Abstract
Recent genome-wide association studies have identified multiple regions at 8q24 that confer susceptibility to many cancers. In our previous work, we showed that the colorectal cancer (CRC) risk variant rs6983267 at 8q24 resides within a TCF4 binding site at the MYC-335 enhancer, with the risk allele G having a stronger binding capacity and Wnt responsiveness. Here, we searched for other potential functional variants within MYC-335. Genetic variation within MYC-335 was determined in samples from individuals of European, African, and Asian descent, with emphasis on variants in putative transcription factor binding sites. A 2-bp GA deletion rs67491583 was found to affect a growth factor independent (GFI) binding site and was present only in individuals with African ancestry. Chromatin immunoprecipitation performed in heterozygous cells showed that the GA deletion had an ability to reduce binding of the transcriptional repressors GFI1 and GFI1b. Screening of 1,027 African American colorectal cancer cases and 1,773 healthy controls did not reveal evidence for association (odds ratio: 1.17, 95% confidence interval: 0.97–1.41, P = 0.095). In this study, rs67491583 was identified as another functional variant in the CRC-associated enhancer MYC-335, but further studies are needed to establish the role of rs67491583 in the colorectal cancer predisposition of African Americans.
Keywords: Enhancer, transcription factor, susceptibility variant, colorectal cancer, association
Colorectal cancer (CRC) is a major public health problem worldwide, accounting for over one million new cases each year. Currently, it is estimated that 147,000 new CRC diagnoses and 50,000 deaths from CRC occur annually in the United States (1). A clear ethnic disparity exists in the occurrence of CRC in the United States; African Americans have the highest incidence and mortality rates for CRC among ethnic populations (2). Genetic and environmental factors are suggested to contribute to the variation in CRC incidence.
Approximately 35% of the variation in CRC risk is attributed to genetic factors (3). Mendelian CRC syndromes account for only 5% of all cases, and the remaining genetic component of CRC is still largely unexplained. Recently, the “common disease–common variant” model of predisposition was addressed by genome-wide association (GWA) studies. To date, GWA studies, conducted in individuals of European descent, have identified 14 common genetic variants (rs6983267, rs4779584, rs4939827, rs16892766, rs10795668, rs3802842, rs4444235, rs9929218, rs10411210, rs961253, rs6691170, rs10936599, rs11169552, rs4925386) that confer increased susceptibility to CRC (4–9). In addition, a recent fine mapping study identified two new independent CRC predisposition single nucleotide polymorphisms (SNPs) near BMP2 (rs4813802) and BMP4 (rs1957636), and suggested that the original CRC-associated SNP rs4779584, close to GREM1, actually tags two functional SNPs. Thus, the original finding may have represented two independent signals, now defined by rs16969681 and rs11632715 (10).
The 8q24 region, marked by SNP rs6983267, is by far the best-studied of the low-penetrance CRC susceptibility loci. rs6983267 has been shown to associate also with prostate cancer (11), and different regions at 8q24 gene desert have been found to confer susceptibility to breast cancer (12), bladder cancer (13), and chronic lymphocytic leukemia (14). It has been suggested that cancer predisposition is mediated by variants in tissue-specific enhancer elements that control the expression of the MYC oncogene. In our previous work, we showed that the CRC predisposition SNP rs6983267 affects a TCF4 binding site in the distal enhancer element, MYC-335, and the risk allele G creates a higher-affinity TCF4 binding than the T-allele, resulting in stronger enhancer activity (15). Pomerantz et al. (2009) demonstrated that the region containing rs6983267 interacts with the MYC promoter, providing evidence that MYC is the target gene (16). The biological mechanism underlying the CRC predisposition at 18q21 (rs4939827) and 8q23 (rs16892766) is similar (17–19). There, the CRC risks are caused by variants that change activity of gene regulatory elements, leading to changes in SMAD7 and EIF3H/UTP23 expression. These studies have created novel insights into the molecular etiology of CRC.
Due to the importance of the 8q24 region in cancer predisposition, it is essential to scrutinize in detail the CRC-associated enhancer MYC-335 at 8q24. Recent fine mapping studies have shown that some of the known CRC loci may contain multiple risk variants, some of which have proposed to be functional (10,19). Moreover, because GWA studies have been performed on Caucasian populations only, little is known about population-specific differences in CRC predisposition. In this study, we characterized the genetic variation within the CRC-associated enhancer MYC-335 at 8q24, aiming at identifying other functional variants that could potentially act as CRC risk alleles. We sequenced the MYC-335 enhancer in African, African American, Asian, and Caucasian samples and investigated the role of rs67491583, which affected a transcription factor binding site, in CRC predisposition in African Americans.
Materials and methods
Subjects
Caucasian control DNA samples used in the study included 188 anonymous Finnish blood donors obtained from the Finnish Red Cross Blood Transfusion Service and 288 U.K. blood donors obtained from Sigma-Aldrich (St. Louis, MO) (human random control DNA panels 1–3). African control samples consisted of 180 Yoruba (YRI) HapMap samples (HAPMAPPT03 and HAPMAPPT04) provided by the Coriell Institute for Medical Research (Camden, NJ). The HAP-MAPPT07 panel comprising 90 samples from people with African ancestry in the southwestern United States was used as a representative of AfricanAmerican population. A series of 36 blood donors from Korea was available as Asian control samples (20).
