Abstract
The Janus kinase/Signal transducers and activators of transcription (JAK/STAT) pathway determines cell fates by regulating gene expression. One example is the specification of the motile cells called border cells during Drosophila oogenesis. It has been established that too much or too little STAT activity disrupts follicle cell identity and cell motility, which suggests the signaling must be precisely regulated. Here, we find that Suppressor of cytokine signaling at 36E (Socs36E) is a necessary negative regulator of JAK/STAT signaling during border cell specification. We find when STAT signaling is too low to induce migration in the presumptive border cell population, nearby follicle cells uncharacteristically become invasive to enable efficient migration of the cluster. We generated a genetic null allele that reveals Socs36E is required in the anterior follicle cells to limit invasive behavior to an optimal number of cells. We further show Socs36E genetically interacts with the required STAT feedback inhibitor apontic (apt) and APT’s downstream target, mir-279, and provide evidence that suggests APT directly regulates Socs36E transcriptionally. Our work shows Socs36E plays a critical role in a genetic circuit that establishes a boundary between the motile border cell cluster and its non-invasive epithelial neighbors through STAT attenuation.
Keywords: Socs36E, Drosophila, Border cells, JAK/STAT, Apontic, Cell motility, Cell fate
Introduction
During normal development, coordinated genetic circuits instruct cells to respond to fate-determining signals. In pathological events, the genes involved in these circuits can become ectopically activated, or regulatory components of endogenous signaling can break down, leading to undesirable outcomes. Thus, decoding how genetic circuits are regulated is important to understanding both development and disease. The Janus kinase (JAK) and Signal transducers and activators of transcription (STAT) proteins are key components in a highly conserved pathway that allows cells to convert extracellular cues into intracellular responses by regulating gene expression (Arbouzova and Zeidler, 2006; Bromberg and Chen, 2001; Levy and Darnell, 2002). Originally discovered for its role in promoting cytokine-induced gene expression, the JAK/STAT pathway has since been implicated in animal development, including regulation of cell proliferation, stem-cell maintenance, cell differentiation, immune system regulation, and cell migration (Arbouzova and Zeidler, 2006; Bromberg and Darnell, 2000; de Cuevas and Matunis, 2011; Hombría and Brown, 2002; Levy and Darnell, 2002; Luo and Dearolf, 2001). Hyper- and hypo-activation of the pathway, however, has been linked to numerous disorders, including various cancers (Bromberg et al., 1999; Chen et al., 2012a; Levy and Darnell, 2002). The requirement for precise JAK/STAT signaling underscores the importance of studying the regulatory components of the pathway.
The border cells of the Drosophila melanogaster ovary require JAK/STAT signaling for their specification and characteristic migration, and have provided some insight into the function and regulation of this pathway (Hombría and Brown, 2002; Montell et al., 2012). The fly genome encodes a single STAT (Stat92E), one JAK (Hopscotch/Hop), and one receptor (Domeless/Dome), as opposed to the numerous orthologs found in mammals; thus the study of the pathway in Drosophila eliminates many issues with redundancy found in vertebrates (Arbouzova and Zeidler, 2006; Devergne et al., 2007; Ghiglione et al., 2002; Hombría and Brown, 2002; Hou et al., 2002; Luo and Dearolf, 2001). The Drosophila ovary is comprised of a procession of egg chambers undergoing oogenesis, which is divided into 14 stages (King, 1970). Each egg chamber is composed of 16 germline cells – one oocyte and 15 nurse cells – surrounded by a monolayer of somatic epithelial cells, the follicle cells (King, 1970; Spradling, 1993). At stage 8, two specialized cells, the anterior polar cells, secrete the cytokine-like molecule Unpaired (Upd), causing graded activation of the JAK/STAT pathway in the 9–12 closest epithelial cells (Montell et al., 2012; Van de Bor et al., 2011). By stage 9, cells that initially had low STAT pathway activation switch it off entirely, thereby reducing the number of follicle cells with STAT activity to 4–6. Cells with high STAT activity assemble around the nonmigratory polar cells to form the border cell cluster. The cluster detaches from the epithelium and migrates along the nurse cells to arrive at the oocyte by stage 10, where it is required to form a fertilizable egg (Montell, 2003; Montell et al., 2012).
STAT controls the specification of border cells through modulation of gene expression. Two essential downstream targets required for normal border cell specification and migration are encoded by the genes slow border cells (slbo) and apontic (apt) (Montell et al., 1992; Silver et al., 2005; Silver and Montell, 2001; Starz-Gaiano et al., 2008). SLBO, the C/EBP transcription factor, promotes border cell specification and represses APT, while APT negatively feeds back on the circuit, repressing STAT and SLBO. APT levels are similar between follicle cells directly adjacent to the polar cells and those more distal at stage 9 (Starz-Gaiano et al., 2008). In contrast, both STAT activity and SLBO expression are graded, decreasing proportionally with distance from the polar cells, and initially detected in a greater number of anterior follicle cells than will eventually become border cells. Mutants that fail to reduce the initial STAT-positive/SLBO-positive population to an appropriate number of cells display abnormal cell invasion and delay, while those disrupting STAT-regulated gene expression result in too few motile cells and loss of migration (Montell et al., 2012; Van de Bor et al., 2011). Thus, optimal border cell migration requires the specification of a precise number of motile cells enclosing the polar cells.
Genetic and expression analyses, along with the finding that loss of apt expands the range and magnitude of SLBO expression, have led to the current genetic circuit paradigm. This states that follicle cells that maintain high levels of STAT activity sustain an above-threshold level of SLBO, which inhibits APT and promotes border cell fate. In contrast, lower levels of activated STAT yield higher signaling via APT than SLBO, establishing cells that remain in the surrounding epithelium as the nurse cell-associated stretch cells, which shut off STAT signaling entirely (Montell et al., 2012; Starz-Gaiano et al., 2009, 2008). In follicle cells with lower STAT activity, APT directs STAT attenuation, in part, by promoting the expression of mir-279, which targets the stat messenger RNA (Yoon et al., 2011). Loss of mir-279, however, results in a less penetrant mutant phenotype than loss of apt, indicating APT is either capable of repressing STAT directly or that it must control the expression of another STAT regulator.
The Suppressor of cytokine signaling (Socs) gene family is composed of well-conserved inhibitors of numerous signal transduction pathways, including JAK/STAT, (Alexander, 2002; Cooney, 2002; Croker et al., 2008; Krebs and Hilton, 2001), making members of this family candidates to be additional regulators in border cell specification. Mammals contain eight Socs genes (Socs 1–7 and CIS), while Drosophila have only three, named after their cytological locations—16D, 36E, and 44A (Arbouzova and Zeidler, 2006; Callus and Mathey-Prevot, 2002; Karsten et al., 2002; Rawlings et al., 2004). While the mammalian SOCS family is divided into two classes – those with a short N-terminus (CIS and SOCS1-3) and those with a long N-terminus (SOCS4-7) – the fly proteins fall only in the latter class (Alexander, 2002; Callus and Mathey-Prevot, 2002; Croker et al., 2008; Karsten et al., 2002; Rawlings et al., 2004). Socs16D and 44A are orthologous to mammalian Socs6 and 7, while Socs36E is most similar to Socs5. A hallmark of SOCS proteins is their conserved architecture near the carboxy terminus – an SH2 domain and a SOCS box – which is essential for their role in ubiquitination (Alexander, 2002; Croker et al., 2008; Rawlings et al., 2004). Through ubiquitin-based attenuation, SOCS proteins are able to fine-tune STAT signaling.
Several studies have demonstrated functional conservation between Drosophila and vertebrate SOCS proteins. In specific contexts, Socs36E has been reported to repress precise receptor tyrosine kinases, including Sevenless during eye development and the epidermal growth factor receptor (EGFR) in the epithelium during wing development (Almudi et al., 2009; Herranz et al., 2012). In the developing wing, Socs36E was also determined to be a negative regulator of the JAK/STAT pathway (Callus and Mathey-Prevot, 2002; Rawlings et al., 2004). These studies also provided evidence that the SH2 and SOCS box domains are essential for Socs36E function in eye and wing development (Almudi et al., 2009; Callus and Mathey-Prevot, 2002). Further, Socs36E has been characterized in the Drosophila testes as an essential negative regulator of JAK/STAT signaling (Issigonis et al., 2009; Singh et al., 2010).
We have determined that Socs36E plays a critical role in specifying the optimal number of border cells. We generated a genetic null allele of Socs36E and found that flies homozygous for this mutation incorrectly specify motile cells, which results in an additional invasive cell phenotype. The phenotypes observed when Socs36E expression was either heightened or lost are consistent with loss of function or gain of function of STAT activity, respectively (Beccari et al., 2002; Silver et al., 2005; Silver and Montell, 2001; Starz-Gaiano et al., 2008; Yoon et al., 2011). We did not observe any phenotypes associated with dorsally-directed migration, which is mediated by EGFR (Duchek and Rørth, 2001; McDonald et al., 2006), suggesting that Socs36E does not regulate EGFR during border cell movement. We determined that Socs36E genetically interacts with apt and its downstream target mir-279, and that APT can bind to a site in the Socs36E enhancer. Our work indicates APT regulates the expression of both Socs36E and mir-279, which are each independently required to limit STAT activity and establish a discrete boundary between the motile border cells and their non-motile neighbors.
