Abstract
Bcl-2-interacting mediator of cell death (Bim) is a pro-apoptotic B-cell lymphoma 2 family member implicated in numerous apoptotic stimuli. In particular, Bim is required for cell death mediated by antimitotic agents, however, mitotic regulation of Bim remains poorly understood. Here, we show that the major splice variant of Bim, BimEL, is regulated during mitosis by the Aurora A kinase and protein phosphatase 2A (PP2A). We observed that BimEL is phosphorylated by Aurora A early in mitosis and reversed by PP2A after mitotic exit. Aurora A phosphorylation stimulated binding of BimEL to the F-box protein beta-transducin repeat containing E3 ubiquitin protein ligase and promoted ubiquitination and degradation of BimEL. These findings describe a novel mechanism by which the oncogenic kinase Aurora A promotes cell survival during mitosis by downregulating proapoptotic signals. Notably, we observed that knockdown of Bim significantly increased resistance of cells to the Aurora A inhibitor MLN8054. Inhibitors of Aurora A are currently under investigation as cancer chemotherapeutics and our findings suggest that efficacy of this class of drugs may function in part by enhancing apoptotic activity of BimEL.
Keywords: Bim, apoptosis, cell cycle, mitosis, aurora kinase
Apoptosis is triggered by many types of cell stress, including DNA damage, oncogene activation, virus infection and metabolic stress. Induction of apoptosis is regulated by the family of B-cell lymphoma 2 (Bcl-2) proteins that have either pro- or anti-apoptotic activities. The family is characterized by the presence of up to four Bcl-2 homology (BH1-4) domains. A subset of the pro-apoptotic Bcl-2 family members containing only the BH3 domain is induced in response to wide range of cell stresses. BH3-only proteins induce apoptosis by activating the pro-apoptotic Bcl-2 family members, Bcl-2-associated X protein (Bax) and Bcl-2 homologous antagonist/killer (Bak), and by antagonizing anti-apoptotic members such as Bcl-2 and induced myeloid leukemia cell differentiation protein 1.1
The Bcl-2-interacting mediator of cell death (Bim) protein is a BH3-only member of the Bcl-2 family that is an important initiator of apoptosis in lymphocytes in response to growth factor withdrawal2, 3, 4 and elimination of autoreactive T cells in the thymus.5 Deletion of a single allele of Bim can accelerate the progression of lymphomas in the Eμ-myc model, suggesting the apoptotic activity of Bim has tumor-suppressor activity in vivo.6 Expression of Bim is regulated by several transcriptional and post-transcriptional mechanisms. At the transcriptional level, the promoter of the Bim gene is activated by stress-inducible transcription factors including activator protein 1 and forkhead box protein O.4, 7, 8 Post-transcriptional mechanisms that have been shown to control Bim expression levels include both mRNA stability9 and protein stability.10, 11, 12
Signaling downstream of receptor tyrosine kinases (RTKs) including epidermal growth factor receptor induce the proteasomal degradation of Bim.13, 14, 15, 16 There are three splice variants of Bim: BimEL, BimL and BimS. The longest form, BimEL, is generally the most highly expressed among the variants.17 Phosphorylation by extracellular signal-regulated kinase 1/2 (Erk1/2) on Serine-69 (S69) of human BimEL has been shown to induce degradation of the protein.12, 18, 19 More recently, phosphorylation at S69 was shown to promote phosphorylation at additional sites on Serines 93/94/98 (S93/94/98) by ribosomal S6 kinase (Rsk1/2) within a conserved phosphodegron motif recognized by the F-box protein beta-transducin repeat containing E3 ubiquitin protein ligase (βTrCP).20
In addition to being phosphorylated in response to growth factor signaling, BimEL is also phosphorylated during mitosis. Early reports suggested that this phosphorylation is catalyzed by ERK1/221 and ERK522 most likely as a pro-survival signal. However, more recent studies have found that mitotic phosphorylation of BimEL is not dependent on ERK1/2 or ERK5 signaling;23 but rather upon cyclin-dependent kinase 1 (CDK1) activity24, 25 and is associated with the poly-ubiquitination and proteasome-dependent turnover of BimEL.25 Although there is now considerable evidence that Bim levels are regulated by phosphorylation and proteasome-mediated proteolysis during mitosis, the mechanism remains unknown.
In the current study, we show that stability of BimEL is regulated during progression through mitosis. BimEL is targeted for proteasome-mediated degradation following phosphorylation of the protein on a conserved phosphodegron recognized by βTrCP1. We further show that the kinase responsible for the mitotic phosphorylation of BimEl at S93/94/98 is Aurora A kinase. As Aurora A inhibitors are being pursued in the clinic as anti-mitotic cancer drugs, these findings have implications in the design of anti-mitotic cancer therapies.
Results
BimEL is phosphorylated and targeted for degradation at mitosis
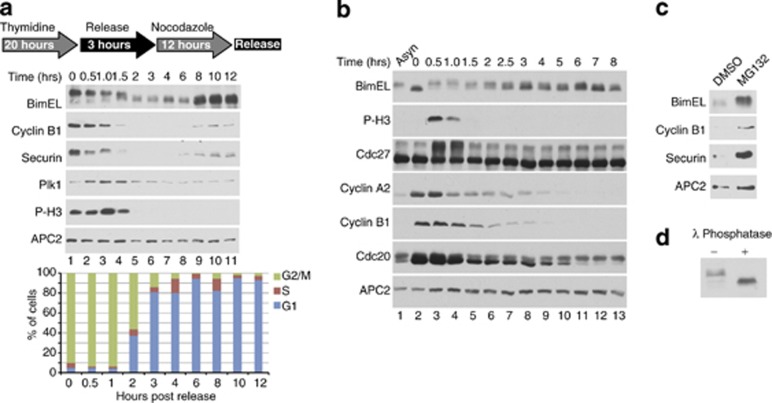
In order to examine expression of BimEL during mitosis, HeLa cells were synchronized using thymidine (Thy) followed by nocodozole (Noc) and analyzed by immunoblot at various times post release. Figure 1a shows that after release of cells from Thy/Noc a slower migrating form of BimEL was apparent, which gradually shifted downward as cells progressed through mitosis (Figure 1a, lanes 1–6). Figure 1b shows that synchronization of HeLa cells using the cdk1 inhibitor RO3306 obtained similar results as with Thy/Noc with one important difference. RO3306 arrests cells at G2/M and BimEL in RO3306-arrested cells shows that the protein is present in a faster migrating form that immediately shifts upward upon release from the drug and entry into mitosis (compare Figure 1b lanes 2 and 3). Interestingly, as cells progressed through mitosis BimEL levels were observed to decrease and reach a low point approximately 90 min following release (compare Figure 1a, lanes 1 and 6). In order to demonstrate that loss of BimEL expression was at the level of protein degradation, HeLa cells were synchronized as in Figure 1a and then treated with the proteasome inhibitor MG132 for 90 min post release. Figure 1c shows that MG132 treatment resulted in the accumulation of BimEL in mitosis and that the stabilized form of BimEL appears as a slower migrating band. In order to demonstrate that the slower migrating form of BimEL was due to phosphorylation, cell extracts were treated with λ phosphatase. Figure 1d shows that phosphatase treatment resulted in a downward shift of BimEL, demonstrating that the protein is phosphorylated in mitosis. These data suggest that BimEL is regulated during mitosis by coupled phosphorylation and proteolytic steps.
Figure 1.