Germline DNA specimens from 1,134 African American CRC cases and 1,732 controls were available from different research centers in the United States. Case–control studies were from University of North Carolina (UNC) (CRC, n = 410; controls, n = 418), Multiethnic Cohort (MEC) (CRC, n = 240; controls, n = 431), University of Chicago (CRC, n = 189; controls, n = 183), Ohio State University (OSU) (CRC, n = 96; controls, n = 192), Cleveland Clinic/Case Western Reserve University (CRC, n = 105; controls, n = 160), and University of Alabama (UAB) (CRC, n = 94; controls, n = 150). DNA samples from UNC, MEC, and OSU were derived from blood (except three OSU cases: one from paraffin block, two from buccal rinse). Samples from the University of Chicago were extracted from formalin-fixed, paraffin-embedded normal tissue sections. Case DNAs from Cleveland Clinic/Case Western Reserve University were from fresh frozen normal colon tissue and controls from blood. DNA from UAB cases was derived from normal tissue surrounding colon tumors, and controls were blood DNAs that were randomly selected from volunteers within the UAB hospital system. In addition, 198 blood DNA controls were received from the Howard University College of Medicine, Washington, DC. Also, 13 tumor DNAs extracted from paraffin-embedded tissue samples were available from OSU. In addition, 44 normal and 26 tumor tissue DNA specimens from Korean colorectal carcinoma patients were studied (20). Normal tissue DNAs from the Finnish CRC patients were selected from a population-based series collected since 1994. All samples were derived with written informed consent and approval from the respective institutions’ ethical review boards.
Cell lines
Epstein-Barr virus transformed B-lymphocyte cell line GM19240 was purchased from the Coriell Institute for Medical Research and maintained in RPMI supplemented with 10% fetal bovine serum (FBS). GM19240 was derived from a HapMap YRI individual. HeLa and HEK293T cells were cultured in DMEM with 10% FBS, and LoVo cells were maintained in DMEM + 1.5 g/L NaHCO3. All plasmid transfections were done with FuGENE HD transfection reagent (Roche). Culture medium was changed 6 hours after transfection.
Sequencing and genotyping
The 1,255-bp MYC-335 enhancer element is located at 128,413,174-128,414,429 bp (GRCh37, Ensemble 56: Sep 2009) in the human chromosome 8. This region was sequenced to determine the genetic variation within the enhancer. The region was polymerase chain reaction (PCR)-amplified from genomic DNA in two overlapping fragments. The primers used in the reactions were as follows: F: 5′-AACTTTCCCAGCCTCGTTCT-3′ and R: 5′-CCATGG GAAAATAGATGGCATA-3′, F: 5′-ATTCCTGACCTACCCCCAAA-3′ and R: 5′-GTTTTCAGGTGCGTGTGTGT-3′. The fragments were sequenced directly using BigDye v3.1 (Applied Biosystems, Carlsbad, CA) sequencing chemistry and the ABI3730 automatic sequencer (Applied Biosystems). Sequence reads were analyzed using Mutation Surveyor v3.24 software (Softgenetics, State College, PA). All sequences were also read manually, and samples with ambiguous results were repeated.
Samples from Cleveland, OSU, and Howard University College of Medicine were genotyped for rs67491583 (GA deletion) and rs6983267 using previously mentioned primers. Case–control series from Chicago, MEC, UNC, and UAB were genotyped for rs67491583 by sequencing, using primers F 5′-TCAATTTCATCTACGTGAAGAGC-3′ and R 5′-TGCAGGATATCTTGGGAATGA-3′.
ChIP
ChIP was performed in HeLa and GM19240 essentially as described in Tuupanen et al. (15) and Turunen et al. (21), using antibodies GFI1B (Rabbit Anti-GFI1B Polyclonal Antibody H-150: sc-22795, Santa Cruz Biotechnology, Santa Cruz, CA), GFI1 (Rabbit Anti-GFI1 Polyclonal Antibody N-20: sc-8558, Santa Cruz Biotechnology), Histone H3 mono methyl K4 (Rabbit Polyclonal Antibody, ab8895, Abcam, Cambridge, MA), or control IgG (normal mouse IgG: sc-2025 or normal rabbit IgG: sc-2027, Santa Cruz Biotechnology).
Allele-specific binding of GFI1 and GFI1B to the rs67491583-containing site was assessed by comparing the height of Sanger sequencing signal peaks from the wild-type allele and the GA deletion-containing alleles in sequencing tracts. Fold changes were calculated relative to input control. Primers targeting the rs67491583-containing fragments were: 5′- GGAGATGCCAAAAAGCCAAT-3′ and 5′- AGAACAGGGGAAGCTGAACA-3′. The primer used for Sanger sequencing was: 5′ - AGAACAGGGGAAGCTGAACA-3′.