Materials and methods
Expression and over-expression assays
We crossed the P[GawB]Socs36ENP5170 and P[GawB]Socs16DNP7149 ((Brand and Perrimon, 1993), Kyoto stock center) lines to w-; UAS-mCD8-GFP ((Lee and Luo, 1999), Bloomington stock center) to determine the expression pattern of Socs36E and Socs16D, respectively. Over-expression experiments were performed at 25 °C to generate the following genotypes: P[GawB]c306 (expressed in anterior follicle cells and referred to as AFC-Gal4 in text, (Manseau et al., 1997)); UAS-mCD8-GFP, c306-Gal4; UAS-Socs36E ((Callus and Mathey-Prevot, 2002), Bloomington stock center)), upd-GAL4 (expressed in polar cells; (Khammari et al., 2011), Bloomington stock center); UAS-mCD8-GFP, upd-GAL4; UAS-Socs36E, Socs36E-Gal4/UAS-mCD8-GFP, Socs16D-Gal4 (P[GawB]Socs16DNP7149); UAS-mCD8-GFP, and Socs16D-Gal4; UAS-Socs36E.
Generation of novel Socs36E alleles
We obtained y1w67c23; P[EPgy2]Socs36EEY06665 ((Bellen et al., 2004; Singh et al., 2010), Bloomington stock center) and out-crossed it to a dominantly marked stock to allow modifiers to be recombined away from the insertional allele. We established a homozygous viable stock, and then isogenized the second chromosome. This generated a “cleaned up” hypomorphic allele of Socs36E EY06665. PCR analysis confirmed the P-element was still present at the Socs36E locus. To excise the P-element, we crossed the cleaned-up Socs36EEY06665 allele to the transposase-bearing stock w*; Sp/CyO; ry506,Dr, Δ2-3/TM6, and re-balanced the chromosome. To check for precise and imprecise excisions in the Socs36E locus, we created primers that flank the P[EPgy2] insertion site (Forward: (5′ TCA CCT TAG CAA GTT CTC AGC ACG C-Exon 2); Reverse: (5′ GAC TGC GGC AGC AAC TGT TGC-Exon 3)). PCR and sequence analysis confirmed Socs36EΔEY06665-A9 and Socs36E-ΔEY06665-B3 were precise excisions. The PCR product from the Socs36E178 line was about 500 base pairs smaller than the wild type product. Sequence analysis using NCBI BLAST (http://blast.ncbi.nlm.nih.gov; (Altschul et al., 1990)) and ClustalW2.1 (http://www.ebi.ac.uk; (Goujon et al., 2010; Larkin et al., 2007)) confirmed an imprecise excision removed approximately 500 base pairs from exon 2 and intron 2–3 of Socs36E (Flybase (flybase.org); (McQuilton et al., 2012)). Using EMBOSS Transeq (http://ebi.ac.uk; (Rice et al., 2000)) we translated our Socs36E178 sequence, which indicated an in-frame TAA stop codon to generate a predicted 178 amino acid polypeptide. All DNA sequencing was performed by Genewiz (South Plainfield. New Jersey). All cytological and sequence information for alleles and genes were acquired using Flybase and Ensembl Genome Browser (ensembl.org; (Kersey et al., 2012)).
Generation of fly stocks for Socs36E mutant rescue
The UAS-Socs36E and UAS-mCD8-GFP transgenes are each on the second chromosome therefore, to co-express either of them with the Socs36E178 allele, we generated recombinant stocks. We utilized PCR analysis to verify the presence of the Socs36E178 allele ((Forward: 5′ GGC GCT GCG ATA AGT ACC ATG ATG-exon 2 at excision site) Reverse: 5′ GGT CAG CTG TGC ACA GCG-intron 2–3)). Independently, we also generated the stock: AFC-Gal4 (P[GawB]c306); Df(2)Exel8038/CyO. We crossed our UAS-Socs36E, Socs36E178 stock and two UAS-mCD8-GFP, Socs36E178 stocks with AFC-Gal4; Df(2)Exel8038/CyO to generate our rescue and control genotypes, respectively, as indicated in the text.
Generation of Socs36E and apt recombinants
To test for a genetic interaction between Socs36E and apt, we generated the following recombinant stocks: the cleaned-up Socs36EEY06665 hypomorphic allele and apt167 (an apt null, (Eulenberg and Schuh, 1997)), or Socs36E178 (genetic null) and P(SUPor-P)aptKG05830 (an apt expression null in egg chambers) (Bellen et al., 2004; Starz-Gaiano et al., 2008). To screen for positive recombination events, we tested for the presence of the apt allele by testing for lethality in trans to a second apt null allele (apt41, (Eulenberg and Schuh, 1997)) and utilized PCR analysis to test for the presence of the P-element in the Socs36E locus in any lines displaying lethality. To test for recombination events between Socs36E178 and P(SUPor-P)aptKG05830, we identified the SUPor-P insertion by the presence of w+, and the Socs36E178 allele by PCR analyses.
Other fly stocks
We compared our Socs36E mutant allele with the previously described alleles: Socs36EPZ1647 (provided by Dr. Erika Matunis; (Issigonis et al., 2009)), Socs36EEY06665 (Singh et al., 2010), and Socs36EEY11 (provided by Dr. Florenci Serras; (Almudi et al., 2009)). To investigate a potential genetic interaction between Socs36E and mir-279, we obtained FRT82B mir-279Δ1.9 and FRT82B mir-279Δ1.2 (provided by Dr. Denise Montell; (Cayirlioglu et al., 2008; Yoon et al., 2011)).
Immunofluorescence
14–16 h prior to dissection, 1–7 day old females were transferred to food with additional yeast at room temperature (22 °C) for the rescue experiments and 25 °C for the remaining experiments. Ovarioles were dissected as previously described (McDonald and Montell, 2005). DAPI (1:1000, Invitrogen: D1306) was applied for 10 min at room temperature.
The following primary antibodies were used: mouse anti-GFP (1:1000, Molecular Probes); rabbit anti-GFP (1:1000, Molecular Probes); rabbit anti-STAT (1:500; provided by Dr. Denise Montell; (Jang et al., 2009)), rat anti-SLBO (1:500; provided by Dr. Pernille Rorth; (Beccari et al., 2002)). The following antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD, and maintained by the University of Iowa, Department of Biology, Iowa City, IA 52242: mouse anti-FasIII (1:75, 7G10: Goodman, C.; (Patel et al., 1987)); rat anti-DCAD2 (1:25, DCAD2: Uemura, T.; (Oda et al., 1994)); mouse anti-Armadillo (1:40, N2 7A1 Armadillo: Wieschaus, E.; (Riggleman et al., 1990)); mouse anti-Eyes Absent (1:1000, EYA10H6: Bonini et al. (1993)). Molecular probes AlexaFluor secondary antibodies (488 nm and 568 nm) were diluted 1:400. Images were acquired on a Zeiss Axioimager microscope equipped with Axiovision software and the Apotome structural interference system for optical sectioning. Adobe Photoshop CS6 was used to process and format images.
Invasive cell analysis and counts
To determine the number of invasive cells in the egg chamber, we used Axiovision software to generate Z-stacks of optical sections spanning the egg chambers in 1.0 μm step sizes. To be considered invasive for the phenotypic analysis, the cell had to be discontinuous with the epithelial layer and predominantly contacting the nurse cells. To be considered phenotypically abnormal in our loss of function analysis, we utilized a stringent criterion: we required more than one additional invasive cell to be present in a stage 10 egg chamber. However, a single additional invasive cell may also be considered phenotypically significant (for example, see (Starz-Gaiano et al., 2008; Yoon et al., 2011)). We, therefore, include separately the penetrance of the presence of a single additional invasive cell.
To quantify SLBO expression and active STAT in anterior follicle cells at stage 8, we generated Z-stacks in the same way as for SLBO cell counts detailed above for post-border cell specification analysis.
Statistical analysis
All statistical analysis for cell count data was performed via a two-tailed t-test. To display cell count data, non-parametric box and whisker plots were utilized. In the box plots, the upper whisker indicates the upper quartile through the maximal value, while the lower whisker is the lower quartile to the minimum value observed. For the boxes, second (lower) and third (upper) quartile bars are separated by the median value. The diamond in each bar indicates the mean.
For statistical analysis of phenotypic penetrances, two-tailed Fischer’s Exact tests were utilized using: http://www.graphpad.com/quickcalcs/contingency1.cfm. For both the two-tailed t-test and Fischer’s Exact tests, we maintained a significance requirement of at least p<0.05.
Electromobility shift assay (EMSA)
We searched the Socs36E enhancer region for putative APT binding sites according to the published consensus binding site (Liu et al., 2003). We identified one site in a Socs36E intron that matched 10 bases of the consensus: ATTCCAATTA. Two 31 bp oligonucleotides (5′-GATCGTTCAGGGAATTCCAATTACCACAATG-3′ and the reverse complement) were used as a 32P-radiolabeled DNA probe. EMSAs were performed as described in (Ueda and Hirose, 1991) with the modification of using a 6% 0.5× TBE gel. His-APT DNA was a gift of S. Hirose. Protein was purified using the Qiagen NTA Ni-Agarose Fast Start Kit. Competition assays were performed with a 50-fold excess of unlabeled oligonucleotides added 30 min prior to addition of the labeled probe.
