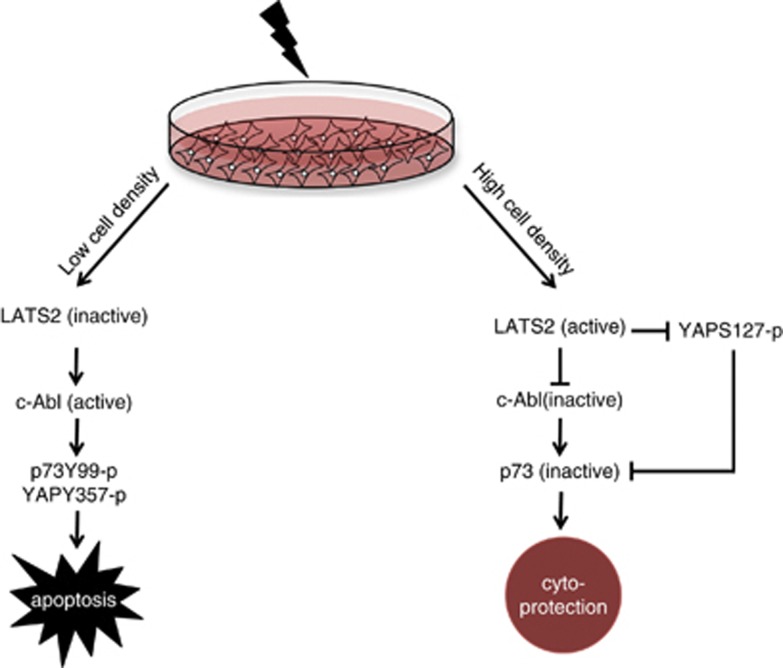
In this issue of CDD, Reuven et al.1 provide an elegant mechanism by which LATS2, a core component of the Hippo pathway, impedes DNA damage-induced apoptosis through inhibition of c-Abl. By dissecting signaling events upon γ-irradiation of sparse and dense cell, the authors revealed a dialogue between Hippo signaling members and DNA damage response (DDR) proteins. The mammalian Hippo tumor suppressor pathway has pivotal roles in regulating organ size, stem cell pluripotency and tumorigenesis.2, 3, 4 The Hippo pathway is composed of a kinase cascade core that includes the MST1/2 serine/threonine kinase, the WW45 scaffold protein, MOB, and the serine/threonine LATS1/2 kinase. Activation of the core cascade through, for example, cell–cell contact due to high cell density leads to phosphorylation of the YAP and TAZ oncoproteins, leading to their sequestration in the cytoplasm and preventing their binding to TEAD transcription factors. This inhibits transcription of downstream target genes implicated in proliferation, anti-apoptosis and epithelial-to-mesenchymal transition. Hippo signaling thus acts through this pathway to suppress tumorigenesis.2, 3, 4
DDR is a complex signaling process that maintains the integrity of the genome in response to DNA damage.5, 6 This signaling process involves a number of factors that either arrest the cell cycle and facilitate DNA repair or, if the DNA damage is too extensive to be repaired, induce apoptosis. A central protein that has been widely associated with various aspects of the DDR is the non-receptor tyrosine (Y) kinase c-Abl (ABL1).7, 8, 9 Upon exposure to γ-irradiation, c-Abl undergoes robust activation leading to phosphorylation of target substrates including p73,7, 8, 9 p63,10 YAP,11 and MDM2.12 c-Abl mediated the phosphorylation of p73 at Y99 and of YAP at Y357 is associated with enhanced apoptosis upon DNA damage, thus implying a tumor suppressive function of YAP.13 This ‘Yin-Yang' function of YAP as an oncogene or tumor suppressor seems to be dependent on its post-translational modification, that is, phosphorylation by LATS1/2 at S127 at high cell density14, 15 or by c-Abl at Y357 upon DNA damage.11, 16 However, the contribution of Hippo signaling in high cell density to apoptosis after DNA damage is largely unknown.
