Abstract
p73, a member of the p53 tumor suppressor family, is involved in neurogenesis, sensory pathways, immunity, inflammation, and tumorigenesis. How p73 is able to participate in such a broad spectrum of different biological processes is still largely unknown. Here, we report a novel role of p73 in regulating lipid metabolism by direct transactivation of the promoter of autophagy-related protein 5 (ATG5), a gene whose product is required for autophagosome formation. Following nutrient deprivation, the livers of p73-deficient mice demonstrate a massive accumulation of lipid droplets, together with a low level of autophagy, suggesting that triglyceride hydrolysis into fatty acids is blocked owing to deficient autophagy (macrolipophagy). Compared with wild-type mice, mice functionally deficient in all the p73 isoforms exhibit decreased ATG5 expression and lower levels of autophagy in multiple organs. We further show that the TAp73α is the critical p73 isoform responsible for inducing ATG5 expression in a p53-independent manner and demonstrate that ATG5 gene transfer can correct autophagy and macrolipophagy defects in p73-deficient hepatocytes. These data strongly suggest that the p73–ATG5 axis represents a novel, key pathway for regulating lipid metabolism through autophagy. The identification of p73 as a major regulator of autophagy suggests that it may have an important role in preventing or delaying disease and aging by maintaining a homeostatic control.
Keywords: ATG5, autophagy, lipid droplets, lipophagy, liver, metabolism, p73, starvation, transcription
p73 belongs to the p53 family, a group of transcription factors that have key roles in the regulation of many cellular processes, such as apoptosis, cell cycle, and senescence, especially following DNA damage.1, 2, 3 Whereas p53 is a tumor suppressor and often either deleted or mutated in tumors, p73 is rarely mutated; however, its expression is often deregulated in cancer.4, 5 In p73-deficient mice, unlike in p53-deficient mice, no increase in spontaneous tumorigenesis is observed.6 It has been demonstrated, however, that p73, in the absence of DNA damage, has a role in neuronal differentiation and development.6, 7, 8, 9 Consequently, p73-deficient mice exhibit neurological defects and, even though no obvious deficiencies in lymphoid or granulocyte populations have been so far detected, such mice show deregulated inflammatory responses and senescence.6, 10
Because of splicing events occurring near the 3′ end of the coding region and because of an alternative promoter located in the third intron of the gene, p73 exists as multiple protein variants.11 The isoforms containing a transactivation (TA) domain (TAp73s) generally behave like p53 with respect to overlapping promoters and biological functions.2, 4 For instance, in contrast to p73-deficient mice, TAp73 knockout mice showed spontaneous as well as carcinogen-induced tumors, indicating that TAp73 is a tumor suppressor.12 The alternative promoter also produces amino-terminal truncated ΔN isoforms, the so-called ΔNp73s, which lack the TA domain and are, therefore, transcriptionally inactive. However, the ΔNp73s can act as negative regulators of p53 and TAp73.2, 4
p53 has recently been linked to another important cellular process, that of autophagy, which controls intracellular homeostasis.13, 14, 15 Various forms of cellular stress induce autophagy. In this process, long-lived proteins and whole organelles are sequestered in double-membraned vesicles, called autophagosomes, and digested.16, 17, 18 Autophagy is a catabolic process through which cells digest their own components to provide energy and building blocks to maintain normal cellular function under unfavorable conditions, but it is also required, although at lower levels, under physiological conditions.16, 17, 19 Interestingly, p53 appears to exhibit a dual role with respect to autophagy: whereas nuclear p53 induces autophagy by activation of the damage-regulated autophagy modulator (DRAM) gene, cytosolic p53 appears to inhibit autophagy by activation of the mTOR pathway.20, 21 It remains unclear in which cellular contexts p53 behaves as an inducer and in which cellular contexts it behaves as an inhibitor of autophagy. In this report, we characterize the involvement of p73 in autophagy.
Results
p73 deficiency results in an accumulation of lipid droplets in liver cells
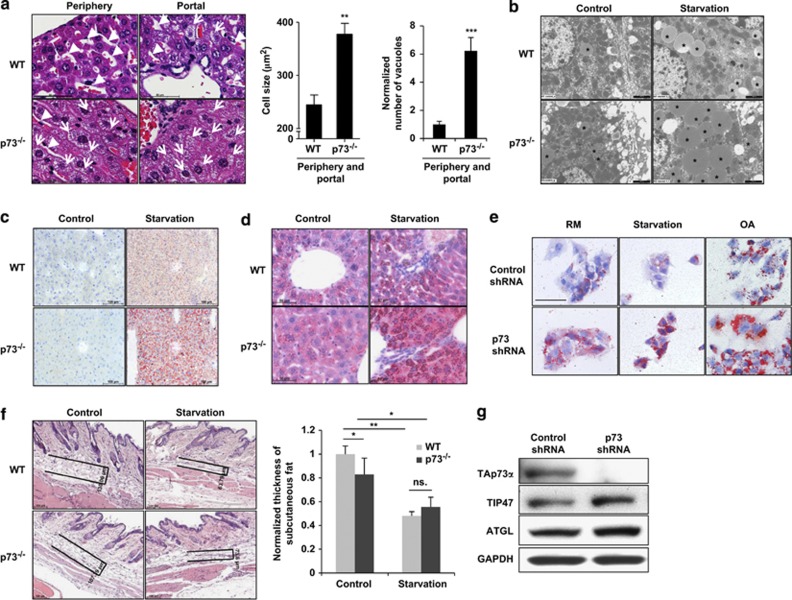
p73-deficient mice display phenotypes associated with dysregulated homeostasis.6, 12 To further characterize possible defects in homeostasis regulation in these mice, we investigated the consequences of starvation and observed massive vacuolization in almost all the regions of the livers of p73-deficient mice, a phenomenon that was seen only in the portal region in wild-type (WT) mice, and then too, to a much lower extent. The vacuoles had a regular shape and largely filled the cytoplasm of affected hepatocytes on account of their huge number and large sizes (Figure 1a and Supplementary Figure S1, white arrows). Interestingly, the vacuolization of hepatocytes upon starvation was more pronounced in aged mice compared with younger mice (Supplementary Figure S2). Moreover, the hepatocytes of p73-deficient mice were hypertrophic compared with those of WT mice, and, consequently, sinusoids were much smaller (Figure 1a and Supplementary Figure S1, white arrow heads). Transmission electron microscopy revealed that the vacuoles lacked any membrane, being homogenous structures with a regular round shape (Figure 1b and Supplementary Figure S3). Such structures have been described previously as lipid droplets (LDs).22 To confirm this assumption, we performed Sudan IV staining, a method for detecting lipids in tissue sections,23 and observed that the vacuoles present in liver cells of p73-deficient mice were Sudan IV-positive (Figure 1c). The accumulation of LDs in p73-deficient mice following starvation was also confirmed by an analysis of the levels of the tail-interacting protein of 47 kDa (TIP47), an LD-associated protein.22 After starvation, TIP47 was concentrated in the vacuoles of hepatocytes, and its presence was much more obvious in the livers of p73-deficient mice, compared with WT mice (Figure 1d).
