Abstract
The Hippo pathway is an evolutionarily conserved pathway that controls cell proliferation, organ size, tissue regeneration and stem cell self-renewal. Here we show that it also regulates the DNA damage response. At high cell density, when the Hippo pathway is active, DNA damage-induced apoptosis and the activation of the tyrosine kinase c-Abl were suppressed. At low cell density, overexpression of the Hippo pathway kinase large tumor suppressor 2 (Lats2) inhibited c-Abl activity. This led to reduced phosphorylation of downstream c-Abl substrates, the transcription coactivator Yes-associated protein (Yap) and the tumor suppressor p73. Inhibition of c-Abl by Lats2 was mediated through Lats2 interaction with and phosphorylation of c-Abl. Lats2 knockdown, or expression of c-Abl mutants that escape inhibition by Lats2, enabled DNA damage-induced apoptosis of densely plated cells, while Lats2 overexpression inhibited apoptosis in sparse cells. These findings explain a long-standing enigma of why densely plated cells are radioresistant. Furthermore, they demonstrate that the Hippo pathway regulates cell fate decisions in response to DNA damage.
Keywords: DNA damage response, high cell density, Hippo pathway, c-Abl inhibition, radioresistant
Severe DNA damage elicits apoptosis, a process mediated by the p53 family of tumor suppressors (reviewed in Vousden and Prives1 and Beckerman and Prives2). However, under certain conditions damaged cells do not undergo apoptosis. DNA damage-induced apoptosis is inhibited at high cell density,3 a behavior that is likely to be due to cell–cell contact. However, the molecular basis of how cell–cell contact inhibits DNA damage-induced cell death has remained obscure.
The Hippo pathway is an evolutionarily conserved pathway that regulates cell proliferation and organ size,4, 5, 6 embryonic stem cell renewal and differentiation7 and cancer stem cells.8 At high cell density, the Hippo signaling cascade is activated, culminating in the activation of the kinases Lats1 and 2 (Lats1/2, large tumor suppressor 1 and 2). Lats1/2, in turn, phosphorylate the transcription coactivators Yap1, Yap2 (Yes-associated protein) and Taz (transcriptional coactivator with PDZ-binding motif), which leads to their cytoplasmic sequestration and proteasomal degradation.9, 10, 11 Under conditions of low cell density, Yap and Taz enter the nucleus, where they coactivate the TEAD (TEA domain) family of transcription factors, and lead to the activation of pro-proliferative, pro-EMT (epithelial to mesenchymal transition) and anti-apoptotic target genes.12, 13, 14, 15, 16 Thus, the Hippo pathway suppresses cellular proliferation at high cell density, and acts as a tumor suppressor by inhibiting oncogenic Yap and Taz.
Although Yap can be a powerful oncogene and pro-proliferative factor, under conditions of DNA damage, Yap functions as a tumor suppressor,17 and coactivates the p53 family member p73 in response to DNA damage. Upon DNA damage, the non-receptor tyrosine kinase c-Abl (ABL1) is activated, and phosphorylates p73 at Tyr99, which supports p73-dependent induction of apoptotic genes.18, 19, 20 Active c-Abl also phosphorylates Yap at Tyr357.21 The tyrosine phosphorylated Yap accumulates and preferentially associates with p73 in targeting apoptotic gene promoters. c-Abl-mediated phosphorylation of Yap therefore switches Yap activity from anti- to pro-apoptotic.21, 22 The involvement of the Hippo-regulated effector Yap in the apoptotic DNA damage response suggested that the Hippo pathway may control apoptosis through Yap. Previous studies have demonstrated that components of the Hippo pathway can have roles in regulation of apoptosis. The Hippo pathway kinases Mst1 and 2 (mammalian STE20-like protein kinase) have pro-apoptotic activity in response to various stimuli (reviewed in Radu and Chernoff23); the tumor suppressor RASSF1A (RAS association domain family 1A) elicits apoptosis through Mst2;24 Lats2 cooperates with the p53 activator ASPP1 (apoptosis-stimulating protein of p53-1) to promote p53-dependent apoptosis;25 and activation of the Hippo pathway is important for anoikis induced by cell detachment.26 However, these studies did not investigate Hippo pathway involvement in DNA damage-induced apoptosis under conditions of high cell density.
We demonstrate here that active Hippo pathway inhibits DNA damage-induced apoptosis. Hippo pathway inhibition of apoptosis was not achieved solely through cytoplasmic retention of Yap, because a constitutively nuclear Yap mutant was not sufficient to enable apoptosis at high cell density. We discovered that the Hippo pathway inhibited the c-Abl-mediated DNA damage response pathway. High cell density or Lats2 activation suppressed activation of c-Abl, and prevented the phosphorylation of downstream substrates p73 and Yap. Overexpression of Lats2 was sufficient to suppress apoptosis in sparse cells in response to DNA damage. Furthermore, knockdown of Lats2, or expression of c-Abl mutants resistant to phosphorylation by or interaction with Lats2, partially restored DNA damage-induced apoptosis in densely plated cells. Our data support a model whereby DNA damage-induced apoptosis is suppressed by the Hippo pathway, through inhibition of c-Abl by Lats2, and explains why dense cultures are resistant to DNA damage-induced apoptosis.
Results
DNA damage-induced apoptosis is inhibited when the Hippo pathway is active
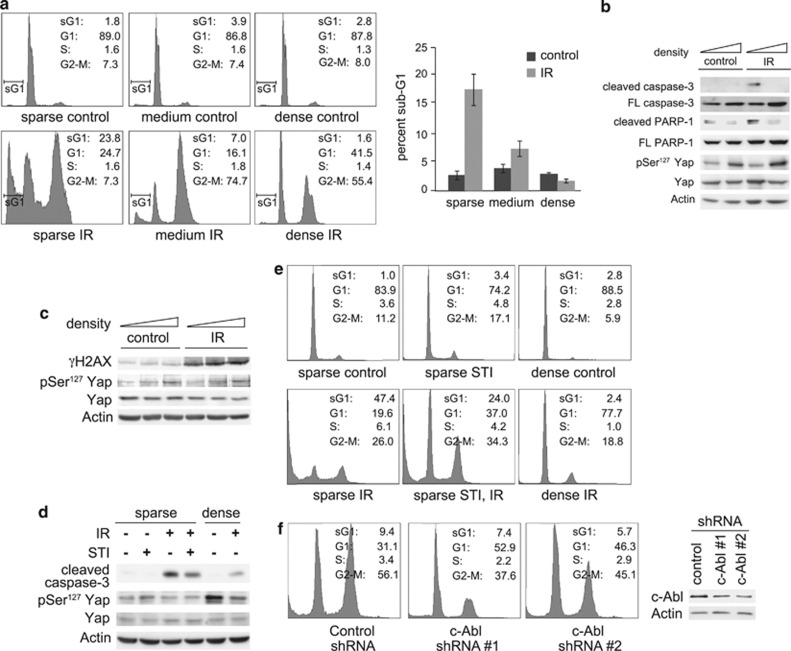
To study the role of the Hippo pathway in regulation of DNA damage-induced apoptosis, we used MCF10A cells, a non-transformed breast epithelial cell line with a functional Hippo pathway.9, 27 We plated the cells at different cell densities and exposed them to γ-irradiation (IR). FACS (fluorescence-activated cell sorting) analysis was performed to determine the percentage of apoptotic cells, defined as those in the sub-G1 fraction. Following IR, sparsely plated cells underwent apoptosis, whereas apoptosis was curtailed in the densely plated cells (Figure 1a). Furthermore, markers of the apoptotic response such as caspase 3 activation and PARP cleavage were detected in the sparsely, but not densely, plated cells (Figure 1b). The lack of cell death at high density was not due to a defect in detecting DNA damage or in the initial DNA damage response. Both densely and sparsely plated cells showed phosphorylation of histone H2AX (γ-H2AX) 2.5 h post-IR (Figure 1c). Upon Hippo pathway activation, Lats phosphorylates Yap at Ser127.9, 28, 29 In the densely plated cells, phosphorylation of Yap at Ser127 was detected, demonstrating that the Hippo pathway was active (Figures 1b–d). To demonstrate that inhibition of apoptosis at high cell density was not cell type specific, we tested other cell lines. We found that DNA damage-induced apoptosis was suppressed at high cell density in NIH3T3 cells (Figures 1d and e), in addition to p53-null H1299 cells (Supplementary Figures 1a and b) and p53-wild-type-containing U2OS cells (Supplementary Figure 1c). Despite the latter two cell lines being tumor derived, they still exhibited Hippo pathway activation at high cell density, as shown by the phosphorylation of Ser127 (Supplementary Figures 1a and d). These cell lines were used to show that the density effect on apoptosis was present in both p53 wild type and p53-null cell lines. Previous studies have demonstrated a central role for c-Abl in induction of DNA damage-induced apoptosis. Yap- and p73-dependent apoptosis requires their phosphorylation by c-Abl.20, 21 c-Abl also contributes to p53-dependent apoptosis through the inhibition of mouse double minute 2 (Mdm2) and Mdm4.30, 31 Consistent with these findings, treatment of the cells with the c-Abl kinase inhibitor STI571, or knockdown of c-Abl with shRNA reduced apoptosis, demonstrating that the process we observed was dependent on c-Abl activity (Figures 1d–f and Supplementary Figures 1a and b).
