC peaks in nevi whose normalized 5-hmC densities were dramatically higher (>5-fold) than in melanomas. These 41,886 peaks are located in 15,240 Refseq genes at either promoters or gene bodies. Taken together, these analyses indicate that loss of 5-hmC is a genome-wide event during melanoma progression.
Figure 3. Genome-wide mapping of 5-mC and 5-hmC in benign nevi and melanomas.
(A) The distribution of 5-mC (green) and 5-hmC (blue) densities in the region of chr16:46,651,039-89,749,255 by MeDIP-seq and hMeDIP-seq. Refseq genes are shown at the bottom.
(B) 5-mC and 5-hmC peak numbers of nevus (red) and melanoma (blue) hMeDIP samples in different genomic regions. Promoters were defined as −2k to +2k relative to TSS.
(C–D) Normalized 5-hmC (C) and 5-mC (D) tag density distribution across the gene body. Each gene body was normalized to 0–100%. Normalized Tag density is plotted from 20% of upstream of TSSs to 20% downstream of TTSs.
(E) Peaks at which 5-hmC is significantly reduced (>5-fold) and 5-mC is significantly increased (>2-fold) in gene bodies in melanomas (Mel) compared to nevi (left panel), and the KEGG pathway analysis results for the associated genes (right panel).
(F–G) MeDIP-seq and hMeDIP-seq results of RAC3, IGF1R and TIMP2 genes (F) and hMeDIP-qPCR verifications (G). The primer targeted regions in panel G are noted by red lines in panel F. Data are shown as mean ± SD (n=3) in panel G.
We next investigated whether the 5-mC genome-wide distribution is also altered in melanomas. We identified 33,374 and 28,830 5-mC peaks in nevi and melanomas (P<10−5, FDR<0.01 and fold enrichment>5), respectively (Figures 3B and S3B). In contrast to 5-hmC, the genome-wide enrichment and distribution pattern of 5-mC is similar between nevi and melanomas, although a slight decrease of 5-mC in melanomas was observed (Figures 3B and 3D), which is consistent with the previously reported global DNA hypomethylation in melanomas (Tellez et al., 2009).
To determine whether gene specific changes of 5-hmC and 5-mC are associated with melanoma progression, we normalized 5-hmC and 5-mC tag densities in nevus and melanoma samples according to each input sequencing read, which permitted quantitative comparison of 5-hmC and 5-mC signal changes at specific genome loci. Since 5-hmC is converted from 5-mC by TET enzymes, we reasoned that the decreased 5-hmC generation in melanomas would result in the accumulation of its substrate, 5-mC, at certain genomic regions compared to nevi. Indeed, we identified 2,144 peaks at which 5-hmC is dramatically higher (>5-fold) and 5-mC is significantly lower (>2-fold) in 3,401 Refseq gene bodies in nevi compared to melanomas (Figure 3E). Importantly, KEGG pathway enrichment analysis for the 3,401 genes revealed that these genes are closely associated with various melanoma related pathways, such as adherens junction (P=6.05×10−9), Wnt signaling (P=1.67×10−7), pathways in cancer (P=8.65×10−7), and melanogenesis pathways (P=4.84×10−4) (Figure 3E). As exemplified in Figure 3F, RAC3, IGF1R and TIMP2 genes show decreased 5-hmC and increased 5-mC in gene bodies in melanomas compared with nevi, and the 5-hmC changes are further verified by conventional hMeDIP-qPCR assays (Figure 3G). Similarly, we also identified 517 peaks at which 5-hmC is dramatically higher (>5-fold) and 5-mC is significantly lower (>2-fold) in 926 gene promoters in nevi compared to melanomas (Figure S3C). Gene Ontology (GO) term and KEGG pathway analyses for the 926 genes also shows that they are involved in the regulation of cell morphogenesis (P=1.18×10−4), cytoskeleton organization (P=0.001), Ras protein signal transduction (P=0.00358), posttranscriptional regulation of gene expression (P=0.0036) (Figure S3C) and Wnt signaling pathway (P=0.009). The 5-mC and 5-hmC densities at representative gene promoters are shown in Figure S3D.
Thus, this study for the first time establishes the genome-wide map of methylome and hydroxylmethylome in nevus and melanoma and reveals the progressive loss of 5-hmC landscape in the epigenome from begin nevus to malignant melanoma. Thus, the demolished 5-hmC landscape is an epigenetic hallmark for melanoma.