Results
Socs36E is expressed in the anterior follicle cells of the egg chamber
Given the essential role of the STAT pathway in border cell specification and motility, we evaluated a potential role of the Socs regulators in these processes. Socs36E had been previously shown to affect border cell movement in over-expression experiments (Silver et al., 2005), but its loss of function phenotype and a detailed characterization of its expression and regulation had not been assayed in this context. At mid-oogenesis, STAT activity can be inferred from expression of its target SLBO, which is initially observed in a gradient in the anterior epithelium, then becomes restricted to the motile border cell population (Fig. 1A–C, (Beccari et al., 2002; Montell et al., 2012; Silver and Montell, 2001; Starz-Gaiano et al., 2008; Xi et al., 2003; Yoon et al., 2011). Using the enhancer trap line P[GawB]Socs36ENP5170 (Almudi et al., 2009), we expressed membrane localized GFP (mGFP), via the Gal4/UAS system (Brand and Perrimon, 1993), as a proxy for Socs36E expression (Fig. 1D–G). We did not observe GFP in the germarium but began to see it localized in the anterior polar and stalk cells at stage 2 (Fig. 1D and D’). As oogenesis progresses, GFP expression is maintained in the anterior follicle cells, as well as a subset of posterior follicle cells, consistent with previous in situ hybridization data for Socs36E at this stage (Fig. 1D–E; (Rawlings et al., 2004)). We observed GFP expression in the border cell cluster and a nearby subset of anterior follicle cells that are not fated as migratory (Fig. 1F–G). By co-staining with the polar cell marker, FasIII, we found GFP expression is also present in the polar cells (data not shown, (Gaziova et al., 2004; Ruohola et al., 1991)).
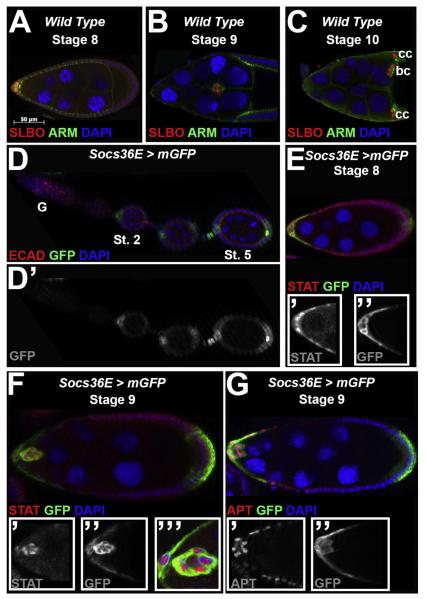
Fig. 1.
Socs36E expression in egg chambers overlaps with STAT activity during border cell specification and migration. For all images, anterior is to the left and the stages of oogenesis are indicated. (A–C) Wild type (Canton-S) egg chambers at mid-oogenesis are stained with antibodies directed against the border cell marker SLBO (red) and beta-cateninprotein (Armadillo, ARM, Green), which is also enriched in the border cells. DAPI (blue) labels DNA. (A) At stage 8, SLBO expression is graded in a subset of anterior-most follicle cells. (B) At stage 9, the border cells migrate between the nurse cells, and maintain SLBO expression. (C) The border cells (bc) reach the oocyte at stage 10, and SLBO is also expressed in centripetal cells (cc). (D–G) Socs36E-Gal4/UAS-mCD8-GFP egg chambersreveal the expression pattern of the Socs36Elocus. (D) GFP is not detected in the germarium (G) but is observed at stage 2 (St. 2) and later (Stage 5, St. 5) in a subset of anterior and posterior follicle cells. The inset (D) shows only GFP expression. (E–F) STAT protein expression, revealed by antibody staining (red), and the Socs36E reporter are both detected in the anterior follicle cells and border cell cluster atstages 8 (E) and 9 (F). The insets show the STAT (’) or GFP (”)staining, alone, or higher magnification at stage 9(’’’). (G) An antibody directed against APT (red) indicatesthis protein is detected in the same cells as GFP at stage 9. The insets show the APT (’) and GFP (”) stainings separately.
The Socs36E expression profile generated by the GFP read-out recapitulated the known pattern of STAT activity in the egg chamber ((Fig. 1A–C and (Harrison et al., 1998; Montell et al., 2012; Silver et al., 2005; Starz-Gaiano et al., 2008; Van de Bor et al., 2011; Xi et al., 2003; Yoon et al., 2011)). To confirm this, we stained Socs36E-Gal4/UAS-mGFP egg chambers with an antibody directed against STAT. At stage 8, there is a strong overlap between cells with activated (nuclear) STAT and mGFP expression (Fig. 1E, see inserts), which continues later in the border cell cluster and a subset of stationary follicle cells from which the cluster detaches (Fig. 1F, see inserts). To characterize Socs36E expression further, we also co-stained with an antibody directed against the STAT downstream target Apontic (APT). We found a similar overlap between cells expressing GFP and APT at the start of cluster formation (Fig. 1G). Since APT expression is also induced by Eyes Absent (EYA) (Starz-Gaiano et al., 2009), APT expression is maintained throughout the anterior follicle cells; thus while at stage 9 the STAT-activity domain becomes restricted, the APT-Socs36E overlap is slightly more expanded in the anterior follicle cells (compare Fig. 1F and G). In addition, a STAT-activity reporter containing sequences from the Socs36E regulatory region requires STAT signaling, is sensitive to APT function, and shows a similar pattern as the enhancer trap ((Bach et al., 2007; Flaherty et al., 2009; Starz-Gaiano et al., 2008), and data not shown). Together, these data suggest that prior to and during border cell specification and migration, Socs36E expression is predominantly superimposed with STAT activity and APT expression.
The two other Socs genes in the fly genome raised the concern that these may be functionally redundant in the egg chamber. It has previously been shown via in situ hybridization that Socs44A expression is restricted to the germline (Rawlings et al., 2004), making it unlikely to affect STAT signaling in follicle cells. To assay Socs16D expression, we utilized the enhancer trap line P[GawB]Socs16DNP7149 to express membrane-bound GFP. We did not observe GFP expression until about stage 6 of oogenesis, in which the GFP was highly expressed in the posterior and main body follicle cells with lower expression in the anterior region (Fig. S1A and B). Socs16D-driven GFP did not significantly overlap with STAT expression (Fig. S1B). Expression data, therefore, support the idea that Socs36E is regulated and functions distinctly from Socs16D and Socs44A.
High levels of Socs36E suppress border cell migration and cluster integrity
Over-expression of Socs36E was previously shown to recapitulate the loss of function stat phenotype and abrogate STAT activation in the border cells (Silver et al., 2005), however, the effects were not described in detail. We examined the effects of Socs36E over-expression more closely utilizing the GAL4/UAS system (Brand and Perrimon, 1993; Callus and Mathey-Prevot, 2002) via an anterior follicle cell driver, AFC-Gal4 (P[GawB]c306), which activates expression at the beginning of stage 8 in the anterior follicle cells and remains active in border and polar cells over the course of migration (Fig. 2A and B; (Silver et al., 2005)). When a full-length Socs36E transgene (Callus and Mathey-Prevot, 2002) was driven by AFC-Gal4, the border cell cluster failed to form properly, and in 53% of the observed stage 10 egg chambers, invasive cells failed to reach the oocyte, compared to 2% of egg chambers ectopically expressing GFP (compare Fig. 2C and D with 2B; (Silver et al., 2005)). The phenotype varied in severity ranging from cases in which a small number of anterior follicle cells clustered around the polar cells (marked by FasIII expression; (Gaziova et al., 2004; Ruohola et al., 1991)) but failed to migrate (Fig. 2C), to the invasive cells being splayed along the migratory path (Fig. 2D) instead of organized into a cohesive cluster. In contrast, over-expression of Socs36E in the polar cells alone by use of the upd-Gal4 driver (Khammari et al., 2011) or in the main body and posterior follicle cells by the Socs16D-Gal4 driver did not cause any border cell defects (data not shown).
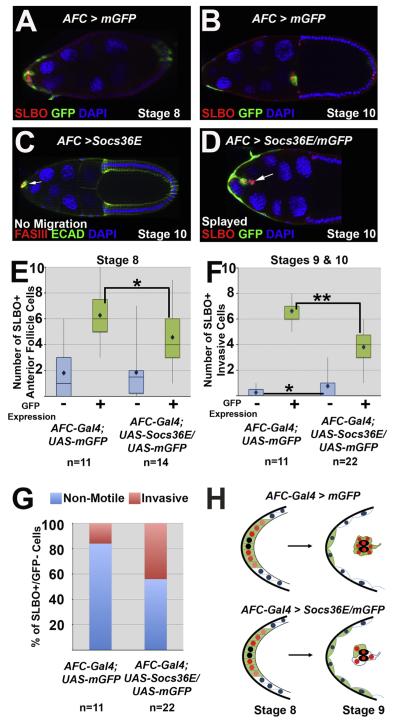
Fig. 2.