BimEL is phosphorylated and targeted for degradation at mitosis. (a) Immnoblot analysis of HeLa cells synchronized using the thymidine/nocodozole (Thy/Noc) protocol as shown (top). Whole-cell extracts were analyzed for expression of BimEL following release from Thy/Noc for the indicated times. Immunoblots for three other proteins known to be degraded during mitosis, cyclin B1, Securin, and Polo-like kinase 1 (Plk-1) were performed for comparison. Anti-phospho histone H3 (P-H3) is included as a marker for onset of mitosis and APC2 as a loading control. Flow cytometry was used to confirm cell cycle phase (bottom). (b) Immunoblot analysis of HeLa cells synchronized with the cdk1 inhibitor RO3306 and released for the indicated length of time or left asynchronous (Asyn). Whole-cell extracts were analyzed by immunoblot analysis for expression of BimEL and three other proteins known to be degraded during mitosis: cyclin B1, cyclin A2, and cdc20. (c) Immunoblot of HeLa cells synchonized as in a and released for 90 min in the presence of DMSO (vehicle control) or MG132. (d) Immunoblot analysis of endogenous BimEL in whole-cell extracts treated with λ phosphatase (+) or control (−) prepared from nocodozole-treated HeLa cells
BimEL is phosphorylated on Serine 69 and Serines 93/94/98 and degraded at mitosis in a mechanism independent of the spindle assembly checkpoint (SAC)
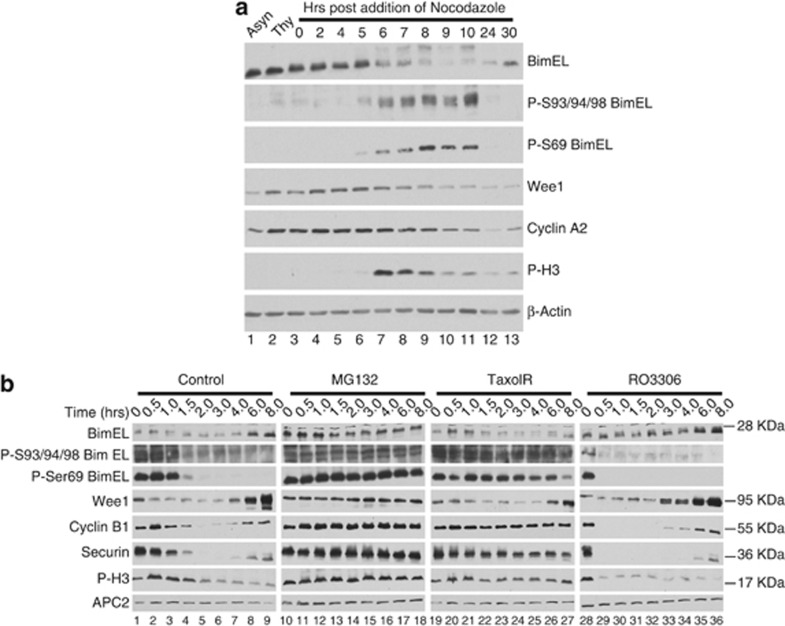
As BimEL degradation occurred concomitantly with phosphorylation, we examined two phosphorylation sites on BimEL previously shown to regulate proteolysis. Erk 1 and 2 were shown to phosphorylate BimEL on S6910, 12, 18 and promote degradation and phosphorylation on S93/94/98 has been shown to induce degradation through the Skp1-Cullin1-F-Box (SCF) complex.20 In order to assess the importance of these sites for BimEL degradation in mitosis, phosphospecific antibodies were used to monitor these modifications during mitosis. 293T cells were synchronized as shown in Figure 1a with the exception that cells were then left in Noc. Figure 2a (lanes 6–10) shows that BimEL becomes phosphorylated on S69 and S93/94/98 in mitosis and is concomitant with degradation of BimEL.
Figure 2.
BimEL is phosphorylated and degraded at mitosis in a mechanism independent of the spindle assembly checkpoint. (a) Immunoblot analysis of 293T cells synchronized as in Figure 1a with the exception that cells were left in nocodozole for the indicated times. Immnoblots were performed for total BimEL and phosphorylated BimEL on serine-69 (P-S69) and serines 93/94/98 (P-S93/94/98). Immunoblots for two other proteins known to be degraded during mitosis, wee1 and cyclin A2, were performed. Anti-phospho histone H3 (P-H3) is included as a marker for onset of mitosis and β-actin as loading control. (b) Immunoblot analysis of HeLa cells synchronized at mitosis as in Figure 1a and released into four different conditions: normal media (control), MG132 (10 μM), Taxol (100 nM), and RO3306 (10 μM). Whole-cell extracts were prepared at the indicated times post release. Immnoblots were performed for total BimEL, P-S69, and P-S93/94/98. Immunoblots for three other proteins known to be degraded during mitosis, Wee1, cyclin B1, and Securin, were performed for comparison. P-H3 is included as a marker for onset of mitosis and APC2 as loading control
To further investigate the mitotic degradation of BimEL, HeLa cells were synchronized as depicted in Figure 1a and released into four different conditions, MG132, Taxol, RO3306, or control (Figure 2b). As expected, release of cells into MG132 stabilized levels of BimEL as well as several other proteins known to undergo proteolysis during mitosis including: cyclin B, securin, and Wee1 (Figure 2b, lanes 10–18). Taxol interferes with tubulin dynamics during mitosis and thereby activates the SAC. SAC activation then inhibits the anaphase-promoting complex/cyclosome (APC/C), which normally mediates degradation of key mitotic substrates such as securin and cyclin B. Figure 2b (lanes 19–27) show that taxol treatment stabilized the APC/C substrates, cyclin B and securin, but did not affect stability of BimEL or Wee1, suggesting that degradation of BimEL was APC/C independent. Interestingly, treatment with RO3306 stabilized BimEL (compare lanes 1–9 with lanes 28–36 of Figure 2b), suggesting that cdk1 activity was required for degradation. Immunoblots using phosphospecific antibodies against S69 and S93/94/98 confirmed that phosphorylation at both of these sites occurred in mitosis and decreased as cell progressed through mitosis (Figure 2b, lanes 1–9). Furthermore, whereas treatment with MG132 and Taxol maintained phosphorylation of both S69 and S93/94/98, RO3306 treatment prevented phosphorylation at these sites. Similar results were obtained using 293T cells (Supplementary Figure 1). Taken together, these data show that phosphorylation sites on BimEL that regulate stability are phosphorylated during mitosis in a cdk1-dependent mechanism. In addition, degradation of BimEL during mitosis was not affected by the SAC, suggesting that the APC/C is not involved in this process.