Figure 1.
p73 deficiency leads to accumulation of LDs in hepatocytes after starvation both in vivo and in vitro. (a) Histology. Liver tissues after starvation were stained using H&E. p73-deficient mice show pathological vacuoles in virtually all hepatocytes (portal region and periphery; white arrows) after starvation. In WT mice, a few vacuoles were detected in the region near to the portal vein only. In addition, the hepatocytes of p73-deficient mice were larger and sinusoids (white arrow heads) were significantly smaller compared with those of WT mice. (Right) The size of the hepatocytes and the numbers of the vacuoles were analyzed in more than 100 cells and presented as the means±S.D.s of three different mice in each group. **P<0.01; ***P<0.001. Bar, 50 μm. Original magnification, × 630. Enlarged images are shown in Supplementary Figure S1. Vacuolization was even more pronounced in aged mice (Supplementary Figure S2). (b) Transmission electron microscopy. Liver sections from WT and p73-deficient mice with and without starvation (24 h) are shown. Vacuoles are homogeneous, lack any membrane, and have a round shape, suggesting that they represent LDs. LDs were indicated with black stars. Bar, 10 μm. Enlarged images are shown in Supplementary Figure S3. (c) Histology. Liver sections from WT and p73-deficient mice in the presence and absence of starvation (24 h) were stained using Sudan IV to detect lipids. Liver cells of p73-deficient mice were strongly Sudan IV-positive after starvation. Bars, 100 μm. (d) Immunohistochemistry. TIP47, a protein associated with LDs, was much more strongly expressed in hepatocytes of p73-deficient mice compared with WT mice after starvation (24 h). Bar, 50 μm. (e) Oil red O staining of HepG2 cells. Cells were infected with lentiviral control shRNA or p73 shRNA. HepG2 cells with normal and reduced p73 levels (g) were kept in a RM, starved or exposed to OA for 24 h before staining. p73-deficient HepG2 cells accumulate more LDs than do control cells under all conditions. Bar, 50 μm. Original magnification, × 630. (f) Histology. Skin sections from the indicated mice were stained using H&E. (Right) The thickness of subcutaneous fat was measured using the AxioVision software and normalized to that of WT mice (n=3 in each group). Bar, 100 μm. (g) Immunoblotting. HepG2 with p73 deficiency exhibit slightly increased levels of TIP47 and ATGL. Results are representative of three independent experiments
Using liver HepG2 cells as a model, we were able to mimic the observations made in p73-deficient mice. The knockdown of p73 using an small hairpin RNA (shRNA)-based approach resulted in increased lipid staining with oil red O, a fat-soluble dye,24 indicative of an accumulation of LDs under cell culture conditions (regular medium; RM), starvation, or following a supply of exogenous oleic acid (OA) (Figure 1e). The same observations were obtained when lipid staining was performed using BODIPY, a lipid-recognizing dye (Supplementary Figure S4). Taken together, we conclude that p73 deficiency leads to the accumulation of LDs both in vitro and in vivo.
During starvation, fat is mobilized from the adipose tissue to the liver in the form of glycerol and free fatty acids, which are then re-esterified and stored in LDs in hepatocytes. The LDs subsequently undergo lipolysis for energy production. As the rate of hepatocellular lipogenesis declines during starvation for up to 24 h,25 the accumulation of LDs in hepatocytes is probably caused by other mechanisms, for example, by excessive lipolysis in the adipose tissue or blockage of lipolysis in the liver. We therefore compared the sizes of subcutaneous fat in the WT and p73-deficient mice. Although the adipose depots were slightly decreased in p73-deficient mice compared with WT mice under conditions of normal food intake, a strong p73-independent reduction was seen upon starvation (Figure 1f). This indicates that there was no excessive lipolysis in the adipose tissue of p73-deficient mice that could explain the massive accumulation of hepatocellular LDs. As cytosolic lipases, such as the adipose triglyceride lipase (ATGL), are mainly responsible for lipolysis in the adipose tissue,26 we also tested ATGL expression, which was even slightly increased in association with p73 deficiency both in vitro (Figure 1g) and in vivo (Supplementary Figure S5). These data suggested that other lipolysis pathway(s) might be impaired in p73-deficient mice.
p73 regulates autophagy independently of p53
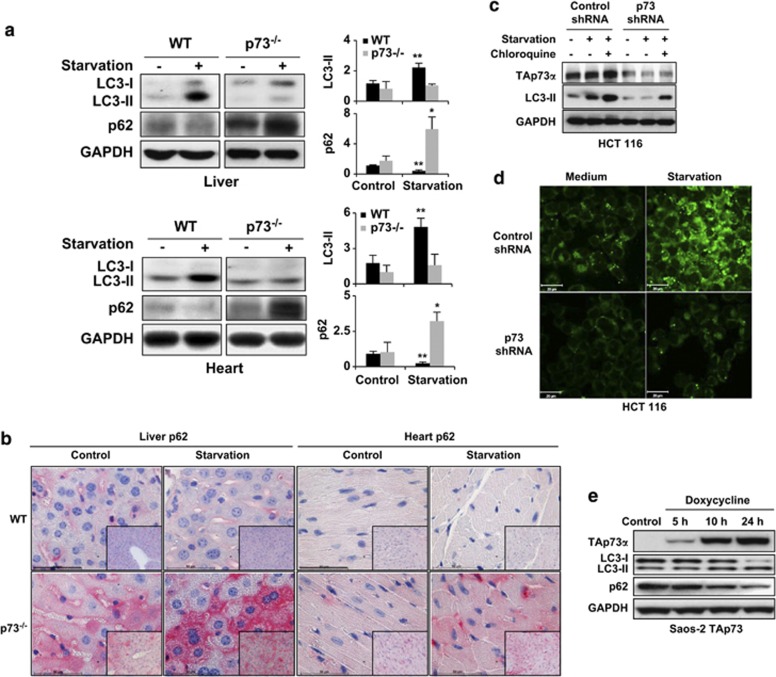