Down-regulation of IDH2 and TET family members in melanomas
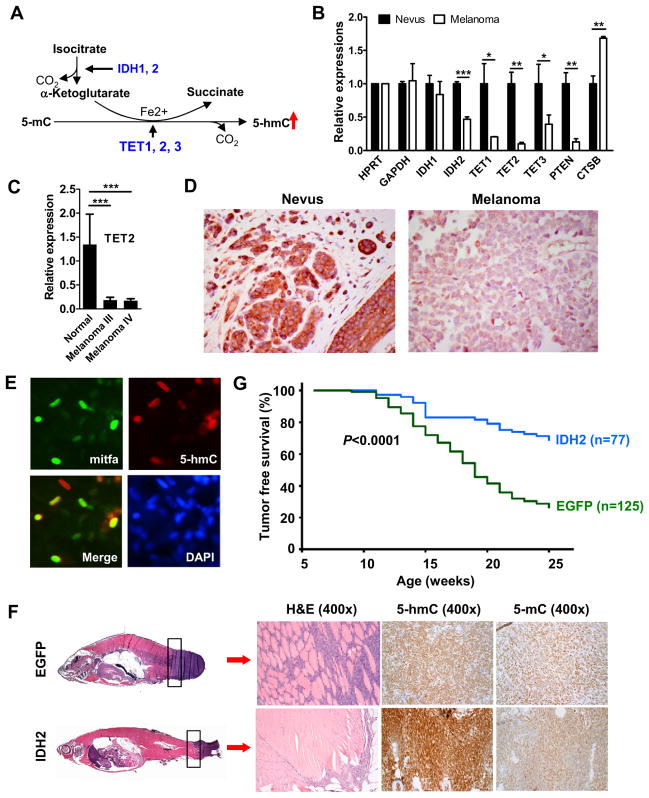
We next sought to determine the imminent cellular factors that are directly responsible for loss of 5-hmC in melanomas. While the TET family of 5-mC DNA hydroxylases are directly responsible for the generation of 5-hmC (Figure 4A), the catalytic reaction requires cofactor α-KG (Ito et al., 2010; Tahiliani et al., 2009) which is mainly controlled by IDHs (Xu et al., 2011a). Therefore, we hypothesized that IDH and/or TET family enzymes play a critical role in the establishment and maintenance of the 5-hmC landscape in the epigenome of melanocytes and their neoplasms, and down-regulation of these key enzymes may be responsible for the loss of 5-hmC in melanomas. To test this hypothesis, we first examined the expression levels of TET and IDH family genes in both nevi and melanomas by RT-qPCR. While IDH1 has a similar expression level between nevi and melanomas, we found that IDH2 is significantly down-regulated in melanomas (Figure 4B). Strikingly, expression of all three TET genes was significantly lower in melanomas than in nevi, with the most dramatic decrease in the TET2 (Figure 4B). As positive controls, decreased expression of tumor suppressor gene PTEN and increased expression of cathepsin B (CTSB) gene was observed in melanomas compared to nevi, while the expression of house keeping gene GAPDH was unchanged (Haqq et al., 2005; Riker et al., 2008; Talantov et al., 2005). Thus, the dramatically decreased expression of IDH2, TET1, TET2 and TET3 is specific and may reflect the down-regulated nature of these genes in melanomas. Furthermore, the decreased expression of TET2 in melanomas was confirmed via melanoma cDNA arrays (Figure 4C), and the significantly decreased expression of IDH2 at the mRNA level was corroborated at the protein level by IHC staining (Figure 4D). These data suggest that the diminished expression of IDH2 and/or TET family genes in melanomas may represent one of the molecular mechanisms underlying global loss of 5-hmC.
Figure 4. Increased 5-hmC level by IDH2 over-expression in a zebrafish melanoma model prolongs tumor free survival.
(A) Schematic diagram of 5-hmC generation by the TET family of 5-mC DNA hydroxylases with cofactors α-ketoglutarate and Fe2+.
(B) Relative expression of genes in nevus and melanoma by RT-qPCR. Each gene expression level was normalized to HPRT house-keeping gene. Data are shown as the mean of three individual patients ± SEM. *P<0.05, **P<0.01, *** P<0.001 by Student’s t-test comparing nevus to melanoma.
(C) Relative TET2 expression in human melanoma cDNA arrays including normal skin (n=3), stage III (n=21) and stage IV melanomas (n=19) by RT-qPCR. Data are shown as mean ± SEM. *** P<0.001 compared the normal skin by Student’s t-test.
(D) Representative IDH2 IHC staining images in nevi (n=4) and melanomas (n=8) at high power (400×).
(E) IF staining of 5-hmC and mitfa in normal zebrafish melanocytes. Green: mitfa; red: 5-hmC; blue: DAPI counterstain of DNA.