BimEL is ubiquitinated at mitosis and requires phosphorylation on Serine 93/94/98
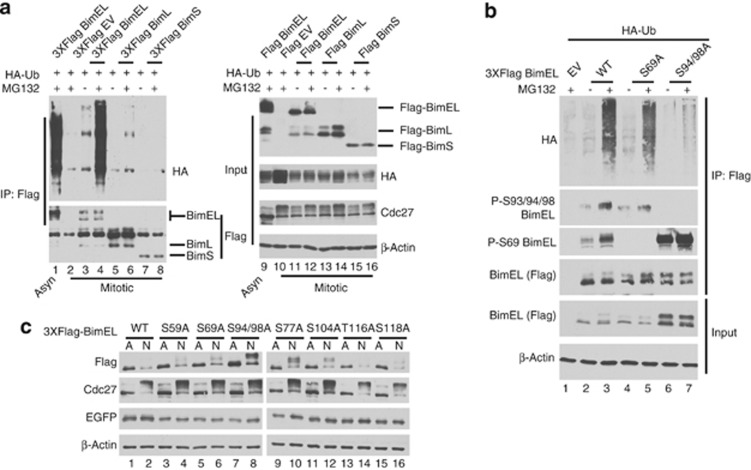
In order to determine whether degradation of BimEL during mitosis is ubiquitination dependent, FLAG-tagged Bim was co-transfected with HA-tagged Ubiqutin (Ub). Bim conjugated with Ub was detected by immunoprecipitation (IP) of Bim using α-FLAG antibody followed by immunoblot for HA. Figure 3a (lane 4) shows that BimEL was ubiquitinated during mitosis and displayed the characteristic laddering in the presence of MG132. Transfection of the two other major splice variants, BimL (lane 6) and BimS (lane 8), did not result in appreciable ubiquitination, suggesting that the degron required for ubiquitination was specific to BimEL. Phosphorylation sites at S69 and S93/94/98 have been previously shown to affect protein stability and are located in the N-terminal region specific to BimEL. We therefore tested ubiquitination of BimEL mutants in which these phosphorylation sites were converted to alanine. Figure 3b shows that mutation of S69 to alanine (S69A) did not affect ubiquitination (lane 5), however, mutation of S94/98 (S94/98A) was severely defective (lane 7). In order to determine whether phosphorylation of BimEL was essential for degradation at mitosis, 293T cells were transfected with FLAG-tagged BimEL and immunoblot performed under asynchronous or mitotic conditions. Figure 3c shows that, as expected, wild-type Flag-BimEL displayed greatly reduced steady-state levels during mitosis (compare lanes 1 and 2). However, the S94/98A mutant was stabilized at mitosis (compare lanes 7 and 8) and the stabilized form of the protein appeared to migrate slower. Serine 104 of BimEL has been previously shown to be phosphorylated by cdk1 during mitosis,24 but mutation of this site had no effect on stability (lanes 11 and 12). The S94/98A mutation is clearly still phosphorylated at other sites in mitosis as the mutated protein migrates slower in mitosis and may be due to phosphorylation by cdk1 as previously reported.24, 25 We further mutated four other potential cdk phosphorylation sites on BimEL but none displayed major effects on mitotic stability of the protein. These data indicate that BimEL is actively ubiquitinated and targeted for degradation during mitosis using a mechanism that requires phosphorylation at S93/94/98.
Figure 3.
BimEL is ubiquitinated at mitosis and requires phosphorylation on Serine 93/94/98. (a) 293T cells were co-transfected with HA-tagged Ubiquitin (Ub) and Flag-tagged BimEL, BimL BimS, or empty vector control (EV). Cells were synchronized as shown in Figure 1a. Asynchrous (Asyn) cells treated with PMA and MG132 was used as a positive control for BimEL ubiqutination (lanes 1 and 9). Cells were treated with MG132 (+) or vehicle control (−) as indicated. (Left) Anti-Flag immunoprecipitation (IP) followed by immunoblot for HA and FLAG epitopes. (Right) Immunoblots were performed on cell extracts used as input for IPs using anti-Flag and HA antibodies to detect Bim and Ub expression. Anti-Cdc27 immunoblot was used to confirm mitotic state of cells and β-actin was monitored as loading control. (b) Nocodozole-treated 293T cells were co-transfected with HA-Ub and Flag-tagged wild-type (WT) BimEL, S69A, S94/98A, or EV control. Anti-Flag IP was performed as in a. Resulting IPs were analyzed by immunoblot using anti-Flag, anti-HA, and the phoshospecific BimEL antibodies (P-S93/94/98 and P-S69). Cells were treated with MG132 (+) or vehicle control (−) as indicated. Immunoblots were performed on cell extracts used as input for IPs using anti-Flag to confirm expression of BimEL constructs and β-actin was monitored as loading control. (c) 293T cells were transfected with WT BimEL or phosphorylation site mutants as indicated. In all cases, EGFP was cotransfected to monitor transfection effeciency. Cell extracts were prepared from either asynchronous (A) or after nocodozole (N; 100 ng/ml) treatment. Immunblots were performed on WCE using anti-Flag to detect BimEL, β-actin as loading control, EGFP to monitor transfection efficiency, and cdc27 to confirm the mitotic state of the cell
Phosphorylation on Serine 93/94/98 of BimEL creates a binding site for βTrCP1
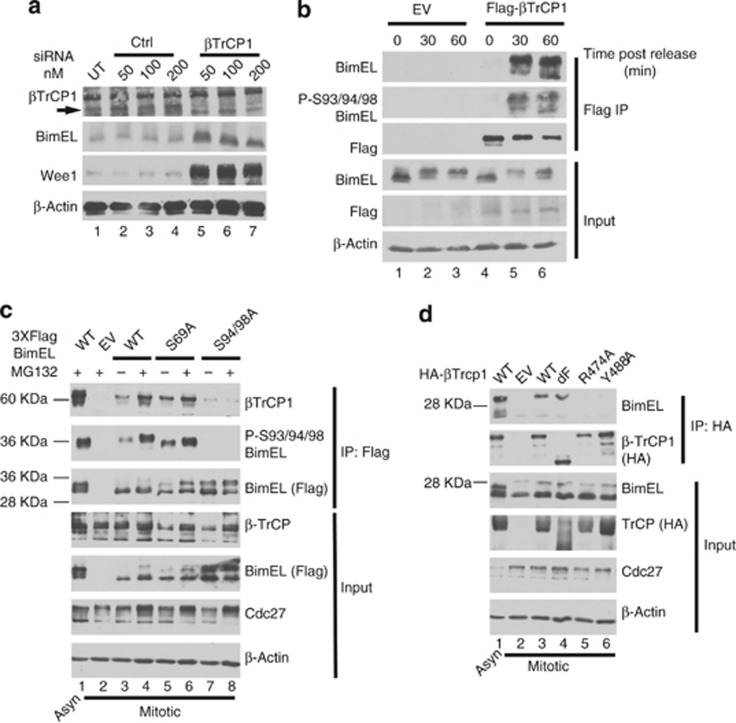
Treatment with phorbol ester has been shown to induce phosphorylation of BimEL on S93/94/98, which creates a binding site for βTrCP1 and targets BimEL for degradation through the SCF complex.20 βTrCP1 is also known to mediate degradation of Wee1 during mitosis.26 As our results in Figure 2 indicated that degradation of BimEL occurred with similar timing and properties as Wee1, we tested if βTrCP1 can also target BimEL during mitosis. Figure 4a shows that siRNA knockdown of βTrCP1 resulted in elevated levels of both Wee1 and BimEL. As binding of βTrCP1 requires phosphorylation of the degron, we determined if binding of BimEL to βTrCP1 occurred specifically during mitosis. Figure 4b shows that HeLa cells synchronized in G2/M using RO3306 showed a dramatic increase in Serine 93/94/98 phosphorylation at 30 min post-release. Furthermore, interaction between BimEL and βTrCP1 occurred only when S93/94/98 phosphorylation was apparent (Figure 4b, lanes 5 and 6). We then determined if S93/94/98 is essential for βTrCP1 interaction with BimEL during mitosis. Figure 4c (lanes 7 and 8) shows that a point mutation of Serines 94/98 to alanines (S94/98A) dramatically reduced binding to βTrCP1. Point mutation of S69 to alanine (S69A), however, had no effect on βTrCP1 binding (lanes 5 and 6). Furthermore, two well-characterized point mutations of βTrCP1, R474A and Y488A,27 that are known to be defective for substrate binding failed to interact to BimEL at mitosis (Figure 4d). Deletion of the F-box, which mediates interaction of βTrCP1 with the SCF was not required for binding to BimEL. Taken together, these data show that activation of the βTrCP1 phosphodegron on BimEL occurs during mitosis and targets the protein for degradation by the SCF complex.
Figure 4.