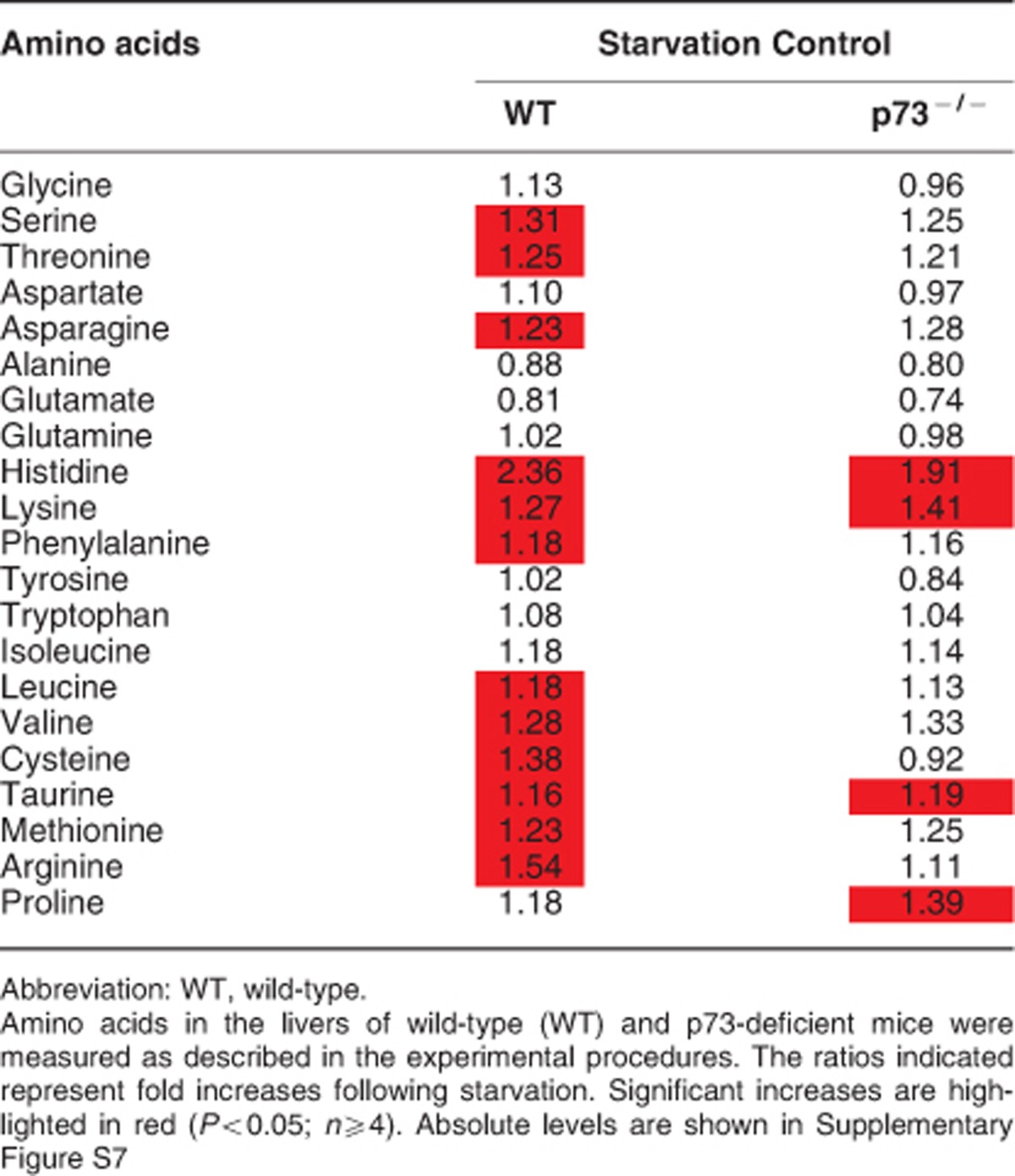
As autophagy has recently been linked to lipolysis,22 we investigated the status of autophagy in vivo by testing the responses in the liver and the heart following starvation in WT and p73-deficient mice. As assessed using immunoblotting, we observed in the liver and heart cells of starved WT mice higher levels of lipidated LC3-II, a protein that associates with newly generated autophagosome membranes and is, therefore, a marker for monitoring autophagy.27 On the other hand, LC3-II levels were essentially unchanged in the liver and heart of starved p73-deficient mice, suggesting that these mice might be deficient in the induction of autophagy (Figure 2a). To verify these findings, we analyzed p62 levels using both immunoblotting and immunohistology. A protein that is degraded upon the induction of autophagy, p62,28 was found in higher levels in the liver and heart both under control and starvation conditions in p73-deficient mice compared with WT mice (Figures 2a and b). Furthermore, we readily detected autophagosomes and autolysosomes in the livers of starved WT mice, but rarely in starved p73-deficient mice (Supplementary Figure S6). In addition, amino-acid levels in the livers of p73-deficient mice were much less elevated following starvation than with WT mice (Table 1, Supplementary Figure S7). These data suggest that p73 deficiency is associated with both lower basal autophagy and defective starvation-induced autophagy.
Figure 2.
p73 regulates autophagy both in vivo and in vitro. (a) Immunoblotting. Livers and hearts were taken from WT and p73-deficient mice under normal feeding conditions and after food deprivation for 24 h (starvation). p73-deficient mice are unable to increase LC3-II levels and accumulate p62 upon starvation. Results are representative of three independent experiments. (Right) The fold changes in expression levels, compared with WT control, are presented. *P<0.05; **P<0.01, n=3. (b) Immunohistochemistry. The same tissues as in panel a were stained with anti-p62 antibody. The p73-deficient mice show slightly increased p62 levels compared with WT mice under normal conditions and a strong p62 accumulation after starvation. Bar, 50 μm. Original magnifications, × 630 and × 200 (lower right corners). Results are representative of three independent experiments. (c) Immunoblotting. HCT 116 cells were infected with lentiviral control shRNA or p73 shRNA. 48 h after infection, cells were starved in an EBSS medium for 1 h. Control (unspecific) shRNA-treated cells had higher LC3-II levels than did p73 shRNA-treated cells. Moreover, whereas the control shRNA-treated cells increased their LC3-II levels upon starvation, p73 shRNA-treated cells exhibited no response. LC3-I was not detectable in this experiment. (d) Autodot staining. Significantly increased staining after starvation was seen in control but not in p73-deficient HCT 116 cells. Bar, 20 μm. Similar results were obtained in HaCaT cells, including using other methods for inducing autophagy (see Supplementary Figure S8). (e) Immunoblotting. Saos-2 cells inducible for TAp73α were treated with doxycycline (2.5 μg/ml) for the indicated time periods. Increased levels of TAp73α resulted in increased LC3-II and reduced p62 levels. Other p73 and p63 isoforms, as well as p53, were without effect in this system (see Supplementary Figure S9)
Table 1. Relative levels of amino acids.
To demonstrate that defects of autophagy in p73-deficient cells occur independently of p53, we used p53-deficient HCT 116 cells to knockdown p73. Cells with reduced levels of p73 had low levels of basal autophagic activity, as reflected by the reduced LC3 lipidation (Figure 2c). Moreover, upon starvation, these cells were unable to induce autophagy, whereas increased levels of LC3-II were seen in unspecific, control shRNA-transduced HCT 116 cells (Figure 2c). In addition, starvation, in combination with chloroquine, which inhibits autophagy by altering lysosomal pH and thereby blocking autophagic flux,29 further increased LC3-lI levels compared with starvation alone (Figure 2c). A lack of autophagy induction upon starvation in p73-deficient HCT 116 cells was also demonstrated by monitoring autophagosome formation using Autodot fluorescence (Figure 2d).30 The same results were obtained in p53-mutated HaCaT cells; following starvation or rapamycin treatment, in contrast to control shRNA-treated cells, p73-deficient HaCaT cells were virtually unable to generate autophagosomes (Supplementary Figure S8).