Ectopic Socs36E causes border cell identity, cohesion, and migration defects. For all egg chambers the stage of oogenesis and the mutant phenotype, if one is present, are indicated. “AFC”stands for Anterior Follicle Cells, where AFC-Gal4 indicates the lineP[GawB]c306. (A–B) AFC-Gal4; UAS-mCD8-GFP egg chambers at stage 8 (A) and stage 10 (B) indicate the expression pattern of the AFCdriver (by GFP antibody staining) in anterior follicle cells and the border cells, marked by SLBO antibody. (C) A stage 10 AFC-Gal4; UAS-Socs36E egg chamber, which over-expresses Socs36E in AFCs, stained with antibodies directed against FasIII (red) and E-Cadherin (green). A cluster of cells forms around the polar cells (indicated by FasIII) at the anterior end (arrow), but no cells migrate. (D) A stage 10 egg chamber co-expressing Socs36E andGFP in the presumptive border cell population(AFC-Gal4; UAS-Socs36E/UAS-mCD8-GFP). SLBO antibody (red) marks a small border cell cluster, which displays a splayed phenotype and fails to detach from the anterior end. The arrow indicates a pair of SLBO-positive/GFP-negative invasive cells. (E–F) Box and whiskers plots quantifyingSLBO-positive cells at stage 8 (E) and stages 9 and 10 (F) in egg chambers expressing GFP alone or with Socs36E under control of the AFC-Gal4 driver. For each genotype, the SLBO-positive cells are categorized as expressing GFP (green bars and +) or not (blue bars and −). The bars represent the range in the second and third quartiles, and mean cell counts are indicated by diamonds;(*) = p<0.05; (**) = p<0.0001. (G) Percent of the SLBO-positive, GFP-negative anterior follicle cells at stage 8 that become either invasive (pink) or remain as non-motile stretch cells (blue). (H) Schematics illustrating altered border cell recruitmentin wild type (top) or when Socs36E is over-expressed (bottom), as quantified in E and F. Polar cells are indicated by black, red nuclei indicate SLBO(+) cells, while blue nuclei are SLBO(−) cells. Green cells indicateGFP (+) cells, which express AFC-Gal4, while white cells do not.
In wild type egg chambers, approximately 6 border cells surround the non-motile polar cells, which is a necessary arrangement for proper cluster movement. In contrast, when Socs36E was over-expressed, small aberrant clusters formed, and some cells were motile independent of association with the polar cells. We defined invasive cells as those that exited the epithelial layer and made contact with the nurse cells, and further classified these as cluster-associated if they were in contact with polar cells, or non-cluster associated if not, and counted each class. We found an average of 3.4 cluster-associated cells and 1.4 non-cluster associated cells in egg chambers over-expressing Socs36E, compared to 5.8 and 0.4 cluster and non-cluster associated cells, respectively, in the GFP control (p<0.0001, p<0.02, respectively). In accord with previous studies, these results suggest high levels of Socs36E in the presumptive border cell population affect cell migration and cluster cohesion, consistent with loss of STAT activity (Montell et al., 2012; Silver et al., 2005; Silver and Montell, 2001; Starz-Gaiano et al., 2008; Van de Bor et al., 2011).
High-levels of Socs36E can alter the range of border cell recruitment
When Socs36E was over-expressed and invasive cells completed migration, the number of cells observed at the oocyte was 1 to 5, compared to 5.8±1.2 in the GFP control. Provided this wide range, we hypothesized that either some cells did not respond to Socs36E, potentially due to very high STAT activation, or that nearby cells not expressing the AFC-Gal4 driver – which are usually maintained as non-invasive epithelial cells – were recruited into a migratory state when the presumptive border cells were incapable of migration. To test this, we co-expressed mGFP and Socs36E in the anterior follicle cells, and assayed protein expression of the STAT target gene slbo. In stage 8 egg chambers, prior to border cell movement, we found a significant decrease in the number of SLBO-positive/GFP-positive anterior follicle cells compared to the controls (4.6 to 6.3, respectively; Fig. 2E). There was, however, no significant difference in the quantity of total GFP-positive cells (Fig. S2A) or the number of SLBO-positive/GFP-negative anterior follicle cells between the two genotypes at stage 8 (average 1.8; Fig. 2E). These data suggest Socs36E can down-regulate a STAT target gene cell autonomously.
Next, we examined invasive cells in stage 9 and 10 egg chambers. In control egg chambers, motility becomes restricted to a smaller number of cells, which maintain expression of AFC-Gal4. All invasive cells are SLBO-positive, with a mean total of 6.9, and in 73% of egg chambers all SLBO-positive invasive cells are also GFP-positive when marked with AFC-Gal4. When Socs36E was over-expressed, though, we found an average of 2.8 fewer SLBO-positive/GFP-positive invasive cells (Fig. 2F) and, surprisingly, we observed many egg chambers (55%) that contained at least one SLBO-positive, GFP-negative invasive cell. On average, 44% of SLBO-positive GFP-negative cells (0.8 out of 1.8) became invasive, compared to 16% (0.3 of 1.8) of these cells acquiring motility in the controls (p<0.05; Fig. 2E–G)). GFP-negative invasive cell(s) could be either cluster-associated or migrating independently. Since some GFP-negative cells were found to be SLBO-positive and motile, it suggests that they originated outside of the AFC-Gal4 expression domain (Fig. 2G and H) and maintained or amplified an initially low STAT activity level. In stages 9–10, we found no significant difference in the total number of GFP-positive cells between egg chambers expressing GFP alone or with Socs36E under the control of AFC-Gal4, which suggests Socs36E over-expression does not affect the expression or activity of the Gal4 driver (Fig. S2B). Together these data suggest Socs36E can function autonomously to attenuate STAT signaling, while it can affect cell identity both autonomously and non-autonomously since cells far from the polar cells became recruited to the motile cluster. Collectively, this supports the hypothesis that over-expression of Socs36E expands the range of border cell recruitment beyond the set of anterior follicle cells normally specified (Fig. 2H).
Socs36E is a necessary STAT regulator in the egg chamber
To identify essential functions of Socs36E in border cell recruitment and migration, we generated novel Socs36E alleles. Socs36EPZ1647 and P(EPgy2)Socs36EEY06665 have been reported as strong loss of function and null alleles respectively in the testes (Issigonis et al., 2009; Singh et al., 2010), while the Socs36EEY11 allele was described as a strong hypomorph in the eye (Almudi et al., 2009). In the ovary, however, each allele carried a stronger phenotype when in trans to a deficiency line than as homozygotes, suggesting neither is a complete loss of function allele (Fig. S3). To generate a null allele, we mobilized the P-element in the P(EPgy2)Socs36EEY06665 line, which is inserted 516 base pairs downstream of the translational start site (Fig. 3A) to create deletions in the Socs36E locus, which we assayed by PCR and sequence analysis (see Materials and Methods). The Socs36E-ΔEY06665-A9 and Socs36EΔEY06665-B3 alleles are precise excisions. We also generated an imprecise excision that removed about 500 base pairs in the Socs36E genomic region between exon 2 and intron 2–3, inducing a frame-shift mutation and generating an in-frame TAA stop codon (Fig. 3B). This premature stop truncates the resultant protein from 633 amino acids (Socs36E-PA isoform) to 178, lacking the well-conserved functional domains. Thus, we named this excision allele Socs36E178. The Socs36E178 allele is homozygous viable and viable in trans to two deficiency lines that lack this cytological region (Df(2)Exel8038 and Df(2)Exel7070 (Parks et al., 2004)).
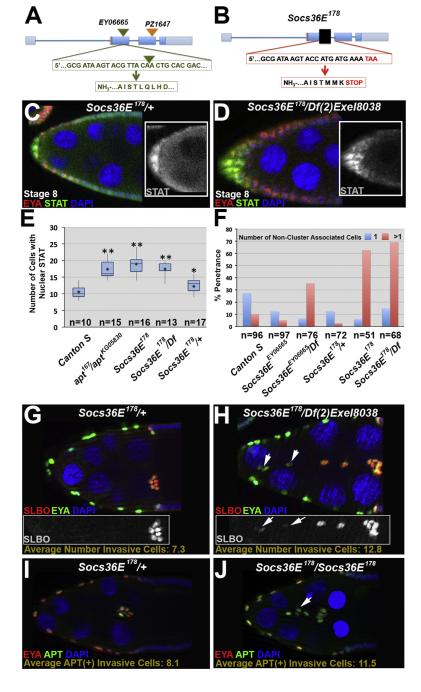
Fig. 3.
Loss of Socs36E increases STAT signaling and allows additional cells to become invasive. (A) Schematic of the Socs36E genomic locus, including previously described insertional alleles: Socs36EEY06665(green triangle) and Socs36EPZ1647(orange triangle). The gene and protein sequences flanking the Socs36EEY06665insertion are shown in green letters; the triangle indicates the insertion site. (B) Schematic of the Socs36E178allele. The black box is scaled to represent the approximately 500 base pairs deleted. The resultant gene and protein sequences are shown in the red boxes. (C–D) Control (C, Socs36E178/CyO) and Socs36E deficient (D) egg chambersstained with antibodies raised against EYA (red), which identifies anterior follicle cells, and STAT (green) depicts the increased range of nuclear STATprior to border cell migration in mutants. Insets: STAT antibody staining alone. (E) Box plotsquantifying nuclear STAT-positive anterior follicle cells at stage 8 in the indicated genotypes. Mean numbers for each are indicated by a diamond. Two-tailed t-tests compared Canton S and each respective genotype, where *=p<0.05; **=p<0.0001. (F) Quantification of the percentage of stage 10 egg chambers withone(blue) or more (pink)non-cluster associated invasive cells when Socs36E is disrupted. (G–H) Stage 10 egg chambers stained with antibodies directed against SLBO (red) and EYA (green). Insets show SLBO expression only. The average number of additional invasive cells is indicated. (G) HeterozygoteSocs36E178/CyOegg chambers have a wild type number of motile cells, which are clustered together at the oocyte. (H) When the Socs36E178allele is in trans to a deficiency for the locus, additional cells become invasive and do not cluster. Arrows indicate reducedSLBO expression in the non-cluster associated cells. (I–J) Stage 9 egg chambers stained with EYA (red) and APT (green) antibodies. The average number of APT-positive invasive cells is given. (I) A Socs36E178/CyO egg chamber displays a wild type number of motile cells. (J)Socs36E deficient egg chambers contain additional APT-positive non-cluster-associated invasive cells (indicated by arrow).