Phosphorylation on Serine 93/94/98 of BimEL creates a binding site for βTrCP1. (a) Knockdown of βTrCP1 expression in HeLa cells using siRNA at the indicated concentrations. A non-silencing siRNA (Ctl) was used as control. Immunoblot analysis was performed against endogenous BimEL and Wee1. β-Actin was monitored as loading control. (b) HeLa cells were transfected with Flag-tagged βTrCP1 or empty vector control (EV). Cells were synchronized in mitosis as in Figure 1a except that cells were released in the presence of MG132. Cell lysates prepared at the indicated times post release. Immunoprecipitation (IP) was performed using anti-Flag antibody and immunoblot analysis used to detect endogenous BimEL, Flag (βTrCP1), and phosphorylated BimEL on serines 93/94/98 (P-S93/94/98). Cell extracts used as input for IP were analyzed by immunoblot to measure expression of total BimEL, Flag (βTrCP1), and β-Actin as loading control. (c) 293T cells were transfected with EV, wild-type (WT) BimEL or phosphorylation site mutants as indicated. Cells were synchronized in mitosis as in Figure 1a. Asynchrous (Asyn) cells treated with PMA and MG132 was used as a positive control (lane 1). Left: asynchronous (Asyn). IP was performed using anti-FLAG antibody and immunoblot analysis used to detect endogenous βTrCP1, Flag (BimEL), and phosphorylated BimEL on serines 93/94/98 (P-S93/94/98). Cell extracts used as input for IP were analyzed by immunoblot to measure expression of total βTrCP1 and Flag (BimEL). Cdc27 immunoblots were used to confirm mitotic state of cells and β-actin as loading control. (d) 293T cells were transfected with EV, wild-type (WT) βTrCP1 or the indicated βTrCP1 mutant. Cells were synchronized as in Figure 1a. Asynchronous cells transfected with HA bTrCP1 WT and treated with PMA and MG132 for 3 h was used as a positive control (lane 1). IP was performed using anti-HA antibody and immunoblot analysis used to detect IPed BimEL and HA (βTrCP1). Cell extracts used as input for IPs were analyzed by immunoblot to measure expression of total Bim and HA (βTrCP1). Cdc27 immunoblots were used to confirm mitotic state of cells and β-actin as loading control
Phosphatase protein phosphatase 2A (PP2A) dephosphorylates BimEL on S93/94/98 and prevents degradation
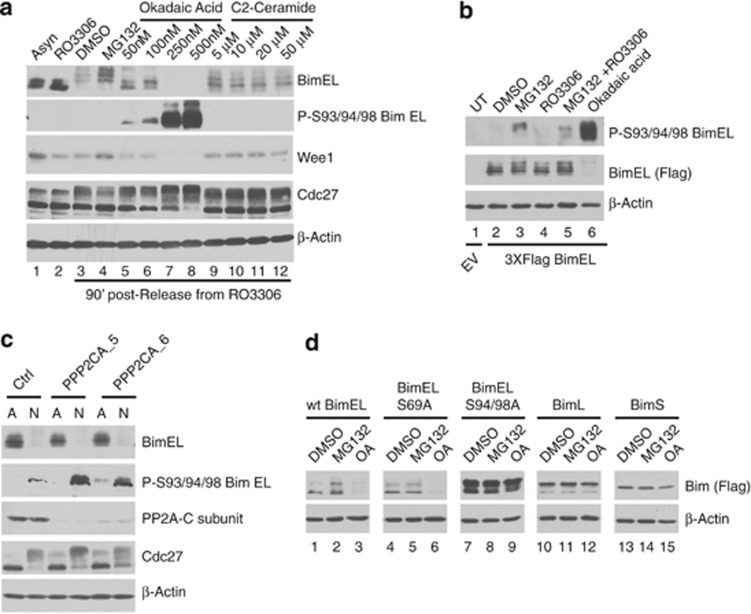
BimEL was previously shown to be dephosphorylated by the phosphatase PP2A and thereby inhibit the ubiquitination and degradation of BimEL, although the mechanism of this effect is not known.28 As shown in Figure 1 BimEL is rapidly phosphorylated as cells enter mitosis and is then gradually dephosphorylated after mitotic exit when BimEL levels increase. We therefore asked if PP2A dephosphorylates S93/94/98 during mitosis and if this regulates mitotic stability of BimEL. Figure 5a (lanes 5–8) shows that treatment of Thy/Noc synchronized HeLa cells with okadaic acid (OA), a specific inhibitor of PP2A, resulted in a dramatic dose-dependent increase of S93/94/98 phosphorylation on endogenous BimEL. Interestingly, levels of BimEL were dramatically reduced in the presence of OA. Similar results were obtained by IP of 293T cells transfected with FLAG-BimEL followed by western blot to detect S93/94/98 (Figure 5b). Conversely, addition of the PP2A activator, C2-Ceremide, resulted in the stabilization of BimEL relative to vehicle controls (Figure 5a, compare lanes 3 and 9). Figure 5c shows that knockdown of PP2A using siRNAs targeting the catalytic subunit also resulted in elevated phosphorylation on S93/94/98. In order to determine if phosphorylation of S93/94/98 was required for OA-induced degradation, wild-type BimEL or phosphorylation site point mutants were transfected into 293T cells synchronized using Thy/Noc. Figure 5d shows that although wild-type BimEL and the mutant at S69 were still degraded in response to OA treatment, mutation of S93/94/98 rendered the protein resistant. Similarly, the two other major splice variants of Bim, BimL and BimS, that lack S93/94/98 were also unaffected by OA (Figure 5d, lanes 10–15). Taken together, these data show that BimEL phosphorylation and stability are dynamically regulated during mitosis with PP2A-mediated dephosphorylation of S93/94/98 stabilizing levels of BimEL after mitosis.
Figure 5.
PP2A dephosphorylates BimEL on Ser93/94/98 and prevents degradation. (a) HeLa cells were synchronized in G2/M with RO3306 and released into fresh media plus vechicle (DMSO), MG132, OA, or C2-ceramide at the indicated concentrations and harvested 90 min post release. Whole-cell extracts were analyzed by immunoblot to detect endogenous BimEL, phosphorylated BimEL on serines 93/94/98 (P-S93/94/98), and Wee1. Cdc27 immunoblots were used to confirm mitotic state of cells and β-actin as loading control. (b) 293T cells were transfected with wild-type (WT) Flag-tagged BimEL or left untransfected (UT). Cells were synchronized in mitosis as in Figure 1 and released into vehicle control (DMSO), MG132 (10 μM), RO3306 (9 μM), and OA (500 nM) as indicated. IP was performed using anti-FLAG antibody (BimEL) and immunoblot analysis used to detect IPed BimEL (FLAG) and P-S93/94/98. β-Actin immunoblot was used as the input control for IPs. (c) Knockdown of the PP2A catalytic subunit in HeLa cells using two different siRNA PPP2CA_5 and PPP2CA_6. A non-silencing siRNA (Ctl) was used as control. Immunoblot analysis was performed against endogenous BimEL, phosphorylated BimEL (P-S93/94/98), and the PP2A-C subunit. Cdc27 immunoblots were used to confirm mitotic state of cells and β-actin as loading control. (d) 293T cells were transfected with WT Flag-tagged BimEL, BimL, BimS, or phosphorylation site mutants as indicated. Transfected cells were synchronized in mitosis as shown in Figure 1a and treated with vehicle control (DMSO), MG132 (10 μM), or OA (500nM) as indicated. Whole-cell extracts were analyzed by immunblot using anti-Flag to detect Bim and β-actin as loading control
Phosphorylation of the βTrCP1 degron in BimEL is mediated by Aurora A kinase during mitosis
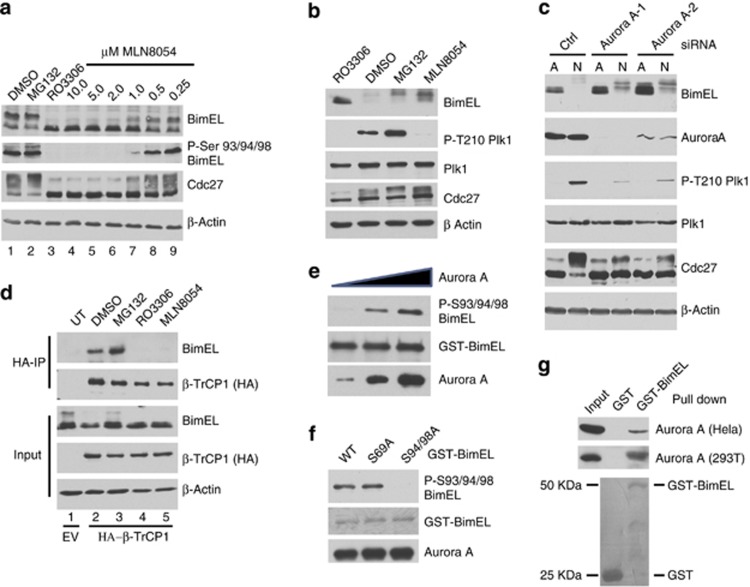