We then used an inducible tet-on system in Saos-2 cells, in which endogenous p53 is absent,31 to induce various p73, p63, and p53 isoforms by adding doxycycline. As assessed using immunoblotting, the induction of TAp73α was associated with higher levels of LC3-II and lower levels of p62, suggesting that TAp73α mediates the induction of autophagy (Figure 2e). In contrast, TAp73β, TAp73γ, and ΔNp73α had no detectable effect on LC3 lipidation or p62 degradation (Supplementary Figure S9a). In addition, TAp63α, but not ΔNp63α, also appeared to induce autophagy, as indicated by the higher levels of LC3-II and the lower levels of p62 (Supplementary Figure S9b). In contrast, slightly reduced levels of LC3-II and an accumulation of p62 were observed after the induction of p53, suggesting that p53, in contrast to p73 and p63, rather acts to counter the onset of autophagy in this system (Supplementary Figure S9c). Taken together, we conclude that TAp73α regulates basal autophagic activity in a p53-independent manner and is required for the induction of autophagy in response to starvation and, perhaps, other autophagy inducers. Therefore, the impairment of lipid metabolism in p73-deficient mice is most likely due to defects in autophagy.
TAp73α regulates ATG5 gene expression
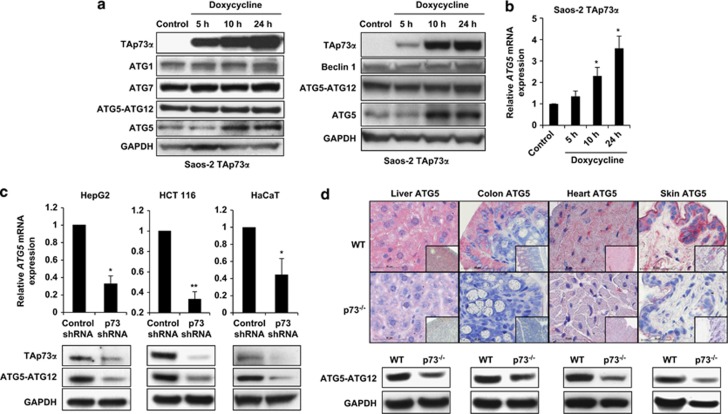
The identification of TAp73α as the critical p73 isoform responsible for regulating autophagy suggested that p73 activates the transcription of one or more genes that are essential in the autophagic pathway. We, therefore, examined the expression levels of several autophagy-related proteins (ATGs) after the induction of TAp73α in Saos-2 cells. Whereas ATG1, ATG7, and Beclin 1 (ATG6) protein expression were unchanged, ATG5 expression was increased both at the mRNA and the protein levels upon the induction of TAp73α (Figures 3a and b). In addition, TAp73β and TAp63α induction was followed by a marginal increase in ATG5 expression. Other p53 family members, including p53, TAp73γ, ΔNp73α, and ΔNp63α, failed to induce ATG5 expression (Supplementary Figure S10).
Figure 3.
p73 regulates ATG5 expression. (a) Immunoblotting. Saos-2 cells inducible for TAp73α were treated with doxycycline (2.5 μg/ml) for the indicated time periods. Expression levels of ATG1, ATG7, Beclin 1, monomeric ATG5, and the conjugated form of ATG5 (ATG5-ATG12) were investigated in two independent experiments. Increased levels of TAp73α resulted in increased monomeric ATG5 levels. TAp73β induction was associated with a slight increases of monomeric ATG5, whereas other members of the p53 family had no effect (see Supplementary Figure S10). (b) Quantitative real-time PCR. Saos-2 cells inducible for TAp73α were treated with doxycycline for the indicated time periods and ATG5 mRNA was measured. Values are means±S.D.s of three independent experiments. *P<0.05. (c) Quantitative real-time PCR (upper panel) and immunoblotting (lower panel). The indicated cells were treated with control shRNA and p73 shRNA. Values in the upper panel are means±S.D.s of three independent experiments. Reduced p73 expression was associated with reduced ATG5 expression at both mRNA and protein levels. p73 shRNA experiments were additionally controlled by real-time PCR (data not shown). (d) Immunohistochemistry (upper panel) and immunoblotting (lower panel). p73-deficient mice show significantly reduced ATG5 expression compared with WT mice. Bar, 50 μm. Original magnifications, × 630 and × 200 (lower right corners). Results are representative of three independent experiments
As these data imply that TAp73 is a transcriptional activator of ATG5, we performed a knockdown of p73 in HepG2, HCT 116, and HaCaT cells to test the influence of such genetic modifications on ATG5 levels. ATG5 expression was significantly decreased in all three cellular systems, both at the mRNA and the protein levels, after the knockdown of p73 (Figure 3c). It should be mentioned that monomeric ATG5 protein was hardly detectable in these cells (data not shown).
To test whether p73 may also regulate ATG5 expression in vivo, we examined tissue sections of several organs of p73-deficient mice. As assessed using immunohistochemistry, we observed reduced levels of ATG5 in the liver, colon, heart, and skin of p73-deficient mice compared with WT mice. In the colon and skin of WT mice, ATG5 was expressed particularly by epithelial cells (Figure 3d, upper panels). Reduced ATG5 expression in p73-deficient organs was also confirmed using immunoblotting (Figure 3d, lower panels). Taken together, these data, both in vivo and in vitro, suggest that ATG5 represents a likely transcriptional target of TAp73, particularly of the α isoform.