In the wild type follicular epithelium in mid-oogenesis, high STAT activation directs cells to become migratory (Montell et al., 2012). We hypothesized that Socs36E mutants would have higher levels or an expanded domain of STAT activation. To investigate this idea, we counted anterior follicle cells with nuclear (activated) STAT in Socs36E deficient egg chambers at stage 8. We found a significant increase in the number of STAT-positive anterior follicle cell nuclei in Socs36E mutants, relative to wild type (18.9±3.1 versus 10.6±1.9, respectively, p<0.0001; Fig. 3C–E). To confirm that STAT activity was expanded, we examined the downstream target and border cell marker SLBO at stage 8 in the Socs36E178/Df(2)Exel8038 genotype. In mutant egg chambers, we observed significantly more SLBO positive cells than in the controls (13.8±3.1 versus 8.1±2.2 (p<0.001)). Together, our results suggest Socs36E limits the range of STAT activation and, when lost, the region of STAT signaling is expanded.
Loss of Socs36E allows additional cells to become motile
Next, we looked at how the loss of Socs36E affects the specification of the migratory border cells. We found that in 63% of stage 10 Socs36E178 homozygous mutant egg chambers, a higher number of cells are motile than in wild type (11.0±2.5 versus 7.2±1.4 in wild type p<0.002), a phenotype similar to that caused by stat pathway gain-of-function (Beccari et al., 2002; Silver et al., 2005; Silver and Montell, 2001). While a single invasive cell that is not cluster-associated occasionally arises in wild type egg chambers, multiple non-cluster trailing cells are observed in only about 10%. However, in 63% of stage 10 egg chambers, abnormally invasive cells in Socs36E178 mutants trail behind the main border cell cluster and are often detached from it; thus, the main cluster migrates to the oocyte normally. The allele in trans to either of two deficiency lines, Df(2)Exel8038 and Df(2)Exel7070 (Parks et al., 2004), also results in a similar phenotypic penetrance and quantity of non-cluster associated additional invasive cells relative to the homozygotes (12.8±2.7 for Socs36E178/Df(2)Exel8038; p<0.0001 (relative to Canton S)): Fig. 3F–H, and data not shown). Furthermore, the previously characterized Socs36E mutant alleles in trans to the Socs36E178 allele or the deficiency lines yields an additional invasive cell phenotype that is increased comparably relative to those homozygous mutants (Fig. S3A–D). In both of our precise excision lines – as homozygotes or when in trans with Socs36E178 – the border cell cluster migrated properly and no additional invasive cells were observed (data not shown). Given that the phenotypic strength is very similar in Socs36E178 homozygotes and in trans to the deficiency lines and that this allele does not have a phenotype when heterozygous, the Socs36E178 allele can be classified as a genetic null allele.
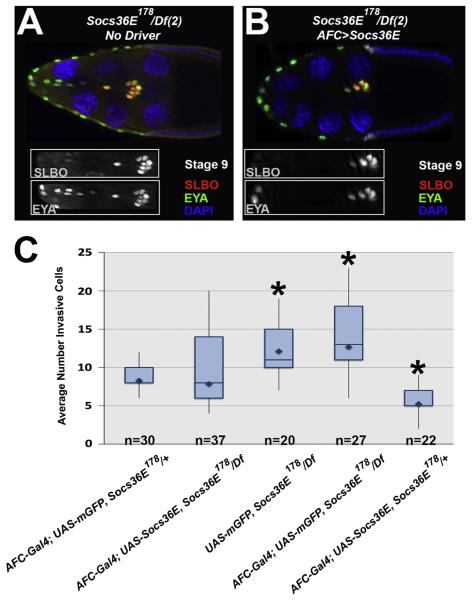
To verify that loss of Socs36E caused the additional invasive cell phenotype observed, we re-expressed Socs36E in the anterior follicle cells in mutant egg chambers. The re-introduction of Socs36E rescued the invasive cell number to close to that in wild type: 7.8±2.5 cells per egg chamber, relative to 12.1±2.8 in the mutants not expressing the Socs36E transgene (Fig. 4A–C, p<0.0001). In contrast, ectopic expression of GFP in the anterior follicle cells was unable to restore proper migratory cell number (12.7±3.2 invasive cells per egg chamber) (Fig. 4C). These data suggest the additional invasive cell phenotype observed in Socs36E178 egg chambers is due specifically to the loss of Socs36E.
Fig. 4.
Re-introduction of Socs36E in Socs36E178mutants restores invasive cell number. (A–B) Egg chambers stained with antibodies directed againstSLBO (red) and EYA (green) atstage 9 or 10of oogenesis, asindicated. The insets show the invasive cells via the indicated antibodyonly. (A) InUAS-Socs36E, Socs36E178/Df(2)Exel8038egg chambers, loss of Socs36E results in additional invasive follicle cells that arenot border cell cluster-associated. (B) InAFC-Gal4; UAS-Socs36E, Socs36E178/Df(2)Exel8038 egg chambers, the re-introduction of Socs36E in anterior follicle cells of a Socs36E mutant restores proper motile cell number. (C) Box plots detail the quantification of the total number of invasive cells observed in the respective genotypes, where the mean is indicated by the diamond. The boxes illustrate the second (lower) and third (upper) quartile bars, separated by the median value. Pairwise comparisons relative to AFC-Gal4; UAS-Socs36E, Socs36E178/Df(rescue genotype) were significant as indicated by the *=P<0.0001.
Once we confirmed the phenotype was due to loss of Socs36E, we examined the expression patterns of the border cell marker SLBO in a Socs36E deficient background. While in wild type, the number of SLBO-positive cells decreases between stages 8 and 10, we found no significant difference between the number of SLBO-positive cells at stage 8 and later stages in Socs36E deficient egg chambers (13.8±3.1 versus 12.8±2.7, respectively; Fig. 3G–H). Furthermore, we observed that SLBO expression varied between and became atypically low in the additional invasive cells in Socs36E deficient egg chambers (Fig. 3H), which indicated partial, but insufficient dampening of STAT activity in the mutants.
Next, we looked at the STAT-downstream target and negative regulator APT (Starz-Gaiano et al., 2009, 2008). APT is broadly expressed in the anterior follicle cells of the egg chamber due to EYA, but has heightened expression in the anterior-most cells due to STAT signaling. While we found a significant increase in the number of APT-positive invasive cells in Socs36E deficient egg chambers (11.5±2.6 versus 8.1±1.0 in wild type, p<0.002, Fig. 3I and J), there was no significant difference between APT-positive invasive cells and total invasive cells in Socs36E mutants (11.5±2.6 versus 11.0±2.5, respectively). In contrast to SLBO expression, we did not observe varied levels of APT expression in the additional invasive cells. Collectively, these results indicate the loss of Socs36E in the egg chamber allows an increased number of follicle cells to reach and maintain the migratory threshold of SLBO expression, while not affecting APT.
Socs36E and apt genetically interact in the anterior follicle cells
Socs36E mutants partially phenocopy egg chambers that have the STAT pathway hyper-activated or have lost the negative regulator apontic (apt) (Fig. 5A, (Beccari et al., 2002; Ghiglione et al., 2002; Silver et al., 2005; Starz-Gaiano et al., 2009, 2008)). APT binds DNA and functions as a transcriptional regulator, and in egg chambers, acts as a feedback inhibitor of STAT signaling (Eulenberg and Schuh, 1997; Gellon et al., 1997; Liu et al., 2003; Starz-Gaiano et al., 2008). One downstream target of APT is mir-279, which negatively regulates STAT post-transcriptionally in the egg chamber (Yoon et al., 2011). The mir-279 loss-of-function phenotype, however, is less severe than that of apt mutants (Starz-Gaiano et al., 2008; Yoon et al., 2011), suggesting APT has another effector, which we hypothesized to be Socs36E.
Fig. 5.