For ChIP-by-sequencing (ChIP-seq), the precipitated DNA was repaired using Klenow and T4 DNA polymerases and T4 polynucleotide kinase (MBI Fermentas, Latvia), and ligated to adapters according to the manufacturer’s instructions (Illumina, San Diego, CA). Subsequently, PCR amplified fragments of approximately 150–200 bp were sequenced using the Illumina Genome Analyzer (University of Helsinki, Helsinki, Finland). Sequencing reads (36 bp) were mapped to the human genome (NCBI36) using Maq software by Heng Li, version 0.6.5. Only high-quality reads that could be reliably mapped (mapping quality score at least 30) were accepted, resulting in a total of 15.1 and 4.8 million reads from GFI1 GM19240 ChIP and IgG control samples. Each read was then extended to a sequence of 170 bp, and height was determined at each position as the number of overlapping sequences.
This analysis yields a maximum peak-width of 250 bp for one occupied GFI1 site (120 bp in both directions from an approximate 10-bp site). The positions with a height of 10 or more were defined as peaks. For each peak, the total number of sequences in the continuous region of four or more overlapping sequences was compared with the number of sequences in the same region in the IgG control. The probability of observing the difference between the sequence counts in the ChIP sample and IgG control by chance was estimated using Winflat program 49. The program was originally developed for digital gene expression analysis, and it can take into account the uncertainty associated with low sequence counts and the difference in the total amount of ChIP and IgG control reads. In total, 16,700 GFI1 peaks in GM19240 with a height of >10 and P < 0.05 were observed.
Luciferase assay
For luciferase reporter assays, a previously generated reporter construct containing MYC-335 enhancer with G allele of rs6983267 was used (15). A reporter construct containing the GA deletion allele was created by site-directed mutagenesis. hGFI1B cDNA was over-expressed in HEK293T, HeLa, and LoVo cells. The 1,406-bp sized, element-containing reporters were transfected into cells together with a Renilla luciferase control reporter (Promega, Madison, WI). Luciferase activities were measured at 32 hours with the DualLuc kit (Promega). Relative luciferase activities were calculated by dividing the firefly luciferase counts with the Renilla controls, and the results were normalized to the wild-type allele.
Analysis of allelic imbalance
Allelic imbalance was analyzed in tumor DNA of 13 CRC cases that were heterozygous for the GA deletion. Normal and tumor DNA from the same patient was sequenced using primers 5′-TCAATTTCATCTACGTGAAGAGC-3′ and 5′-TGCAGGATATCTTGGGAATGA-3′, and AI was scored by comparing the allelic peak ratios in tumor DNA respective to the normal tissue.
Statistical analyses
We expected heterogeneity across study groups; therefore, we used boxplots to identify possible outliers in the allele frequency data (Table 2, Supplementary Figure S1) (22). Case series UAB was identified as an outlier and was excluded from further analysis. African American populations are highly heterogenous; therefore, genotype counts were calculated separately for different case–control series. Association analyses were performed by combining genotyped counts from all the sample series. Association analyses were performed using R software (v2.10.1). Pearson’s chi-square test was used to calculate P-values, odds ratios (ORs), and the associated confidence intervals (CIs). Power calculations were performed at http://statpages.org/proppowr.html.
Table 2.
Frequency of rs6749158 in the African American CRC cases and controls
| Sample series |
Genotype | HWE test P-valuea |
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Wt | Het | Hom | Frequency | ||||
| UNC | ||||||||
| CRC | 408 | 331 | 74 | 3 | 12.1 | 0.78 | ||
| Controls | 410 | 338 | 70 | 2 | 10.9 | 0.56 | ||
| MEC | ||||||||
| CRC | 240 | 200 | 38 | 2 | 8.8 | 1.00 | ||
| Controls | 431 | 368 | 61 | 2 | 7.5 | 1.00 | ||
| Chicago | ||||||||
| CRC | 182 | 147 | 30 | 5 | 11 | 0.05 | ||
| Controls | 176 | 137 | 36 | 3 | 11.9 | 1.00 | ||
| OSU | ||||||||
| CRC | 95 | 81 | 13 | 1 | 7.9 | 0.62 | ||
| Controls | 188 | 161 | 26 | 1 | 7.4 | 1.00 | ||
| Cleveland | ||||||||
| CRC | 102 | 76 | 25 | 1 | 13.2 | 0.69 | ||
| Controls | 153 | 125 | 27 | 1 | 9.5 | 1.00 | ||
| Washington | ||||||||
| Controls | 189 | 156 | 31 | 2 | 9.3 | 0.70 | ||
| UAB | ||||||||
| Controls | 136 | 115 | 20 | 1 | 8.1 | 1.00 | ||
| HAPMAPPT07 | ||||||||
| Controls | 90 | 80 | 10 | 0 | 5.6 | 1.00 | ||
| Total | ||||||||
| CRC | 1027 | 835 | 180 | 12 | 9.9 | 0.60 | P = 0.095 | |
| Controls | 1773 | 1480 | 281 | 12 | 8.6 | 0.77 | OR = 1.17 | 95% c.i 0.97–1.41 |
Pearson’s Chi-squared test with simulated P-value (based on 1e+06 replicates).
