Abstract
Actively transcribed genes are enriched with the histone variant H3.3. Although H3.3 deposition has been linked to transcription, mechanisms controlling this process remain elusive. We investigated the role of the histone methyltransferase Wolf–Hirschhorn syndrome candidate 1 (WHSC1) (NSD2/MMSET) in H3.3 deposition into interferon (IFN) response genes. IFN treatment triggered robust H3.3 incorporation into activated genes, which continued even after cessation of transcription. Likewise, UV radiation caused H3.3 deposition in UV-activated genes. However, in Whsc1−/− cells IFN- or UV-triggered H3.3 deposition was absent, along with a marked reduction in IFN- or UV-induced transcription. We found that WHSC1 interacted with the bromodomain protein 4 (BRD4) and the positive transcription elongation factor b (P-TEFb) and facilitated transcriptional elongation. WHSC1 also associated with HIRA, the H3.3-specific histone chaperone, independent of BRD4 and P-TEFb. WHSC1 and HIRA co-occupied IFN-stimulated genes and supported prolonged H3.3 incorporation, leaving a lasting transcriptional mark. Our results reveal a previously unrecognized role of WHSC1, which links transcriptional elongation and H3.3 deposition into activated genes through two molecularly distinct pathways.
Keywords: BRD4, HIRA, histone H3.3 exchange, transcription elongation, WHSC1
Introduction
The histone variant, H3.3, is expressed throughout the cell cycle and deposited along with transcription in a replication-independent manner (Elsaesser et al, 2010; Talbert and Henikoff, 2010). Replication-independent histone replacement is a process conserved from yeast to humans, and is likely of great biological importance. H3.3 generally localizes to the regions of active gene expression, and is suggested to leave an epigenetic mark relevant to transcriptional memory (Hake and Allis, 2006; Henikoff, 2008; Ng and Gurdon, 2008). Genome-wide analyses showed that H3.3 is enriched in transcriptionally active genes, in the promoters near the transcription start sites (TSSs), as well as the gene body and transcription end sites (TESs) in mammalian and Drosophila cells (Mito et al, 2005; Wirbelauer et al, 2005; Daury et al, 2006; Jin et al, 2009; Goldberg et al, 2010). H3.3 deposition is induced in some genes upon transcriptional activation (Janicki et al, 2004; Schwartz and Ahmad, 2005; Sutcliffe et al, 2009). However, H3.3 also occupies telomeres and pericentric heterochromatin, indicating its diverse presence and the function beyond transcription (Jin et al, 2009; Drane et al, 2010; Goldberg et al, 2010). Consistent with its assumed broad activities, H3.3 can substitute for the canonical H3.1 in replication-coupled histone deposition, although H3.1 cannot substitute for H3.3 in replication-independent deposition (Ray-Gallet et al, 2011). Further supporting the biological importance of H3.3, mutations in the H3.3A gene and those in the H3.3 deposition pathways were reported in malignant brain tumours (Schwartzentruber et al, 2012; Wu et al, 2012).
H3.3 deposition is mediated by multiple factors, including HIRA, ATRX/DAXX, DEK, and CHD2 (Tagami et al, 2004; Drane et al, 2010; Goldberg et al, 2010; Lewis et al, 2010; Sawatsubashi et al, 2010; Harada et al, 2012). The histone chaperon HIRA plays a pivotal role in H3.3 incorporation in transcriptionally active genes (Goldberg et al, 2010). In agreement with a role in transcription-linked H3.3 deposition, HIRA is bound to both the initiating and elongating forms of RNA polymerase II (Pol II) (Ray-Gallet et al, 2011).
Despite intense efforts towards understanding the process of replication-independent H3.3 deposition, molecular mechanisms underlying the events remain incompletely understood. In this study, we investigated transcription-coupled H3.3 deposition mainly focussing on the interferon (IFN)-stimulated genes (ISGs). We previously reported that IFN treatment triggers rapid H3.3 deposition in ISGs, exhibiting a distinct spatial gradient clearly biased for the TES. Moreover, IFN-induced H3.3 deposition continued well after the cessation of ISG transcription (Tamura et al, 2009). In this system, H3.3 deposition correlated well with the trimethylation of H3K36 (H3K36me3), as it is accumulated in ISGs after IFN treatment with a strong bias towards the TES. H3K36me3 is a mark for active gene expression that increases upon transcriptional activation (Edmunds et al, 2008; Suganuma and Workman, 2011; Wagner and Carpenter, 2012). In yeast, H3K36me3 is mediated by the Set2 methyltransferase (Strahl et al, 2002; Li et al, 2003; Du and Briggs, 2010). Wolf–Hirschhorn syndrome candidate 1 (WHSC1, also known as NSD2 or MMSET) is a putative mammalian Set2 homologue (Stec et al, 1998; Lachner and Jenuwein, 2002). WHSC1 possesses a methyltransferase activity for histone H3K27, H3K36, and H4K20 (Kim et al, 2008; Marango et al, 2008; Kuo et al, 2011; Pei et al, 2011). WHSC1 is associated with diseases affecting growth and development, and plays a role in DNA damage response (Bergemann et al, 2005; Pei et al, 2011). Recently, Nimura et al (2009) generated Whsc1-deficient mice and showed that WHSC1 is essential for embryonic development.
To delineate a possible mechanistic link between H3K36me3 and induced H3.3 deposition, we asked the role of WHSC1 and tested Whsc1−/− mouse embryonic fibroblasts (MEFs) for IFN-induced H3.3 incorporation. We found that both H3.3 deposition and ISG transcription were markedly diminished in Whsc1−/− cells. Immunoprecipitation analysis showed that WHSC1 interacts with the bromodomain protein 4 (BRD4), and is recruited to the ISGs, thus contributing to ISG elongation. BRD4 is a BET family protein that binds to acetylated histones (Dey et al, 2003). BRD4 recruits the positive transcription elongation factor b (P-TEFb) and stimulates transcription elongation from many genes (Jang et al, 2005; Yang et al, 2005; Mochizuki et al, 2008; Hargreaves et al, 2009). After recruitment, WHSC1 interacted with the H3.3-specific chromatin assembly factor HIRA and drove prolonged H3.3 deposition in ISGs well beyond transcription. We found that inhibition of P-TEFb did not completely prevent IFN-induced H3.3 deposition, suggesting that WHSC1 directs P-TEFb-mediated elongation and HIRA-dependent H3.3 deposition through molecularly separable mechanisms.
Results
WHSC1 is required for IFN-induced transcription and H3.3 incorporation
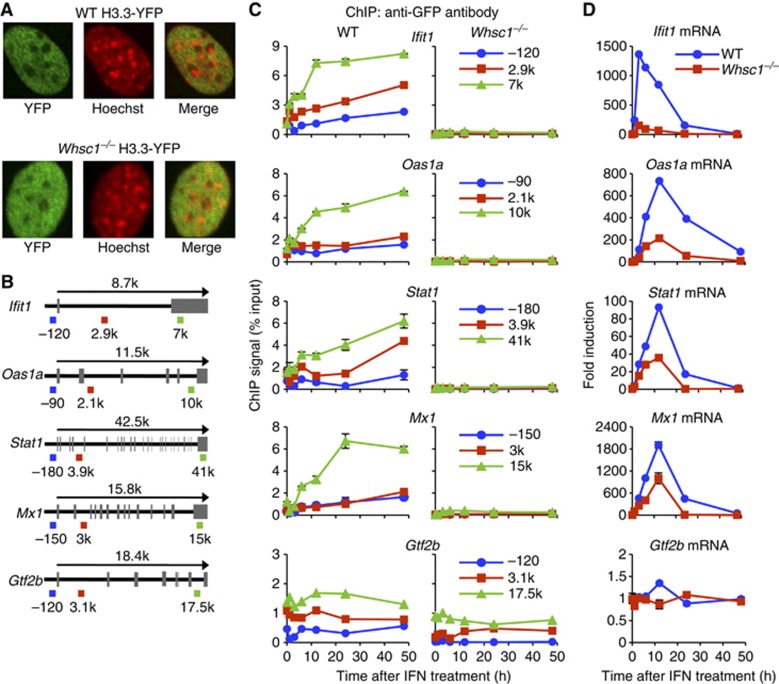
To study the role of WHSC1 in H3.3 deposition, we tested Whsc1−/− and wild-type (WT) MEFs stably expressing H3.3 fused to the yellow fluorescent protein (H3.3-YFP) (Figure 1A). Immunoblot analyses in Supplementary Figure S1A verified that H3.3-YFP was expressed at similar levels as H3.1-YFP, both of which were <10% of total H3 in these cells. As might be expected of the WHSC1 methyltransferase activity, partial reduction in H3K36me3 was observed in Whsc1−/− cells as compared with WT cells. Reduced H3K36me3 expression in Whsc1−/− cells was confirmed at three different extract concentrations (Supplementary Figure S1B). In addition, H3.3-YFP had higher levels of K36me3 than H3.1-YFP in WT cells, consistent with the enrichment of active histone marks in H3.3 (Hake et al, 2006; Loyola et al, 2006). Whsc1−/− and WT cells were stimulated with IFNβ (hereafter IFN) at 100 U/ml, an optimal concentration for ISG induction (Darnell et al, 1994). Chromatin immunoprecipitation (ChIP) assays were performed using anti-GFP antibody to detect H3.3 incorporation in four typical ISGs, Ifit1, Oas1a, Stat1, and Mx1, each at three different regions, near the TSS (blue), a mid coding region (gene body, red), and near or at the TES (green, see gene maps in Figure 1B). In WT cells, H3.3-YFP incorporation began immediately after IFN stimulation in all ISGs, showing a notable TES-preferred accumulation, which lasted for 48 h, long past ISG mRNA expression, consistent with our earlier report (Tamura et al, 2009). In contrast, in Whsc1−/− cells, H3.3-YFP incorporation was virtually undetectable in ISGs at all regions examined (Figure 1C). It should be noted here that H3.3-YFP was expressed at comparable levels in WT and Whsc1−/− cells, and their levels were not altered after IFN treatment (Supplementary Figure S2). ChIP analysis for the total histone H3 revealed similar levels of deposition in WT and Whsc1−/− cells in all ISGs, thus indicating that Whsc1−/− cells are fully competent in general histone deposition, but are selectively deficient in induced H3.3 incorporation (Supplementary Figure S3). It should be also noted that IFN stimulation did not increase H3.3-YFP and total H3 incorporation in Gtf2b, a housekeeping gene not induced by IFN, both in WT and Whsc1−/− cells (Figure 1B; Supplementary Figure S3).