Previous studies have shown that the βTrCP1 phosphodegron at S93/94/98 is phosphorylated by Rsk1/2 in response to treatment with phorbol ester. Rsk1/2 is activated downstream of receptor tyrosine signaling but does not explain how this site becomes phosphorylated during mitosis in a cdk1-dependent manner. Moreover, we show in Supplementary Figure 1 that addition of the Rsk1/2 inhibitor BI-D1870 did not significantly affect phosphorylation of S94/98 during mitosis. Inhibition of several other kinases downstream of RTKs also had little or no effect on S93/94/98 (Supplementary Figure 2), suggesting that RTK signaling is dispensable for S93/94/98 phosphorylation during mitosis. Although the cdk1 inhibitor RO3306 completely prevented S93/94/98 phosphorylation, these sites do not have the required context of a cdk1 site suggesting another mitotic kinase downstream of cdk1 is required. The S93/94/98 phosphodegron includes the amino-acid sequence 91-RSSSG-95, which contains the consensus for Aurora A kinase (R-X-X-S/T-φ, where φ is any hydrophobic amino acid29). Intriguingly, mitotic activation of Aurora A has been shown to be dependent upon cdk1 activity.30 We therefore determined if S93/94/98 is phosphorylated during mitosis by Aurora A. 293T cells were arrested using Thy/Noc and released into increasing concentrations of the Aurora kinase inhibitor MLN8054. Figure 6a (lanes 4–9) shows that MLN8054 prevented mitotic phosphorylation of S93/94/98 in a dose-dependent manner. Furthermore, Figure 6b shows that addition of MLN8054 to mitotic HeLa cells stabilized levels of BimEL during mitosis. Under these conditions treatment with MLN8054 also prevented phosphorylation on a well-characterized Aurora A site on Polo-like kinase (T210).31, 32 Genetic inhibition of Aurora A activity using siRNAs also resulted in elevated levels of BimEL in mitosis (Figure 6c). As phosphorylation of the βTrCP1 degron is required for interaction with BimEL, we determined if MLN8054 affects this interaction. Figure 6d shows that BimEL failed to co-IP with βTrCP1 in cells treated with MLN805.
Figure 6.
Phosphorylation of the BimEL degron is mediated by Aurora A kinase during mitosis. (a) 293T cells were synchronized in mitosis as in Figure 1a and released into vehicle control (DMSO), MG132, RO3306, or the indicated concentration of MLN8054. Whole-cell extracts were analyzed by immunoblot to detect endogenous BimEL and phosphorylated BimEL on serines 93/94/98 (P-S93/94/98). Cdc27 immunoblots were used to monitor mitotic state of cells and β-actin as loading control. (b) HeLa cells were synchronized in G2/M with RO3306 and released into fresh media plus vechicle (DMSO), MG132 (10 μM), or MLN8054 (5 μM) and harvested 90 min post release. Whole-cell extracts were analyzed by immunoblot to detect endogenous BimEL. P-T210 Plk1 and total Plk1 were used as controls to confirm the downstream effects of the aurora A inhibitor MLN8054. (c) Knockdown of Aurora A expression in HeLa cells using two different siRNA. A non-silencing siRNA (Ctl) was used as control. Immunoblot analysis was performed against endogenous BimEL and Aurora A. P-T210 Plk1 and total Plk1 were used as a control to confirm the downstream effects of the knockdown. Cdc27 immunoblots were used to confirm mitotic state of cells and β-actin as loading control. (d) 293T cells were transfected with HA-tagged βTrCP1 or empty vector control (EV). Cells were synchronized in mitosis as in Figure 1a and released into vehicle control (DMSO), MG132 (10 μM), RO3306 (9 μM), MLN3306 (5 μM), or left untreated (UT). Immunoprecipitation (IP) was performed using anti-HA antibody. Immunoblot analysis was perfored on IPs to detect endogenous BimEL and transfected βTrCP1 (HA). Cell extracts used as input for IP were analyzed by immunoblot to measure expression of total BimEL, HA (βTrCP1), and β-Actin as loading control. (e and f) Recombinant GST-BimEL or phosphorylation site mutants produced in E. coli were incubated with increasing amounts of purified Aurora A kinase (e) or 500 ng purified Aurora A kinase (f). Reactions were analyzed by immunoblot to detect GST-Bim, P-S93/94/98 GST-BimEL, and Aurora A. (g) Recombinant GST or GST-BimEL produced in E. coli were incubated with either mitotic Hela or 293T extracts prepared using the standard thymidine/nocodazole protocol. Immunoblot analysis was performed to detect co-precipetated endogenous Aurora A. Mitotic Hela or 293T cell extracts were used as an input to measure expression of endogenous Aurora A. Image of Red Ponceau-stained membrane used to show GST proteins
In order to determine if Aurora A is able to directly phosphorylate BimEL, we combined purified Aurora A and glutathione S-transferase (GST)-BimEL in vitro. Figure 6e shows that Aurora A was indeed able to directly phosphorylate BimEL at S93/94/98 in vitro. It has been suggested that phosphorylation at S69 is required for effective phosphorylation at S93/94/98, however, a S69A mutant was still effectively phosphorylated by Aurora A in vitro (Figure 6f). BimEL has been shown to form complexes with kinases that phosphorylate it.19, 20, 24 Similarly, in Figure 6g we show that that GST-BimEL was able to pull down Aurora A from mitotic extracts derived from HeLa or 293T cells.
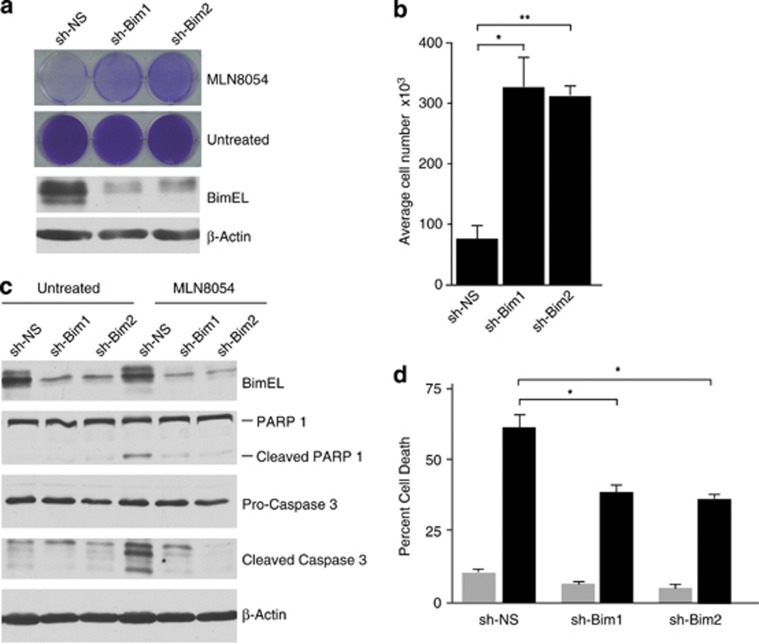
Numerous clinical trials are ongoing testing Aurora kinase inhibitors as cancer therapeutics.33 As our data show that Aurora A phosphorylates BimEL on S93/94/98, triggering the proteolysis of the protein, the efficacy of Aurora A inhibitors may be dependent on stabilization of BimEL. In order to test this hypothesis, we determined if levels of Bim expression could affect response to MLN8054. Figure 7a shows that HeLa cells with stable knockdown of Bim were significantly more resistant to treatment with MLN805 relative to non-silencing control (sh-NS). Quantitation of this effect shows that after 3 days of treatment with MLN8054, HeLa cultures with Bim knockdown survived the treatment and contained nearly fourfold the number of cells compared with sh-NS (Figure 7b). MLN8054 has been previously shown to induce apoptosis in cancer cells34 so we therefore determined if treatment with MLN8054 induces death in a Bim-dependent mechanism. Figure 7c shows that treatment of sh-NS cells with MLN8054 resulted in caspase-3 and poly (ADP-ribose) polymerase 1 (PARP1) cleavage that was inhibited by Bim knockdown. Moreover, NLN8054 treatment of sh-NS cells induced significant levels of apoptosis that was partially reversed in cell lines with stable knockdown of Bim (Figure 7d).