Relationship between rs67491583 genotype and MYC expression
To examine a relationship between the rs67491583 genotype and the expression level of MYC, publicly available exon array gene expression data from 89 YRI HapMap Epstein-Barr virus–transformed lymphoblastoid cell lines were used (GEO GSE7761) (23). To discard erroneous probes, probe set intensities were generated using the Brainarray (v.12.1.0, ENSG) custom CDF file (24). The data was normalized using RMA normalization provided in exonmap package (v1.0.07) for R (v2.9.0). A two-sided t test assuming equal variance in the groups was used to compare MYC expression (ENS-G00000136997_AT) between the wild-type (n = 75) and heterozygous (n = 14) cell lines.
Another sample set was obtained from OSU and included B-lymphocyte cell line pellets in Trizol from 40 healthy individuals. Twenty of 40 were wild-type for rs67491583, 19 were heterozygotes, and 1 was a homozygote. Total RNA was extracted with standard Trizol-based protocol, and 800 ng of total RNA was reverse transcribed to cDNA using the Promega MMLV enzyme. Relative expression of MYC was determined with TaqMan chemistry and the ABI Prism 7500 sequence detection system using assays for MYC (Hs00153408_m1) and endogenous control PGK (Hs99999906_m1) (both from Applied Biosystems).
Results
Sequencing the MYC-335 element (Supplementary Table S1) in 727 control samples (247 U.K., 174 Finnish, 180 HapMap YRI, 90 HapMap African Americans, 36 Koreans) revealed 13 variants, of which 6 were known (rs6983267, rs34835043, rs73706717, rs67180956, rs7008058, rs67491583) and 7 were new variants (Table 1). Three of the novel variants (novel 3, 4, 5) were present in only one sample each. Allele frequencies of the variants varied between populations. Our previous notion that the CRC predisposition SNP rs6983267 affects the TCF4 binding site at the MYC-335 prompted us to particularly look for variants that affected any of the 24 other enhancer element locator (EEL)-predicted transcription factor (TF) binding sites within the MYC-335 enhancer (14). Indeed, two such variants were identified (Table 1, Supplementary Table S1). rs67180956 was located at the first nucleotide of the broad complex 3 binding site TGTTTTGTTTA; however, this site is not conserved in mouse (GTTATAGTTTT) and, therefore, was not studied further. rs67491583, which is a 2-bp deletion, mapped to a growth factor independent (GFI) binding site CAGAGATTGC (GRCh37: chr8: 128414228-128414237). The deletion affected GA nucleotides at the fifth and sixth position of the binding sequence (CAGAGATTGC). rs67491583 has been previously found in the genome of Craig Venter; however, no population genetics were available for this variant, and it has not, to our knowledge, been reported elsewhere. In our data, the deletion was found only in samples with African origin and was absent in 421 Caucasians and 36 Koreans. Allele frequency of the deletion was 8.9% in HapMap YRI samples and 5.6% in HapMap African American samples (Table 1). Due to the low number of control samples representing Asian population, we screened 70 Korean CRC patients (44 normal and 26 tumor DNAs) for the GA deletion. None of these displayed the GA deletion. In addition, no GA deletions were identified among 79 Finnish CRC patients.
Table 1.
Variants within MYC-335 revealed by sequencing 727 control samples
| Minor allele frequency |
||||||||
|---|---|---|---|---|---|---|---|---|
| Variation | Type | Location chr8 | TF binding site | U.K. | Finland | YRI | SW AFAM | Korea |
| rs6983267a | G>T | 128413305 | TCF4 | 0.541 | 0.535 | 0.977 | 0.822 | 0.389 |
| rs34835043 | A>G | 128413389 | 0.038 | 0.056 | 0.069 | 0.028 | 0.069 | |
| rs73706717 | T>C | 128413510 | 0 | 0 | 0.117 | 0.128 | 0 | |
| Novel 1 | G>A | 128413593 | 0 | 0 | 0.017 | 0.017 | 0 | |
| Novel 2 | G>T | 128413783 | 0.081 | 0.115 | 0 | 0.022 | 0 | |
| rs67180956 | T>C | 128413809 | Broad Complex 3 | 0.041 | 0.057 | 0.138 | 0.094 | 0.069 |
| Novel 3 | G>A | 128413829 | 0 | 0 | 0.003 | 0 | 0 | |
| rs7008058 | T>C | 128413978 | 0 | 0 | 0.117 | 0.128 | 0 | |
| Novel 4 | G>A | 128413985 | 0 | 0.003 | 0 | 0 | 0 | |
| Novel 5 | C>T | 128414065 | 0 | 0.003 | 0 | 0 | 0 | |
| rs67491583 | Del GA | 128414232e128414233 | GFI1 | 0 | 0 | 0.089 | 0.056 | 0 |
| Novel 6 | C>T | 128414371 | 0.067 | 0.075 | 0.028 | 0.017 | 0.028 | |
| Novel 7 | G>C | 128414372 | 0 | 0 | 0.006 | 0 | 0 | |
Frequency of the risk allele G is presented.