Figure 1.
WHSC1 is required for induction of ISG transcription and H3.3 incorporation. (A) Intranuclear localization of H3.3-YFP was visualized by immunostaining of WT and Whsc1−/− MEFs with anti-GFP antibody (green), counterstained with Hoechst 33342 for DNA (red). (B) Schematic map of ISGs and Gtf2b. The arrow indicates the transcribed region. Exons are marked by grey boxes. Positions of primers used in ChIP analysis are shown underneath and colour-coded. (C) IFN induced H3.3 incorporation. H3.3-YFP expressing WT and Whsc1−/− cells were treated with IFN for 1, 3, 6, 12, 24, and 48 h, and ChIP assays were performed using anti-GFP antibody at the indicated sites of ISGs. Gtf2b was tested as a control. Values represent the average of duplicate determinations ±s.d. (D) Induction of ISG mRNA in the above cells was detected by qRT–PCR, normalized by Gapdh and expressed as fold induction. Values represent the average of two determinations ±s.d.
To correlate IFN-induced H3.3 deposition with ISG transcription, quantitative real-time PCR (qRT-PCR) was performed to measure ISG mRNA levels in WT and Whsc1−/− cells. In WT cells, IFN led to rapid mRNA induction for all ISGs, with a nearly 100- to 2000-fold increase in transcript levels that peaked at 3–12 h followed by a rapid decline at 24 h (blue). In contrast, the ISG mRNA levels were strikingly lower in Whsc1−/− cells (red), whose levels approached only 10–50% of those in WT cells at all time points (Figure 1D). Gtf2b mRNA expression, however, was similar in Whsc1−/− and WT cells. These results indicate that WHSC1 is critically required for IFN-induced transcription and H3.3 deposition in ISGs, but not for housekeeping genes. To test whether the defects in Whsc1−/− cells are attributed to impaired IFN signalling, we examined IFN-induced phosphorylation of STAT1. STAT1 is a DNA sequence-specific transcription factor, which controls ISG induction upon its phosphorylation mediated by JAK/STAT pathway activation (Darnell et al, 1994). Immunoblot analyses showed that STAT1 was phosphorylated after IFN stimulation both in WT and Whsc1−/− cells at comparable levels, confirming that IFN signalling is intact in Whsc1−/− cells (Supplementary Figure S1C). Thus, the defects observed in Whsc1−/− cells lie downstream of STAT1 activation.
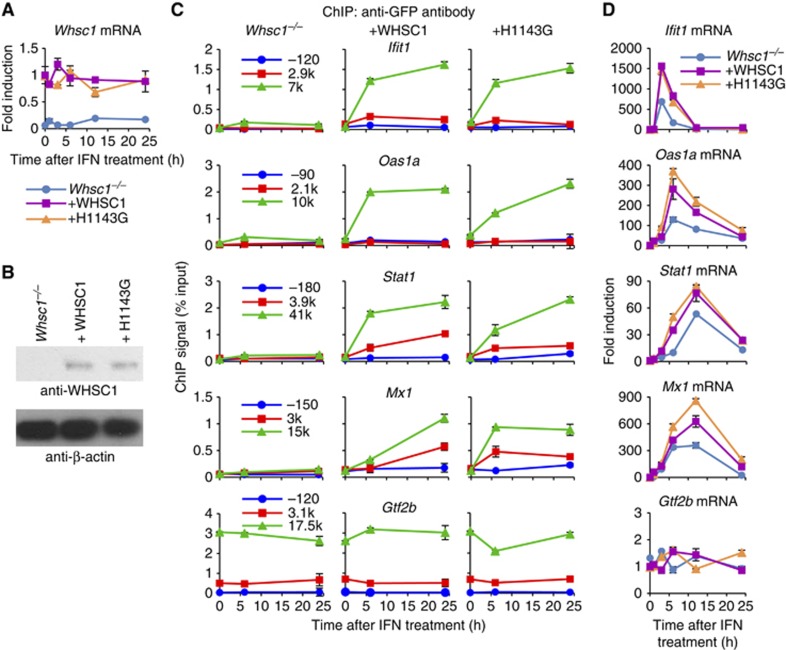
To corroborate that the lack of H3.3 incorporation in Whsc1−/− cells is accounted for by the absence of WHSC1, rather than a secondary change(s), we reintroduced WT WHSC1 and the catalytically inactive mutant WHSC1 (H1143G) into Whsc1−/− cells (Nimura et al, 2009). Data in Figure 2A and B confirmed that upon reintroduction, Whsc1 mRNA and the protein were expressed from these constructs in Whsc1−/− cells. As shown in Figure 2C, WT WHSC1 reintroduction restored IFN-induced H3.3-YFP incorporation in all ISGs. Surprisingly, the catalytically inactive mutant, H1143G, also rescued H3.3 deposition. Correlating with these results, ISG mRNA induction was rescued by both WT WHSC1 and the mutant (Figure 2D). Together, these results show that WHSC1 plays a critical role in both ISG transcription and H3.3 deposition, the role achieved without histone methyltransferase activity. To further substantiate the role of WHSC1, we knocked down Whsc1 gene expression in WT cells by small-interfering RNA (siRNA) (Supplementary Figure S4). Induction of all four ISGs was consistently reduced in Whsc1 knockdown cells by up to 50%, reinforcing the idea that WHSC1 itself participates in ISG transcription and H3.3 deposition.
Figure 2.
WHSC1 reintroduction rescues ISG transcription and H3.3 incorporation in Whsc1−/− cells. (A) Whsc1−/− cells expressing H3.3-YFP were transduced with WT WHSC1 or the catalytically defective mutant (H1143G) vectors. Cells were treated with IFN for indicated times, and Whsc1 mRNA was detected by qRT–PCR. Values represent the average of two determinations ±s.d. Untransduced Whsc1−/− cells were tested as a negative control. (B) WHSC1 protein expression in above cells was detected by immunoblotting with anti-WHSC1 antibody. β-actin was tested as a loading control. (C) Above cells were tested for IFN-induced H3.3 incorporation by ChIP assays as in Figure 1C. Values represent the average of duplicate determinations ±s.d. (D) Above cells were tested for ISG mRNA expression by qRT-PCR. Values represent the average of two determinations ±s.d.
Source data for this figure is available on the online supplementary information page.
WHSC1 is recruited to the ISGs to facilitate HIRA recruitment
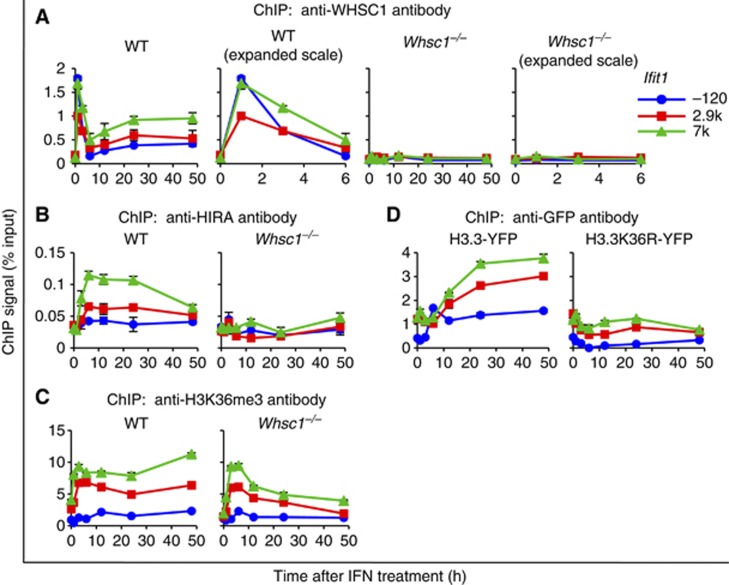
To assess how WHSC1 regulates ISG transcription and H3.3 deposition, we examined whether WHSC1 is recruited to the ISGs. ChIP analysis with anti-WHSC1 antibody showed that after IFN stimulation, WHSC1 rapidly accumulated at all three regions of the ISGs, which peaked at 1–3 h (Figure 3A; Supplementary Figure S5A, left). WHSC1 occupancy declined thereafter at 6 h, but rose again in the subsequent, post-ISG transcription period and remained roughly constant thereafter, showing a clear TES preference. WHSC1 accumulation in Gtf2b was at near background level throughout the period. As expected, no WHSC1 binding was detected in Whsc1−/− cells (Figure 3A; Supplementary Figure S5A, right).