An interesting observation made by the Oren group in 2004 showed that the p53 response to DNA damage, mostly apoptosis, was attenuated by high cell density,17 a phenomena likely caused by cell–cell contact, yet the molecular basis of this behavior has remained elusive. Reuven et al.1 propose a functional crosstalk between Hippo pathway and DDR to explain this behavior. Authors used several cell types to show that DNA damage-induced apoptosis, as assessed by percentage of the subG1 population, caspase-3 activation and PARP cleavage, is inhibited when Hippo pathway is active, that is, at high cell density. The authors found no differences between p53 wild-type and p53 mutant cell lines, suggesting that p53 is not the main player in this context. Importantly, a c-Abl kinase inhibitor STI571 or c-Abl knockdown phenocopied the reduced apoptotic phenotype in dense cells, supporting c-Abl involvement in mediating apoptosis. Examining the downstream effector of Hippo signaling under these conditions revealed inactivation of YAP, as assessed by its phosphorylation at S127, thus confirming that the Hippo pathway is active. Notably, enforced nuclear YAP localization by using YAPS127A mutant did not rescue inhibition of apoptosis, implying additional members of the Hippo pathway are involved.
Interestingly, under conditions of DNA damage, dense cells displayed attenuated c-Abl activation. Proper autophosphorylation of c-Abl as well as phosphorylation of its downstream substrates YAP (Y357) and p73 (Y99) was markedly reduced suggesting that activation of Hippo pathway inhibits c-Abl even following DNA damage. The authors venture further to show that LATS2 physically interacts with c-Abl and functionally phosphorylates c-Abl, leading to reduced c-Abl kinase function. Remarkably, a point mutation in the c-Abl SH3 domain rescued LATS2 inhibition of c-Abl, implying that the SH3 domain is involved in mediating this interaction. The authors further demonstrated that by mimicking active Hippo signaling, through overexpression of LATS2, in low-density cell culture, DNA damage-induced apoptosis is reduced. On the other hand, LATS2 depletion increased apoptosis, even in densely plated cells. Finally, overexpression of mutant c-Abl constructs, which lack LATS2 phosphosite or harbor a point mutation in SH3 domain, overcame LATS2 inhibition and resulted in apoptosis in densely plated cells upon DNA damage. Taken together, these observations by Reuven et al.1 suggest that when DNA damage is induced in dense cells, activated LATS2 binds and inhibits c-Abl-mediating phosphorylation of p73Y99 and YAPY357, resulting in reduced apoptosis and cytoprotection (Figure 1). Importantly, under these conditions, LATS2 can also bind and phosphorylate YAP at S127, resulting in its cytoplasmic sequestration, thus YAP becomes unable to bind and co-activate p73 to enhance its apoptotic function.18 These findings suggest that under certain circumstances, LATS2 can function as anti-apoptotic protein.
Figure 1.
In late response to DNA damage, c-Abl is activated leading to phosphorylation and activation of p73 (p73Y99-p)8, 9, 10 and YAP (YAPY357-p)12 resulting in cell apoptosis. Reuven et al.1 demonstrate that upon exposure of dense cells to ionizing radiation, activated LATS2 phosphorylates c-Abl, hence inhibiting its kinase function and resulting in reduced p73Y99-p and YAPY357-p-mediated apoptosis leading to cytoprotection. According to this model, LATS2-mediated phosphorylation of YAP at S127 sequesters YAP in the cytoplasm, thus YAP could not function as co-activator of p73 to enhance apoptosis,18 thereby contributing also to cytoprotection
Notably, previous findings have strongly suggested that LATS proteins function as positive regulators of apoptosis. In fact, both LATS1 and LATS2 are established as known tumor suppressors that regulate cell proliferation, cell cycle progression and apoptosis (reviewed in Visser and Yang19). Although LATS1 and LATS2 possess minor differences in expression patterns and mechanisms of action, both kinases share many functions although it remains to be determined whether active LATS1 is also capable of inhibiting c-Abl activity and apoptosis upon DNA damage of dense cells. Besides their role in Hippo signaling,20 LATS1/2 are coupled to several well-established cell signaling pathways. For example, constitutively active RAS induces LATS2 expression, and, as such, loss of LATS2 cooperates with oncogenic Ras to promote tumorigenesis.21 In addition, LATS2 augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1 promoting the death of polyploid cells.22 Furthermore, loss of function of either LATS1 or LATS2, largely due to epigenetic changes, is associated with a variety of tumor types, including soft tissue sarcomas, leukemia as well as breast, prostate, lung and esophageal cancers (reviewed in Visser and Yang19). Altogether, these features are widely associated with tumor suppressor roles of LATS1/2. However, the context in Reuven et al.1 study is different. One possibility that rises from the current work is that exposure of high culture density to DNA damage alters the target substrate specificity of LATS2, perhaps favoring the inhibition of proapoptotic proteins and activation of antiapoptotic proteins, thereby conferring increased resistance to DNA damage. The effects presented by this study are robust and might suggest that the Hippo pathway inhibits other apoptotic factors, other than c-Abl, which still need to be uncovered. Such effects could account for radio and chemoresistance of aggressive cancer cells buried within a tumor and exhibit extensive cell–cell interactions. Careful dissection of LATS2/YAP/cAbl pattern and apoptotic markers in such tumor cells might further confirm the clinical implication of these observations.