Figure 7.
Knockdown of Bim protects cells from apoptosis induced by Aurora A inhibition. (a) Bim expression was knocked down in HeLa cells using two different lentivirus delivered shRNAs (sh-Bim1 and shBim2). A non-silencing shRNA (sh-NS) was used as control. (Top) Knockdown and control cells were treated with 5 μM MLN8054 for 72 h. Resulting tissue culture plates were stained with crystal violet. (Bottom) Immunoblots of whole-cell extracts confirming knockdown in Bim shRNA-expressing cells. β-Actin is shown as loading control. (b) Quantitation of cell numbers from experiment in a. Student's t-test was used to determine statistical significance (*P≤0.05, **P≤0.01). (c) Control (sh-NS) or sh-Bim1 and sh-Bim2 HeLa cells were treated with 5 μM MLN8054 for 48 h. Immunoblots of whole-cell extracts were performed to confirm Bim knockdown and cleaved PARP-1 and Caspase 3 were used to demonstrate apoptotic effect of MLN8054. β-Actin is shown as a loading control. (d) HeLa cells were treated as described for c, stained with AnnexinV and 7AAD, and analyzed by flow cytometry. The percentage of AnnexinV-positive cells is indicated for each treatment. Grey bars indicate untreated and black bars are treatment with MLN8054
Taken together, these data show that S93/94/98 on BimEL is phosphorylated by Aurora A, creating a binding site for βTrCP1 and inducing degradation of the protein. Furthermore, inhibition of cell growth and induction of apoptosis by Aurora A inhibition is at least partially dependent upon Bim expression levels.
Discussion
The levels of Bim protein are tightly regulated at the level of protein stability and are responsive to many cues including growth factor and stress signaling. Previous studies have suggested that Bim is regulated by phosphorylation during mitosis but the molecular details of these observations have not been elucidated. In the current study, we show that proteolysis of BimEL is induced during mitosis following phosphorylation by Aurora A kinase. Phosphorylation of BimEL on the phosphodegron at S93/94/98 creates a binding site for the F-box protein βTrCP. This mechanism of BimEL inactivation is similar to that observed after growth factor stimulation with the exception that the βTrCP phosphodegron is activated by Rsk1/2 downstream of growth factors rather than Aurora A.20 We observed that the phosphorylation of S93/94/98 is dynamic and reversed by the activity of PP2A leading to the stabilization of BimEL proteins levels after mitosis. Aurora A is itself degraded late in mitosis by the APC/C and may also explain how BimEL levels return to normal state.35, 36
The biological functions of BimEL degradation during mitosis need further study but are likely due to the mechanism of how the apoptotic activity of Bim is normally kept in check. In healthy cells, BimEL remains sequestered to the microtubule network, which neutralizes its apoptotic activity.37, 38 Stress-activated signaling such as the c-Jun N-terminal kinase 1 pathway can phosphorylate Bim, resulting in release from microtubules and induction of apoptosis.39 As the onset of mitosis triggers the global disassembly of the microtubule network in preparation for assembly of the mitotic spindle, this would also result in widespread release of sequestered Bim. Induction of Bim degradation may therefore be a mechanism to cope with the sudden release of cytoskeleton-associated Bim during mitosis.
Downregulation of Bim and other core components of the apoptotic machinery may be a general property of cells entering mitosis. Caspase-2, -8, and -9 are phosphorylated by Cdk1/Cyclin B1 during mitosis and these modifications inhibit pro-apoptotic activity.40, 41, 42 A general feature of mitotic cells is the loss of attachment to extracellular matrix as the cytoskeleton is remodeled in preparation for spindle formation. When anchorage-dependent cells remain in a detached state for extended periods, they undergo a process termed anoikis.43 Interestingly, Bim is required for cell death induced by anoikis,44 suggesting that downregulation of Bim during mitosis may prevent cell death in response to extracellular matrix detachment.
Our data show that Aurora A phosphorylates BimEL during mitosis and promotes degradation of the protein. The Aurora kinase family consists of three paralogues that control key events during mitosis including mitotic entry, centrosome regulation and cytokinesis.45 The Aurora kinases become activated during mitosis and are subsequently shut down late in mitosis by being targeted for degradation by the APC/C. As would be expected, we observe that levels of BimEL begin to increase late in mitosis when activity of Aurora A is inhibited. Aurora A and B are each required for multiple events during mitosis and inhibition of either activity through chemical or genetic methods leads to severe mitotic defects.45 Although we cannot completely rule out the possibility that Aurora B may also target BimEL, several lines of evidence suggest that Aurora A is the major kinase-targeting BimEL in mitosis. First, we observe that mitotic phosphorylation of BimEL on S93/94/98 is dependent upon Cdk1/Cyclin B. Only Aurora A has been shown to definitively require Cdk1/Cyclin B for activation during mitosis.30 Second, Aurora A has been shown to act as a bona fide oncogene and induction of Bim degradation is consistent with this activity. Third, we observe that the Aurora A inhibitor MLN8054 prevents mitotic phosphorylation of BimEL on S93/94/98 and stabilizes the protein. MLN8054 inhibits Aurora A with 40-fold selectivity over Aurora B,46 indicating that Aurora A is the main kinase being affected in our experiments.
Aurora A is overexpressed in a wide range of tumor types and has been demonstrated to enhance tumor cell growth both in vivo and in vitro.45 Aurora A has been shown to downregulate several tumor-suppressor mechanisms. The p53 tumor suppressor was shown to be phosphorylated by Aurora A leading to destabilization and degradation of the p53 protein in a mechanism reminiscent of what we have observed for BimEL.47, 48 Similarly, Aurora A was shown to phosphorylate BRCA1 and promote G2/M cell cycle progression.49 Thus, in addition to the wide range of substrates targeted by Aurora A regulating chromosome and spindle dynamics during mitosis, it can also phosphorylate and deactivate tumor-suppressor pathways including Bim.
Numerous Aurora kinase inhibitors are currently in clinical trials for a wide range of cancer types.33 Our results suggest that Bim expression is required for efficacy of Aurora A inhibition. Several reports have shown that pharmacologic inhibition of Aurora A leads to apoptosis and our results suggest that this may due in part to mitotic stabilization of Bim.34, 50, 51 Interestingly, Li et al. conclusively demonstrated that apoptosis induced by Aurora A inhibition occurs through the intrinsic mitochondrial pathway requiring the activity of either Bax or Bak.34 This is consistent with the activation of a BH3-only protein such as Bim, which functions in part by activating pro-apoptotic Bcl-2 family members Bax and Bak.1 As Aurora A is upregulated in many tumor types, it may contribute to the transformed phenotype by maintaining low levels of BimEL.