Figure 3.
WHSC1 is recruited to ISGs upon IFN stimulation. (A–C) Recruitment of WHSC1 (A), HIRA (B), and H3K36me3 (C) on indicated position of Ifit1 was tested in WT and Whsc1−/− cells treated with IFN for indicated times by ChIP analysis using respective antibodies. In (A), binding of WHSC1 was plotted in an expanded X axis to better depict early recruitment kinetics. Values represent the average of duplicate determinations ±s.d. (D) NIH3T3 cells were transduced with WT H3.3-YFP or mutant H3.3K36R-YFP, treated with IFN for indicated times, and their deposition into Ifit1 was detected by ChIP using anti-GFP antibody. Values represent the average of duplicate determinations ±s.d.
We next tested whether the histone chaperon HIRA is recruited to ISGs, since HIRA is shown to be involved in H3.3 incorporation in transcriptionally active genes (Goldberg et al, 2010). In WT cells, HIRA accumulated in the ISGs after IFN stimulation with a pattern very similar to that of H3.3-YFP: HIRA accumulation was highest at the TES and it continued well after ISG transcription (Figure 3B; Supplementary Figure S5B). In contrast, HIRA failed to accumulate in ISGs in Whsc1−/− cells, indicating that WHSC1 is required for driving HIRA recruitment. Immunoblot analysis verified that levels of HIRA expression were similar in WT and Whsc1−/− cells before and after IFN stimulation. Similarly, WHSC1 expression in WT cells was not affected after IFN treatment (Supplementary Figure S2).
Since H3.3 deposition and H3K36 trimethylation correlated well in our previous work, and as WHSC1 is a methytransferase for H3K36, we next tested whether H3K36 trimethylation is defective in Whsc1−/− cells (Nimura et al, 2009; Tamura et al, 2009). H3K36me3 increased after ISG stimulation in WT cells as expected (Figure 3C; Supplementary Figure S5C, left). Somewhat unexpectedly, H3K36me3 also increased in Whsc1−/− cells, although with kinetics different from WT cells: in Whsc1−/− cells, H3K36me3 increased only in the initial 3–6 h and then declined, whereas it remained high in WT cells (Figure 3C; Supplementary Figure S5C, right). Because H3K36me3 accumulated even in Whsc1−/− cells after IFN treatment, we next asked whether other histone methyltransferases are recruited to ISGs. To this end, we tested SETD2, a mammalian Set2 homologue suggested to be a major enzyme for H3K36me3 (Edmunds et al, 2008). As shown in Supplementary Figure S6A, SETD2 rapidly accumulated at the TSS of ISGs and peaked at 3–6 h upon IFN stimulation in WT cells. SETD2 also accumulated in Whsc1−/− cells, although slightly lower in amounts relative to WT cells. These results suggest that SETD2 may mediate methylation of H3K36 after 3–6 h of IFN stimulation in both WT and Whsc1−/− cells.
As WHSC1 is reportedly involved in both di- and tri-methylation of H3K36, we also examined accumulation of H3K36 dimethylation (H3K36me2) (Kuo et al, 2011). In WT cells, H3K36me2 increased in ISG coding region and rapidly diminished after IFN treatment. However, in Whsc1−/− cells, H3K36me2 failed to accumulate in the ISGs at detectable levels (Supplementary Figure S6B). To further study the relationship between H3.3 deposition and H3K36 methylation, we tested a mutant H3.3-YFP, in which K36 was mutated to arginine (H3.3K36R), a mutation that would prevent methylation. Consistent with previous reports that H3K36R can be incorporated into the histone octamer (Tanaka et al, 2007; Du and Briggs, 2010), H3.3K36R-YFP was expressed and localized to the nucleus at similar levels as WT H3.3-YFP (Supplementary Figure S7A and B). However, this mutant, unlike WT H3.3-YFP, was not incorporated into ISGs after IFN stimulation, although a low level of incorporation was observed before stimulation (Figure 3D; Supplementary Figure S5D). These data support the notion that H3.3K36 methylation is required, but not sufficient for induced H3.3 deposition.
WHSC1 plays a role in ISG elongation
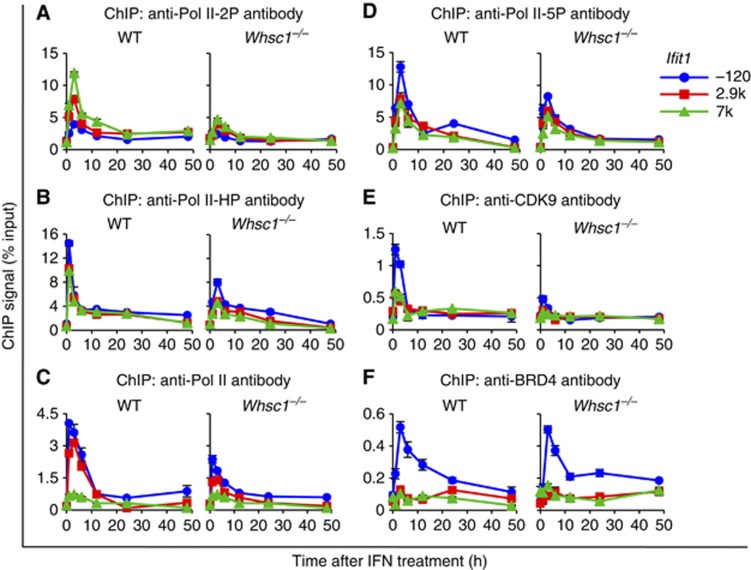
The transition from transcription initiation to elongation requires phosphorylation of Pol II at serine 2 in the C-terminal domain by P-TEFb, composed of CDK9 and CYCLIN T1 (Core and Lis, 2008; Nechaev and Adelman, 2011). P-TEFb is recruited to many genes by binding to the BRD4 (Jang et al, 2005; Yang et al, 2005; Dey et al, 2009; Hargreaves et al, 2009). We have recently shown that BRD4 is recruited to ISGs upon IFN stimulation primarily by binding to acetylated histones, and that BRD4 recruitment is obligatory for productive ISG elongation as it recruits P-TEFb and other elongation factors (Patel et al, 2013). Here, we investigated whether WHSC1 plays a role in ISG elongation. As shown in Figure 4A and Supplementary Figure S8A, the elongating form of Pol II, phosphorylated at serine 2 (Pol II-2P), rapidly accumulated upon IFN stimulation in ISGs. Pol II-2P accumulation peaked at 3 h, and was higher in the coding region than in the TSS. Thereafter, Pol II-2P binding declined to basal levels, correlating with the kinetics of ISG mRNA induction. Pol II-2P accumulation was, however, drastically reduced in Whsc1−/− cells in all ISGs, pointing to a defect in elongation. Supporting defective elongation rather than defective Pol II assembly, accumulation of hypophosphorylated Pol II (Pol II-HP), which markedly increased in the TSS upon IFN addition in WT cells, was only moderately affected in Whsc1−/− cells (Figure 4B; Supplementary Figure S8B). Accumulation of total Pol II was also tested with another antibody that recognizes both phosphorylated and hypophosphorylated Pol II, and was essentially the same as that of hypophosphorylated Pol II (Figure 4C; Supplementary Figure S8C). Accumulation of total Pol II and Pol II phosphorylated at serine 5 (Pol II-5P), the initiation form of Pol II, was also tested with additional respective antibodies (Figure 4C and D; Supplementary Figure S8C and D). We found that total Pol II and Pol II-5P accumulated in ISGs with a pattern very similar to that of Pol II-HP in both WT and Whsc1−/− cells. These results favour the possibility that the initial Pol II recruitment and initiation occurred, but the subsequent elongation was inhibited in Whsc1−/− cells. The partial reduction in total Pol II, Pol II-HP, and Pol II-5P binding in Whsc1−/− cells is likely due to reduced transcription reinitiation, associated with multiple cycles of Pol II recruitment following elongation (Yudkovsky et al, 2000; Mapendano et al, 2010; Patel et al, 2013). Further supporting defective elongation, P-TEFb recruitment, tested with anti-CDK9 antibody, was 2- to 3-fold lower in Whsc1−/− cells than in WT cells, revealing a role of WHSC1 in P-TEFb recruitment (Figure 4E; Supplementary Figure S9A). As P-TEFb recruitment and ISG elongation depends on BRD4, we next asked whether BRD4 recruitment is affected in Whsc1−/− cells. Data in Figure 4F and Supplementary Figure S9B showed that BRD4 recruitment was unaffected in Whsc1−/− cells. BRD4 bound to all ISGs in both WT and Whsc1−/− cells at comparable levels. Thus, WHSC1 acts downstream of BRD4 and facilitates P-TEFb recruitment and ISG elongation. It should be noted here that accumulation of P-TEFb, Pol II, Pol II-HP, Pol II-2P, and Pol II-5P to Gtf2b was not affected in Whsc1−/− cells, reinforcing the view that WHSC1 impacts on rapidly activated transcription, but not on constitutive transcription (Supplementary Figures S8 and S9). The expression levels of BRD4, P-TEFb, Pol II, Pol II-HP, and Pol II-5P were not affected by IFN stimulation in both WT and Whsc1−/− cells, although the levels of Pol II-2P were increased after IFN stimulation, probably as a result of transcription activation of numerous ISGs (Supplementary Figure S2).