Figure 1.
DNA damage-induced apoptosis is inhibited at high cell density. (a) MCF10A cells were plated at different cell densities. Cells were irradiated at 20 Gy, and were harvested for fluorescence-activated cell sorting (FACS) analysis 72 h post-irradiation (IR). Left panels, profile plots of representative FACS analysis. Percentages of cells in the different fractions (sub-G1, G1, S and G2-M) are indicated. Right panel, graph showing quantification of three independent experiments. (b) Cells were treated as in (a), and analyzed by immunoblot with the indicated antibodies. (c) Cells were plated and irradiated as in (a), but were harvested 2.5 h post-IR for immunoblot analysis. (d) NIH3T3 cells were plated sparse or dense and were treated with 10 μM STI571 or dimethyl sulfoxide (solvent control) as indicated. Cells were irradiated 2.5 h later at 40 Gy, and were harvested 96 h post-IR, and analyzed by immunoblotting. (e) NIH3T3 cells were treated as in (d), and were analyzed by FACS. Percentages of cells in the different fractions are indicated. (f) MCF10A stably expressing small hairpin RNA (shRNA) to c-Abl or non-targeting control were plated sparse, and were irradiated at 25 Gy. Left: cells were harvested for FACS analysis 96 h post-IR. Percentages of cells in the different fractions are indicated. Right: the expression of c-Abl in the c-Abl-shRNA-expressing cells was analyzed by immunoblotting
Yap nuclear localization is not sufficient to enable apoptosis at high cell density
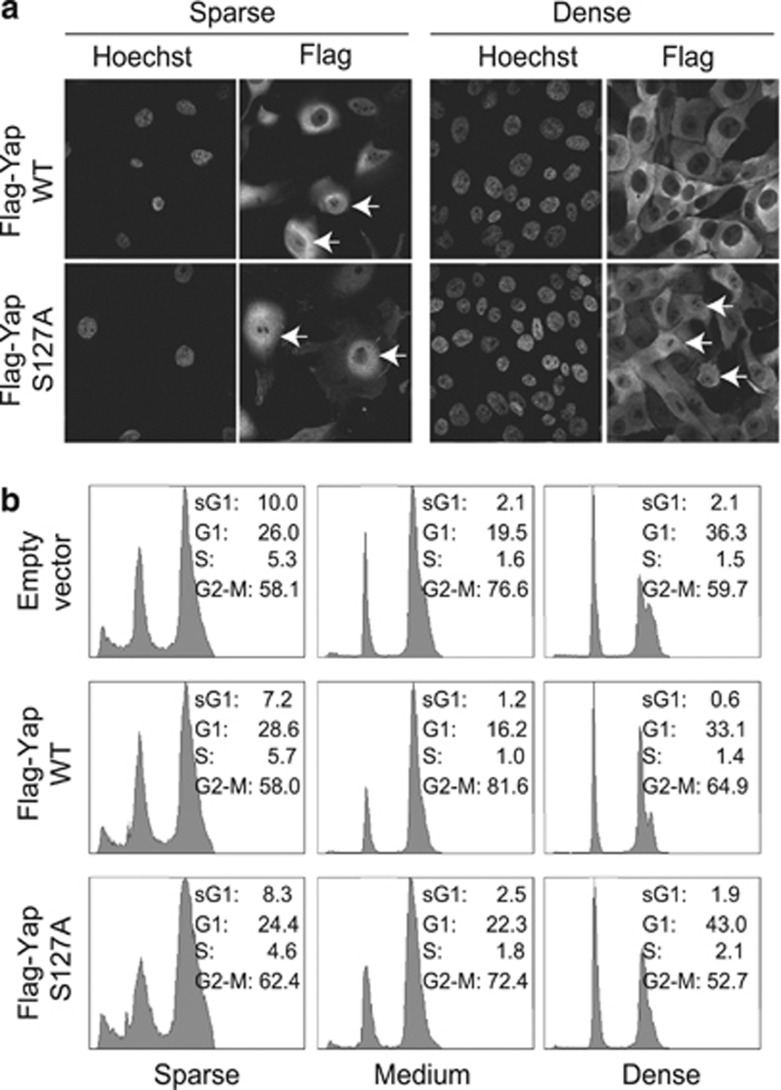
Yap is an important coactivator of p73 in the apoptotic response. At high cell density, Yap is mostly cytoplasmic, therefore this could be a way by which the Hippo pathway inhibits apoptosis at high cell density. To test this possibility, we used MCF10A cells expressing Flag-Yap, or the Flag-Yap S127A mutant, which is nuclear localized.9 Yap is important for apoptosis in these cells, as knockdown of Yap1 in MCF10A cells reduced DNA damage-induced apoptosis (Supplementary Figure 2). Under DNA damage, at low cell density, Flag-Yap and the S127A mutant were nuclear localized (Figure 2a). In contrast, at high cell density, Flag-Yap was, by large, nuclear excluded whereas the Flag-Yap S127A mutant still showed nuclear staining. We found that even in cells expressing nuclear Yap S127A, apoptosis was inhibited by high cell density (Figure 2b). As nuclear Yap could not rescue apoptosis, this implied that the Hippo pathway suppresses apoptosis through an additional mechanism.
Figure 2.