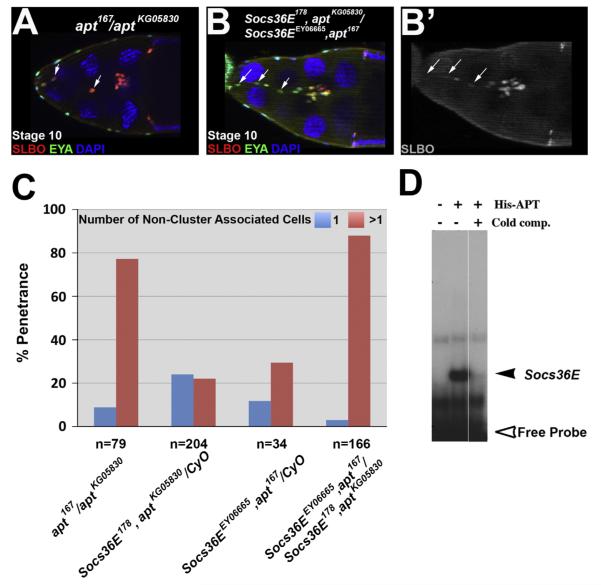
Socs36E and apt genetically interact to limit cell invasiveness. (A–B) Egg chambers stained with antibodies directed against SLBO (red) and EYA (green) at stage 10 of oogenesis. (A) Anapt loss of function egg chamber contains additional invasive cells, which are non-cluster associated, indicated by arrows. (B) The border cell cluster remains tethered to the anterior end by additional invasive cells, resulting in incomplete migrationin a Socs36E, apt double mutant egg chamber; (B) shows only the SLBO staining. Arrows indicate reducedSLBO expression in the additional invasive cells. (C) Quantification of the phenotypic penetrance of stage 10 egg chambers displaying one (blue) or more (pink) non-cluster associated invasive cells in mutant egg chambers of the indicated genotypes. (D) An electromobility shift assay (EMSA) reveals purified His-tagged APT protein binds to a site from the regulatory region of Socs36E, shown by the radioactively-labeled DNA detained in the gel in the second lane (black arrowhead), compared to free probe (open arrowhead). Binding is competed away by addition of excess unlabeled probe (cold competitor, third lane).
To test this idea, we performed expression and genetic interaction experiments between Socs36E and apt. By use of the Socs36E-Gal4 line, we established that there is overlap between Socs36E-driven GFP and APT expression in the anterior follicle cells (Fig. 1G). For genetic analysis, we compared the phenotypes in Socs36EEY06665/+ and apt167/+ to those in egg chambers from the double heterozygous females (Socs36EEY06665,+/+, apt167). We found a more than additive increase in the penetrance of the additional invasive cell phenotype when a single copy of Socs36E or apt was removed from the egg chamber, relative to a reduction of both: 55% of the double heterozygous egg chambers contained more than one additional invasive cell relative to 10% and 15% in Socs36EEY06665/+ and apt167/+, respectively. Next, we created Socs36E, apt recombinant mutant fly stocks using multiple apt and Socs36E loss of function alleles (see Materials and methods). Stage 10 egg chambers homozygous mutant for apt have an average of 12.6±3.0 total invasive cells (Fig. 5A and (Starz-Gaiano et al., 2008)). By comparison, the Socs36E, apt double homozygous mutants have an average of 13.5±3.0 total invasive cells per egg chamber, which was significantly greater than the heterozygous controls (which ranged between 7.3 and 8.4 invasive cells, depending on the genotype, with a standard deviation from the mean of ±1.5 and p<0.0001; Fig. 5B), and not significantly greater than apt/apt mutants (Fig. 5B and C, compare to Fig. 3G and A). These genetic results suggest Socs36E and APT have over-lapping roles in motile cell specification.
apt mutant flies have delayed border cell migration due to an impediment in detachment (Starz-Gaiano et al., 2008), however, in Socs36E mutant egg chambers, although there are additional motile cells, the main border cell cluster is able to complete migration. Thus, we examined border cell detachment in the double mutants. In 44% of stage 10 Socs36E, apt double homozygous mutant egg chambers, the border cells are tethered to the anterior end of the egg chamber by the non-cluster associated cells (Fig. 5B and B’). The inability to detach from neighboring follicle cells induced a migration delay in 31% of stage 10 double mutant egg chambers, comparable to the 32% observed in apt loss of function (Starz-Gaiano et al., 2008). However, trailing cells in Socs36E, apt/Socs36E, apt egg chambers displayed uneven levels of SLBO expression, which we observed in Socs36E but not apt deficient egg chambers (Fig. 5B and B’). These data suggest that APT functions independently in regulating cell detachment, but both APT and Socs36E are needed to convert the initial graded STAT activity into a binary ON/OFF signal for proper specification of cell identity.
In many cases, Socs36E expression is turned on by STAT activity, triggering a negative feedback loop (Alexander, 2002; Alexander and Hilton, 2004; Croker et al., 2008). To determine if APT may also directly contribute to Socs36E expression, we performed an Electromobility Shift Assay (EMSA). We tested for direct binding of purified His-tagged APT protein to radiolabeled oligonucleotides matching the APT consensus site (Liu et al., 2003) that we identified within the Socs36E enhancer region. We found that APT can bind this sequence (Fig. 5D). In genetic support of this finding, we determined that the penetrance of the additional invasive cell phenotype is only slightly higher between loss of function of apt and Socs36E, apt double homozygous mutant egg chambers (Fig. 5A–C), which is consistent with a model in which APT is upstream of Socs36E. These data combined with the observed expression patterns and genetic interaction support the idea that APT functions as a transcriptional regulator to induce Socs36E expression directly. Collectively, our data suggest a model in which Socs36E is co-regulated by APT and STAT to limit the migratory population to the border cell cluster by reducing STAT activity.
Socs36E and mir-279 function together to limit cell invasion
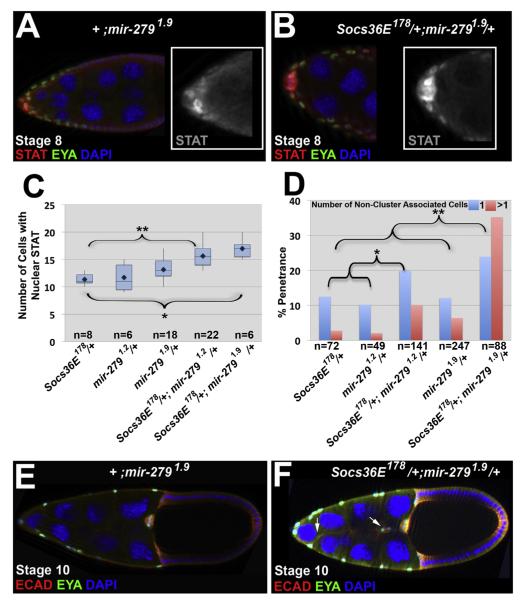
mir-279 is a downstream target of APT that negatively regulates STAT at the transcript level (Yoon et al., 2011). Since our data suggest Socs36E is also a downstream target of APT, we predicted there would be a synergistic relationship between Socs36E and mir-279. For genetic interaction tests, we looked at a single reduction of each gene (Socs36E/+; mir-279/+) using our Socs36E178 allele and two mir-279 null alleles (Cayirlioglu et al., 2008; Yoon et al., 2011). We counted the number of anterior follicle cells containing nuclear localized (activated) STAT at stage 8. We found a significant increase in anterior follicle cells with STAT activation when a single copy of both genes was removed (15.6±2.1 and 17.0±1.9), relative to loss of a single copy of either individual gene (11.4±1.2 Socs36E and 11.8±2.1 or 13.2±1.9 mir-279 alleles; p<0.005 for all genotypes; Fig. 6A–C). Further, when we removed a single copy of both genes there was a more than additive increase in the percentage of egg chambers containing more than one additional invasive cell, relative to loss of a single copy of only one of the genes (Fig. 6D–F). This penetrance varied depending on the mir-279 allele used, however, a synergistic relationship with Socs36E178 was observed in each case. These results suggest Socs36E and mir-279 function downstream of APT to limit the range of STAT activation prior to border cell cluster formation, which is necessary to optimize the number of motile cells.
Fig. 6.
Socs36E and mir-279 genetically interact to limit cell invasiveness. (A–B), (E–F) Egg chambers from following genotypes are shown: Control genotype, +/CyO; mir-2791.9/+(A and E) and Socs36E178/+; mir-2791.9/+ (B and F), at the stage indicated. (A–B) Staining with antibodies directed against EYA (green) and STAT (red)reveals the range of STAT activation in control egg chambers (A) compared tothose deficient for one copy of Socs36E andmir-279 (B). (C) Box plot quantification of nuclear STAT-positive anterior follicle cells in the genotypes indicated; means are indicated by diamonds. Statistically significant results relative to Socs36E178/+are indicted by *P<0.0003; **P<0.0001. (D) Phenotypic penetrance of one (blue) or more (pink) non-cluster associated cells in stage 10 egg chambers bearing mutations inSocs36E or mir-279 as indicated.Statistical analysis was performed using a two-tailed Fischer Exact test to compare the total penetrance of Socs36E178/+; mir-279/+ egg chambers exhibiting at least one additional invasive cell to their respective controls: Socs36E178/+; +/+ and CyO/+; mir-279/+. *P<0.02; **P<0.0001. (E–F) Egg chambers stained with antibodies directed against EYA (green) and E-Cadherin(red)toreveal invasive follicle cells. (E) A mir-279 heterozygote displays wild-type border cell numbers and movement. (F) A single copy reduction of both Socs36E and mir-279 results in additional follicle cells with invasive abilities (indicated by arrows).