(F) Tumors were smaller and less invasive and had higher 5-hmC levels in miniCoopR IDH2 zebrafish than mini-CoopR EGFP control zebrafish. Histology of the melanomas from miniCoopR-EGFP control zebrafish and miniCoopR-IDH2 zebrafish are shown in the left panels. The H&E staining of tumor sections shows an infiltrative pattern of tumor in control miniCoopR-EGFP zebrafish at the body and tail junction, while the tumor shows much less infiltrative borders in IDH2 over-expressing zebrafish.
(G) Significant prolongation of tumor-free survival in miniCoopR-IDH2 zebrafish (n=77) compared with miniCoopR-EGFP control zebrafish (n=125).
Over-expression of IDH2 in a zebrafish melanoma model increases 5-hmC levels and prolongs tumor free survival
To test the hypothesis that over-expression of IDH2 may result in increased 5-hmC levels in melanoma and suppress tumor growth, we employed a recently developed transgenic (Tg) zebrafish model for melanoma, in which BRAFV600E is expressed under the control of the mitfa gene melanocyte-specific promoter on a p53 mutant background (p53−/−) (Ceol et al., 2011). While the 5-hmC level is high in normal zebrafish melanocytes shown by co-stained with melanocyte-specific mitfa (Figure 4E), 5-hmC is barely detected in melanomas lesion of EGFP control animals (Figure 4F upper panel). IDH2 over-expression greatly increases the 5-hmC level in melanoma lesions compared to that of EGFP control animals (Figure 4F lower panel), while over-expression of IDH2 R172K mutant does not shown any significant effects on the 5-hmC level (data not shown). Strikingly, tumor incidence curve analysis revealed that IDH2 wt over-expressing group, but not IDH2 R172K over-expressing group, showed significantly increased tumor-free survival compared to the EGFP control group (P=7.8×10−9 by logrank test) (Figures 4G and S4). Collectively, these data suggest that the isocitrate dehydrogenase activity of IDH2 plays an important role in maintaining proper levels of 5-hmC in melanocytes and may function as a putative tumor suppressor for melanoma progression.
Reestablishing the 5-hmC landscape in the epigenome of human melanoma cells by reintroducing TET2
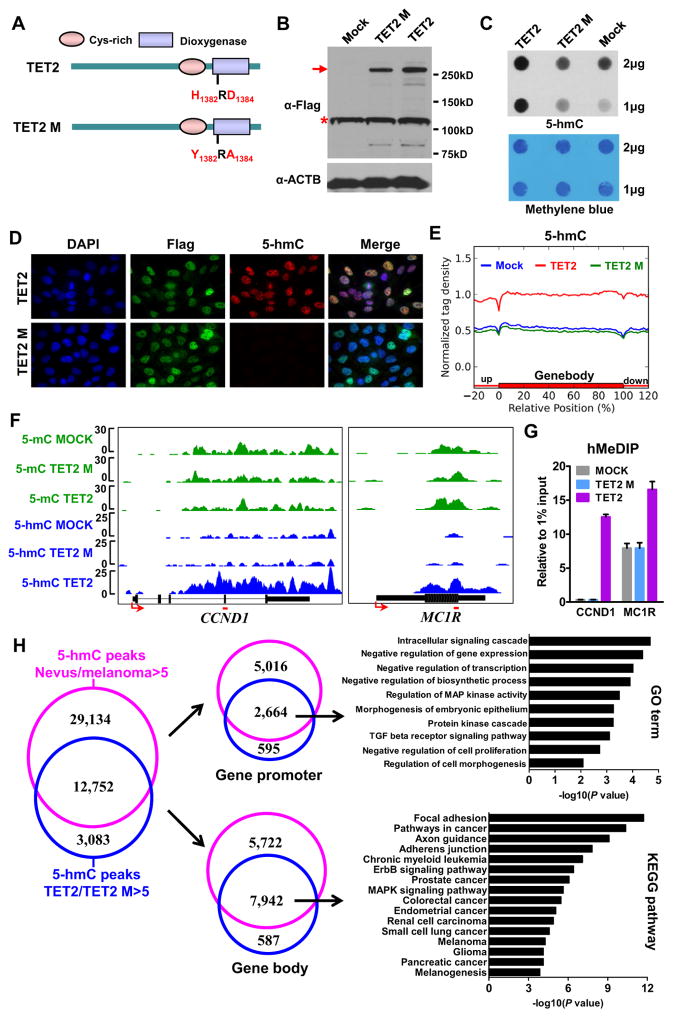
We next asked whether reintroducing TET2, the most dramatically decreased TET family gene in human melanomas, can rescue the demolished 5-hmC landscape in melanoma cells. To exclude the 5-mC hydroxylase-independent function of TET2, we generated pure monoclonal A2058 stable cell lines over-expressing flag-tagged full length wt TET2 or the iron-binding site (H1382RD1384) disrupted catalytically-inactive mutant (TET2 M) (Figure 5A), as well as the vector only control (Mock). The over-expression of full-length TET2 and TET2 M was verified by Western blot and RT-qPCR assays (Figures 5B and S5A). We confirmed the global increase of 5-hmC levels in TET2 over-expressing cells, but not TET2 M over-expressing cells, compared to Mock cells by dot-blot and IF assays (Figures 5C and 5D).