Nuclear Yes-associated protein (Yap) does not rescue apoptosis at high cell density. (a) MCF10A cells expressing Flag-Yap1, Flag-Yap1 S127A or vector control were plated sparse and dense, and irradiated at 20 Gy. After 24 h, cells were fixed for immunofluorescence analysis with the anti-Flag antibody. Nuclear staining is denoted by Hoechst 33342. Panels show representative fields of three independent experiments. Arrows indicate some of the cells with nuclear-localized Yap1. (b) The cells used in (a) were plated sparse and dense, irradiated at 20 Gy and harvested for FACS analysis 120 h post-IR. Percentages of cells in the different fractions (sub-G1, G1, S and G2-M) are indicated
Lats2 overexpression inhibits Yap in sparse cells
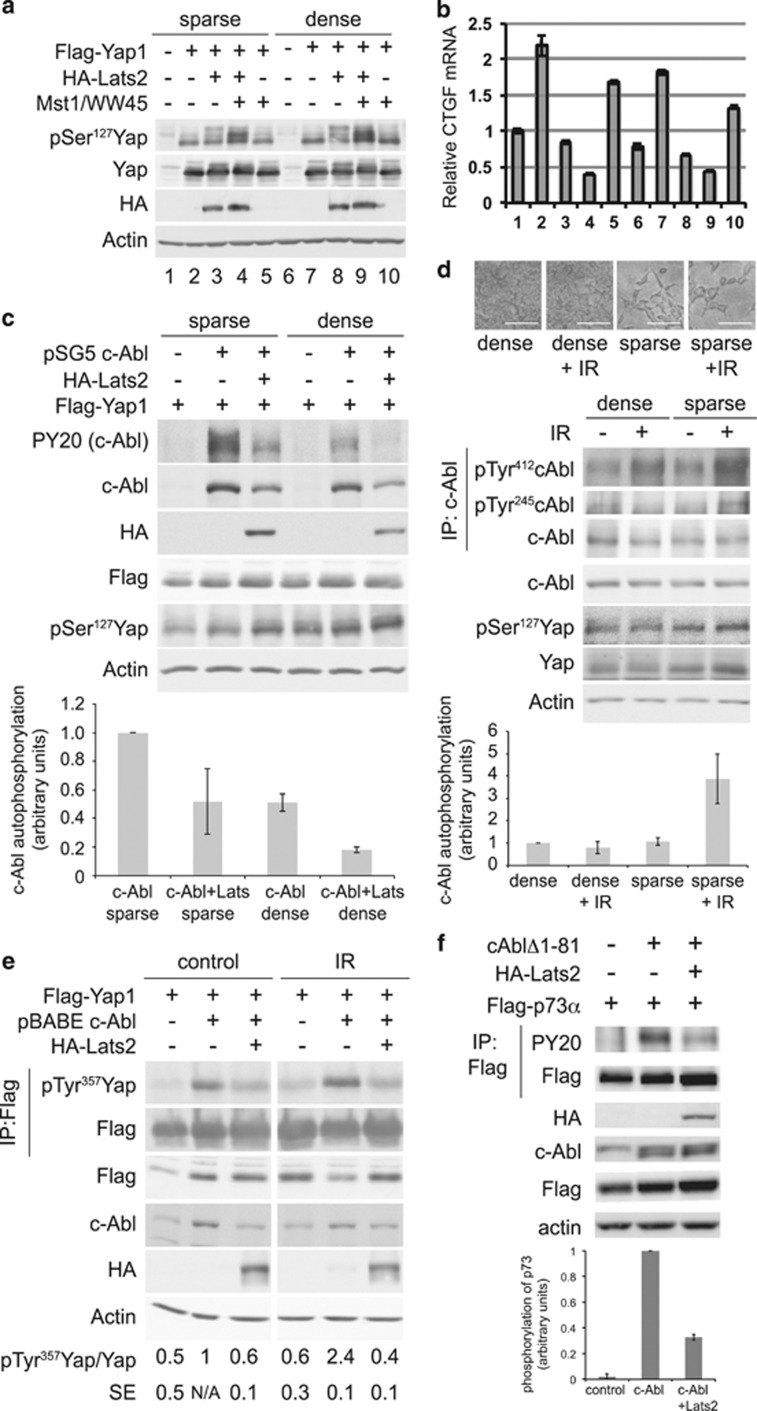
To investigate high cell density inhibition of apoptosis and Hippo pathway involvement, we used overexpression of Hippo pathway core proteins to recapitulate the effect of high cell density. To validate this system, we overexpressed Flag-Yap1, along with combinations of HA-Lats2 and untagged Mst1 and WW45 in HEK293 cells. Following transfection, cells were replated under sparse and dense conditions. Lats2 was active when overexpressed, even under sparse conditions, leading to phosphorylation of Yap at Ser127 (Figure 3a). Co-expression of Mst1 and WW45, activators of Lats2, led to a further increase in pSer127. These effects were intensified at high cell density. Phospho-Yap appeared with several upshifted bands, likely representing incremental phosphorylation of the five different Lats2 sites on Yap.9, 28 Analysis of mRNA by qPCR confirmed expression of the untagged Mst1 and WW45 (Supplementary Figures 3a and b). Furthermore, qPCR showed that expression of endogenous CTGF, a target gene of Yap, increased with Yap overexpression, but was inhibited with Lats2, and was inhibited further when Lats2 was expressed together with Mst1 and WW45 (Figure 3b). These results indicate that overexpression of Lats2 is sufficient to activate Lats2, leading to Yap inhibition similar to what is found with Hippo activation at high cell density.
Figure 3.
High-cell density inhibits c-Abl autophosphorylation. (a–b) Activation of Lats by overexpression and high-cell density. HEK293 cells were transfected with the indicated and green fluorescent protein (GFP)-expressing plasmids. Cells were replated 28 h post-transfection to produce sparse and dense plates. Sixteen hours later, cells were harvested and immunoblotted with the indicated antibodies (a). (b) CTGF mRNA levels were analyzed by qPCR and normalized by GFP mRNA. (c) High cell density and Lats2 overexpression inhibit c-Abl autophosphorylation. HEK293 cells were transfected with Flag-Yap1 (all lanes), and indicated plasmids. Transfected cells were replated sparse and dense, harvested 11 h later, and analyzed by immunoblotting. PY20 detects phosphotyrosine. Lower panel: Quantification of c-Abl autophosphorylation, N=2. (d) c-Abl DNA damage-induced activation is inhibited at high cell density. HEK293 cells were plated at high and low cell density. Cells were irradiated at 20 Gy, and were photographed (upper panel, scale bar=0.1 mm) and then harvested 24 h post-IR. Middle panel: c-Abl was immunoprecipitated and samples were analyzed by immunoblotting with the indicated antibodies. Lower panel: Quantification of c-Abl autophosphorylation, N=6. (e) Lats2 inhibits IR-induced c-Abl phosphorylation of Yap1. HEK293 cells were transfected with the plasmids indicated, and 24 h post-transfection, cells were irradiated at 20 Gy. Cells were harvested 4 h post-IR, and Flag-Yap1 was immunoprecipitated. Samples were analyzed by immunoblotting with the indicated antibodies. Quantification of pTyr357 Yap, N=3. (f) Lats2 inhibits c-Abl phosphorylation of p73. Proteins were immunoprecipitated from transfected HEK293 cells with anti-Flag. Upper panel: Samples were analyzed by immunoblotting with the indicated antibodies. Lower panel: Quantification of p73 phosphorylation, N=3
c-Abl activation under DNA damage is compromised in dense cells
c-Abl is an important regulator of apoptosis, therefore we next asked whether c-Abl can be activated at high cell density. c-Abl is autophosphorylated upon activation at several tyrosine residues, including Tyr245 and Tyr412,32 thus autophosphorylation was used as an indicator of c-Abl activation. To test whether the Hippo pathway could inhibit c-Abl, wild-type c-Abl was transfected with or without Lats2 into HEK293 cells. The following day, transfected cells were replated under sparse and dense conditions. Figure 3c shows that basal c-Abl autophosphorylation due to overexpression could be detected most strongly in the sparsely plated cells. Overexpression of Lats2 or plating of the cells at high density reduced c-Abl autophosphorylation. The inhibition of c-Abl at high cell density was intensified with Lats2 overexpression. Activation of endogenous Lats2 by high cell density and activation of exogenous Lats2 by overexpression was validated by the increase in Yap phosphorylated at Ser127. This demonstrated that high cell density, or Lats2 overexpression, could inhibit c-Abl autophosphorylation. To test whether c-Abl activated by DNA damage is inhibited at high cell density where the endogenous Lats2 is active, HEK293 (Figure 3d), NIH3T3 (Supplementary Figure 3c) and MCF10A (Supplementary Figure 3d) cells were plated under sparse and dense conditions, and irradiated. Activation of endogenous c-Abl following IR was evident in sparsely plated cells, but was inhibited at high cell density. Under IR, Yap is an important c-Abl substrate, therefore, we next tested whether Lats2 overexpression could inhibit the DNA damage-induced phosphorylation of Yap on Tyr357 by c-Abl. Yap was phosphorylated by wild-type c-Abl in response to IR, but when Lats was coexpressed, Yap phosphorylation was reduced (Figure 3e). Together these results show that the Hippo pathway kinase, Lats2, prevented the damage-induced activation of c-Abl and its phosphorylation of Yap.