Discussion
We have determined, through both gain-of-function and loss-of-function analysis, that Socs36E is necessary to limit STAT-directed migration in egg chambers. When we over-expressed Socs36E in the presumptive border cell population, most of these cells failed to reach the STAT/SLBO-migratory threshold and migrate efficiently to the oocyte (this study and (Silver et al., 2005)). These observations are consistent with reduction of STAT activation (Beccari et al., 2002; Ghiglione et al., 2002; Silver et al., 2005; Silver and Montell, 2001). Surprisingly, though, we found that nearby cells not over-expressing Socs36E could become abnormally motile, taking the place of those with high levels of Socs36E (Fig. 2F and G). We propose these data suggest Socs36E functions cell autonomously to attenuate STAT activation, but can also influence cell identity non-autonomously. SOCS proteins are able to function as the substrate recognition component of E3 ligase complexes in the ubiquitination pathway (Babon et al., 2009; Croker et al., 2008; Kile et al., 2002; Piessevaux et al., 2008). Therefore, it is possible that heightened Socs36E disrupts the expression of DOME on the apical side of the follicle cells, which could allow the activating UPD signal to disperse along a greater range than normal. The cell autonomous (STAT attenuation) and non-autonomous (UPD dispersion) hypotheses are not mutually exclusive and both could provide a mechanism to support our data. These results imply there is a compensatory mechanism when the presumptive border cells are incapable of migration to establish enough motile cells to escort the non-motile polar cells.
We determined that previously described loss of function alleles for Socs36E were not genetic nulls in egg chambers, so we generated new mutant alleles. Our loss of function mutant analyses with a genetic null revealed that Socs36E is required to limit the domain of STAT-activation and establish an optimal number of motile cells. Consistent with this idea, mutants homozygous for our allele, Socs36E178, had a significant increase in the number of STAT-positive anterior follicle cells prior to migration and showed invasive behavior in an excess number of follicle cells, similar to constitutive activation of JAK (Beccari et al., 2002; Silver et al., 2005; Silver and Montell, 2001). Our data suggest Socs36E is needed to convert the graded STAT-activity observed during stage 8 to a distinct ON/OFF cell fate decision prior to migration to restrict the number of motile cells.
Furthermore, we found Socs36E genetically interacts with the STAT-negative regulator apt: combined single-copy reduction of both genes caused a synergistic increase in the additional invasive cell phenotype relative to a reduction of each gene alone. We also found that APT is able to bind to the regulatory region of Socs36E, suggesting APT may activate Socs36E expression. Our Socs36E, apt double homozygous mutant phenotype supports an epistasis model in which APT is upstream of Socs36E, however, the double mutants displayed a slightly elevated penetrance of the additional invasive cell phenotype relative to loss of function apt alone (~90% versus 75%, respectively). This increase of penetrance suggests Socs36E may also be regulated by another factor, which is consistent with previous studies that found Socs family members are downstream targets of STAT (Alexander, 2002; Croker et al., 2008; Rawlings et al., 2004; Yoshimura et al., 2007). In the egg chamber, STAT-directed Socs36E expression is further supported by observations that reporters with Socs36E regulatory regions are influenced by activation levels of STAT and expressed in the anterior follicle cells of the egg chamber (this study and (Bach et al., 2007; Flaherty et al., 2009; Starz-Gaiano et al., 2008)). We, therefore, suggest Socs36E is positively regulated by STAT in a direct manner and indirectly through the STAT target APT (Starz-Gaiano et al., 2008). Consistent with this, we found overlap between APT and Socs36E expression. While APT expression expands beyond the range of Socs36E, we observed Socs36E is expressed in anterior follicle cells that receive both APT and STAT activity. We propose APT functions to potentiate Socs36E expression in regions of low STAT signaling. In further support of this, we observed APT binding to a sequence in the Socs36E regulatory region, suggesting it directly activates Socs36E expression.
We also determined Socs36E and mir-279 genetically interact in the egg chamber, which is consistent with each being independent downstream targets of APT and negative regulators of STAT (Montell et al., 2012; Yoon et al., 2011). While we would expect Socs36E; mir-279 double mutants to recapitulate the full apt phenotype, technical details of this experiment hindered its completion. However, reduction of each gene by a single copy was sufficient to increase significantly the number of anterior follicle cells with activated STAT and synergistically increased the penetrance of egg chambers with more than one additional invasive cell, relative to controls. Thus, coinciding with our Socs36E mutant analysis, our data suggest both negative regulators are necessary to limit the range of STAT activity prior to border cell recruitment and migration. These results support proposals that feedback loops and multiple repressors provide the accuracy and robustness necessary to distinguish specific cell fates (Chen et al., 2012b; Freeman, 2000).
In contrast to apt mutants, the border cell cluster is able to reach the oocyte and complete its migration in egg chambers deficient of Socs36E, because the additional invasive cells detach from the main cluster. This implies APT regulates aspects of migration independently of Socs36E. This phenotypic difference suggests that APT acts through miR-279 to facilitate detachment and therefore timely migration (Yoon et al., 2011), while Socs36E and miR-279 are crucial for limiting the number of invasive cells. We propose Socs36E and miR-279 are required in follicle cells surrounding the border cells to inhibit STAT at the protein and transcript level, respectively. This negative-feedback regulation produces a sharp boundary between the motile border cells and non-motile epithelial cells (Fig. 7).
Fig. 7.
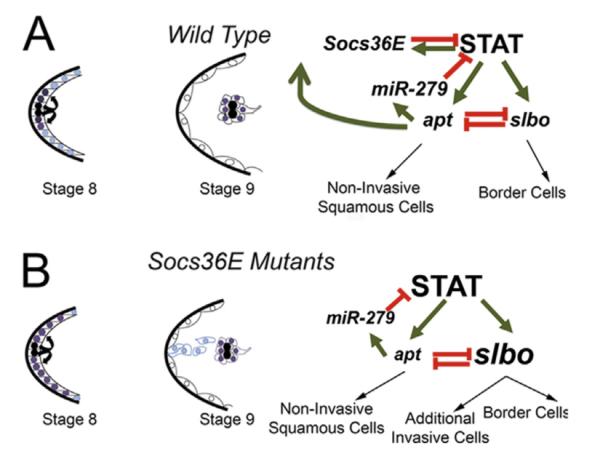
Proposed model:Socs36E is integral to a genetic circuit that attenu STAT activity to optimize the number of motile cells in the egg chamber. (A) In wild-type anterior follicle cells at stage 8, signal from the polar cells (indicated with black circles) activates STAT highly in neighboring cells (purple) and to a lower extent in farther cells (blue). At stage 9 as the border cells move away, STAT activity is maintained in motile cells and shut down in non-invasive cells that remain in the squamous epithelium (white). The system of determining the invasive cells from the epithelium requires the genetic regulatory circuit drawn on the right. (B) When Socs36E function is lost, STAT is highly activated across a larger field at stage 8 (indicated by purple circles). At stage 9, STAT activity remains high in border cells, but is also maintained in other cells, which become inappropriately invasive (blue). The impact of loss of Socs36E on the genetic circuit is shown on the right. See text for details.
Socs36E can negatively regulate Epidermal Growth Factor Receptor (EGFR) in Drosophila (Callus and Mathey-Prevot, 2002; Herranz et al., 2012; Rawlings et al., 2004). The EGFR pathway plays a key role in guiding the dorsal movement of the border cell cluster, and over-expression of EGFR ligands in the egg chamber or expression of a constitutively active EGFR in the border cells allows proper cluster formation but causes severe migration defects (Bianco et al., 2007; Duchek and Rørth, 2001; Inaki et al., 2012; McDonald et al., 2006; Poukkula et al., 2011; Prasad and Montell, 2007). These phenotypes are distinct from the additional invasive cell phenotype observed in loss of function Socs36E egg chambers. Thus, our data are consistent with Socs36E functioning predominantly as a STAT attenuator in the egg chamber, and not by regulating EGFR. It is, however, likely that Socs36E, as well as STAT and mir-279, have additional downstream targets, adding further complexity to the regulation of cell motility.
In mammals, SOCS proteins can perform tumor suppressive activities. For example, Socs expression levels are severely decreased in several cancers, including gastric (SOCS1 and 3) (Deng et al., 2010; To et al., 2004), hepatocellular carcinoma (SOCS3) (Niwa et al., 2005), and breast and ovarian carcinomas (SOCS1 and 2) (Nakagawa et al., 2008; Sutherland et al., 2004). A recent study suggested a tumor-secreted miRNA targets Socs5 in endothelial cells allowing STAT-driven angiogenesis and invasion, linking the closest mammalian ortholog of Socs36E to the regulation of STAT and invasive behavior (Zhuang et al., 2012). These studies implicate the SOCS family as important regulators of cell migration through attenuation of STAT activity. We have found this to be a conserved role of the family between vertebrates and Drosophila, as Socs36E is a necessary negative regulator of cell invasion through attenuation of STAT. Thus, work in this system can further our understanding of complex genetic pathways, and provide insight into what may go wrong in abnormal or disease contexts.
Supplementary Material
Acknowledgements
We appreciate the generosity extended by members of the fly community, particularly Dr. Denise Montell, Dr. Erika Matunis, Dr. S. Hirose, Dr. Reinhard Schuh, Dr. Florenci Serras, Dr. Steven Hou, and Dr. Pernille Rorth, for reagents and protocols. We thank the Bloomington Drosophila Stock Center, the Drosophila Genetic Resource Center in Kyoto Institute of Technology, and Developmental Studies Hybridoma Bank for providing reagents. We would also like to thank Dr. Nicholas Gaiano and Steven A. Szebenyi for critical comments and discussion on the manuscript. AJM is supported by the NIH Chemistry-Biology Interface Training Fellowship (NIH T32 GM066706-06) and DOE #P200A090094. This research was supported in part by a NSF Career Award (1054422) and a Basil O’Connor Starter Scholar Award from the March of Dimes awarded to MSG.