Figure 5. TET2 over-expression re-establishes the 5-hmC landscape in the epigenome of human melanoma cells.
(A) Schematic diagram of TET2 wt and TET2 catalytically inactive mutant (TET2 M) proteins.
(B) The expression of Flag-tagged TET2 and Flag-tagged TET2 M proteins by Western blot. Red arrow denotes the full length TET2 and TET2 M bands, and red star denotes non-specific bands. ACTB was used as a loading control.
(C) Global 5-hmC levels in MOCK, A2058 TET2 and A2058 TET2 M stable cell lines by dot-blot assay. The Methylene blue staining was used as total genomic DNA loading control.
(D) IF analysis of A2058 TET2 and A2058 TET2 M stable cell lines. The Flag antibody was used to detect Flag-tagged TET2 and Flag-tagged TET2 M. Blue: DAPI counterstain of DNA; green: Flag; red: 5-hmC.
(E) Normalized 5-hmC tag density distribution across the gene body. Each gene body was normalized to 0–100%. Normalized tag density is plotted from 20% of upstream of TSSs to 20% downstream of TTSs.
(F–G) MeDIP-seq and hMeDIP-seq results of CCND1 and MC1R genes (F) and hMeDIP-qPCR verifications (G). The primer targeted regions in panel G are noted by red lines in panel F. Data are shown as mean ± SD (n=3) in panel G.
(H) Venn diagrams showing the overlap between 5-hmC peaks which are dramatically higher (>5-fold) in nevi than melanomas (pink) and 5-hmC peaks which are dramatically higher (>5-fold) in TET2 over-expression cells compared to TET2 M over-expression cells (blue) (left panel), and the associated genes according the peak location either at gene promoter (middle upper panel) or in gene body (middle lower panel). The GO term and KEGG pathway analyses results are shown in the right panels.
See also Figures S3 and S5 and Table S7.
We next generated the genome-wide maps of 5-mC and 5-hmC in Mock, TET2- and TET2 M-over-expressing melanoma cells by MeDIP-seq and hMeDIP-seq as described above (Figure S3A). While only marginal changes in 5-mC levels among Mock, TET2- and TET2 Mover-expressing melanoma cells were observed (Figures S3A and S5B), TET2 over-expressing melanoma cells showed re-establishment of the 5-hmC landscape in their epigenome (Figure 5E). No significant 5-hmC level differences were found between Mock and TET2 M cells. As exemplified by CCND1 and MC1R genes (Figure 5F), deep sequencing data were confirmed by conventional hMeDIP-qPCR assays (Figure 5G).
Further bioinformatic analyses identified 15,835 peaks in TET2 over-expressing cells whose 5-hmC densities are dramatically higher (>5-fold) than in TET2 M cells. 80% (12,752/15,835) of these peaks overlap with a significant portion (30%, 12,752/41,886) of 5-hmC peaks whose 5-hmC levels are dramatically lower (>5-fold) in melanomas than nevi (Figure 5H, left panel), suggesting that the 5-hmC landscape in those genomic regions can be reestablished by reintroducing TET2 in human melanoma cells. These overlapping 5-hmC peaks were further analyzed according to their locations (promoter or gene body) at associated genes. We identified 2,664 Refseq genes whose promoters have consistently higher 5-hmC densities in nevi and TET2 over-expressing cells compared to melanomas and TET2 M cells, respectively (Figure 5H, upper middle panel). GO term analysis reveals that these 2,664 genes are mainly associated with intracellular signaling cascade, regulation of gene transcription, cell proliferation and morphogenesis (Figure 5H, upper right panel). Similarly, we identified 7,942 Refseq genes whose gene bodies have consistently higher 5-hmC densities in nevi and TET2 over-expressing cells compared to melanomas and TET2 M cells, respectively (Figure 5H, lower middle panel). Importantly, KEGG pathway analysis shows an impressive functional association of these genes with various cancer-related pathways, such as focal adhesion (P=1.9×10−12), pathways in cancer (P=4.32×10−11), adherens junction (P=7.94×10−10), melanoma (P=5.39×10−5) as well as ErbB (P=3.86×10−7) and MAPK (P=2.29×10−6) signaling pathways (Figure 5H, lower right panel).
In sum, these biochemical and genome-wide analyses suggest that reintroducing wt TET2, but not the TET2 catalytically inactive mutant, is able to significantly increase the global 5-hmC level and reestablish, at least in part, the 5-hmC landscape in the epigenome of human melanoma cells, which may subsequently affect several biological processes, such as cancer progression.