To further investigate Lats2 inhibition of c-Abl, we tested Lats2 effect on phosphorylation of p73 by constitutively active c-Abl (Δ1-81).33 Co-expression of Lats2 reduced the phosphorylation of p73 by active c-Abl (Figure 3f). As Lats2 inhibited constitutively active c-Abl, it implied that Lats2 was acting directly, and not via an upstream activator of c-Abl. Furthermore, when Lats2 was coexpressed with its upstream activators Mst1 and WW45, p73 phosphorylation and c-Abl autophosphorylation were further reduced (Supplementary Figure 3e). This indicates that Hippo pathway activation leads to inhibition of c-Abl, and prevents phosphorylation of its downstream substrates.
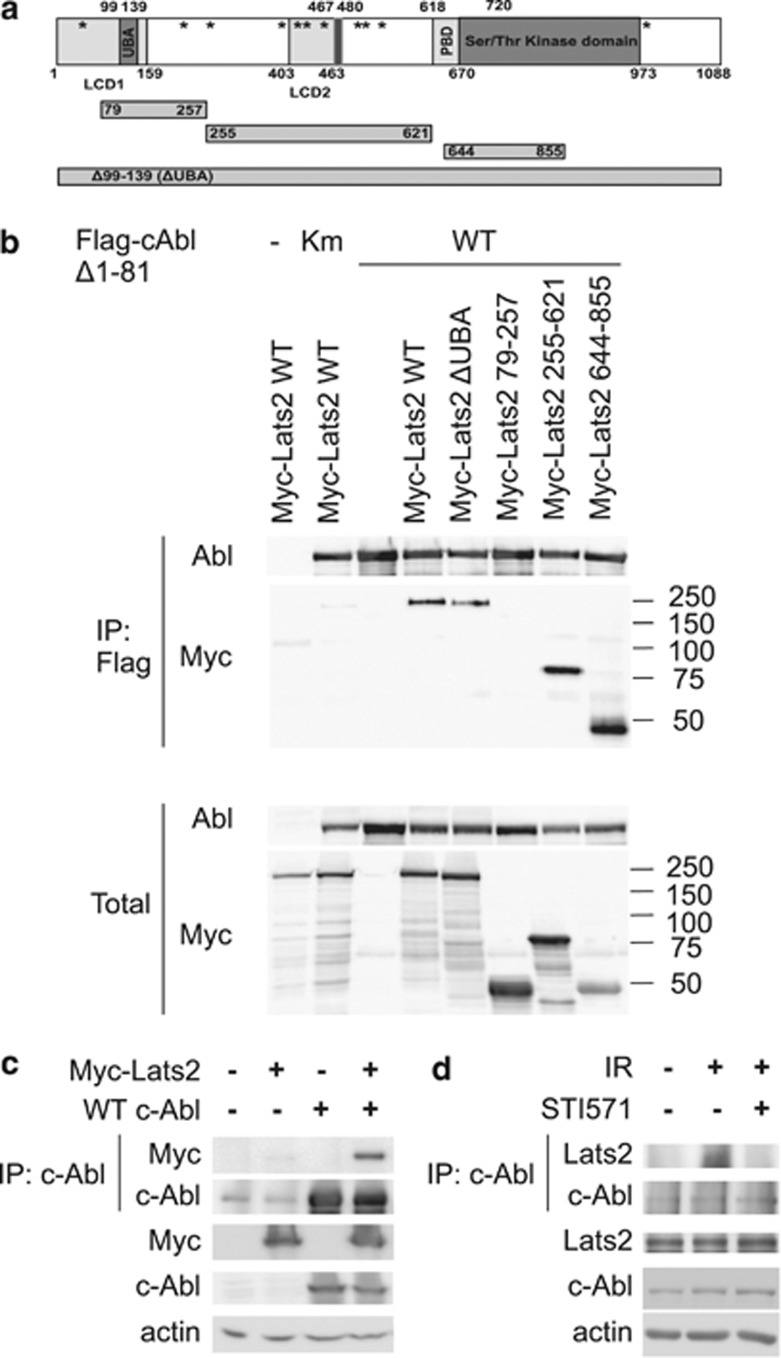
Hippo kinase Lats2 binds c-Abl
The effect of Lats2 on constitutively active c-Abl suggested that Lats is acting directly, through interaction with c-Abl. Lats2 possesses several PxxP motifs, the consensus for binding to SH3 domains34 (Figure 4a). We tested and mapped the interaction between c-Abl Δ1-81 and Lats2 by using co-immunoprecipitation (IP) of transfected constructs. Lats2 was capable of binding to c-Abl through several domains (Figure 4b). Interestingly, Lats2 did not interact with kinase-dead c-Abl. All of the Lats2 constructs tested, with the exception of the aa79–257 fragment, co-immunoprecipitated with constitutively active c-Abl. In addition, full-length wild type c-Abl also interacted with Lats2 (Figure 4c). Endogenous Lats2 co-immunoprecipitated with endogenous c-Abl under DNA damage conditions, but not in the presence of the c-Abl inhibitor STI571 (Figure 4d). These results suggest that Lats2 interacts with active c-Abl via multiple domains.
Figure 4.
Interaction of c-Abl and Lats2. (a) Schematic representation of human Lats2 and truncation mutants. LCD, Lats Conserved Domain (aa1–160, 403–463; UBA, ubiquitin-binding domain (aa99–139), Ser/Thr kinase domain (aa670–973), PAPA repeat (aa467–480). Consensus SH3 binding motifs (PXXP) are indicated by asterisks. (b) Lats2 interacts with c-Abl through multiple sites. HEK293 cells were transfected with pCDNA Flag Δ1-81Abl or Km (kinase mutant) and pCDNA Myc-Lats2 variants (WT, ΔUBA; 79–257; 255–621; 644–855). Cells were harvested and Flag-Abl was immunoprecipitated by anti-Flag agarose. IP and total extract samples were analyzed by immunoblotting. (c) Lats2 interacts with WT c-Abl. HEK293 cells were transfected with pSLX c-Abl and pCDNA Myc-Lats2. Cells were harvested and c-Abl was immunoprecipitated by anti-c-Abl antibodies and protein A/G agarose. IP and total extract samples were analyzed as in (b). (d) Lats2 interacts with c-Abl following DNA damage. HEK293 cells were treated with 20 μM STI571, where indicated, and irradiated at 40 Gy 1 h later. Cells were harvested 72 h post-IR, and endogenous c-Abl was immunoprecipitated. IP and total extracts were analyzed by SDS-PAGE and immunoblotting
c-Abl is a direct substrate of the Hippo kinase Lats2
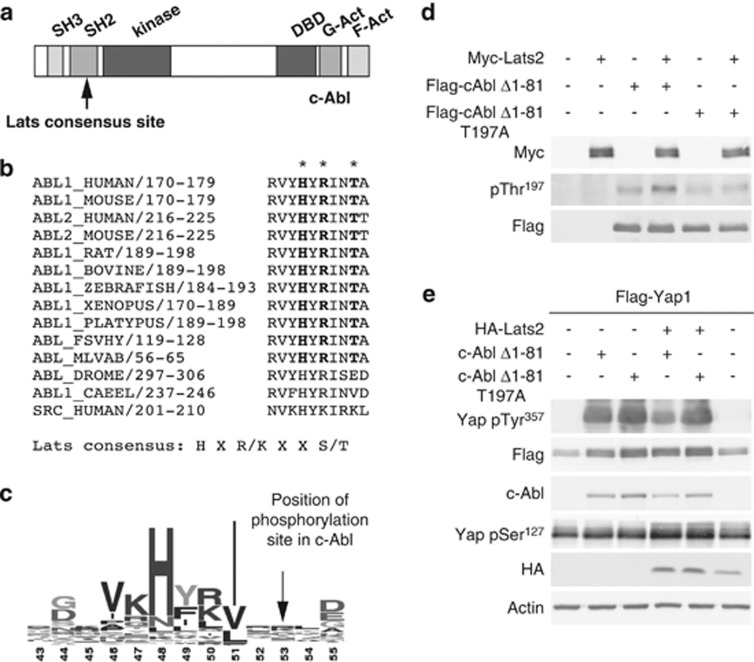
We observed that c-Abl possesses a consensus site for Lats phosphorylation in its SH2 domain, the domain responsible for binding to phosphotyrosine (Figure 5a). The consensus, HxR/H/KxxS/T,9, 28 is present in c-Abl (ABL1) and Arg (Abl-related gene, ABL2) from higher organisms (Figure 5b) and in v-Abl, but is not conserved in worms or flies. Likewise, it is not conserved in the related Src family kinase c-Src. A consensus sequence logo compiled by Prosite35 from 444 SH2 domain sequences shows that the phosphorylation site S or T is not conserved among general SH2 domains (Figure 5c). We prepared a phospho-specific antibody to detect c-Abl phosphorylated at Thr197, the consensus phosphorylation site. This antibody recognized wild-type c-Abl phosphorylated in vitro by activated Lats2, but not the T197A mutant, suggesting that Lats2 phosphorylates c-Abl at the Thr197 site (Figure 5d). The activity of the myc-Lats2 used in the in vitro assay was confirmed by its ability to phosphorylate Yap in vitro (Supplementary Figure 4a). Furthermore, a myc-Lats2 kinase mutant did not phosphorylate c-Abl in vitro (Supplementary Figure 4b). To determine whether the Thr197 phosphorylation site is important for the inhibition of c-Abl by Lats2, we tested the ability of wild type constitutively active c-Abl (Δ1-81), and the T197A c-Abl mutant to phosphorylate Yap when transfected together with Lats2. The T197A mutant was more resistant to Lats2 inhibition than the wild type, and phosphorylated Yap even in the presence of Lats2 (Figure 5e). These results suggest that phosphorylation of c-Abl by Lats2 has a role in the inhibition of c-Abl.