Materials and Methods
Antibodies
Mouse monclonal antibodies to the following proteins were purchased from the indicated manufacturers and used for immunoblotting according to standard protocols: Cdc20, cyclin B1, cyclin A2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Cdc27 (BD Biosciences, San Jose, CA, USA), EGFP (Clontech, Mountain View, CA, USA), Plk-1 (Life Technologies, Burlington, ON, Canada), Securin (Abcam, Toronto, ON, Canada), anti-Flag M2, (Sigma-Aldrich, Oakville, ON, Canada), anti-HA (Covance, Princeton, NJ, USA), PP2A-C subunit (Cell Signaling, Danvers, MA, USA), Phospho Ser-Thr-Pro MPM2 (EMD Millipore, Billerica, MA, USA). The following rabbit polyclonal antibodies were purchased from the indicated manufacturers: Bim (EMD Millipore), β-actin (Sigma-Aldrich), APC2 (Biolegend, San Diego, CA, USA), Phospho BimEL(S69), and Phospho-(Ser) CDK substrates (Cell Signaling) PARP-1 and Caspase 3 (Santa Cruz Biotechnology). Rabbit monoclonal antibodies were from Cell Signaling (βTrCP, Phospho Plk-1-T210, and Aurora A). The phospho Histone H3 Ser10 rat polyclonal was purchased from Sigma-Aldrich. Anti-Phospho-BimEL (S93/94/98) has been described previously20 and was a kind gift from Dr. Michele Pagano. Mouse TrueBlot, secondary anti-mouse IgG antibody, was from eBioscience (San Diego, CA, USA).
Plasmids and cloning
The three major splice variants of Bim were obtained by RT-PCR from form HeLa total RNA and cloned into p3XFLAG-myc-CMV-26 Expression Vector (Sigma-Aldrich). Stop sequences were generated to exclude the C-terminal myc tag. HA βTrCP1 was a kind gift from Dr. Michele Pagano. pRK5-HA-Ubiquitin-WT was from Addgene (Cambridge, MA, USA) (plasmid 17608).52 BimEL and βTrCP1 point mutations were generated using site-directed mutagenesis. To generate the GST-BimEL fusion protein, BimEL WT or phosphorylation mutants were sub-cloned into pGEX-6P1 (GE Healthcare, Baie-D'Urfé, QC, Canada). All constructs were verified by restriction digest analysis and DNA sequencing.
Cells and drug treatments
Hela and 293T cells were obtained from ATCC. All cells were maintained in Dulbecco's modified Eagle medium (Wisent Inc., St-Bruno, QC, Canada) supplemented with 10% fetal bovine serum (HyClone/Thermo Scientific) and 0.1% gentamicin (Wisent Inc.). Mitotic cells were prepared by incubating with 2 mM Thy for 20 h followed by 3 h release into normal medium before the addition of 100 ng/ml Noc (Sigma). Rounded cells were collected after treatment for 12 h. RO3306 (Enzo Life Sciences, Farmingdale, NY, USA) synchronization was performed by treating Hela cells for 19 h with 9 μM RO3306. The drug was then washed out and cells released into fresh media for the specified time points. For proteasome inhibition studies, cells were incubated with 10 μM MG132 (EMD Millipore) for 3 h before further treatments. Other drugs that were used were: PMA (10 ng/ml) for 3 h, MLN8054 (Selleck Chemicals, Houston, TX, USA), OA (EMD Millipore), C2-Ceramide (Sigma-Aldrich) for the indicated times and concentrations. Treatment of cell extracts with λ phosphatase (New England BioLabs, Whitby, ON, Canada) was performed by adding 50 μg of total protein extract to the 1 × phosphatase buffer (supplied by manufacturer) supplemented with 2 mM MnCl2 and incubated with 10 U of λ phosphatase at 30 °C for 30 min. The reactions were stopped by the addition of 1 × Laemmli sample buffer.
Extract preparation, IP, and immunoblotting
For HA βTrCP 1 or 3XFlag BimEL IPs, transfected 293T or Hela cells were lysed in 1 ml IP lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40, 1 mM EDTA, 1 mM Na3VO4, 50 mM NaF, 10 mM β-glycerophosphate, 1 mM PMSF and protease inhibitor (Roche Diagnostics, Laval, QC, Canada) per 107 cells on ice for 20 min. Cell debris was pelleted by centrifugation at maximum speed for 15 min at 4 °C. The supernatant was then incubated with 30 μl anti-Flag or anti-HA affinity gel (Sigma-Aldrich) for 2 h at 4 °C. The beads were then pelleted and washed four times with the same buffer. Following the washes, the beads were pelleted and resuspended in 1 × Laemmli sample buffer, boiled for 5 min, and stored at −20 °C until further use. Immunoblots were performed using standard protocols and visualized by enhanced chemiluminescence (Perkin Elmer, Waltham, MA, USA).
In vivo ubiquitination assay
In vivo ubiqutination assays were performed as previously described.53 Briefly, 293T cells were co-transfected with pRK5-HA ubiquitin (Addgene) and 3Xflag Bim constructs using the Lipofectamine 2000 protocol. 24 h post-transfection the standard Thy/Noc protocol was applied. Drug treatment was performed in the last 2–3 h of the experiment. Cells were washed twice with PBS before re-suspending the in 4–5 volumes of 50 mM Tris-HCl, pH 7.4, 0.25 M NaCl, 0.1% Triton X-100, 1 mM EDTA, 50 mM NaF, 1 mM DTT, 0.1 mM Na3VO4, protease inhibitor tablet and 2 mM N-ethylmaleimide. The regular IP protocol was followed as described above.
Gene silencing using RNAi and shRNA
The siRNA oligonucleotide sequence targeting βTRCP1/2 has been described previously54 and corresponds to nt 515–535 of human βTRCP1 and nt 262–282 of human βTRCP2. The following siRNA sequences were used to knock down Aurora A:55 5′-725AUGCCCUGUCUUACUGUCA743-3′ and 5′-155AUUCUUCCCAGCGCGUUCC173-3′. To knock down PP2A catalytic subunit, we used the following siRNA sequences Hs_PPP2CA_5 5′-ATGGAACTTGACGATACTCTA-3′ and Hs_PPP2CA_6 5′-CAAACAATCATTGGAGCTTAA-3′.56 The sh-NS siRNA sequence used was 5′-AATTCTCCGAACGTGTCACGT-3′ (Qiagen, Germantown, MD, USA). Cells were transfected with 50 nM siRNAs using the Lipofectamine 2000 protocol as provided by the manufacturer. Cells were subjected to two rounds of transfection (24 and 48 h after plating) before Thy/Noc or RO3306 synchronization as described above. Lentivirus shRNA constructs targeting Bim were obtained from Sigma (Mission shRNA pLKO.1-puro, NM_138621.x-541s1c1 and NM_138621.x-522s1c1). SHC002 MISSION Non-Target shRNA was used as the sh-NS. Lentivirus were packaged as previously described.57
Flow cytometry
Cells were collected by scraping, washed twice with cold PBS and resupended in Nuclear Isolation and Stain (NIM-DAPI 10) from Beckman Coulter Canada (Mississauga, ON, Canada) to obtain an average concentration of 1 × 106 cells/ml. DNA content was determined by flow cytometry on a Cell Lab Quanta SC flow cytometer (Beckman Coulter, Canada). Apoptosis assays were performed in Hela cells treated with MLN8054 or vehicle control (DMSO). Cells were collected by scraping and washed twice with ice-cold PBS then resupended in 100 μl of AnnexinV binding buffer (2.5 mM CaCl2, 140 mM NaCl, 7.75 mM HEPES (pH 7.4)) and then incubated with PE-AnnexinV (BD Biosciences) and 7-amino-actinomycin D (7AAD; A.G. Scientific, San Diego, CA, USA) according to the manufacturing protocol. Cells were then analyzed on the same Flow cytometer as above for staining with PE-Annexin alone or PE-Annexin plus 7AAD staining. At least 10 000 cells were analyzed per sample.