Tumor invasion and growth is suppressed by TET2-mediated reestablishment of the 5-hmC landscape in melanoma cells
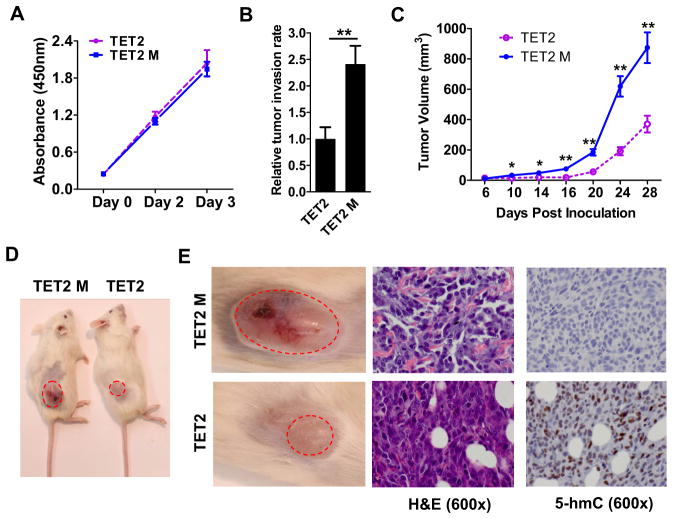
To determine the biological consequence of TET2-mediated reestablishment of the 5-hmC landscape in melanoma cells, we first compared the invasive ability of A2058 cells over-expressing TET2 and TET2 M by in vitro Matrigel assay. While these two cell lines have similar in vitro proliferation rates (Figure 6A), TET2 over-expressing cells show a significantly lower invasion rate than TET2 M cells (Figure 6B). Next, we investigated the putative tumor suppressor role of 5-hmC and TET2 using in vivo xenograft assays. TET2- and TET2 M- over-expressing melanoma cells were injected into NSG mice and tumor growth was monitored. TET2 over-expressing melanoma cells gave rise to significantly smaller tumors compared to TET2 M melanoma cells (Figures 6C and 6D). NSG mice injected with Mock melanoma cells showed no significant differences in tumor growth compared to TET2 M over-expressing melanoma cells (data not shown). Importantly, 5-hmC IHC staining of the xenografted tumor sections showed that smaller tumors derived from TET2 over-expressing cells retained relatively high levels of 5-hmC (Figure 6E, right panels). Thus, these data highlight a potential tumor suppressor role of increased 5-hmC levels meditated by the enzymatic activity of TET2 in melanoma.
Figure 6. Over-expression of TET2 in human melanoma cells suppresses tumor growth in NSG xenograft mice.
(A) The proliferation curves of A2058 TET2 and A2058 TET2 M stable cell lines.
(B) A2058 TET2 melanoma cells show less in vitro invasion than A2058 TET2 M melanoma cells by Matrigel tumor invasion assay. Data are shown as mean ± SD (n=3). ** P<0.01 by Student’s t-test.
(C) Tumor growth curves of A2058 TET2 and A2058 TET2 M cells xenografted to NSG mice. Data are shown as mean± SEM (n=10). *P<0.05, **P<0.01 by Student’s t-test.
(D) Representative images of tumor-bearing NSG mice xenografted with A2058 TET2 M (left) or A2058 TET2 cells (right) at 4 weeks post inoculation.
(E) H&E and 5-hmC IHC staining of TET2 M (upper) and TET2 (lower) xenografts. The regions shown in left panels are noted by red dash circles in panel D.
DISCUSSION
Here we report that “loss of 5-hmC” is a distinctive epigenetic event of neoplastic progression in melanoma that correlates with clinical relapse-free survival and melanoma staging parameters. Thus, 5-hmC holds promise as a putative molecular biomarker with predictive and prognostic value. The present study also for the first time illustrates the genome-wide 5-hmC landscape of benign nevi and melanomas and reveals the strikingly demolished 5-hmC levels and distribution along the epigenome of melanomas in comparison with benign nevi. Furthermore, loss of 5-hmC in melanoma is caused, at least in part, by the decreased expression of key enzymes, IDH2 and TET family proteins, controlling 5-hmC production. In relevant animal models, increase in 5-hmC levels via either IDH2 or TET2 overexpression is shown to suppress tumor invasion and growth and improve tumor free survival. Taken together, the present study provides multiple layers of evidence to support that genome-wide “loss of 5-hmC” is a new epigenetic hallmark of melanoma with diagnostic and prognostic advantages over global DNA hypomethylation, a recognized epigenetic mark of cancer. Of clinical and therapeutic significance, the present study also opens a new avenue for cancer prevention by targeting the cellular and biochemical pathways that can re-establish 5-hmC levels and landscape in melanoma.