Figure 4.
Defective ISG elongation in Whsc1−/− cells. WT and Whsc1−/− cells were treated with IFN for indicated times, and the recruitment of Pol II-2P (A), Pol II-HP (B), Pol II (C), Pol II-5P (D), CDK9 (E), and BRD4 (F) to indicated position of Ifit1 was tested by ChIP using respective antibodies. Values represent the average of duplicate determinations ±s.d.
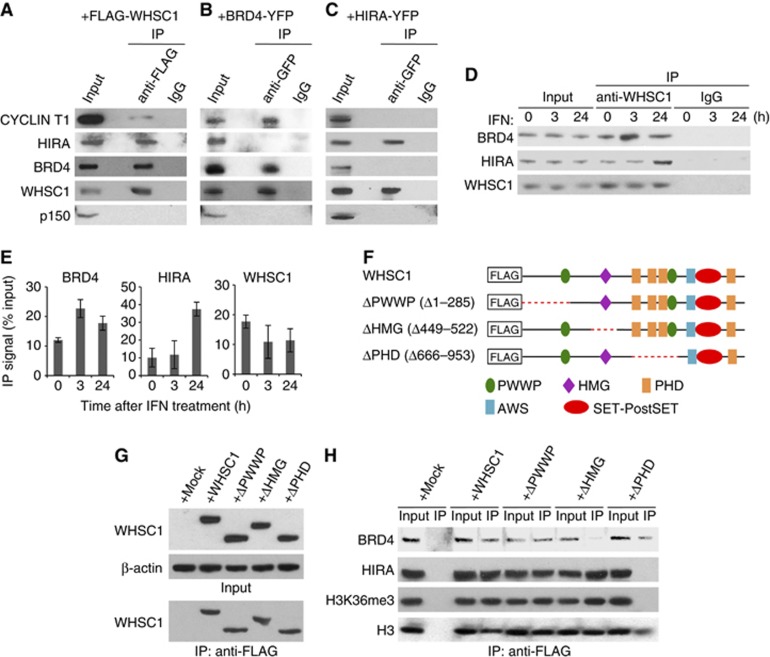
WHSC1 interacts with BRD4 and HIRA independently on the ISG chromatin
The above data raised the possibility that WHSC1 has two roles: one, recruiting P-TEFb to stimulate ISG elongation and two, recruiting HIRA to drive H3.3 deposition. To test this possibility, co-immunoprecipitation (Co-IP) assays were performed with Whsc1−/− cells expressing FLAG-tagged WHSC1. As shown in Figure 5A, the FLAG-WHSC1 precipitates contained BRD4, HIRA, and P-TEFb, the latter detected by anti-CYCLIN T1 antibody. To determine whether WHSC1 recruits these factors together or separately, two reciprocal assays were performed. First, using NIH3T3 cells stably expressing YFP-tagged BRD4, Co-IP assays were carried out. Immunoblot data in Figure 5B showed that the BRD4-YFP precipitates contained WHSC1 and P-TEFb, but not HIRA. Second, Co-IP assays were performed with WT cells expressing YFP-tagged HIRA. As shown in Figure 5C, the immunoprecipitates brought down by anti-GFP antibody contained only WHSC1. These results indicate that HIRA does not interact with BRD4 and P-TEFb, and that WHSC1 interacts with BRD4/P-TEFb and HIRA separately. Notably, these immunocomplexes did not contain p150, a subunit of histone chaperone CAF-1, which is involved in replication-coupled H3.1 deposition (Figure 5A–C, lower panels) (Tagami et al, 2004). Co-IP experiments with anti-WHSC1 antibody showed that endogenous WHSC1 was co-precipitated with HIRA and BRD4, and that the levels of both precipitates increased after IFN treatment but with varying kinetics (Figure 5D and E). The amounts of BRD4 co-precipitated with WHSC1 peaked at 3 h, whereas HIRA co-precipitated with WHSC1 peaked at around 24 h.
Figure 5.
WHSC1 interacts with BRD4 and HIRA independently. (A) Whole cell extracts from Whsc1−/− cells transiently expressing FLAG-WHSC1 were immunoprecipitated with anti-FLAG antibody, and tested for co-precipitation of indicated proteins by immunoblotting. Normal IgG was used as a control. Input represents 10% of total protein. (B) Extracts from NIH3T3 cells expressing BRD4-YFP were immunoprecipitated with anti-GFP antibody, and tested for co-precipitation of indicated proteins by immunoblotting. (C) Whole cell extracts from WT cells transiently expressing HIRA-YFP were immunoprecipitated with anti-GFP antibody, and tested for co-precipitation of indicated proteins by immunoblotting. (D) Whole cell extracts from WT cells treated with IFN for indicated times were immunoprecipitated with anti-WHSC1 antibody, and tested for co-precipitation of BRD4 and HIRA. (E) Graphic representation of the experiments shown in (D). The amounts of precipitated proteins were quantified and normalized by input protein. Values are the average of two assays ±s.d. (F) A diagram of FLAG-tagged WHSC1 deletion mutants. The red dots indicate deleted regions. (G) Whole cell extracts from Whsc1−/− cells transiently expressing above deletion constructs were immunoblotted with anti-WHSC1 antibody (left) or immunoprecipitated with anti-FLAG antibody, followed by immunoblotting with anti-WHSC1 antibody (right). (H) Proteins immunoprecipitated from above cells with anti-FLAG antibody were immunoblotted for BRD4, HIRA, H3K36me3, or total H3.
Source data for this figure is available on the online supplementary information page.
To assess the domain(s) of WHSC1 necessary for the interaction with BRD4 and HIRA, a series of FLAG-tagged WHSC1 deletion constructs were tested (a diagram in Figure 5D). WHSC1 consists of PWWP, high-mobility group (HMG), plant homeotic domain (PHD), associated with SET (AWS), and SET-PostSET domains. The AWS and SET-PostSET domains are conserved among Set2 homologues. Some of these domains are implicated in certain activities, for example, HMG in DNA binding, PHD in the interaction with H3, and PWWP in the interaction with H3K36me3 (Taverna et al, 2006; Vezzoli et al, 2010). Immunoblot data in Figure 5E showed that the deletions of expected sizes were expressed and precipitated by anti-FLAG antibody. As seen in Figure 5F Co-IP experiments, ΔHMG failed to precipitate BRD4, whereas WT WHSC1, ΔPWWP, and ΔPHD precipitated BRD4, although ΔPHD was less effective in precipitating BRD4 than others. On the other hand, ΔHMG precipitated HIRA and H3K36me3 at levels similar to WT WHSC1 and ΔPWWP. However, ΔPHD did not precipitate either protein. These results indicate that the HMG domain is important for the interaction with BRD4, and the PHD domain is critical for the interaction with HIRA and H3K36me3. We noted that all WHSC1 deletions tested precipitated total H3, although the amounts of H3 precipitates by ΔPHD were somewhat less than those by others (Figure 5H, lower panel). These results are consistent with a recent report that WHSC1 binds to non-methylated H3 through its C-terminal domain (He et al, 2013). These results suggest that all of the WHSC1 deletions are recruited to chromatin. To test whether any of these deletions rescue ISG mRNA induction, the deletions and WT WHSC1 were transiently expressed in Whsc1−/− cells and ISG mRNA induction was examined (Supplementary Figure S10). ΔPWWP and ΔPHD partially rescued ISG mRNA induction. However, ΔHMG did not, indicating that the interaction with BRD4 is required for WHSC1 to stimulate ISG induction.