The GFI binding sequence CAGAGATTGC at the MYC-335 element contains sequence GATT (AATC) that is a core binding motif for GFI proteins, GFI1 and GFI1b (25). According to the affinity matrix available in the JASPAR database, the identified binding sequence (CAGAGATTGC) differs slightly from the consensus binding sequence of GFI (CAGTGATTTG). The GA deletion at the EEL-predicted binding site results in generation of a weaker GFI site that has a 5.5-fold lower affinity than the GA-containing sequence. The identified GFI1 binding sequence is highly conserved in evolution. According to Ensemble multispecies alignment, exactly the same sequence is present in chimpanzee, orangutan, rhesus macaque, cow, rat, and mouse genomes. Dog and horse show one base pair difference in the binding sequence (data not shown).
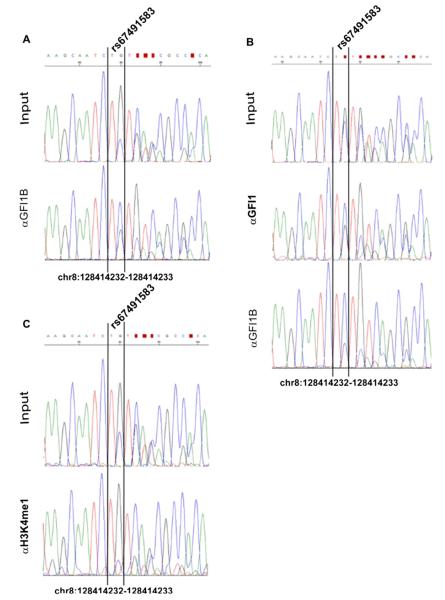
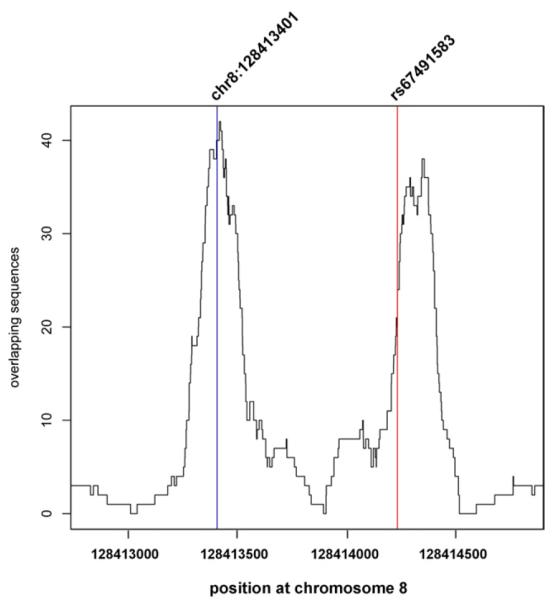
We assessed whether the GA deletion has an effect on TF binding in cells. We genotyped CRC cell lines RKO, GP5D, HCT8, HUTU80, HCA7, LS174T, LS180, HCT116, VACO5, SW480, LoVo, and CCL231 as well as cervical adenocarcinoma cell line HeLa for the GA deletion rs67491583. All the CRC cell lines were wild-type for rs67491583. HeLa cells that originate from a 31-year-old African American woman were found to be heterozygous for rs67491583. We tested the effect of the deletion on GFI binding by conducting chromatin immunoprecipitation (ChIP) in heterozygous cell lines HeLa and GM19240, using antibodies for GFI1, GFI1B, H3K4me1, and p300 (Figure 1, Supplementary Figure S1). GFI1 proteins are not expressed in HeLa; therefore, GFI1B was transiently over-expressed on these cells before the experiments. Sanger sequencing of the input and immunoprecipitated DNA indicated that GFI1B bound more strongly to the wild-type allele than to the mutant allele (Figure 1A). Similarly, in the lymphoblast cell line GM19240, endogenous GFI1 and GFI1B preferred to bind to the wild-type allele (Figure 1B). In turn, H3 monomethyl Lysine 4 (H3K4me1), a correlate of enhancer activity (26), was preferentially found in the GA deletion-containing allele of GFI1B-expressing HeLa cells (Figure 1C). We observed an approximately twofold decrease in H3K4 monomethylation in the allele that binds to GFI, consistent with the action of GFI as a repressor. Although the effect is not all-or-nothing, it is quite large considering that only a single TF binding site is affected in a regulatory element that binds to many TFs. H3K4me1 was also tested in GM19240; however, the binding was balanced between the alleles. This is probably because GM19240 is a lymphoblastoid cell line, and it is likely that this tissue-specific enhancer (15) is not functional in lymphoblasts. However, p300, another correlate of enhancer activity, preferred to bind to the wide-type allele in GFI1B-expressing HeLa cells (Supplementary Figure S2), suggesting that the GA deletion may also decrease transcriptional co-activator binding. To gain a comprehensive picture of GFI1 occupancy at MYC-335, we performed ChIP-seq in GM19240 cells (Figure 2). ChIP-seq indicated two peaks within the 2,000-bp genomic fragment containing MYC-335 (Figure 2).