The IHC staining approach to detect 5-hmC may have practical applications clinically. We and others have observed similar loss of 5-hmC in other solid tumors such as breast, ovarian, and colon carcinoma (data not shown) using the same methods (Haffner et al., 2011; Yang et al., 2012). The anti-5-hmC antibody-based IHC strategy could lead to the development of a new, simple, sensitive and practical adjuvant diagnostic assay. Future studies focusing on borderline lesions and with sufficient clinical outcome annotation are now indicated to evaluate the utility of “loss of 5-hmC” as a novel diagnostic and prognostic tool.
The high level of 5-hmC in differentiated and benign nevomelanocytes raises an intriguing question as to the nature of the biological role of 5-hmC in melanocyte differentiation, self-renewal and malignant transformation. Studies of embryonic stem (ES) cells indicate that 5-hmC may be involved in the regulation of cell differentiation and lineage commitment (Ito et al., 2010; Xu et al., 2011b). Until now, skin tissue-specific 5-hmC distribution and genome-wide mapping of 5-hmC in cancer have not been well studied. Our findings of a high level of 5-hmC in mature melanocytes and benign nevi, as well as a significantly lower level of 5-hmC associated with melanoma, provide new insights supporting a role of 5-hmC in pathways fundamental to cellular differentiation and de-differentiation. We postulate that the well-controlled dynamic level of 5-hmC during transition from ES cell to melanocyte progenitor to terminally-differentiated melanocyte is a novel epigenetic signature of melanocyte differentiation, the perturbation of which may lead to the induction of oncogenic pathways underlying melanoma progression.
There are several ways to influence 5-hmC levels in cells (Figure 4A). Presumably, dysfunction of TET and/or IDH enzymes, two key factors involved in 5-hmC generation, would greatly reduce 5-hmC generation. 10% of melanomas (4/39) harbor an IDH1 or IDH2 mutation Shibata et al., 2011), whereas no TET mutations have been reported in melanoma. The low penetration of IDH and TET mutations in melanoma constitutes robust evidence that other cancer pathways inactivating these 5-hmC-generating enzymes must play a major role in down-regulation of 5-hmC. Herein, we demonstrate the significant decrease in TET1, TET2, TET3 and IDH2 gene expression in melanomas compared to benign nevi, which suggests that insufficient enzymes required for the conversion of 5-mC to 5-hmC may account for one of the molecular mechanisms underlying global “loss of 5-hmC” in melanomas.
In support of this hypothesis, we show that increasing 5-hmC levels and partially reestablishing the 5-hmC landscape in melanoma cells by restoring expression of active TET2 enzyme but not the catalytically inactive TET2 mutant, significantly suppresses tumor growth in a murine human melanoma xenograft model. Importantly, this study excludes the possibility that the tumor-suppressive effect is due to the over-expression of TET2 itself. Rather, such an effect is due to elevated levels of 5-hmC on the genes important for key cellular processes. Moreover, we demonstrated that a forced increase in 5-hmC in an established zebrafish melanoma model via over-expression of IDH2 wt, but not IDH2 R172K mutant, significantly suppressed tumor growth and prolonged tumor-free survival. These data suggest that IDH2, but not IDH1, is specifically down-regulated in melanomas and that the wt IDH2 acts as a putative tumor suppressor in the zebrafish melanoma model although the IDH family of enzymes have been considered candidate oncogenes in various tumors (Ward et al., 2010; Wrzeszczynski et al., 2011). Nonetheless, while the nature of the putative tumor suppressor function of IDH2 in melanoma and other tumor types warrants future investigations, a dramatic increase in the global 5-hmC level is the most pronounced epigenetic alteration observed both in the TET2 and IDH2 over-expressing melanoma animal models. Thus, these data demonstrate that high levels of 5-hmC and the appropriate 5-hmC landscape in the epigenome of melanocytes and nevus cells, both potential melanoma progenitors, play a role in preserving the integrity of these indolent cells and in preventing melanoma initiation and progression. Our study supports the novel concept that an elevated level of 5-hmC can serve as a distinctive epigenetic molecular beacon for the reversal of an aggressive melanoma phenotype. It further indicates that particular TET and IDH family enzymes have putative tumor suppressor functions in melanoma progression, and spontaneously targeted down-regulation or inactivation of multiple key enzymes in 5-hmC generating pathway is one of the epigenetic mechanisms underlying melanoma development.