Figure 5.
Lats2 inhibits c-Abl activity, and phosphorylates c-Abl at its SH2 domain. (a) Schematic representation of c-Abl. The SH3, SH2, kinase, DNA binding (DBD) and G and F-actin (G-Act, F-Act) binding domains are indicated. The putative Lats phosphorylation site is indicated by an arrow. (b) Lats consensus phosphorylation site is conserved in Abl from higher organisms. Alignment of region of putative Lats phosphorylation sites in Abl from different organisms. The Lats consensus site is shown in bold and its position in the aligned regions is marked by asterisks. The corresponding region of human c-Src is shown for comparison. (c) Lats phosphorylation site is not conserved in all SH2 domains. A consensus sequence logo compiled by Prosite from 444 SH2 domain sequences showing the region containing the Lats phosphorylation site in c-Abl. (d) Lats2 phosphorylates c-Abl at Thr197 in vitro. In vitro kinase assays were performed and analyzed by immunoblot. The anti-phospho c-Abl (Thr197) antibody was used to detect c-Abl phosphorylated by Lats2. (e) c-Abl T197A mutant is resistant to inhibition by Lats2. HEK293 cells were transfected with Flag-Yap1 (all lanes) and the plasmids indicated. Cell extracts were analyzed by immunoblotting
The Hippo kinase Lats2 binds the c-Abl SH3 domain
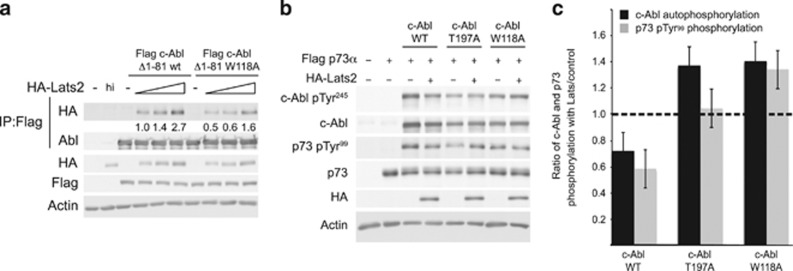
Our results (Figure 4b) indicating that multiple Lats2 domains were capable of interacting with c-Abl, and that Lats2 possesses a number of consensus motifs for SH3 domain binding (Figure 4a), suggested that Lats2 inhibition of c-Abl could be mediated through Lats2 binding to the SH3 domain of c-Abl. To validate this, we made a point mutation in the SH3 domain of c-Abl (W118A) at a position shown to reduce SH3 domain binding to PXXP motifs,36 and tested the interaction between c-Abl and Lats2 by co-IP. Lats2 co-immunoprecipitated approximately 2-fold less efficiently with the W118A mutant c-Abl than with the wild type, indicating lower binding with this mutant (Figure 6a). We next compared the sensitivity of the constitutively active c-Abl Δ1-81, and the T197A and W118A mutants, to inhibition by Lats2. Lats2 inhibited wild-type c-Abl Δ1-81, as indicated by a decrease in phosphorylation of p73 (pTyr99) and c-Abl autophosphorylation (pTyr245), but the T197A and W118A mutants escaped inhibition (Figures 6b and c). These results indicate that Lats2 inhibits c-Abl, and the SH3 domain is involved in mediating the interaction between Lats2 and c-Abl.
Figure 6.
c-Abl SH3 mutants are resistant to inhibition by Lats2. (a) Mutation of c-Abl at W118 impairs binding to Lats2. HEK293 cells were transfected with the constructs indicated, with 0.25, 0.5, and 1 μg of HA-Lats2 plasmids used. Flag-c-Abl constructs were immunoprecipitated and IP and total extracts were analyzed by immunoblotting. Quantification of the amount of HA-Lats2 that co-immunoprecipitated, normalized to the amount of immunoprecipitated c-Abl, is presented. (b,c) c-Abl T197A and W118A mutants are not inhibited by Lats2. HEK293 cells were transfected with plasmids expressing the proteins indicated. (b) Extracts were analyzed by immunoblotting. (c) The ratio of c-Abl autophosphorylation in the presence of Lats2 versus control (black bars), and the ratio of p73 pTyr99 in the presence of Lats2 versus control (gray bars) is presented. The level of c-Abl autophosphorylation was normalized to the amount of total c-Abl, and the level of phosphorylated p73 was normalized to total p73. Ratios were calculated from three independent experiments. The dashed line represents a ratio of 1, where phosphorylation of c-Abl and p73 is the same in the presence or absence of Lats2. Ratios less than 1 indicate inhibition of phosphorylation in the presence of Lats2
Hippo pathway inhibits apoptosis at high cell density through inhibition of c-Abl by Lats2
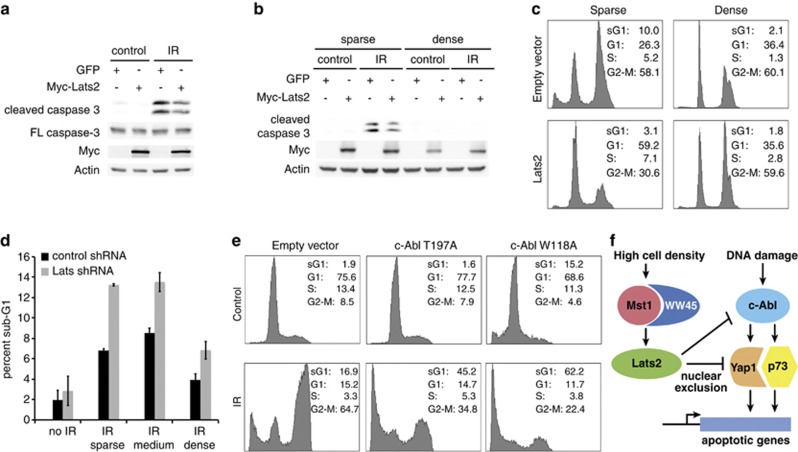
Our initial results indicated that high cell density inhibited apoptosis, and we hypothesized that this inhibition is mediated by the Hippo pathway, and specifically, through the downstream kinase Lats2. Thus, our model predicts that overexpression of Lats2 in sparse cells should reduce DNA damage-induced apoptosis, and conversely, knockdown of Lats2 should enable apoptosis in densely plated cells. To challenge this prediction, we analyzed the effect of overexpression or knockdown of Lats2 on DNA damage-induced apoptosis at different cell densities. Overexpression of Lats2 in H1299 and MCF10A cells led to a reduction in DNA damage-induced apoptosis in sparsely plated cells (Figures 7a–c). In contrast, knockdown of Lats2 increased apoptosis, including in densely plated cells (Figure 7d, Supplementary Figure 5a). Knockdown of Lats2 was confirmed by immunoblot and by qPCR (Supplementary Figures 5b and c). This demonstrates that Lats2 is involved in the inhibition of DNA damage-induced apoptosis.