Figure 1.
ChIP followed by PCR and Sanger sequencing for rs67491583. (A) In HeLa cells, ectopically over-expressed GFI1B favors binding to the wild-type allele of rs67491583. Sanger-sequencing of anti-GFI1B precipitated DNA (αGFI1B) reveals that the wild-type allele (TC) is enriched by the GFI1B precipitation when compared with the input DNA (Input). (B) In the GM19240 lymphoblast, endogenous GFI1 (αGFI1) and GFI1B (αGFI1B) both prefer to bind to the wild-type allele (TC). (C) In HeLa cells over-expressing GFI1B, Sanger-sequencing of anti-histone H3 monomethyl Lysine 4 precipitated DNA (αH3K4me1) reveals that the risk-allele (TG) is enriched by the H3K4me1 precipitation when compared with the input DNA (Input).
Figure 2.
GFI1 occupancy in this genomic region in GM19240 lymphoblast cells. The 2 kb region at chromosome 8q24 with the highest number of overlapping sequences as determined by ChIP-seq. rs67491583, indicated by a red line, resides near one of the two peaks. The blue line marks the other EEL-predicted GFI binding site in this region.
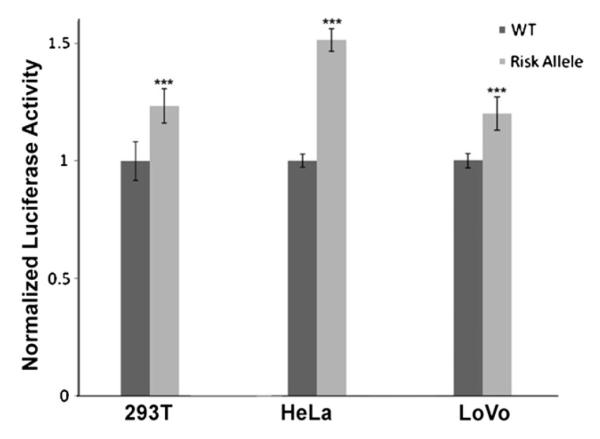
To evaluate the effect of rs67491583 on MYC-335 enhancer activity, we generated a luciferase reporter construct containing the GA deletion. Wild-type and GA deletion-containing constructs were transfected into HEK293T, HeLa, and LoVo cells with or without GFI1B. cDNA and luciferase activity was measured after 32 hours. Consistent with weaker co-activator binding by the GA-deletion allele (Supplementary Figure S2), in the absence of GFI1B co-expression, the MYC-335 reporter with the GA deletion showed weaker luciferase activity than the wild-type reporter in LoVo cells (P = 9.28 × 10−5) (Supplementary Figure S3). When GFI1B was co-expressed, the GA deletion-containing enhancer displayed significantly increased reporter activity in HEK293T (P = 9.7 × 10−6), HeLa (P = 1.8 × 10−8), and LoVo cells (P = 0.0005) (Figure 3). These results suggest that in the absence of GFI, the GA deletion is a somewhat weaker enhancer, but in the presence of GFI, the GA deletion is more active as it binds less repressor.
Figure 3.
Enhancer activity is changed when GA nucleotides were absent at rs67491583. The GA deletion allele-containing enhancer fragment (risk allele) displays a slightly but statistically significantly higher enhancer activity than wild-type allele-containing enhancer fragment (WT) in luciferase assay in GFI1B cDNA transfected HEK293T, HeLa, and LoVo CRC cells. Error bars indicate one standard deviation (n = 6).
The GA deletion was present only in samples from African descent; therefore, we tested whether it could influence CRC susceptibility in African American individuals. A total of 1,027 of 1,134 (90.6%) CRC cases and 1,683 of 1,732 (97.2%) controls were included in the association analyses after genotyping and outlier detection. Including the 90 African American HapMap samples, the total number of genotyped controls was 1,773. After combining the data from all the different sample series that could be made available for the study (Table 2), the allele frequency of the deletion was higher in CRC cases (9.9%) than in controls (8.6%); however, this difference did not reach statistical significance (P = 0.095; OR: 1.17, 95% CI: 0.97–1.41). Table 2 shows that allele frequencies varied between different sample series, suggesting heterogeneity across study groups. It was estimated that the study had 80% power to detect variants with minor allele frequencies of 10% and ORs of 1.36 at significance level of 0.05, but only 40% power to capture variants with frequencies of 10% and ORs of 1.2. GA deletion homozygotes were found to be more common among cases (12/1,027 = 1.7%) than controls (12/1,773 = 0.7%) (Table 3), suggesting that CRC risk may be further increased in GA deletion homozygotes (OR: 1.73, 95% CI: 0.76–3.95).
Table 3.