The present study also attempts to address the molecular mechanisms directly linking the gene specific 5-hmC loss to melanoma formation. Genome-wide mapping and comparative analyses of 5-mC and 5-hmC landscape in benign nevi, primary melanomas, MOCK, TET2- and TET2 M-over-expressing melanoma cells indicated that a program of genes involving various cancer pathways display significant reduction of 5-hmC in comparisons between benign nevi and melanoma, which can be reversed by over-expression of active TET2 but not inactive TET2 M. However, we did not find simple correlations between significant loss of 5-hmC and expression of associated genes because a reduced 5-hmC level is associated with both up- and down-regulated genes in melanoma compared with nevi. This is not surprising since we and others have shown complex roles of 5-hmC in gene transcription regulation in mouse ES cells (Ficz et al., 2011; Xu et al., 2011b). Thus, understanding the intricate relationship between the regulation of 5-hmC and associated gene transcription remains a challenge in 5-hmC biology (Cimmino et al., 2011; Wu and Zhang, 2011). Of note, we identified a subset of genes showing significant 5-hmC level decreases and simultaneous 5-mC level increases in melanomas compared to nevi. The strong association of this subset of genes with various cancer pathways suggests that gene-specific 5-hmC loss may partially contribute to the abnormal DNA methylation pattern in the epigenome of melanoma that has been linked to the progression of various cancers. However, there are a portion of genes showing significant reduction in 5-hmC levels but no obvious 5-mC level changes in melanomas compared to nevi, suggesting that other independent molecular mechanisms, such as 5-hmC-mediated regulation of cell division/replication, cell differentiation/senescence and/or genome/epigenome instability, may also be involved in linking the loss of 5-hmC to melanoma progression. Future studies aimed at identifying these potential 5-hmC-related mechanisms should enhance insights into 5-hmC function in melanoma formation and progression.
Finally, with melanoma as a paradigm of aggressive cancer, our study provides important insight for future functional studies of 5-hmC in cancer biology. Increasing 5-hmC levels via over-expressing TET2 reversed the genome-wide 5-hmC distribution from the global 5-hmC loss pattern in melanoma toward a benign nevus-like pattern. More importantly, the phenotype of melanoma was rescued by increasing 5-hmC levels via over-expressing either TET2 or IDH2 in animal models. Thus, “loss of 5-hmC” in melanoma progression is a fundamental epigenetic event that provides proof-of-principle that key factors in the 5-hmC generating pathway can be therapeutically targeted to restore 5-hmC in human melanoma, thus revealing new strategies for the design of melanoma treatment.
EXPERIMENTAL PROCEDURES
Immunohistochemical (IHC) Staining
Immunohistochemical studies employed 5-μm sections of formalin-fixed, paraffin-embedded tissue. Slides were de-paraffinized and rehydrated, and after antigen retrieval, were placed in 2N HCl for 30 minutes, rinsed in distilled water and placed in 100mM Tris-HCl pH8.5 for 10 minutes. All were stained on the Dako Autostainer (Dako Corporation, Carpinteria, CA) using the EnVision (Dako) staining reagents. Sections were incubated for 60 minutes with either rabbit-anti-5-hmC at 1:10,000 (Active motif) or mouse-anti-5-mC at 1:500 (Eurogentec) and then incubated with the EnVision+ Dual Link (Dako) detection reagent for 30 minutes. Sections were washed, treated with a solution of diaminobenzidine and hydrogen peroxide (Dako) for 10 minutes, and after rinsing, a toning solution (DAB Enhancer, Dako) was used for 2 minutes to enrich the final color.
Glucosylation of genomic 5-hmC
Genomic DNA (700ng) purified from benign nevi or melanomas was incubated with 1μl of T4 Phage β-glucosyltransferase (NEB) and 1μl of UDP-glucose [6–3H] (ARC) in 1 × NEB buffer 4 at 37°C overnight, followed by protease K digestion. DNA was purified and radioactivity was measured by Beckman Coulter scintillation counter LS6500.
MeDIP-seq and hMeDIP-seq
Genomic DNA of human melanomas, nevi and human melanoma cell lines was purified and sonicated. Illumina barcode adapters were ligated before hMeDIP. Adaptor-ligated DNA (5 μg) was denatured and incubated with 3 μl of 5-hmC antibody (Active Motif) or 5 μg of 5-mC antibody (Eurogentec) at 4°C overnight. Antibody-DNA complexes were captured by protein A/G beads. The immunoprecipitated DNA was purified and sequenced followed by standard Illumina protocols (Xu et al., 2011b). Read sequences were mapped to the human genome (hg19) using ELAND v2 in the CASAVA (Illumina, v1.6) package. Significantly enriched regions were determined by Model-based Analysis of ChIP-Seq (MACS) package (Zhang et al., 2008). GO term and KEGG pathway analyses were performed by the database for annotation, visualization and integrated discovery (DAVID) programs (Huang et al., 2009).