Figure 7.
Lats2 regulates DNA damage-induced apoptosis. (a-c) Overexpression of Lats2 inhibits DNA damage-induced apoptosis in sparse cells. (a) H1299 cells stably overexpressing myc-Lats2 or GFP control were plated sparse, and were irradiated at 25 Gy. Cells were harvested 2 days post-IR, and were analyzed by immunoblotting. (b) MCF10A cells stably overexpressing myc-Lats2 or GFP control were plated at low and high density, and were irradiated at 20 Gy. Cells were harvested 62 h post-IR, and analyzed as in (a). (c) MCF10A cells stably overexpressing myc-Lats2 or control were plated and irradiated as in (b). Cells were harvested for FACS analysis 120 h post-IR. Percentages of cells in the different fractions (sub-G1, G1, S, and G2-M) are indicated. (d) Knockdown of Lats2 leads to increase in DNA damage-induced apoptosis. H1299 cells stably expressing shRNA to Lats2 or control were plated at different densities, and were irradiated at 25 Gy. Cells were harvested for FACS analysis 24 h post-IR. The percentage of cells in the sub-G1 fraction for control (black bars) and Lats2 knockdown (gray bars) is shown. (e) Expression of c-Abl T197A and W118A mutants rescues DNA damage-induced apoptosis at high cell density. H1299 cells stably overexpressing full-length c-Abl W118A or T197A mutants, or the empty plasmid were plated dense and irradiated at 25 Gy. Cells were analyzed by FACS 72 h post-IR, as in (c). (f) High cell density inhibits DNA damage-induced apoptosis through inhibition of c-Abl by the Hippo pathway; a model. DNA damage induces activation of c-Abl, which phosphorylates Yap and p73, leading to transcription of pro-apoptotic genes. At high cell density, the Hippo pathway is activated, leading to the activation of Mst and WW45. Mst and WW45 in turn activate Lats, which inhibits Yap and c-Abl. Inhibition of c-Abl prevents the phosphorylation of Yap and p73, which inhibits apoptosis
Our results have shown that Lats2 inhibits c-Abl activity. Previous studies, and our results (Figures 1d–f and Supplementary Figures 1a and b) have shown a dependence on c-Abl for mediating DNA damage-induced apoptosis. Thus, our model predicts that expression of c-Abl mutants that are not inhibited by Lats2 could enable apoptosis of cells at high cell density. To test this, H1299 cells were transduced to overexpress wild-type c-Abl, the W118A and T197A mutants, or pBabe empty vector. DNA damage-induced apoptosis was inhibited at high cell density in the control or wild-type c-Abl expressing cells, but apoptosis was induced by IR at high cell density in the cells expressing c-Abl W118A or T198A, which are not inhibited by Lats2 (Figure 7e and Supplementary Figure 6a, expression of constructs shown in Supplementary Figure 6b) Taken together, these results show that the Hippo pathway inhibits DNA damage-induced apoptosis at high cell density, and that this is mediated through inhibition of c-Abl by Lats2 (Figure 7f).
Discussion
Our findings show that inhibition of DNA damage-induced apoptosis at high cell density is mediated through the inhibition of the tyrosine kinase c-Abl, an important inducer of cell death under DNA damage, by the Hippo pathway kinase Lats2 (Figure 7f). Hippo pathway inhibition of DNA damage-induced apoptosis appears to be a robust process, and therefore we would predict that the Hippo pathway may also be inhibiting other pro-apoptotic factors in order to achieve the effect. Nevertheless, the fact that manipulation of Lats2 or c-Abl alone was able to induce or reduce the inhibition indicates that they have an important role in the process. Overexpression of Lats2 in sparse cells reduced DNA damage-induced apoptosis, and knockdown of Lats2 increased DNA damage-induced apoptosis at high cell density, despite the fact that other Hippo pathway elements (such as Lats1 or other pathway components) may also be involved. Furthermore, expression of c-Abl mutants resistant to Lats inhibition restored apoptosis at high cell density. In contrast, apoptosis could not be rescued at high cell density by overexpressing nuclear Yap, indicating that cytoplasmic sequestration of Yap likely contributes to, but is not sufficient for the prevention of apoptosis. c-Abl is required for p73-dependent apoptosis, through direct phosphorylation of p7318, 19, 20 and through phosphorylation of Yap.21 While c-Abl does not directly phosphorylate p53,37 it does lead to increased p53 levels and activity through the phosphorylation of the p53 ubiquitin E3 ligase Mdm2 and the related inhibitor of p53 Mdmx.30, 31 Thus, by inhibiting c-Abl, the Hippo pathway targets both p73- and p53-dependent apoptosis.
The fact that densely plated cells are radio- and chemoresistant was previously noted, but the underlying molecular mechanisms remained obscure. A previous study3 showed that apoptotic activation of p53 is suppressed in densely plated cells, but the mechanism was not resolved. Previous studies have demonstrated differential p53 activation depending on cell type and developmental state. Following whole-body irradiation, p53 accumulates and causes apoptosis in certain cell types, such as thymocytes, but not in others, such as hepatocytes.38 Other studies have demonstrated differences in p53 accumulation and apoptosis depending on developmental state and cell type.39, 40, 41, 42 The general finding is that maximal induction of p53 is found in less-differentiated cells.40 In a study that examined radiation-induced expression of p53 in the mouse intestine,42 it was found that the highest p53 levels are at the base of the crypt in the small intestine in the stem cell population, with high levels also found in the proliferating transit cell population. Low levels are found in the differentiated cells of the villi. This study showed that the cells expressing high levels of p53 undergo apoptosis, whereas the cells expressing low p53 express the inducer of cell-cycle arrest, p21. Interestingly, this pattern of p53 response to DNA damage correlates with Hippo pathway activation in the intestinal crypt. Nuclear Yap (indicating inactive Hippo) is found in the progenitor/stem cell compartment, however, as the cells differentiate, the Hippo pathway is activated and Yap is reduced.43 Thus, there is a correlation between Hippo activation in differentiated cells and lack of DNA damage-induced apoptosis.
Our findings predict that other damage-induced cell fate pathways, such as senescence, should be favored in cells with active Hippo pathway, where apoptosis is suppressed. Recent studies have shown links between the Hippo pathway and the pRB (retinoblastoma) pathway in the context of cellular senescence.44, 45, 46, 47 The latter studies show a role for Lats2 in mediating repression of E2F target genes by pRB in senescence. These studies induced senescence through oncogene expression or serum withdrawal. It will be interesting to determine whether DNA damage-induced senescence also depends upon Lats2.