Genotype associations of rs6749158 in African American samples
| CRC | Controls | P-value | OR | 95% CI | |
|---|---|---|---|---|---|
| Hom vs. het/wt | 12/1015 | 12/1761 | 0.17 | 1.73 | 0.76–3.95 |
| Hom/het vs. wt | 192/835 | 293/1480 | 0.14 | 1.16 | 0.95–1.42 |
| Hom vs. wt | 12/835 | 12/1480 | 0.16 | 1.77 | 0.77–4.04 |
| Het vs. wt | 180/835 | 281/1480 | 0.23 | 1.14 | 0.92–1.39 |
In our previous reports, we have shown that during CRC tumor evolution, the CRC risk allele G of variant rs6983267 is selected for through increases in copy number (15,27). Similarly, allelic imbalance at the GA deletion was analyzed in the tumor DNA of 13 heterozygous cases. Copy number increase was detected in 4 of 13 tumors (31%), and 3 of 4 AI events affected the mutant allele.
Finally, we studied the relationship between the rs67491583 genotype and MYC expression. We used gene expression data determined on 89 YRI HapMap lymphoblastoid cell lines (23), as well as MYC expressions measured with real-time quantitative PCR (qPCR) in lymphoblastoid cell lines of 40 African American control individuals. We could not detect an association between the rs67491583 genotype and MYC expression either in the microarray gene expression data (P = 0.17) or in the qPCR data (P = 0.95).
Discussion
In our previous work, we used a computational tool, enhancer element locator, and identified an evolutionary conserved enhancer element MYC-335 that contains CRC predisposition SNP rs6983267 (15). rs6983267 resides at a TCF4 binding site, and the CRC risk allele G has the ability to enhance TCF4 binding and promote Wnt signaling. Here, we have sequenced through the MYC-335 enhancer in individuals with European and African descent and identified a GA deletion (rs67491583) at a putative GFI binding site. Interestingly, rs67491583 occurred only in samples from individuals of African origin.
The GA deletion rs67491583 resides 927 bp downstream from the SNP rs6983267. The functional data presented in this study show that the GA deletion has an effect on transcription factor binding; it reduces the binding of GFI proteins to the MYC-335 enhancer. GFI1 and GFI1b are zinc finger transcriptional repressors that function by binding to DNA elements containing a core AATC sequence (25). GFI1 and GFI1b are functionally very similar and both play an important role in hematopoietic development and lymphomagenesis. Gfi1b is required for the development of erythroid and megakaryocytic lineages (28). Mice lacking functional Gfi1 are neutropenic, and heritable GFI1 mutations in humans cause severe congenital neutropenia (29,30). GFI proteins can also function as proto-oncogenes. Gfi1 is a frequent target of retroviral integration in T and B cell lymphomas, leading to transcriptional activation of the Gfi1 gene. Gfi1b also plays an essential role in erythroleukemia and megakaryocytic leukemia (31). Additionally, Gfi1 has been shown to regulate differentiation of nonhematopoietic tissues, such as inner ear hair cells, lung neuroendocrine cells, and intestinal epithelial cells (32–34). In the mouse intestine, expression of Gfi1 is found in cells throughout the developing intestine (33). In mature small and large intestines, expression is detected mainly in the crypts. Gfi1 functions downstream of Math1 in early progenitors, contributing to lineage determination of intestinal epithelial progenitors. Because Gfi1 is important in intestinal epithelial cell proliferation, a role in intestinal malignancies is feasible. Variants rs6983267 and rs67491583 might contribute to colorectal neoplasia by increasing the activity of the MYC-335 enhancer and MYC expression and, in that way, disturb the intestinal homeostasis. Consistently, MYC has been described as a GFI1 and GFI1b target gene in myeloid cells (35,36).
We failed to detect an association between rs67491583 and CRC risk in African Americans. Due to low frequency of the deletion in the African American population (~10%), an even larger sample size would have been needed to obtain statistical power to detect significant association. Moreover, African Americans are genetically a highly heterogenous group and, without adjustment for population stratification association, analyses might give spurious results. Previous studies have shown conflicting results about the contribution of rs6983267 to CRC risk in African Americans (37,38).
The hypothesis that rs67491583 affects CRC predisposition is intriguing. First, both MYC-335 variants rs6983267 and rs67491583 affect transcription factor binding sites and are highly differentially distributed in different ethnic populations. Based on the HapMap data, the frequency of G allele of rs6983267 is highest in Africa (~97%), whereas moving on from Africa the frequency is reduced (~82% in African Americans, 50% in Caucasian populations, 39% in Han Chinese, and 29% in Japanese). In this study, a similar phenomenon was detected for the GA deletion at the GFI site. The allele frequency is approximately 10% in African Americans, but in individuals of European descent the GA deletion is very rare. These observations suggest that both alleles are functional and under selective pressure from environmental exposure. Second, the predicted effect of the G allele at the TCF4 binding site and the GA deletion at the GFI binding site on MYC expression is similar. The CRC risk allele G of rs6983267 enhances TCF4 binding and the GA deletion reduces repressor binding, which should lead to increased expression of a target gene. Evidence obtained from this and previous works indicate that the MYC-335–bearing G allele of rs6983267 in concert with the GA deletion at the GFI1 binding site is a stronger enhancer than the “wild-type” element. The stronger form of the enhancer—potentially leading to increased expression of MYC and increased proliferation—could provide protection in certain environmental challenges, such as tolerance to gastrointestinal infections.