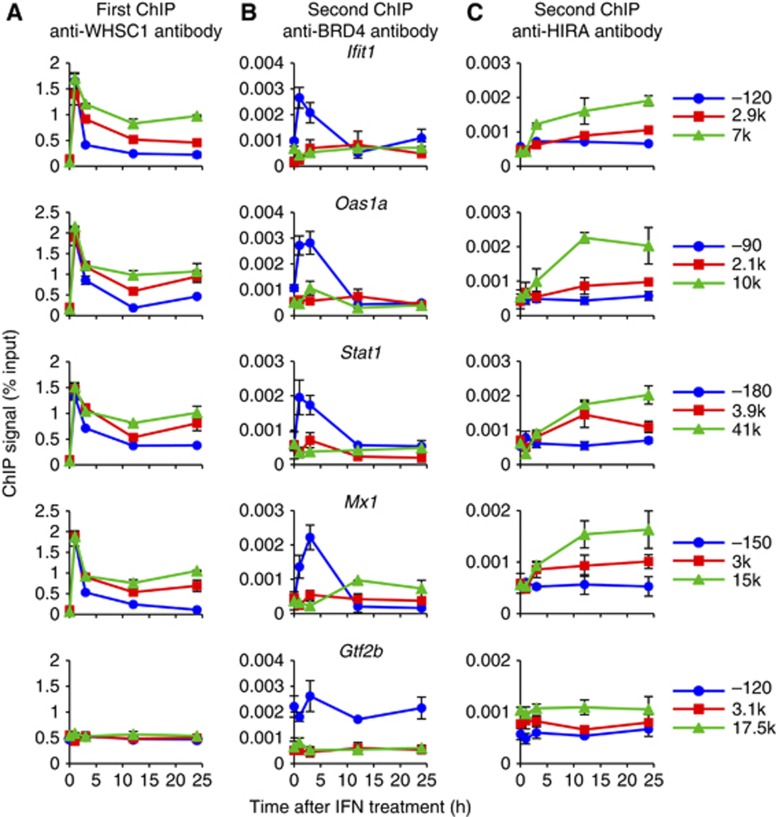
We then asked whether WHSC1 binds to the ISG chromatin along with BRD4/P-TEFb or HIRA. To this end, sequential ChIP assays were carried out. Chromatin was first precipitated with anti-WHSC1 antibody, followed by the second ChIP performed with anti-BRD4 or anti-HIRA antibody. The first ChIP with anti-WHSC1 antibody revealed recruitment of WHSC1 to ISGs and background binding to Gtf2b as expected (Figure 6A). The second ChIP found a considerable amount of BRD4 and HIRA on the ISG chromatin that had been precipitated with WHSC1, indicating that these proteins and WHSC1 co-occupied ISGs (Figure 6B and C). Significantly, BRD4 and HIRA showed spatially and temporally distinct distribution patterns. BRD4 was present almost exclusively at the TSS and only for the initial 3 h. In contrast, HIRA accumulated more slowly, but lasted as long as 24 h, and displayed a clear TES preference. These results further support the possibility that BRD4 and HIRA interact with WHSC1 independently, occupying separate spaces of ISGs with different time courses. A possible scenario is that WHSC1 initially associates with BRD4/P-TEFb at the TSS, and subsequently travels across the ISG coding regions to the TES along with HIRA, but without BRD4. It is of note that the kinetics and distribution patterns observed for BRD4 and HIRA in the second ChIP were consistent with Co-IP results (see Figure 5C) and similar to the conventional ChIP pattern seen by anti-BRD4 or anti-HIRA antibody alone (see Figures 4F and 3B), suggesting that the bulk of BRD4 and HIRA bind to ISGs together with WHSC1.
Figure 6.
WHSC1 co-occupies the ISG chromatin with BRD4 and HIRA independently. WT cells were treated with IFN for indicated times, and sequential ChIP assays were performed first with anti-WHSC1 antibody (A), then with anti-BRD4 (B) or anti-HIRA (C) antibody. Values represent the average of duplicate determinations ±s.d.
As above data indicate that HIRA recruitment is directed by WHSC1, we examined whether HIRA reciprocally affects WHSC1 recruitment by testing cells stably expressing Hira short-hairpin RNA (shRNA). In the Hira knockdown cells, endogenous Hira transcript levels were reduced by 50 to 70% (Supplementary Figure S11A). Further, H3.3-YFP deposition was markedly reduced in Hira knockdown cells relative to cells expressing control shRNA (Supplementary Figure S11B). Nevertheless, WHSC1 was recruited to all ISGs in Hira knockdown cells and control shRNA cells at comparable levels (Supplementary Figure S11C). These results indicate that HIRA acts downstream of WHSC1, and is required for H3.3 deposition in the ISGs. The slight reduction in ISG mRNA induction seen by Hira knockdown is reminiscent of the reduced ISG induction by H3.3 knockdown reported earlier, consistent with the contribution of H3.3 deposition to ISG transcription (Tamura et al, 2009).
WHSC1 directs ISG elongation and H3.3 deposition partly through different molecular processes
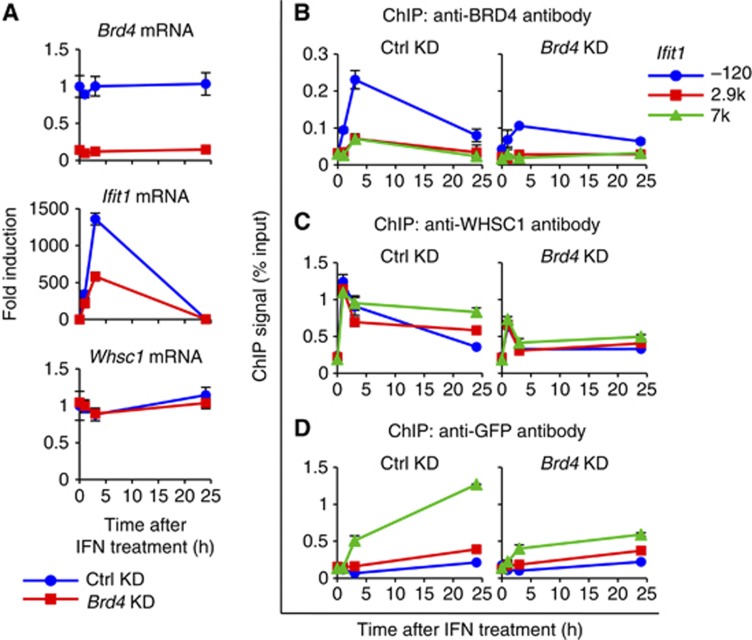
In light of the above findings that BRD4 recruitment was intact in Whsc1−/− cells (see Figure 4F and Supplementary Figure S9B), we next investigated whether BRD4 is responsible for WHSC1 recruitment. We knocked down the expression of BRD4 in WT cells, which lead to >80% reduction in BRD4 transcript expression without affecting WHSC1 transcript levels, as reported in our previous works (Figure 7A) (Jang et al, 2005; Dey et al, 2009). Data in Figure 7A and Supplementary Figure S12A showed that mRNA levels of all four ISGs tested were lower in Brd4 knockdown cells compared with control cells throughout the IFN treatment. In contrast, Brd4 knockdown had virtually no effect on Whsc1 and Gtf2b transcript levels. As expected, recruitment of BRD4 was consistently reduced in Brd4 knockdown cells (Figure 7B; Supplementary Figure S12B). We next tested whether Brd4 knockdown impacted on the recruitment of WHSC1. ChIP analysis in Figure 7C and Supplementary Figure S12C showed that IFN-stimulated WHSC1 recruitment to all ISGs was inhibited in Brd4 knockdown cells by 50% compared with control cells. These data led us to predict that Brd4 knockdown would also inhibit H3.3 deposition. Indeed, incorporation of H3.3-YFP into ISGs after IFN stimulation was reduced in Brd4 knockdown cells (Figure 7D; Supplementary Figure S12D).
Figure 7.
BRD4 is required for H3.3 deposition and ISG induction. (A) WT cells expressing Brd4 shRNA or control shRNA were treated with IFN for indicated times, and mRNA levels of Brd4, Ifit1, and Whsc1 were measured by qRT-PCR. Values represent the average of two determinations ±s.d. (B–D) ChIP analysis was performed to detect recruitment of BRD4 (B), WHSC1 (C), and H3.3-YFP (D) into Ifit1 using respective antibodies. Values represent the average of duplicate determinations ±s.d.