TAp73α and TAp73β bind to and transactivate the ATG5 gene promoter
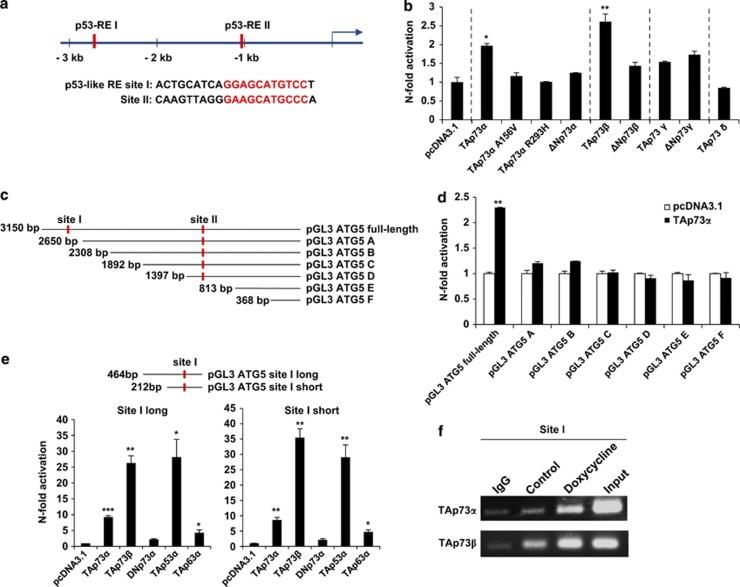
To directly test whether p73 binds to and activates the promoter of ATG5, we first analyzed the ATG5 promoter sequence using the MatInspector promoter analysis tool. As the binding sequence specific to p73 has not been identified, and because p73 and p53 often share the same DNA-binding sequence,2 we asked whether p53-specific binding sites exist in the ATG5 promoter region. We discovered two p53 response elements (p53-RE) (Figure 4a). We subsequently cloned the ATG5 promoter sequence in the firefly luciferase vector, pGL3, and co-transfected H1299 cells with this construct, together with the pRL-TK Renilla luciferase vector as an internal control and pcDNA3.1 constructs expressing different p53 family members or empty vectors. Lipofectamine transfection was employed, after which dual-luciferase assays were performed. TAp73α and TAp73β activated the ATG5 promoter, whereas TAp73γ and other p73 isoforms, which lack the N-terminal TA domain, had no effect (Figure 4b). Two mutant forms of TAp73α (TAp73αA156V and TAp73αR293H) that do not bind to p53-RE32 served as negative controls in these experiments. As we had identified two p53-REs in the ATG5 promoter, we tested whether one of them alone would be sufficient for TAp73α activation of the ATG5 gene. To answer this question, the full-length ATG5 promoter was truncated and cloned in the pGL3 plasmid (pGL3 ATG5 A-F) to perform dual-luciferase assays (Figure 4c). However, only the full-length ATG5 promoter, and not the truncated promoter lacking the binding site I, was activated by TAp73α (Figure 4d). Thus, site I, but not site II, is required for TAp73α activation.
Figure 4.
p73 transactivates the ATG5 gene promoter. (a) Two p53 response elements (red bars) were found in the ATG5 promoter. The core sequence of p53 response elements are indicated in red below. The blue arrow indicates the transcriptional start site. (b) The effect of different isoforms or mutant forms of p73 on the ATG5 promoter was assessed using a dual-luciferase assay in H1299 cells. Values are means±S.D.s of three independent experiments. (c) and (d) Full-length and truncated forms of the ATG5 promoter were generated (c) and dual-luciferase assays were performed in H1299 cells (d). Values are means±S.D.s of three independent experiments. (e) Dual-luciferase assays of p73 isoforms were performed in H1299 cell using a plasmid containing only the p53-RE I (site I). Values are means±S.D.s of three independent experiments. (f) ChIP assay. Saos-2 cells inducible for TAp73α and TAp73β were treated with doxycycline for 24 h. The eluted DNA was assessed by PCR using primers specific to the site I of the ATG5 promoter sequence
We cloned, therefore, a truncated promoter containing only site I in the pGL3 plasmid for dual-luciferase assays. With two different constructs of site I, differing just in length, we detected strong increases in luciferase activity for both TAp73α and TAp73β. Interestingly, the luciferase activity was ∼5- and 10-fold higher, respectively, compared with the full-length ATG5 promoter, pointing to the possibility that other regions of the ATG5 promoter may limit its activity (Figure 4e). Finally, we performed chromatin immunoprecipitation (ChIP) assays in which, after induction of TAp73α or TAp73β, we detected enrichment in the DNA fragments to which p73 bind. Among these fragments, for both TAp73α and TAp73β, were the ATG5 promoter sequences with site I (Figure 4f).
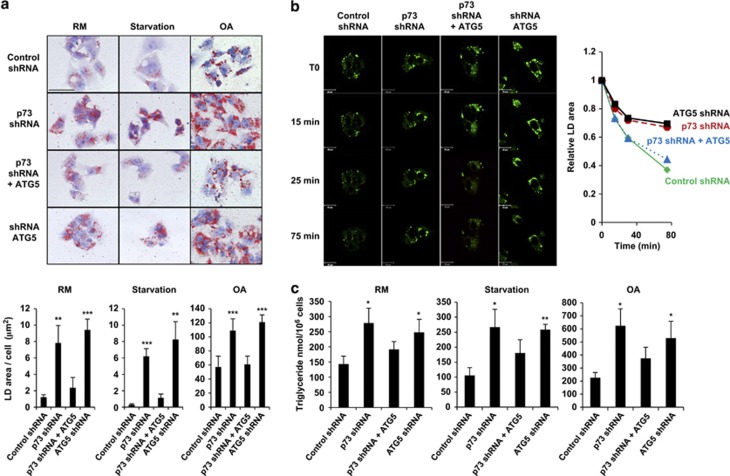
Impaired degradation of LDs in p73-deficient liver cells can be corrected by ATG5 gene transfer
To investigate whether the defect in hepatocellular lipid metabolism in p73-deficient mice was the consequence of deficient ATG5 expression, liver HepG2 cells were stained using Lysotracker in combination with BODIPY to monitor the possible degradation of LDs via the lysosomal machinery. When these cells were either starved or fed with OA to stimulate lipid accumulation, we observed LDs being engulfed by lysosomes, whereas few LDs were seen under control conditions (Supplementary Figure S11). Thus, HepG2 cells represent a suitable in vitro model to investigate the role of p73 and ATG5 in hepatocellular lipid metabolism and autophagy.
Following the modification of p73 or ATG5 expression (Supplementary Figure S12), we stained HepG2 cells with oil red O to monitor the degradation of LDs under normal conditions or upon starvation or following OA supply. The accumulation of LDs was observed not only in p73- but also in ATG5-deficient cells (Figure 5a). Moreover, enforced expression of ATG5 in p73-deficient cells restored LD degradation and presented a pattern similar to those of control cells (Figure 5a). We also analyzed in a time-dependent manner the degradation of LDs upon starvation. Control HepG2 cells degraded LDs within 30 min, whereas cells with decreased p73 or ATG5 expression were unable to do so even at later time points (Figure 5b). Again, enforced expression of ATG5 was able to restore LD degradation in p73-deficient cells (Figure 5b). Moreover, an accumulation of LDs in p73- and ATG5-deficient HepG2 cells was associated with a significantly increased triglyceride (TG) content under normal conditions or upon starvation or following OA supply. ATG5 gene transfer into p73-deficient cells could reverse this effect (Figure 5c). In sum, these data strongly suggest that the defect in lipid metabolism observed in p73-deficient mice is owing to a defect in Atg5 gene expression, and, consequently in autophagy.
Figure 5.