Taken together, we have characterized in detail the genetic variation within the MYC-335 regulatory element and have identified a population-specific functional variant at the GFI binding site. We were unable to detect significant association between rs67491583 and CRC risk in African Americans. However, our study power was limited and more extensive studies are needed to detect the putative association. In theory, the strong predominance of the G allele in individuals with African origin together with the disrupted GFI site could, in part, explain the high CRC incidence in African Americans. Importantly, the low incidence of CRC in Africa provides hope that the effects of these two candidate risk alleles can be controlled by environmental and lifestyle factors, such as diet. Further work to investigate the contribution of MYC-335 variants in neoplasia, as well as normal homeostasis, is highly warranted.
Supplementary Material
Acknowledgments
We thank Mairi Kuris, Inga-Lill Svedberg, Iina Vuoristo, Maarit Ohranen, and Sini Miettinen for technical assistance. This work was supported by the grants from Academy of Finland (Finnish Center of Excellence Program 2006–2011); the Finnish Cancer Society; the Sigrid Juselius Foundation; and the National Institutes of Health [R01 CA66635 to R.S.S, CA67941 and CA16058 to A.d.l.C].
Footnotes
Supplementary data Supplementary data related to this article can be found online at doi:10.1016/j.cancergen.2012.01.005.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal S, Bhupinderjit A, Bhutani MS, et al. Colorectal cancer in African Americans. Am J Gastroenterol. 2005;100:515–523. doi: 10.1111/j.1572-0241.2005.41829.x. discussion 514. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 5.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genomewide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 6.Jaeger E, Webb E, Howarth K, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 8.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 12.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiemeney LA, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 16.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pittman AM, Naranjo S, Webb E, et al. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19:987–993. doi: 10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittman AM, Naranjo S, Jalava SE, et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 2010;16. 6(9):ii, e1001126. doi: 10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvajal-Carmona LG, Cazier JB, Jones AM, et al. Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet. 2011;20:2879–2888. doi: 10.1093/hmg/ddr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Launonen V, Avizienyte E, Loukola A, et al. No evidence of Peutz-Jeghers syndrome gene LKB1 involvement in left-sided colorectal carcinomas. Cancer Res. 2000;60:546–548. [PubMed] [Google Scholar]
- 21.Turunen MM, Dunlop TW, Carlberg C, et al. Selective use of multiple vitamin D response elements underlies the 1 alpha,25-dihydroxyvitamin D3-mediated negative regulation of the human CYP27B1 gene. Nucleic Acids Res. 2007;35:2734–2747. doi: 10.1093/nar/gkm179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGill R, Tukey JW, Larsen WA. Variations of box plots. The American Statistician. 1978;32:12–16. [Google Scholar]
- 23.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zweidler-Mckay PA, Grimes HL, Flubacher MM, et al. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 27.Tuupanen S, Niittymaki I, Nousiainen K, et al. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res. 2008;68:14–17. doi: 10.1158/0008-5472.CAN-07-5766. [DOI] [PubMed] [Google Scholar]
- 28.Saleque S, Cameron S, Orkin SH. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Person RE, Li FQ, Duan Z, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karsunky H, Zeng H, Schmidt T, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 31.Elmaagacli AH, Koldehoff M, Zakrzewski JL, et al. Growth factor-independent 1B gene (GFI1B) is overexpressed in erythropoietic and megakaryocytic malignancies and increases their proliferation rate. Br J Haematol. 2007;136:212–219. doi: 10.1111/j.1365-2141.2006.06407.x. [DOI] [PubMed] [Google Scholar]
- 32.Wallis D, Hamblen M, Zhou Y, et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- 33.Shroyer NF, Wallis D, Venken KJ, et al. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazanjian A, Wallis D, Au N, et al. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res. 2004;64:6874–6882. doi: 10.1158/0008-5472.CAN-04-0633. [DOI] [PubMed] [Google Scholar]
- 35.Duan Z, Horwitz M. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc Natl Acad Sci U S A. 2003;100:5932–5937. doi: 10.1073/pnas.1031694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurent B, Randrianarison-Huetz V, Kadri Z, et al. Gfi-1B promoter remains associated with active chromatin marks throughout erythroid differentiation of human primary progenitor cells. Stem Cells. 2009;27:2153–2162. doi: 10.1002/stem.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J, Wilkens LR, Stram DO, et al. Generalizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populations. Cancer Epidemiol Biomarkers Prev. 2011;20:70–81. doi: 10.1158/1055-9965.EPI-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kupfer SS, Anderson JR, Hooker S, et al. Genetic heterogeneity in colorectal cancer associations between African and European Americans. Gastroenterology. 139:1677–1685. 1685.e1–8. doi: 10.1053/j.gastro.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.