NSG mice melanoma xenograft assay
NOD/SCID interleukin-2 receptor (IL-2R) γ-chain null (NSG) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained under defined conditions in accordance with institutional guidelines. Experiments were performed according to approved experimental protocols. For tumorigenicity studies, MOCK, wt TET2 or TET2 M A2058 melanoma cells were injected subcutaneously into the flanks of NSG mice (1×106/injection). Tumor growth was assayed as a time course (Schatton et al., 2008) for the duration of the experiment or until excessive tumour burden or disease state required protocol-stipulated euthanasia.
IDH2 over-expression in a zebrafish melanoma model
The miniCoopR assay was performed as previously described (Ceol et al., 2011). Transgenes were expressed in zebrafish melanocytes in the background of a stably-integrated BRAFV600E transgene and a p53 loss-of-function mutation. The background also contained a mitfa loss-of-function mutation, which blocked melanocyte development. Transgenes were coupled, via the miniCoopR vector, to a rescuing mitfa gene, ensuring that rescued melanocytes also expressed the transgene being tested. IDH2 was cloned into the miniCoopR vector, and the resulting miniCoopR-IDH2 construct was injected into single-cell zebrafish embryos. Transgenic animals were selected and melanoma onset measured weekly as compared to miniCoopR-EGFP control animals. Animals with melanomas were isolated and tumors allowed to progress for two weeks prior to being sacrificed. Tumors were formalin fixed, embedded and sectioned transversely to assess invasion.
Supplementary Material
HIGHLIGHTS.
Loss of 5-hmC is an epigenetic hallmark of melanoma with diagnostic/prognostic value
Genome-wide mapping reveals a demolished 5-hmC landscape in human melanoma epigenome
Down-regulating IDH2 and TETs suggests a mechanism underlying 5-hmC loss in melanoma
TET2 and IDH2 set the 5-hmC landscape, suppress melanoma growth and increase survival
Acknowledgments
We thank Dr. David E. Fisher for his critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) grant GM078458 to Y.G.S, SPORE Core: NIH-NCI SPORE in Skin Cancer 5P50CA093683-08, and partly by the Chinese Ministry of Education project 985. Y.G.S. is a PEW scholar.
Footnotes
ACCESSION NUMBERS
Sequencing data have been deposited to GEO (accession number GSE38231).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine Ross L, Aifantis I. TET Family Proteins and Their Role in Stem Cell Differentiation and Transformation. Cell stem cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway K, Edmiston SN, Khondker ZS, Groben PA, Zhou X, Chu H, Kuan PF, Hao H, Carson C, Berwick M, et al. DNA-methylation profiling distinguishes malignant melanomas from benign nevi. Pigment cell & melanoma research. 2011;24:352–360. doi: 10.1111/j.1755-148X.2011.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011 doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, Allen RE, Singer MI, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Devesa SS, Hartge P, Tucker MA. Recent trends in cutaneous melanoma incidence among whites in the United States. J Natl Cancer Inst. 2001;93:678–683. doi: 10.1093/jnci/93.9.678. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA: a cancer journal for clinicians. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, et al. Integrative genome comparison of primary and metastatic melanomas. PloS one. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell D, Assoku M, Galloway M, Mulholland P, Tomlinson I, Bardella C. Screen for IDH1, IDH2, IDH3, D2HGDH and L2HGDH mutations in glioblastoma. PloS one. 2011;6:e19868. doi: 10.1371/journal.pone.0019868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nature genetics. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Liu S, Ren S, Howell P, Fodstad O, Riker AI. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment cell & melanoma research. 2008;21:545–558. doi: 10.1111/j.1755-148X.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- Marks R. Epidemiology of melanoma. Clin Exp Dermatol. 2000;25:459–463. doi: 10.1046/j.1365-2230.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer research. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riker A, Enkemann S, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant R, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Medical Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel Genes Associated with Malignant Melanoma but not Benign Melanocytic Lesions. Clinical Cancer Research. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- Tellez CS, Shen L, Estecio MR, Jelinek J, Gershenwald JE, Issa JPJ. CpG island methylation profiling in human melanoma cell lines. Melanoma Research. 2009;19:146–155. doi: 10.1097/cmr.0b013e32832b274e. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting α-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzeszczynski KO, Varadan V, Byrnes J, Lum E, Kamalakaran S, Levine DA, Dimitrova N, Zhang MQ, Lucito R. Identification of Tumor Suppressors and Oncogenes from Genomic and Epigenetic Features in Ovarian Cancer. PLoS ONE. 2011;6:e28503. doi: 10.1371/journal.pone.0028503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & Development. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of alpha-Ketoglutarate-Dependent Dioxygenases. Cancer cell. 2011a;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell. 2011b;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2012 doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.