Downregulation of p73 results in the accumulation of LDs, which disappear following the ATG5 gene transfer. (a) Oil red O staining. HepG2 cells with modulated p73 and/or ATG5 levels (see immunoblots in Supplementary Figure S12) were kept in a RM, starved or exposed to OA for 24 h, and stained with oil red O to visualize LDs. LDs were further quantified using the Image Pro Plus software and data are presented below. p73- and ATG5-deficient cells were virtually unable to degrade LDs, whereas an ATG5 gene transfer corrected the defect in p73-deficient cells. Bar, 50 μm. Original magnification, x630. Values are means±S.D.s of three independent experiments **P<0.01; ***P<0.001. (b) BODIPY staining. The degradation of LDs in HepG2 cells was followed within 75 min upon starvation and quantified using the Imaris software and is presented on the right. ATG5 gene transfer helps to degrade LDs in p73-deficient cells with similar kinetics as is seen in control cells. Bar, 20 μm. (c) TG measurements. HepG2 cells with reduced p73 and/or ATG5 levels were kept in a RM, and starved or exposed to OA for 24 h before analysis. Cells with reduced p73 or ATG5 levels exhibited significantly increased TG contents. The increased TG content in p73-deficient cells was reversible following the ATG5 gene transfer. Values are means±S.D.s of three independent experiments *P<0.05; **P<0.01
Discussion
Our results show that p73 regulates the transcription of ATG5, a gene essential for autophagosome formation.33 Thus, p73-deficient mice demonstrate reduced levels of Atg5, resulting in lower basal levels of autophagy and the inability to induce autophagy upon starvation. The deficient autophagy has dramatic consequences for the lipid metabolism in the liver, which seems to be unable to metabolize LDs for energy. Differently from the lipolysis in the adipose tissue, where cytosolic lipolysis mainly takes place,34 other organs, such as the liver, use additional mechanisms, such as autophagy-related lysosomal lipolysis, to digest fat.22 This process seems to be particularly important during starvation, as the defect in autophagy-related lysosomal lipolysis due to p73 deficiency was morphologically reflected by the massive accumulation of LDs in hepatocytes. Moreover, the finding that p73-deficient hepatocytes were also unable to prevent lipid accumulation following an exogenous supply of OA suggests that p73 has a general role in hepatocellular lipid metabolism. A disturbance in lipid metabolism is a feature of multiple human diseases, such as metabolic syndrome, fatty liver disease, and hepatocellular carcinoma. Although p73-based syndromes have not yet been identified, it is possible that the p73–ATG5 pathway is either absolutely or relatively insufficient in these diseases, contributing to their pathogenesis.
The transcription-dependent regulation of autophagy has been described.35, 36, 37 The p73–ATG5 axis reported here complements these studies and provides a significant step forward in understanding the regulation of autophagy. Although previous reports have already suggested a potential link between p73 and autophagy,38, 39 we provide here, for the first time, a clear molecular link between p73 and autophagy using a significant series of analyses in vitro and in vivo. As a consequence, it might be possible to explain the diverse phenotypes of p73-deficient mice. For instance, the severe neuronal defects of p73-deficient mice might be the consequence of defective autophagy, which is known to have a role during development.40 Moreover, p73-deficient mice suffer from chronic infections and inflammatory diseases, which may also be, at least partially, the consequence of insufficient autophagy. In Crohn's disease, for example, defects of the epithelial barrier function, due to abnormalities in autophagy associated with secretory dysfunction of Paneth cells, have been described.41
The definition of cell death pathways has been pivotal in medical research.42, 43, 44 The regulatory link between p73 and ATG5 implies that ultimatively p73 is involved in multiple additional functions previously reported to be autophagy-dependent. For instance, p73 may subsume crucial roles in innate immunity, as autophagy has a key role in digesting intracellular pathogens.45 Moreover, autophagy is essential for adaptive immunity on account of its role in antigen presentation.46 In addition, as ATG5 has been shown to be essential for positive selection in the thymus,47 p73 might be central to the prevention of autoimmunity. Finally, autophagy seems to regulate aging,48, 49 and p73 could represent a master regulator in this process. Our observation that the accumulation of LDs after starvation increases with age supports such an assumption. Future studies will be required to test these hypotheses and to formally establish the key role of p73 in the maintenance of homeostasis in organisms following environmental stress.
Materials and Methods
Mice
The p73-deficient mice were generated as previously described.6 Mice were bred and subjected to procedures listed under the project license issued by the United Kingdom Home Office. Experiments were performed on 8–12 week-old mice. In selected experiments, we used 20-week-old mice. Starvation was induced by food deprivation over a period of 24 h, but with free access to drinking water.
Cells and cell cultures
H1299 cells (ATTC, Rockville, MD, USA) were cultured in RPMI 1640+GlutaMAX-I, Saos-2 cells31 in Dulbecco's modified Eagle's medium (DMEM)-F12 (1 : 1), HCT 116 cells (DSMZ, Braunschweig, Germany) in McCoy's 5a, and HaCaT (CLS, Eppelheim, Germany) in DMEM (Invitrogen, LuBioScience GmbH, Lucerne, Switzerland). All media were supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen).
Lentiviral gene transfer
Lentiviral plasmids of p73 and ATG5 shRNAs were purchased from Sigma-Aldrich (Buchs, Switzerland). The full-length ATG5 coding sequence was subcloned into the HIV-derived vector pWPT, which contains a cytomegalovirus promoter.50 T293 cells were transfected with the plasmid of interest together with the envelope vector pMD2G and the packaging vector psPAX2 (kindly provided by Dr. D Trono, EPFL, Lausanne, Switzerland) using the calcium phosphate transfection method for lentivirus production.50 Lentivirus was collected after 24 h after transfection, centrifuged briefly, and filtered through a 0.22-μM cellulose nitrate filter (Millipore AG, Volketswil, Switzerland).
Immunoblotting
Cells were collected, washed using cold PBS and lysed using a lysis buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X 100, 2 mM EDTA, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, and 200 μM sodium orthovanadate with a freshly prepared protease inhibitor cocktail and PMSF (both Sigma-Aldrich). Protein concentrations were measured using the BCA Protein Assay kit (Thermo Fisher Scientific, Perbio Science Switzerland SA, Lausanne, Switzerland). Thirty micrograms of protein were loaded on NuPage gels (Invitrogen) and transferred onto PVDF membranes (Immobilion-P; Millipore, Zug, Switzerland). The membranes were blocked using the Tris buffered saline (TBS) buffer with 0.1% Tween 20 (TBST) containing 5% non-fat dry milk and incubated with primary antibodies at 4 °C overnight. The primary antibodies were: monoclonal mouse anti-ATG5 (1 : 1000; Nanotools, Teningen, Germany), monoclonal mouse anti-LC3 (1 : 1000; Nanotools), polyclonal rabbit anti-Beclin 1 (1 : 1000; Abgent, Oxfordshire, UK), monoclonal mouse anti-p62 (1 : 1000; Santa Cruz, Labforce AG, Nunningen, Switzerland), rabbit anti-human ATG1 (1 : 1000; Sigma-Aldrich), rabbit anti-human ATG7 (1 : 1000; Medical and Biological Laboratories, LabForce AG), rabbit anti-TIP47 (1 : 100; Santa Cruz, Labforce AG), rabbit anti-ATGL (1 : 100, Santa Cruz, Labforce AG), rabbit anti-human p73-FL (1 : 1000, ref. Sayan et al., 2005), mouse monoclonal anti-human p53 (1 : 1000; Santa Cruz, Labforce AG), anti-HA (1 : 1000; Santa Cruz, Labforce AG), and monoclonal mouse anti-GAPDH (1 : 10 000; Chemicon International, Zug, Switzerland). Membranes were washed using TBST and then incubated with the corresponding HRP-conjugated secondary antibody (Amersham, Glattbrugg, Switzerland) and visualized using an ECL-based approach (ECL-Kit, Amersham). Intensity of the protein bands were quantified using a Licor/odyssey scanner and the ImageJ software. The intensity of each band of interest was normalized to the loading control GAPDH and subsequently compared with the control conditions.