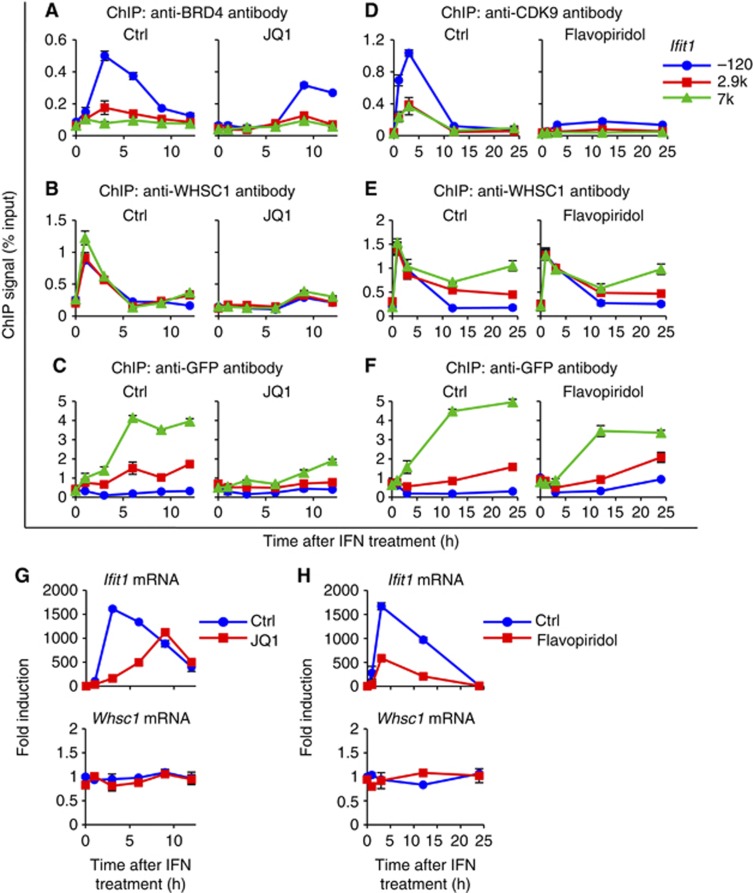
To further confirm that BRD4 is required for the recruitment of WHSC1, we tested JQ1, a small molecule inhibitor specific for the bromodomains of the BET family (Filippakopoulos et al, 2010; Nicodeme et al, 2010). We have previously shown that JQ1 blocks recruitment of BRD4 and P-TEFb to the ISGs, which led to strong inhibition of ISG transcription (Patel et al, 2013). As shown in Figure 8A and Supplementary Figure S13A, treatment of WT cells with JQ1 and IFN inhibited recruitment of BRD4 to all ISGs. In contrast, the inactive, isomeric compound tested as a negative control had no effect on BRD4 recruitment. Recruitment of WHSC1 was almost completely abolished upon JQ1 treatment in all ISGs (Figure 8B; Supplementary Figure S13B). Binding of WHSC1 to Gtf2b remained at background levels, irrespective of JQ1 treatment. In addition, JQ1 markedly inhibited H3.3-YFP deposition in all ISGs (Figure 8C; Supplementary Figure S13C). The inhibition was essentially complete in the early 6 h and extended to 9 and 12 h. As expected, JQ1 also inhibited ISG mRNA expression in these cells without affecting Whsc1 and Gtf2b expression (Figure 8G; Supplementary Figure S13D). Together, these results indicate that binding of BRD4 to the ISGs is critically required for the recruitment of WHSC1, which is in turn required for the subsequent ISG induction and H3.3 incorporation.
Figure 8.
WHSC1 directs ISG elongation and H3.3 deposition through different molecular processes. (A, B) 10 μM of (+)-JQ1 or the inactive stereoisomer (−)-JQ1 (Ctrl) was added to the cells 1 h prior to IFN treatment for indicated times. ChIP assays were performed for recruitment of BRD4 (A) and WHSC1 (B) into Ifit1 using respective antibodies. Values represent the average of duplicate determinations ±s.d. (C) ChIP analysis was performed for WT cells expressing H3.3-YFP after IFN and JQ1 treatment using anti-GFP antibody. Values represent the average of duplicate determinations ±s.d. (D, E) 100 nM of Flavopiridol or vehicle (Ctrl) was added to the cells simultaneously with IFN treatment for indicated times. ChIP assays were performed for recruitment of CDK9 (D) and WHSC1 (E) into Ifit1 using respective antibodies. Values represent the average of duplicate determinations ±s.d. (F) ChIP analysis was performed for WT cells expressing H3.3-YFP after IFN and Flavopiridol treatment using anti-GFP antibody. (G, H) WT cells were treated with IFN in the presence of JQ1 (G) or Flavopiridol (H), and mRNA levels of Ifit1 and Whsc1 were measured by qRT-PCR. Values represent the average of two determinations ±s.d.
The above data highlighted a tight linkage between ISG elongation and H3.3 deposition. We felt it is important to study further whether H3.3 deposition was an automatic consequence of ISG elongation, or mediated by a mechanism independent of elongation. To this end, we tested Flavopiridol, a CDK9-specific kinase inhibitor that blocks the activity of P-TEFb and resultant elongation (Chao et al, 2000; Chen et al, 2012). In the presence of Flavopiridol, IFN-induced recruitment of CDK9 to ISGs was markedly inhibited (Figure 8D; Supplementary Figure S14A). However, Flavopiridol did not affect recruitment of WHSC1 and BRD4 (Figure 8E; Supplementary Figures S14B and S15A). Furthermore, recruitment of HIRA was unaffected in the presence of Flavopiridol, and H3.3-YFP accumulation still took place, although the accumulation was modestly reduced (Figure 8F; Supplementary Figures S14C and S15B). As expected, Flavopiridol caused a marked reduction in ISG mRNA induction without affecting Whsc1 and Gtf2b transcript levels, consistent with our previous report (Figure 8H; Supplementary Figure S14D) (Patel et al, 2013). Thus, the blockade of P-TEFb-mediated ISG elongation did not result in a blockade of H3.3 deposition. These results are consistent with the view that ISG elongation and H3.3 deposition, while linked by WHSC1, are executed partly through molecularly different processes. It may be envisaged that in the presence of Flavopiridol, WHSC1 still travelled across the ISG by a mechanism independent of P-TEFb (Bartkowiak et al, 2010; Devaiah et al, 2012). It should be noted that JQ1 and Flavopiridol did not affect expression of the factors tested in these experiments (Supplementary Figure S16A and B).
WHSC1 is essential for UV-induced transcription and H3.3 deposition
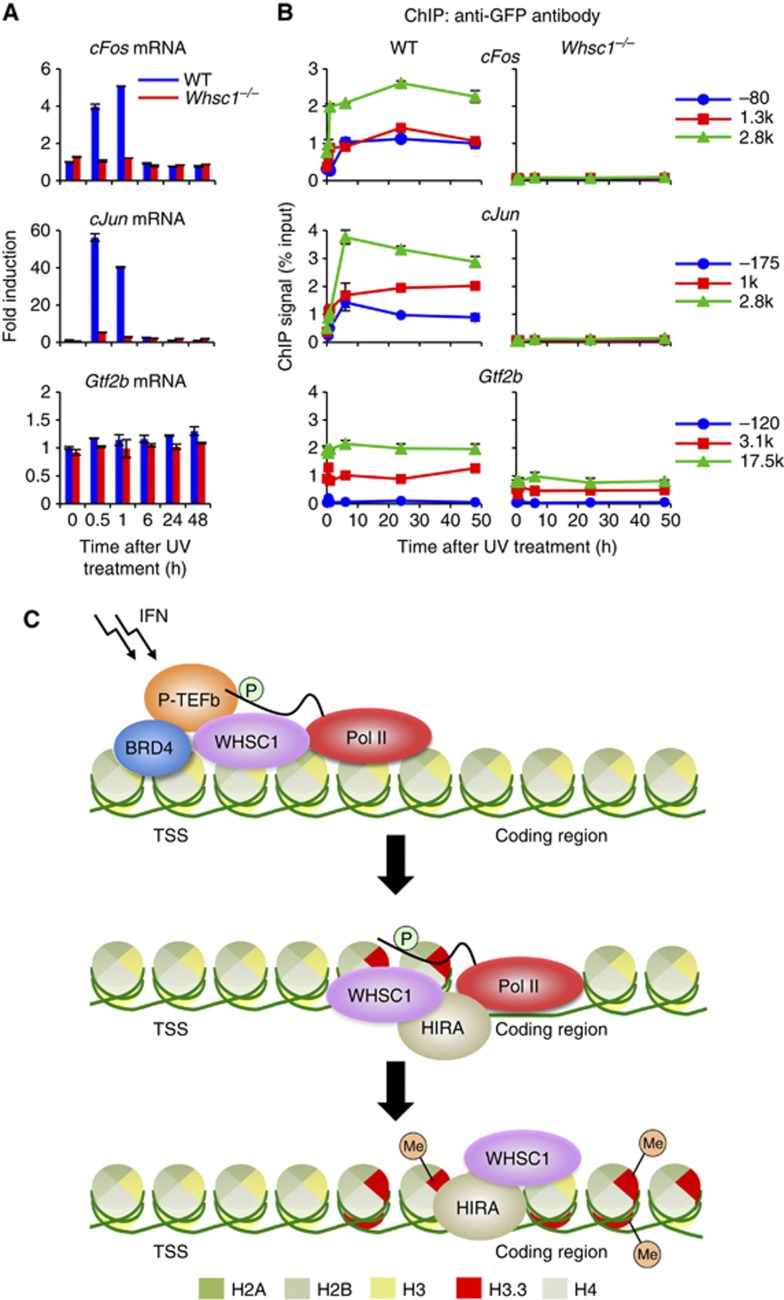
Finally, to assess whether WHSC1 is generally required for H3.3 incorporation in activated transcription, we investigated another activation model in which transcription is induced in response to UV radiation (Cooper and Bowden, 2007). In Figure 9A, WT and Whsc1−/− cells were irradiated with UV-B and induction of c-Fos and c-Jun, typical UV activated genes, was tested by qRT-PCR. As expected, mRNAs for both genes were induced in WT cells at 30 min and 1 h after UV radiation, followed by a sharp fall at 6 h. In contrast, neither gene was induced in Whsc1−/− cells. As shown in Figure 9B, UV treatment triggered rapid incorporation of H3.3-YFP into both genes in WT cells. Similarly to the ISGs, UV-induced H3.3 accumulation persisted up to 48 h, long past c-Fos and c-Jun transcription, and exhibited a clear TES preference. In contrast, H3.3-YFP deposition was essentially undetectable in Whsc1−/− cells for both genes. UV treatment, however, did not change H3.3-YFP levels in Gtf2b. The protein expression levels of H3.3-YFP were identical between WT and Whsc1−/− cells (Supplementary Figure S16C). Thus, the requirement of WHSC1 for induced H3.3 deposition is not limited to ISGs, rather, it may represent a common feature among activated transcription.