The Hippo pathway inhibits proliferation by keeping the downstream effectors, Yap and Taz, in check. However, it is possible that the Hippo pathway prevents proliferation through other mechanisms as well. Although this study focused on the apoptotic role of c-Abl, c-Abl is also a pro-proliferative factor (reviewed in Sirvent et al.48 and Colicelli49). Activation of c-Abl in the cytoplasm is involved in the response to growth factors50, 51, 52 and F-actin assembly.53 Furthermore, as shown by the example of CML (chronic myeloid leukemia), uncontrolled activation of ABL is tumorigenic.54 Our study shows that Lats2 can inhibit c-Abl. It is possible that this inhibition is relevant to the anti-proliferative effect of Lats at high cell density. In addition, as a tumor suppressor, Lats2 may be one of the controls that normally prevent over-activation of the proto-oncogene c-Abl. It will be of interest to examine tumors with low Lats activity to determine whether c-Abl activity is increased, and whether the tumor depends on c-Abl activity for its growth.
Materials and Methods
Cells and cell culture
The cell lines used were human embryonic kidney cells HEK293 and HEK 293T, the non-transformed human breast epithelial cell line MCF10A, human non–small-cell lung carcinoma H1299 p53-null cells, human osteosarcoma U2OS cells and mouse NIH3T3 cells. Cells were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO (Life Technologies), Grand Island, NY, USA) supplemented with 8% fetal bovine serum (GIBCO), 100 units/ml penicillin, 100 μg/ml streptomycin and cultured at 37 °C in a humidified incubator with 5.6% CO2. H1299 cells were cultured under the same conditions with Roswell Park Memorial Institute (RPMI; GIBCO) medium. MCF10A were grown in DMEM/F12 (Biological Industries, Kibbutz Beit Haemek, Israel) supplemented with 5% donor horse serum (GIBCO), 2 mM glutamine (Biological Industries), 20 ng/ml epidermal growth factor (EGF), 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin (all from Sigma, St Louis, MO, USA), and antibiotics, as above. Plating densities are defined as <40% confluent for ‘sparse' and confluent for ‘dense'. After plating, cells were allowed to attach for at least 16 h before treatment (such as irradiation) or harvesting. Light microscopy photographs of cells were performed using an Olympus IX70 microscope (Olympus, Tokyo, Japan) connected to a DVC camera. Flow cytometry analyses were done as previously described.55 For flow cytometry analysis, at least 20 000 cells were collected for each sample.
Plasmids, transfection and mRNA analysis
The plasmids used were: pCDNA Flag-Yap1,21 pSLX c-Abl WT and pSLX c-Abl Δ1-81, kindly provided by G Superti-Furga.33 pCDNA c-Abl Δ1-81 was sub-cloned from pSLX c-Abl Δ1-81, and Flag-c-Abl Δ1-81 was constructed by inserting a Flag tag at the N-terminus of the construct. pSG5-c-Abl wt and kinase mutant are described in Agami et al.20 The 1b isoform was used for generation of full-length c-Abl constructs, and the numbering of c-Abl residues refers to positions in the 1b isoform. The EcoRI–BamHI (blunted) c-Abl fragment from pSG5-c-Abl wt was sub-cloned into pBABE puro at EcoRI-SalI (blunted) to make pBABE c-Abl used in this project. The T197A, W118A and K290H (kinase-dead) mutations in pCDNA cAbl Δ1-81, pCDNA Flag-c-Abl Δ1-81 and pBABE c-Abl were generated by site-directed mutagenesis. Myc-Lats2 constructs were provided by M Oren and Y Aylon,56 and HA-Lats2, Mst1 and WW45 constructs were provided by XJ Yang. To generate pBabe Flag-Yap, the coding region of human Yap21 was cloned into pBabe puro expression vector with the addition of the Flag tag at the N-terminus. Yap S127A mutant was constructed by site-directed mutagenesis. To generate cell lines stably expressing Abl or Lats2, retrovirus infection was performed by transfecting 293 Phoenix retrovirus packaging cells with pBabe puro c-Abl or pWZL blast Myc-Lats2 or the respective empty vectors. For stable knocking down of Lats2 or c-Abl, 293T cells were transfected with the mission pLKO.1 shRNA vectors targeting Lats2, c-Abl, GFP or harboring the non-targeting sequence (Sigma) together with pLP1, pLP2 and pLP-VSVG packaging plasmids (Invitrogen, Life Technologies, Carlsbad, CA, USA). For knockdown of Yap1, the pRetroSuper plasmid targeting Yap155 was used. Forty-eight hours after transfection, viral supernatant was filtered through a 0.45-μm filter, supplemented with 8 μg/ml polybrene, and used to infect MCF10A or H1299. Twenty-four hours after infection, cells were selected with 2 μg/ml puromycin (Sigma) or 5 μg/ml blasticidin (Calbiochem, Merck Millipore, Billerica, MA, USA) in the culture medium. All transfections were done by the calcium phosphate method as described.55 Total RNA was extracted using the TRI-Reagent solution (Molecular Research Center, Cincinnati, OH, USA), DNase treated using the RNAse-free DNAse set (Qiagen, Hilden, Germany) and then reverse-transcribed by iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Quantitative RT-PCR was performed with SYBR Green PCR Master Mix (Kapa Biosystems, Woburn, MA, USA) using the LightCycler 480 Instrument (Roche Diagnostics, Basel, Switzerland).
Immunoblot and co-immunoprecipitation studies
Immunoblots and IPs were done as previously described.55 The antibodies used were: anti-HA, monoclonal anti-β-actin, anti-phospho-Yap (Tyr357), and anti-FLAG M5 (Sigma); anti-Yap1 H125, anti-c-Abl K12, anti-c-Abl 8E9, anti-full-length caspase-3 (H-277), anti-general phosphotyrosine (PY20), anti-phospho-p73 (Tyr99) (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-p73 BL906 (Bethyl Laboratories, Montgomery, TX, USA); anti-phospho c-Abl (Tyr412) (Abcam, Cambridge, UK); anti-phospho c-Abl (Tyr245), anti-cleaved caspase-3 and anti-phospho-Yap (Ser127) (Cell Signaling, Beverly, MA, USA); anti-PARP (Enzo Life Sciences, Ann Arbor, MI, USA), anti-Lats2 (Novus Biologicals) and anti-phospho-histone H2A.X(Ser139) (Upstate Biotechnology, Lake Placid, NY, USA). Anti-myc monoclonal antibodies were generated by the antibody unit, Weizmann Institute (Rehovot, Israel). Anti-c-Abl phospho Thr197 polyclonal antibody was raised by immunization of rabbits with a synthetic phosphopeptide (CEGRVYHYRIN(pT)ASD), corresponding to c-Abl 1b aa187–200, with the N-terminal cysteine added for conjugation, conjugated to KLH (keyhole limpet hemocyanin). Antibodies were generated and affinity purified by Biomatik (Cambridge, ON, Canada). For IP of HA- and Flag-tagged proteins, anti-Flag M2 agarose and anti-HA agarose (Sigma) were used. For other IP, protein A/G agarose (Santa Cruz Biotechnology) was used. Horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoReasearch Laboratories (West Grove, PA, USA). Enhanced chemiluminescence was performed with the EZ-ECL kit (Biological Industries) and signals were detected by the ImageQuant LAS 4000 (GE Healthcare, Piscataway, NJ, USA) or by exposure to film. Intensities of bands were quantified by the ImageQuant TL software. Quantification of band intensities in numerical or chart form was added to aid in presentation of immunoblot results. For comparison of multiple experiments, values within one experiment were normalized to a standard set at 1. ‘SE' refers to standard error.
Immunofluorescence staining
MCF10A cells stably expressing Flag-Yap were seeded on glass coverslips, γ-irradiated (20 Gy) the next day, and 24 h following IR fixed in 4% paraformaldehyde for 30 min, permeabilized with 0.5% (v/v) Triton X-100 for 25 min, and blocked with FCS containing 10% skim milk and 0.2% Tween 20. Cells were incubated with mouse monoclonal anti-Flag M5 antibody followed by the Cy3-conjugated donkey anti-mouse antibody (Jackson Immunoresearch Laboratories), and the nuclei were stained with Hoechst 33342 (Molecular Probes (Life Technologies), Carlsbad, CA, USA). Images were acquired using Zeiss LSM 710 confocal (Zeiss, Munich, Germany) scanning system using a × 60/1.4 NA oil objective, and processed by Zen 2009 software (Zeiss). Photoshop (Adobe, San Jose, CA, USA) was used to make combinatory figures.