Quantitative real-time PCR
RNA was isolated using the SV Total RNA Isolation System (Promega, Duebendorf, Switzerland) according to the technical manual provided. RNA was reverse transcribed to cDNA, and real-time PCR was performed using the iQ SYBR Green Supermix (Bio-Rad, Reinach BL, Switzerland) with a real-time PCR machine (iQ5 Multicolor Real-Time PCR Detection System, Bio-Rad). The primers used were human ATG5: 5′-CAC AAG CAA CTC TGG ATG GGA TTG-3′ and 5′-GCA GCC ACA GGA CGA AAC AG-3′ human p73: 5′-GGC TGC GAC GGC TGC AGA GC-3′ and 5′-GCT CAG CAG ATT GAA CTG GGC CAT G-3′, human 18S: 5′-ATC CCT GAA AAG TTC CAG CA-3′ and 5′-CCC TCT TGG TGA GGT CAA TG-3′ (Microsynth, Balgach, Switzerland).
Dual-luciferase assay
H1299 cells were co-transfected with the pGL3 firefly luciferase vector containing the ATG5 promoter sequence, pRL-TK Renilla luciferase vector, and a pcDNA3 construct expressing the indicated p53 family members32, 51 or empty vector using the Lipofectamine method (Lipofectamine 2000, LuBioScience GmbH). Twenty-four hours after transfection, cells were lysed in the luciferase lysis buffer according to the manufacturer's instructions (Dual-Luciferase Reporter Assay System, Promega) and luminescence was measured using a Chameleon reader (HIDEX, Turku, Finland). Luciferase activity was normalized to that of Renilla luciferase activity.
ChIP assay
ChIP was performed using a ‘ChIP-IT Express Enzymatic' kit (Active Motif, Rixensart, Belgium). To preserve the intact protein/DNA interactions, the intact cells were first fixed with formaldehyde for cross-linking. DNA was then sheared into small fragments with the ready-to-use enzymatic cocktail. The protein/DNA complexes were immunoprecipitated, and the contaminating proteins were then removed by treatment with proteinase K. The DNA fragments were collected and then analyzed using 39 cycles of PCR to amplify ATG5 promoter sequences. Primers used for PCR were 5′-TGA AGG GAT GGG CCG AAT TAA AGG-3′ and 5′-GAA GGT TGT GGG CTT ACC GGA AT-3′.
Histology
Mouse tissue sections were deparaffinized and rehydrated for H&E staining according to the protocol provided (Sigma-Aldrich). Vacuoles were counted in at least 100 cells of liver sections from three mice. The cell size was measured using the Image Pro Plus software (Media Cybernetics, Rockville, MD, USA) for at least 100 hepatocytes from three mice. Frozen sections of livers from WT and p73-deficient mice were also assessed after Sudan IV staining to visualize lipids as previously described.52
Immunohistochemistry
Paraffin-embedded tissue sections were first deparaffinized and rehydrated. Heat-induced antigen retrieval was performed in a sodium citrate buffer (10 mM, pH 6.0). Staining was performed by incubating the sections with monoclonal anti-ATG5 antibody (1 : 100; Nanotools), monoclonal anti-p62 antibody (1 : 100; Santa Cruz), or polyclonal anti-TIP47 antibody (1 : 100; Santa Cruz) in a blocking buffer. Proteins were visualized using the Dako REAL Detection System, Alkaline Phosphatase/RED kit (Dako, Glostrup, Denmark), showing a red color in the tissue sections. Micrographs were taken with the help of an Axiovert 35 microscope (Carl Zeiss, Jena, Germany) using the AxioVision Rel. 1.8 software (Carl Zeiss).
Immunofluorescence
Paraffin-embedded tissue sections were first deparaffinized and rehydrated, and antigen retrieval performed as described under immunohistochemistry. Staining was performed by incubating the sections with rabbit anti-p73 antibody (1 : 100, Lifespan BioSciences, Seattle, WA, USA), rabbit anti-p53 antibody (1 : 100, Lifespan BioSciences), or rabbit anti-ATGL antibody (1 : 100; Santa Cruz), followed by a secondary goat anti-rabbit antibody (488; Invitrogen). The slides were mounted using a mounting medium (Dako) and subsequently analyzed using confocal laser scanning microscopy (LSM 510).
Autodot staining
Cells were first seeded on coverslips in a 24-well plate to allow adherence. Autodot (stock concentration: 0.1 M; Abgent, Lucerne, Switzerland) was diluted 1 to 1000 in PBS, applied to cells in the presence or absence of the stimuli and incubated at 33 °C for 15 min. Cells were then washed using PBS, fixed with 4% paraformaldehyde and mounted using Dako Mounting Media (Dako). Autophagosomes were detected as a punctate pattern using confocal laser scanning microscopy (LSM 510). The excitation filter of Autodot is 380–420 nm, and the barrier filter is 450 nm.
Electron microscopy
Tissue blocks that were 1 mm3 in size were fixed with 5% glutaraldehyde in PBS (pH 7.3) and immersed further in 2% osmium tetroxide. Samples were dehydrated and infiltrated with increasing ethanol concentrations and Spurr's resin, respectively. Ultrathin sections (60 nm) were prepared using a diamond knife, placed onto grids, and stained with uranyl acetate and lead citrate for imaging.
BODIPY staining
shRNA-treated HepG2 cells were seeded on coverslips 24 h before staining using BODIPY (10 μg/ml) (Sigma-Aldrich) as previously described.22 Live cell images were acquired using confocal laser scanning microscopy (LSM 510). LDs were analyzed using the Imaris software. Total areas of LDs were measured and then normalized to those of control conditions.
Oil red O staining
shRNA-treated HepG2 cells were seeded on coverslips for 24 h and subsequently treated as indicated. Oil red O staining (125 μM) was performed as previously described.24, 53 Images were acquired with the help of an Axiovert 35 microscope using the AxioVision Rel. 1.8 software. LDs stained in red were analyzed using the Image Pro Plus software. The red color was selected in HSI color modus and the total LD areas were calculated for at least 100 cells from three independent experiments.