In vitro kinase and in vitro binding assays
Five hundred nanograms of recombinant GST-BimEL (WT or mutant) purified from E. coli were incubated at 30 °C for 30 min with 500 ng recombinant His Aurora A (Millipore) in a 30 μl of kinase buffer (50 mM Tris pH 7.5, 10 mM MgCl2, 1 mM DTT) plus 100 μM ATP. Reaction was stopped by the addition of 4 × laemmli buffer. For in vitro binding assays, 5 mg of Hela or 293T lysates were incubated with 1 μg of GST alone or GST BimEL for 2 h at 4 °C. Samples were then processed as described above for IPs.
Acknowledgments
This work was supported by a grant (MOP-179122) from the Canadian Institutes of Health Research (CIHR) and a Discovery grant from the Natural Sciences and Engineering Research Council (NSERC to JGT). JGT is a CIHR New Investigator and FRSQ Research Scholar. MM-K was supported by a studentship from the Egyptian Ministry of Higher Education and Scientific Research.
Glossary
- APC/C
anaphase promoting complex/cyclosome
- Bak
Bcl-2 homologous antagonist/killer
- Bax
Bcl-2-associated X protein
- Bcl2
B-cell lymphoma 2
- Bim
Bcl-2-interacting mediator of cell death
- βTrCP
beta-transducin repeat containing E3 ubiquitin protein ligase
- CDK1
cyclin-dependent kinase 1
- ERK
extracellular signal-regulated kinase
- GST
glutathione S-transferase
- IP
immunoprecipitation
- NOC
nocodazole
- sh-NS
non-silencing control
- OA
okadaic acid
- PARP-1
poly (ADP-ribose) polymerase 1
- Plk-1
Polo-like kinase 1
- PP2A
protein phosphatase 2A
- RSK
ribosomal S6 kinase
- RTK
receptor tyrosine kinase
- SAC
spindle assembly checkpoint
- SCF
Skp1-Cullin1-F-Box
- Thy
Thymidine
- Ub
ubiquitin
- 7AAD
7-amino-actinomycin D
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Happo L, Strasser A, Cory S. BH3-only proteins in apoptosis at a glance. J Cell Sci. 2012;125 (Pt 5:1081–1087. doi: 10.1242/jcs.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science (New York, NY) 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, et al. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron. 2001;29:615–628. doi: 10.1016/s0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29:629–643. doi: 10.1016/s0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- Matsui H, Asou H, Inaba T. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell. 2007;25:99–112. doi: 10.1016/j.molcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003;22:6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations PLoS Med 200741669–1679.discussion 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A.Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics PLoS Med 200741681–1689.discussion 1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–11875. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly LA, Cullen L, Visvader J, Lindeman GJ, Print C, Bath ML, et al. The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am J Pathol. 2000;157:449–461. doi: 10.1016/S0002-9440(10)64557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Barrett T, Flavell RA, Davis RJ. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell. 2008;30:415–425. doi: 10.1016/j.molcel.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Howes E, Balmanno K, Cook SJ. Extracellular signal-regulated kinases 1/2 are serum-stimulated "Bim(EL) kinases" that bind to the BH3-only protein Bim(EL) causing its phosphorylation and turnover. J Biol Chem. 2004;279:8837–8847. doi: 10.1074/jbc.M311578200. [DOI] [PubMed] [Google Scholar]
- Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, et al. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graos M, Almeida AD, Chatterjee S. Growth-factor-dependent phosphorylation of Bim n mitosis. Biochem J. 2005;388 (Pt 1:185–194. doi: 10.1042/BJ20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girio A, Montero JC, Pandiella A, Chatterjee S. Erk5 is activated and acts as a survival factor in mitosis. Cell Signal. 2007;19:1964–1972. doi: 10.1016/j.cellsig.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Czernick M, Rieger A, Goping IS. Bim is reversibly phosphorylated but plays a limited role in paclitaxel cytotoxicity of breast cancer cell lines. Biochem Biophys Res Commun. 2009;379:145–150. doi: 10.1016/j.bbrc.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Mac Fhearraigh S, Mc Gee MM. Cyclin B1 interacts with the BH3-only protein Bim and mediates its phosphorylation by Cdk1 during mitosis. Cell Cycle. 2011;10:3886–3896. doi: 10.4161/cc.10.22.18020. [DOI] [PubMed] [Google Scholar]
- Gilley R, Lochhead PA, Balmanno K, Oxley D, Clark J, Cook SJ. CDK1, not ERK1/2 or ERK5, is required for mitotic phosphorylation of BIMEL. Cell Signal. 2012;24:170–180. doi: 10.1016/j.cellsig.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, III, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Van Horn RD, Chu S, Fan L, Yin T, Du J, Beckmann R, et al. Cdk1 activity is required for mitotic activation of aurora A during G2/M transition of human cells. J Biol Chem. 2010;285:21849–21857. doi: 10.1074/jbc.M110.141010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science (New York, NY) 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss DS, Beijnen JH, Schellens JH. Clinical experience with aurora kinase inhibitors: a review. Oncologist. 2009;14:780–793. doi: 10.1634/theoncologist.2009-0019. [DOI] [PubMed] [Google Scholar]
- Li M, Jung A, Ganswindt U, Marini P, Friedl A, Daniel PT, et al. Aurora kinase inhibitor ZM447439 induces apoptosis via mitochondrial pathways. Biochem Pharmacol. 2010;79:122–129. doi: 10.1016/j.bcp.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Castro A, Arlot-Bonnemains Y, Vigneron S, Labbe JC, Prigent C, Lorca T. APC/Fizzy-related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi S, Honda K, Sugiura K, Yamaguchi A, Furukawa K, Urano T. Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS Lett. 2002;519:59–65. doi: 10.1016/s0014-5793(02)02711-4. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- Day CL, Puthalakath H, Skea G, Strasser A, Barsukov I, Lian LY, et al. Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands. Biocheml J. 2004;377 (Pt 3:597–605. doi: 10.1042/BJ20031251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Davis RJ. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci USA. 2003;100:2432–2437. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, et al. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J. 2009;28:3216–3227. doi: 10.1038/emboj.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan LA, Clarke PR. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell. 2007;26:301–310. doi: 10.1016/j.molcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Matthess Y, Raab M, Sanhaji M, Lavrik IN, Strebhardt K. Cdk1/cyclin B1 controls Fas-mediated apoptosis by regulating caspase-8 activity. Mol Cell Biol. 2010;30:5726–5740. doi: 10.1128/MCB.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP. Anoikis. Cell Death Differ. 2005;12 (Suppl 2:1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochimica et biophysica acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci USA. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, et al. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- Ouchi M, Fujiuchi N, Sasai K, Katayama H, Minamishima YA, Ongusaha PP, et al. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- Huang XF, Luo SK, Xu J, Li J, Xu DR, Wang LH, et al. Aurora kinase inhibitory VX-680 increases Bax/Bcl-2 ratio and induces apoptosis in Aurora-A-high acute myeloid leukemia. Blood. 2008;111:2854–2865. doi: 10.1182/blood-2007-07-099325. [DOI] [PubMed] [Google Scholar]
- Lin ZZ, Hsu HC, Hsu CH, Yeh PY, Huang CY, Huang YF, et al. The Aurora kinase inhibitor VE-465 has anticancer effects in pre-clinical studies of human hepatocellular carcinoma. J Hepatol. 2009;50:518–527. doi: 10.1016/j.jhep.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Pagano M. Experimental tests to definitively determine ubiquitylation of a substrate. Methods Enzymol. 2005;399:249–266. doi: 10.1016/S0076-6879(05)99017-4. [DOI] [PubMed] [Google Scholar]
- Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, et al. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA (New York, NY) 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.