γ-Ray irradiation
Cells were subjected to a γ-radiation in a Millenium 870LC Irradiator with a 137Cs source (Maintenance International Ltd, UK).
In vitro kinase assays
HEK293 cells were transfected separately with Flag-c-Abl constructs or Flag-Yap1, with myc-Lats2 transfected alone or together with untagged Mst1 and WW45, or transfected with control plasmids. Cells were harvested 24–30 h post-transfection, and were lysed in Buffer A (10 mM Tris-HCl pH 7.5, 250 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT, protease inhibitor cocktail (Sigma) and phosphatase inhibitor cocktails II and III (Sigma, for Tyrosine and Serine/Threonine phosphatases). Cells were incubated for 20 min on ice and centrifuged at 13 000g for 10 min. Proteins were immunoprecipitated using anti-Flag agarose beads (Sigma), or anti-myc antibody with Protein A/G sepharose (Santa Cruz). Immunoprecipitates (IPs) were washed four times with Buffer A, then twice with kinase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, and 0.01% Brij 35). Flag-tagged proteins were eluted by incubating beads in kinase buffer with 0.1 mg/ml Flag peptide for 10 min at 30 °C. Flag beads were centrifuged at 3000 × g, and eluted proteins were transferred to another tube. For the assay, eluted Flag-tagged proteins or control were added to the tubes containing the immunoprecipitated control or myc-Lats2 (not eluted from the beads). BSA was added to 200 μg/ml, and ATP to 200 μM. Reactions were incubated at 30 °C for 30 min, and stopped by the addition of Laemmli protein gel sample buffer, and boiling for 1 min. Samples were analyzed by SDS-PAGE and immunoblotting.
Acknowledgments
We thank M Oren and Y Aylon for helpful discussion and for the myc-Lats2 constructs and reagents. We thank XJ Yang for the HA-Lats2, Mst1 and WW45 constructs. We thank G Superti-Furga for the pSLX c-Abl constructs. STI571 was kindly provided by Novartis Pharmaceuticals. We thank R Keshet for providing the pBabe Flag-Yap constructs, and M Elbaz for her technical assistance. This work was supported by grants from the Israel Science Foundation (grant no. 551/11), Israel Cancer Research Fund, the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel's Ministry of Science and Technology (MOST) and from the Minerva Foundation with funding from the Federal German Ministry for Education and Research.
Glossary
- ASPP
apoptosis-stimulating protein of p53
- c-Abl (ABL1)
Abelson tyrosine protein kinase 1
- Arg (ABL2)
Abl-related gene
- EMT
epithelial to mesenchymal transition
- FACS
fluorescence-activated cell sorting
- Lats
large tumor suppressor
- Mdm
mouse double minute
- Mst
mammalian STE20-like protein kinase
- PARP
poly ADP-ribose polymerase
- RASSF
RAS association domain family
- Rb
retinoblastoma
- Taz
transcription coactivator with PDZ-binding motif
- WW45
45 kDa WW domain protein
- Yap
Yes-associated protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by B Zhivotovsky
Supplementary Material
References
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harbor Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar J, Cohen-Noyman E, Geiger B, Oren M. Attenuation of the p53 response to DNA damage by high cell density. Oncogene. 2004;23:2128–2137. doi: 10.1038/sj.onc.1207325. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr., Levrero M, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Levy D, Reuven N, Shaul Y. A regulatory circuit controlling Itch-mediated p73 degradation by Runx. J Biol Chem. 2008;283:27462–27468. doi: 10.1074/jbc.M803941200. [DOI] [PubMed] [Google Scholar]
- Radu M, Chernoff J. The DeMSTification of mammalian Ste20 kinases. Curr Biol. 2009;19:R421–R425. doi: 10.1016/j.cub.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Molecular cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Ofir-Rosenfeld Y, Yabuta N, Lapi E, Nojima H, Lu X, et al. The Lats2 tumor suppressor augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1. Genes Dev. 2010;24:2420–2429. doi: 10.1101/gad.1954410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J. 2002;21:3715–3727. doi: 10.1093/emboj/cdf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman V, Lenos K, Popowicz GM, Silberman I, Grossman T, Marine JC, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem. 2009;284:4031–4039. doi: 10.1074/jbc.M809211200. [DOI] [PubMed] [Google Scholar]
- Brasher BB, Van Etten RA. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- Pluk H, Dorey K, Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasher BB, Roumiantsev S, Van Etten RA. Mutational analysis of the regulatory function of the c-Abl Src homology 3 domain. Oncogene. 2001;20:7744–7752. doi: 10.1038/sj.onc.1204978. [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Ben-Dor I, Shaul Y. c-Abl tyrosine kinase selectively regulates p73 nuclear matrix association. J Biol Chem. 2003;278:34475–34482. doi: 10.1074/jbc.M301051200. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Owens B, Briscoe CV, Thomas DB, Lane DP, Hall PA.Coupling between gamma irradiation, p53 induction and the apoptotic response depends upon cell type in vivo J Cell Sci 1995108Pt 51843–1848. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Hupp TR, Midgley CA, Stuart D, Campbell SJ, Harper A, et al. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- Gottlieb E, Haffner R, King A, Asher G, Gruss P, Lonai P, et al. Transgenic mouse model for studying the transcriptional activity of the p53 protein: age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 1997;16:1381–1390. doi: 10.1093/emboj/16.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova EA, Chernov MV, Franks R, Wang K, Armin G, Zelnick CR, et al. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 1997;16:1391–1400. doi: 10.1093/emboj/16.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JW, Pritchard DM, Hickman JA, Potten CS. Radiation-induced p53 and p21WAF-1/CIP1 expression in the murine intestinal epithelium: apoptosis and cell cycle arrest. Am J Pathol. 1998;153:899–909. doi: 10.1016/S0002-9440(10)65631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Dick FA, Mymryk JS. Sweet DREAMs for Hippo. Genes Dev. 2011;25:889–894. doi: 10.1101/gad.2050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop K, Conery AR, Litovchick L, Decaprio JA, Settleman J, Harlow E, et al. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011;25:814–830. doi: 10.1101/gad.2000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–813. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirvent A, Benistant C, Roche S. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell. 2008;100:617–631. doi: 10.1042/BC20080020. [DOI] [PubMed] [Google Scholar]
- Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan D, Sims JT, Plattner R. Aggressive breast cancer cells are dependent on activated Abl kinases for proliferation, anchorage-independent growth and survival. Oncogene. 2008;27:1095–1105. doi: 10.1038/sj.onc.1210714. [DOI] [PubMed] [Google Scholar]
- Tanos B, Pendergast AM. Abl tyrosine kinase regulates endocytosis of the epidermal growth factor receptor. J Biol Chem. 2006;281:32714–32723. doi: 10.1074/jbc.M603126200. [DOI] [PubMed] [Google Scholar]
- Woodring PJ, Hunter T, Wang JY. Regulation of F-actin-dependent processes by the Abl family of tyrosine kinases. J Cell Sci. 2003;116 (Pt 13:2613–2626. doi: 10.1242/jcs.00622. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. Chronic myeloid leukemia. New Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell and Differentiation. 2007;14:743–751. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- Okada N, Yabuta N, Suzuki H, Aylon Y, Oren M, Nojima H. A novel Chk1/2-Lats2-14-3-3 signaling pathway regulates P-body formation in response to UV damage. J Cell Sci. 2011;124 (Pt 1:57–67. doi: 10.1242/jcs.072918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.