TG measurements
The TG contents of HepG2 cells under the indicated conditions were measured using a Triglyceride Quantification Kit (Abcam, Cambridge, UK) and performed according to the manufacturer's instructions.
Analyses of liver amino-acid concentrations
Liver samples from control and starved mice were extracted and prepared for analysis using a standard solvent extraction method. Briefly, sample preparation was conducted using a proprietary series of organic and aqueous extractions to remove the protein fraction, while allowing maximum recovery of small molecules. The resulting extract was divided into two fractions: one for analysis using liquid chromatography and one for analysis using gas chromatography. Samples were placed briefly on a TurboVap (Zymark, London, UK) to remove the organic solvent. Each sample was then frozen and dried under vacuum. Samples were then prepared for the appropriate instrument, either liquid chromatography and mass spectrometry (LC/MS) or gas chromatography and mass spectrometry (GC/MS). Following log transformation and imputation with minimum observed values for each compound, contrasts resulting from the two-way ANOVA were used to identify amino acids that differed significantly between experimental groups.
Statistics
Data were expressed as mean ±S.D.s and the differences between values were analyzed using the Mann–Whitney U-test. A P<0.05 was considered statistically significant.
Acknowledgments
This study was funded by the Swiss National Science Foundation (310030_129640 and 310030_146181) for HUS as well as by the Medical Research Council, UK, and Grants from ‘Alleanza contro il Cancro' (ACC12), AIRC (2008-2010_33-08) (no. 5471) (2011-IG11955), AIRC 5xmille (no. 9979), Telethon Grant GGPO9133, and IDI-IRCCS (RF06 c.73, RF07 c.57, RF08 c.15, RF07 c.57) for GM.
Glossary
- ATG5
autophagy-related protein 5
- ATGL
adipose triglyceride lipase
- chIP
chromatin immunoprecipitation
- DMEM
Dulbecco's modified Eagle's medium
- LD
lipid droplet
- OA
oleic acid
- RM
regular medium
- shRNA
small hairpin RNA
- TA
transactivation
- TG
triglyceride
- TIP
tail-interacting protein
- WT
wild-type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function. Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ. One billion years of p53/p63/p73 evolution. Proc Natl Acad Sci USA. 2009;106:17609–17610. doi: 10.1073/pnas.0910634106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol. 2011;12:259–265. doi: 10.1038/nrm3086. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA. 2009;106:797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, et al. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci USA. 2011;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, et al. MicroRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26:2009–2014. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Petit I, Murray-Zmijewski F, Goullet de Rugy T, Fernandes K, Meuray V, et al. Diverse p63 and p73 isoforms regulate Δ133p53 expression through modulation of the internal TP53 promoter activity. Cell Death Differ. 2012;19:816–826. doi: 10.1038/cdd.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–1824. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak G, Cooper GM. p53 is a major component of the transcriptional and apoptotic program regulated by PI 3-kinase/Akt/GSK3 signaling. Cell Death Dis. 2012;3:e400. doi: 10.1038/cddis.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse I, Barbollat-Boutrand L, Molina M, Berthier-Vergnes O, Joly-Tonetti N, Martin MT, et al. Functional interplay between p63 and p53 controls RUNX1 function in the transition from proliferation to differentiation in human keratinocytes. Cell Death Dis. 2012;3:e318. doi: 10.1038/cddis.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Dirat B, Laurent K, Puissant A, Auberger P, Budanov A, et al. Sestrin2 integrates Akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ. 2013;20:611–619. doi: 10.1038/cdd.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold A, Vervoort SJ, Martin BP, Bots M, Johnstone RW. Induction of autophagy does not alter the anti-tumor effects of HDAC inhibitors. Cell Death Dis. 2012;3:e387. doi: 10.1038/cddis.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelle M, Perez VM, Rojkind M. Lipid quantitation in formalin-fixed liver sections. J Histochem Cytochem. 1998;36:1471–1474. doi: 10.1177/36.12.2461410. [DOI] [PubMed] [Google Scholar]
- Ljungberg O, Tibblin S. Peroperative fat staining of frozen sections in primary hyperparathyroidism. Am J Pathol. 1979;95:633–641. [PMC free article] [PubMed] [Google Scholar]
- Gibbons GF, Pullinger CR, Björnsson OG. Changes in the sensitivity of lipogenesis in rat hepatocytes to hormones and precursors over the diurnal cycle and during longer-term starvation of donor animals. J Lipid Res. 1984;25:1358–1367. [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Huang Q, Kim YS, Pan N, Ong WY, et al. Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy. 2008;4:457–466. doi: 10.4161/auto.5662. [DOI] [PubMed] [Google Scholar]
- Munafó DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, et al. Two new p73 splice variants, gamma and delta, with different transcriptionally activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischhusen J, Melino G, Weller M. p53 and its family members—reporter genes may not see the difference. Cell Death Differ. 2004;11:1150–1152. doi: 10.1038/sj.cdd.4401477. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledelius C, Jacobsen P, et al. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;52:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, O'Prey J, Bell HS, Ryan KM. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ. 2007;14:1071–1079. doi: 10.1038/sj.cdd.4402108. [DOI] [PubMed] [Google Scholar]
- Rosenbluth JM, Pietenpol JA. mTOR regulates autophagy-associated genes downstream of p73. Autophagy. 2009;5:114–116. doi: 10.4161/auto.5.1.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Blumberg RS. Autophagy, microbial sensing, endoplasmic reticulum stress, and epithelial function in inflammatory bowel disease. Gastroenterology. 2011;140:1738–1747. doi: 10.1053/j.gastro.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 2012. 19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert EK, Levine AJ, Vazquez A. Proliferation and tissue remodeling in cancer: the hallmarks revisited. Cell Death Dis. 2012;3:e397. doi: 10.1038/cddis.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Vander-Heiden MG, Munoz-Pinedo C, Emerling BM. Emerging concepts: linking hypoxic signaling and cancer metabolism. Cell Death Dis. 2012;3:e303. doi: 10.1038/cddis.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannage M, Münz C. MHC presentation via autophagy and how viruses escape from it. Semin Immunopathol. 2010;32:373–381. doi: 10.1007/s00281-010-0227-7. [DOI] [PubMed] [Google Scholar]
- Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemicki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A, et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol Cell Biol. 2004;24:10593–10610. doi: 10.1128/MCB.24.24.10593-10610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Xu L, Ren S, Pandak WM, Chen S, Yin L. StAR overexpression decreases serum and tissue lipids in apolipoprotein E-deficient mice. Lipids. 2009;44:511–519. doi: 10.1007/s11745-009-3299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Huang HC, Lin JK. Theaflavins attenuate hepatic lipid accumulation through activating AMPK in human HepG2 cells. J Lipid Res. 2007;48:2334–2343. doi: 10.1194/jlr.M700128-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.