Figure 9.
WHSC1 is required for UV-induced transcription and H3.3 incorporation. (A) WT and Whsc1−/− cells expressing H3.3-YFP were irradiated with UV-B (4 mJ/cm2) and induction of c-Fos and c-Jun mRNAs was detected at indicated times by qRT–PCR. Values represent the average of two determinations ±s.d. Gtf2b mRNA was tested as a control. (B) H3.3 incorporation into the indicated regions within the c-Fos (top), c-Jun (middle), and Gtf2b (bottom) genes was tested by ChIP analysis using anti-GFP antibody. Values represent the average of duplicate determinations ±s.d. (C) A model for WHSC1-dependent ISG elongation and H3.3 deposition. Upon IFN stimulation, BRD4 is recruited to ISGs, which in turn recruits WHSC1 and P-TEFb at the TSS (top). P-TEFb phosphorylates Pol II at serine 2 in the C-terminal domain to start elongation. WHSC1 recruits HIRA and these proteins travel together along with Pol II-2 P across the ISG coding region, launching elongation-coupled H3.3 deposition (middle). WHSC1 and HIRA continue to stay on the ISG coding region even after completion of ISG transcription to support sustained, post-elongation phase of H3.3 deposition (bottom).
Discussion
Our study shows that WHSC1 directs H3.3 deposition in IFN- and UV-activated genes. Remarkably, induced H3.3 deposition continued for an extended period even after the ending of transcription, and left a lasting chromatin mark on ISGs and UV-induced genes. In addition, induced H3.3 deposition exhibited a characteristic positional preference for the TES. Our subsequent analysis revealed that WHSC1 drives H3.3 deposition in two phases likely through different molecular mechanisms, the first, elongation-coupled phase, which then shifts to the second, post-elongation phase of deposition.
WHSC1 links ISG elongation and H3.3 deposition
Our conclusion that WHSC1 is required for H3.3 deposition comes from the following observations: (1) IFN- and UV-induced H3.3 deposition was abolished in Whsc1−/− cells, (2) reintroduction of WHSC1 into Whsc1−/− cells rescued H3.3 deposition in ISGs, (3) blockage of WHSC1 recruitment by the bromodomain inhibitor, JQ1 blocked H3.3 deposition, and (4) recruitment of H3.3-specific histone chaperon HIRA was absent in Whsc1−/− cells. In addition to H3.3 deposition, WHSC1 played an important role in ISG elongation, as ISG mRNA induction was severely inhibited in Whsc1−/− cells, but this condition was rescued by WT WHSC1 and the catalytically inactive WHSC1 mutant. Transcription initiation starts with the assembly of Pol II and other factors on the TSS (Roeder, 2005). Although Pol II is preassembled and paused at the promoter proximal region in some genes (Core and Lis, 2008; Rahl et al, 2010; Nechaev and Adelman, 2011). ISGs did not have paused Pol II prior to transcription, rather, Pol II was assembled de novo after IFN stimulation, leading to the recruitment of BRD4 and P-TEFb (Patel et al, 2013). BRD4 is a bromodomain protein of the BET family that interacts with P-TEFb and stimulates transcription of many genes (Jang et al, 2005; Yang et al, 2005; Mochizuki et al, 2008; Hargreaves et al, 2009). Further supporting the role of WHSC1 in elongation, WHSC1 interacted with P-TEFb along with BRD4, and in Whsc1−/− cells, accumulation of the elongating form of Pol II (Pol II-2P) was inhibited more strongly than that of total Pol II. Furthermore, JQ1, an inhibitor for BRD4 and BET family of bromodomain proteins, inhibited not only BRD4/P-TEFb recruitment, but also WHSC1 recruitment, indicating that WHSC1 acts downstream of BRD4 and helps to facilitate P-TEFb recruitment (Figure 9C). It should be noted that inhibition of ISG transcription in Whsc1−/− cells was incomplete, suggesting that a low level of elongation occurred in some ISGs without requiring WHSC1.
Corroborating the role in elongation and H3.3 deposition, WHSC1 was recruited to ISGs upon IFN stimulation, and interacted with BRD4, P-TEFb, and HIRA. The interaction of WHSC1 with BRD4-P-TEFb required mainly the HMG domain and that with HIRA the PHD domain, respectively. Sequential ChIP analyses showed that the two WHSC1 complexes occupied the ISG chromatin in a spatially and temporally distinct fashion. While BRD4 and WHSC1 co-occupied the TSS briefly during transcription, HIRA and WHSC1 co-occupied the coding regions and the TES of the ISGs for extended periods beyond transcription. We provide a first line of evidence that while WHSC1 links elongation and H3.3 deposition, the two events are directed in part by molecularly separable mechanisms. That is, inhibition of P-TEFb-mediated elongation by Flavopiridol did not inhibit H3.3 deposition as much, although it drastically reduced P-TEFb recruitment and ISG mRNA expression. As phosphorylation of Pol II at serine 2 is mediated by multiple factors, P-TEFb recruitment to ISGs may not be indispensable for H3.3 deposition (Bartkowiak et al, 2010; Devaiah et al, 2012).
On the basis of these findings, we envisage that WHSC1 is initially recruited at the TSS through BRD4, and helps recruit P-TEFb initiating elongation. WHSC1 then moves across the ISG coding regions possibly along with the Pol II, recruiting HIRA on route (model in Figure 9C). A recent study reported that HIRA interacts with Pol II-2P (Ray-Gallet et al, 2011). Thus, it is possible that WHSC1 may move along with the elongating Pol II and launches H3.3 deposition by interacting with HIRA. In a similar context, Set2 was shown to bind to Pol II-2P during transcription (Li et al, 2002, 2003). The results suggest that WHSC1 and HIRA remained on the ISGs long after ISG transcription, the pattern well correlated with the sustained, TES-biased H3.3 deposition.
The role of K36me3 in H3.3 deposition
The close concordance between H3K36me3 and H3.3 deposition pointed to a possible causal link for the two events (Tamura et al, 2009). Analysis of the H3.3K36R mutant showed that H3K36 methylation is indispensable for H3.3 incorporation in ISGs. Nonetheless, H3K36me3 accumulated in ISGs in Whsc1−/− cells. These data lead us to suggest that H3K36me3 is required, but not sufficient for H3.3 deposition. Since trimethylation of H3K36 can be catalysed by multiple Set2-related histone methyltransferases, it is likely that Set2-related enzymes other than WHSC1 are involved in H3K36 methylation (Edmunds et al, 2008; Wang et al, 2008; Nimura et al, 2009; Rahman et al, 2011). Indeed, SETD2 was recruited to ISGs in both WT and Whsc1−/− cells. In yeast, repression of internal initiation is dependent on the histone deacetylation, for which Set2-mediated H3K36me3 is required, and this process is important for the regulation of mRNA synthesis (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005; Li et al, 2007; Du and Briggs, 2010; Venkatesh et al, 2012). As H3.3 deposition particularly in the gene body correlates with H3K36 trimethylation, and as WHSC1 reportedly associates with the histone deacetylase HDAC1, WHSC1 may link histone deacetylation, regulation of mRNA synthesis, and H3.3 deposition (Nimura et al, 2009; Tamura et al, 2009).
Generality of induced H3.3 deposition
In this study, H3.3 deposition was found in two unrelated activation models, one with IFN and the other with UV stimulation. In both cases, H3.3 deposition displayed characteristic long duration and the TES preference, and was dependent on WHSC1. Thus, the requirement of WHSC1 for transcription elongation and H3.3 deposition may be a fairly common feature among many models of rapidly activated transcription. However, it is noteworthy that WHSC1 was not required for transcription of housekeeping genes such as Gtf2b. Occupancies of BRD4, Pol II, and P-TEFb on the Gtf2b gene were unaffected in Whsc1−/− cells, although H3.3 deposition was slightly lower in these cells. In light of the report that the pace of transcription could influence the degree of nucleosomal destabilization, it may be reasonable to suggest that WHSC1 is dispensable for slow, constitutive transcription, but is essential for a rapidly induced transcription (Kulaeva et al, 2010).
It is possible that H3.3 deposition is regulated by multiple factors, depending on the types of transcription. A recent report showing that the chromatin remodelling factor CHD2 is required for H3.3 deposition in the myogenic genes that are transcriptionally activated during C2Cl2 cell differentiation, but not for housekeeping genes supports this possibility (Harada et al, 2012).