Footnotes
Appendix A. Supporting information Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.03.022.
References
- Alexander W. Suppressors of cytokine signalling (SOCS) in the immune system. Nat. Rev. Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Almudi I, Stocker H, Hafen E, Corominas M, Serras F. SOCS36E specifically interferes with Sevenless signaling during Drosophila eye development. Dev. Biol. 2009;326:212–223. doi: 10.1016/j.ydbio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arbouzova N, Zeidler M. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Babon J, Sabo J, Zhang J, Nicola N, Norton R. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J. Mol. Biol. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene. Express Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Beccari S, Teixeira L, Rørth P. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech. Dev. 2002;111:115–123. doi: 10.1016/s0925-4773(01)00615-3. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rørth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Chen X. STAT proteins: signal tranducers and activators of transcription. Methods Enzymol. 2001;333:138–151. doi: 10.1016/s0076-6879(01)33052-5. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Darnell JJ. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Wrzeszczynska M, Devgan G, Zhao Y, Pestell R, Albanese C, Darnell JJ. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Callus B, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GS, Gunning D, Lai EC, Zipursky SL. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008;319:1256–1260. doi: 10.1126/science.1149483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012a;36:529–541. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu Z, Mei C, Yu D, Small S. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012b;149:618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R. Suppressors of cytokine signaling (SOCS): inhibitors of the JAK/STAT pathway. Shock. 2002;17:83–90. doi: 10.1097/00024382-200202000-00001. [DOI] [PubMed] [Google Scholar]
- Croker BA, Kiu H, Nicholson SE. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008;19:414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas M, Matunis EL. The stem cell niche: lessons from the Drosophila testis. Development. 2011;138:2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JY, Sun D, Liu XY, Pan Y, Liang H. STAT-3 correlates with lymph node metastasis and cell survival in gastric cancer. World J. Gastroenterol. 2010;16:5380–5387. doi: 10.3748/wjg.v16.i42.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devergne O, Ghiglione C, Noselli S. The endocytic control of JAK/STAT signalling in Drosophila. J. Cell Sci. 2007;120:3457–3464. doi: 10.1242/jcs.005926. [DOI] [PubMed] [Google Scholar]
- Duchek P, Rørth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291:131–133. doi: 10.1126/science.291.5501.131. [DOI] [PubMed] [Google Scholar]
- Eulenberg K, Schuh R. The tracheae defective gene encodes a bZIP protein that controls tracheal cell movement during Drosophila embryogenesis. EMBO J. 1997;16:7156–7165. doi: 10.1093/emboj/16.23.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Zavadil J, Ekas LA, Bach EA. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev. Dyn. 2009;238:2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- Gaziova I, Bonnette PC, Henrich VC, Jindra M. Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development. 2004;131:2715–2725. doi: 10.1242/dev.01143. [DOI] [PubMed] [Google Scholar]
- Gellon G, Harding K, McGinnis N, Martin M, McGinnis W. A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development. 1997;124:3321–3331. doi: 10.1242/dev.124.17.3321. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Georgenthum E, Carballès F, Médioni C, Cerezo D, Noselli S. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129:5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Hong X, Hung NT, Voorhoeve PM, Cohen SM. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev. 2012;26:1602–1611. doi: 10.1101/gad.192021.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombría J, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr. Biol. 2002;12:R569–575. doi: 10.1016/s0960-9822(02)01057-6. [DOI] [PubMed] [Google Scholar]
- Hou SX, Zheng Z, Chen X, Perrimon N. The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev. Cell. 2002;3:765–778. doi: 10.1016/s1534-5807(02)00376-3. [DOI] [PubMed] [Google Scholar]
- Inaki M, Vishnu S, Cliffe A, Rørth P. Effective guidance of collective migration based on differences in cell states. Proc. Nat. Acad. Sci. U.S.A. 2012;109:2027–2032. doi: 10.1073/pnas.1115260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang A, Chang Y, Bai J, Montell D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat. Cell Biol. 2009;11:569–579. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten P, Häder S, Zeidler M. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DS, Keenan S, Kerhornou A, Koscielny G, Langridge N, McDowall MD, Megy K, Maheswari U, Nuhn M, Paulini M, Pedro H, Toneva I, Wilson D, Yates A, Birney E. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khammari A, Agnès F, Gandille P, Pret AM. Physiological apoptosis of polar cells during Drosophila oogenesis is mediated by Hid-dependent regulation of Diap1. Cell Death Differ. 2011;18:793–805. doi: 10.1038/cdd.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile B, Schulman B, Alexander W, Nicola N, Martin H, Hilton D. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- King RC. Development in Drosophila melanogaster. Academic Press; New York: 1970. [Google Scholar]
- Krebs D, Hilton D. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Levy D, Darnell JJ. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Liu Q, Jindra M, Ueda H, Hiromi Y, Hirose S. Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development. 2003;130:719–728. doi: 10.1242/dev.00297. [DOI] [PubMed] [Google Scholar]
- Luo H, Dearolf CR. The JAK/STAT pathway and Drosophila development. Bioessays. 2001;23:1138–1147. doi: 10.1002/bies.10016. [DOI] [PubMed] [Google Scholar]
- Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K, Palter K, Selleck S. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Montell DJ. Analysis of cell migration using Drosophila as a model system. Methods Mol. Biol. 2005;294:175–202. doi: 10.1385/1-59259-860-9:175. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, Montell DJ. Multiple EGFR ligands participate in guiding migrating border cells. Dev. Biol. 2006;296:94–103. doi: 10.1016/j.ydbio.2006.04.438. [DOI] [PubMed] [Google Scholar]
- McQuilton St P, Pierre SE, Thurmond J, Consortium F. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell DJ. Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, Spradling AC. Slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nat. Rev. Mol. Cell Biol. 2012;13:631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Iida S, Osanai T, Uetake H, Aruga T, Toriya Y, Takagi Y, Kawachi H, Sugihara K. Decreased expression of SOCS-3 mRNA in breast cancer with lymph node metastasis. Oncol. Rep. 2008;19:33–39. [PubMed] [Google Scholar]
- Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T, Yamamoto J, Kubo T, Yoshikawa H. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406–6417. doi: 10.1038/sj.onc.1208788. [DOI] [PubMed] [Google Scholar]
- Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell–cell adhesion. Dev. Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: A glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Piessevaux J, Lavens D, Peelman F, Tavernier J. The many faces of the SOCS box. Cytokine Growth Factor Rev. 2008;19:371–381. doi: 10.1016/j.cytogfr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Poukkula M, Cliffe A, Changede R, Rørth P. Cell behaviors regulated by guidance cues in collective migration of border cells. J. Cell Biol. 2011;192:513–524. doi: 10.1083/jcb.201010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev. Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Rawlings J, Rennebeck G, Harrison S, Xi R, Harrison D. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 2004;5:38. doi: 10.1186/1471-2121-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Riggleman B, Schedl P, Wieschaus E. Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- Silver DL, Geisbrecht ER, Montell DJ. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Singh S, Zheng Z, Wang H, Oh S, Chen X, Hou S. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J. Cell. Physiol. 2010;223:500–510. doi: 10.1002/jcp.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. Developmental Genetics of Oogenesis. Cold Spring Harbor Laboratory Press; New York: 1993. [Google Scholar]
- Starz-Gaiano M, Melani M, Meinhardt H, Montell D. Interpretation of the UPD/JAK/STAT morphogen gradient in Drosophila follicle cells. Cell Cycle. 2009;8:2917–2925. doi: 10.4161/cc.8.18.9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell. 2008;14:726–738. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Lindeman GJ, Choong DY, Wittlin S, Brentzell L, Phillips W, Campbell IG, Visvader JE. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726–7733. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- To KF, Chan MW, Leung WK, Ng EK, Yu J, Bai AH, Lo AW, Chu SH, Tong JH, Lo KW, Sung JJ, Chan FK. Constitutional activation of IL-6-mediated JAK/STAT pathway through hypermethylation of SOCS-1 in human gastric cancer cell line. Br. J. Cancer. 2004;91:1335–1341. doi: 10.1038/sj.bjc.6602133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Hirose S. Defining the sequence recognized with BmFTZ-F1, a sequence specific DNA binding factor in the silkworm, Bombyx mori, as revealed by direct sequencing of bound oligonucleotides and gel mobility shift competition analysis. Nucleic Acids Res. 1991;19:3689–3693. doi: 10.1093/nar/19.13.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Bor V, Zimniak G, Cérézo D, Schaub S, Noselli S. Asymmetric localisation of cytokine mRNA is essential for JAK/STAT activation during cell invasiveness. Development. 2011;138:1383–1393. doi: 10.1242/dev.056184. [DOI] [PubMed] [Google Scholar]
- Xi R, McGregor J, Harrison D. A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev. Cell. 2003;4:167–177. doi: 10.1016/s1534-5807(02)00412-4. [DOI] [PubMed] [Google Scholar]
- Yoon WH, Meinhardt H, Montell DJ. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat. Cell Biol. 2011 doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, Oeh J, Modrusan Z, Bais C, Sampath D, Ferrara N. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012 doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.