Altogether, the current work by Reuven et al.1 introduces an interesting twist into the function of Hippo signaling members in apoptosis, suggesting that activated LATS2 may have a significant role in inhibiting apoptosis upon DNA damage. Whether other cellular behaviors are favored under these conditions, such as senescence, remained to be explored. This may have tremendous consequences(s) on, for example, cellular response to treatment. Although the study advances our understanding of the involvement of LATS2 in the cellular response to DNA damage-induced apoptosis in dense cells, additional work is required to obtain a comprehensive understanding on the physiological outcomes of this involvement. Research on the Hippo pathway is relatively new but has expanded quickly in recent years. The connection between DDR and the Hippo pathway would provide new insights for both fields.
Acknowledgments
I am grateful to Dr. Ittai Ben-Porath for critical reading of the manuscript. The Aqeilan lab is supported, in part, by funds from the Israel Science Foundation (ISF, #12-542).
The author declares no conflict of interest.
References
- Reuven N, et al. Cell Death Differ 2013. e-pub ahead of print 12 July 2013; doi: 10.1038/cdd.2013.83 [DOI]
- Pan D. Dev Cell. 2010. pp. 491–505. [DOI] [PMC free article] [PubMed]
- Salah Z, Aqeilan RI. Cell Death Dis. 2011. p. e172. [DOI] [PMC free article] [PubMed]
- Zhao B, Tumaneng K, Guan KL. Nat Cell Biol. 2011. pp. 877–883. [DOI] [PMC free article] [PubMed]
- Jackson SP, Bartek J. Nature. 2009. pp. 1071–1078. [DOI] [PMC free article] [PubMed]
- Shiloh Y, Ziv Y. Nat Rev Mol Cell Biol. 2013. pp. 197–210. [DOI] [PubMed]
- Agami R, et al. Nature. 1999. pp. 809–813. [DOI] [PubMed]
- Gong JG, et al. Nature. 1999. pp. 806–809. [DOI] [PubMed]
- Yuan ZM, et al. Nature. 1999. pp. 814–817. [DOI] [PubMed]
- Gonfloni S,C. Nat Med. 2009. pp. 1179–1185. [DOI] [PubMed]
- Levy D, et al. Mol Cell. 2008. pp. 350–361. [DOI] [PubMed]
- Goldberg Z, et al. EMBO J. 2002. pp. 3715–3727. [DOI] [PMC free article] [PubMed]
- Bertini E, et al. Cell Cycle. 2009. pp. 49–57. [DOI] [PubMed]
- Hao Y, et al. J Biol Chem. 2008. pp. 5496–5509. [DOI] [PubMed]
- Oka T, Mazack V, Sudol M. J Biol Chem. 2008. pp. 27534–27546. [DOI] [PubMed]
- Levy D, Reuven N, Shaul Y. J Biol Chem. 2008. pp. 27462–27468. [DOI] [PubMed]
- Bar J, et al. Oncogene. 2004. pp. 2128–2137. [DOI] [PubMed]
- Strano S, et al. J Biol Chem. 2001. pp. 15164–15173. [DOI] [PubMed]
- Visser S, Yang X. Cell Cycle. 2010. pp. 3892–3903. [DOI] [PubMed]
- Zhao B, Lei QY, Guan KL. Curr Opin Cell Biol. 2008. pp. 638–646. [DOI] [PMC free article] [PubMed]
- Aylon Y, et al. Oncogene. 2009. pp. 4469–4479. [DOI] [PMC free article] [PubMed]
- Aylon Y, et al. Genes Dev. 2010. pp. 2420–2429. [DOI] [PMC free article] [PubMed]