In conclusion, this work identifies WHSC1 as a factor essential for inducible H3.3 deposition, a process governed by two distinct phases, that leads to the formation of a long-lasting chromatin mark on activated genes. Finally, Wolf–Hirschhorn syndrome may be associated with defects in H3.3 deposition.
Materials and methods
Cells
WT and Whsc1−/− MEFs were prepared from day 13.5 embryos, and maintained in Dulbecco’s minimal essential medium (DMEM) plus 10% fetal bovine serum (Atlantic Biologicals) and antibiotics. NIH3T3 cells (ATCC) were grown in DMEM containing 10% donor bovine serum. Cells were treated with mouse recombinant IFNβ (100 units/ml, PBL Interferon Source) or UV (4 mJ/cm2) for indicated times. The bromodomain small molecule inhibitor (+)-JQ1 or the stereoisomer (−)-JQ1 (10 μM) was added to the culture 1 h before IFN treatment (Filippakopoulos et al, 2010). Flavopiridol (100 nM, Sigma-Aldrich) or vehicle (DMSO) was added to the cell culture simultaneously with IFN.
Constructs and siRNA
Retrovirus vectors expressing H3.1-YFP and H3.3-YFP, and the viral transduction procedures were described (Tamura et al, 2009). H3.3K36R-YFP was constructed by site-directed mutagenesis. A retrovirus vector for BRD4-YFP was constructed by a procedure similar to that used above for H3-YFP. Retroviral vectors for WHSC1 and WHSC1H1143G were constructed by cloning full-length WHSC1 cDNA from pCAGIP-WHSC1 into pMSCVpuro (Clontech) (Nimura et al, 2009). WHSC1 and its deletion mutants, ΔPWWP, ΔHMG, and ΔPHD (lacking amino acid residues 1–285, 449–522, and 666–953, respectively), were cloned in a CMV-driven expression vector to generate pFLAG-CMV-WHSC1 (Sigma-Aldrich). The human HIRA cDNA (Origenes) was subcloned into pEYFP-N1 (Clontech) to generate a vector expressing HIRA as a fusion protein with YFP at the C-terminus. WHSC1 siRNA and scrambled control siRNA were purchased from Santa Cruz Biotechnology. The template of HIRA shRNA was as follows (5′-GATCCCCGATGGTCGGAGGAGAATCATTCAAGAGATGATTCTCCTCCGACCATCTTTTTA-3′ and 5′-AGCTTAAAAAGATGGTCGGAGGAGAATCATCTCTTGAATGATTCTCCTCC GACCATCGGG-3′), and was ligated into pSUPERretro (OligoEngine) to generate the shRNA expressing retroviral vector. A retroviral BRD4 shRNA vector and viral transduction procedures were described (Jang et al, 2005; Dey et al, 2009).
Antibodies
Rabbit antibodies for BRD4 and WHSC1 were described (Dey et al, 2009; Nimura et al, 2009). Antibodies for the following proteins were purchased from indicated vendors: antibodies against histone H3 (ab1791), H3K36me2 (ab9049), and H3K36me3 (ab9050) were from Abcam; those against CDK9 (sc-484), CYCLIN T1 (sc-10750), Pol II (N-20, sc-899), phosphorylated STAT1 (sc-417), HIRA (sc-48774), SETD2 (sc-99451), p150 (sc-10772), and β-actin (sc-7210) were from Santa Cruz Biotechnology; antibodies for Pol II-HP (8WG16, MMS-126R), Pol II-2P (H5, MMS-129R), and Pol II-5P (H14, MMS-134R) were from Covance Research Products; and anti-GFP antibody (11814460001) was from Roche Applied Science; and anti-FLAG antibody (2368) was from Cell Signaling.
Reverse transcription and qPCR
Total RNA was extracted with the TRIzol reagent (Invitrogen), and cDNA was prepared with Superscript II (Invitrogen). qPCR was performed using the SYBR Green Master Mix (Applied Biosystems) in combination with the ABI Prism 7500 sequence detection system (Applied Biosystems). Transcript levels were normalized by Gapdh and expressed as relative to those in unstimulated cells. Primer sequences used for qPCR are available upon request.
ChIP and sequential ChIP assays
ChIP assays were performed as described (Tamura et al, 2009) with a modification. Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and disrupted by sonication. Indicated antibodies were incubated with Dynabeads protein G (Invitrogen) for 2 h, followed by incubation with precleared chromatin corresponding to 4 × 105 cells overnight at 4°C. Immunoprecipitated chromatin was de-cross-linked, and purified DNA was subjected to qPCR. For sequential ChIP assays, anti-WHSC1-conjugated beads were first incubated with chromatin preparations corresponding to 1 × 107 cells, and chromatin immunocomplex was eluted with 10 mM DTT by incubating at 37°C for 30 min, followed by second ChIP with antibody for BRD4 or HIRA. Eluted DNA was then purified and analysed with the standard qPCR procedure.
Immunoprecipitation and immunoblot
WT FLAG-WHSC1 and FLAG-WHSC1 deletions were transiently transfected into Whsc1−/− MEFs. BRD4-YFP was transduced into NIH3T3 cells. HIRA-YFP was transiently transfected into WT MEFs. Whole cell extracts were immunoprecipitated with ANTI-FLAG M2 affinity gel (Sigma-Aldrich) or anti-GFP antibody-conjugated Dynabeads protein G. Immunoprecipitates were fractionated by 4–20% gradient SDS–PAGE, and analysed by immunoblotting using indicated antibodies. For detection of WHSC1, HIRA, SETD2, BRD4, CYCLIN T1, Pol II, Pol II-HP, Pol II-2P, and Pol II-5P, whole cell extracts from WT and Whsc1−/− MEFs were immunoblotted with indicated antibodies. For detection of phosphorylated STAT1, nuclear extracts from WT and Whsc1−/− MEFs were immunoblotted with indicated antibodies. Levels of YFP-tagged H3 and methyl modifications were assessed by immunoblotting of acid-extracted total histones using respective antibodies (Tamura et al, 2009).
Supplementary Material
Acknowledgments
We thank Drs D Singer, K Zhao, and J Zhu for critical reading of the manuscript; Mr W Huynh, Drs A Dey, and H Kurumizaka for advice on experiments. This work was supported by the Intramural Program of NICHD and the NIH Intramural AIDS Targeted Antiviral Program, National Institutes of Health. NS was partially supported by the NIH-Japan Society for Promotion of Science (JSPS) Fellowship.
Author contributions: KN and TT made preliminary observations and gave suggestions to experiments; NS, TK, and MCP designed and performed experiments, and analysed data; KU provided Whsc1−/− cells and comments on the project; TDH provided JQ1 and comments on the project; KO conceived and steered the project; NS and KO wrote the manuscript. All authors read and approved the submitted manuscript.
Footnotes
The authors declared that they have no conflict of interests.
References
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL (2010) CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann AD, Cole F, Hirschhorn K (2005) The etiology of Wolf-Hirschhorn syndrome. Trends Genet 21: 188–195 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH (2000) Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem 275: 28345–28348 [DOI] [PubMed] [Google Scholar]
- Chen S, Dai Y, Pei XY, Myers J, Wang L, Kramer LB, Garnett M, Schwartz DM, Su F, Simmons GL, Richey JD, Larsen DG, Dent P, Orlowski RZ, Grant S (2012) CDK inhibitors upregulate BH3-only proteins to sensitize human myeloma cells to BH3 mimetic therapies. Cancer Res 72: 4225–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper SJ, Bowden GT (2007) Ultraviolet B regulation of transcription factor families: roles of nuclear factor-kappa B (NF-kappaB) and activator protein-1 (AP-1) in UVB-induced skin carcinogenesis. Curr Cancer Drug Targets 7: 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319: 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE Jr, Kerr IM, Stark GR (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421 [DOI] [PubMed] [Google Scholar]
- Daury L, Chailleux C, Bonvallet J, Trouche D (2006) Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep 7: 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ 3rd, Singer DS (2012) BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA 109: 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K (2003) The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA 100: 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K (2009) Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 20: 4899–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A (2010) The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 24: 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du HN, Briggs SD (2010) A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 Lys36 methylation, histone acetylation, and repression of cryptic transcription. J Biol Chem 285: 11704–11713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL (2008) Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27: 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaesser SJ, Goldberg AD, Allis CD (2010) New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev 20: 110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N et al. (2010) Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD (2006) Histone H3 variants and their potential role in indexing mammalian genomes: the "H3 barcode hypothesis". Proc Natl Acad Sci USA 103: 6428–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF (2006) Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem 281: 559–568 [DOI] [PubMed] [Google Scholar]
- Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S, Kumamaru H, Saiwai H, Tsubota T, Kurumizaka H, Akashi K, Tachibana T, Imbalzano AN, Ohkawa Y (2012) Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J 31: 2994–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R (2009) Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Li F, Zhang J, Wu J, Shi Y (2013) The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (Nuclear Receptor SET Domain) family members in their histone H3 recognition. J Biol Chem 288: 4692–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S (2008) Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9: 15–26 [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534 [DOI] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL (2004) From silencing to gene expression: real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Kim JY, Kee HJ, Choe NW, Kim SM, Eom GH, Baek HJ, Kook H, Seo SB (2008) Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol 28: 2023–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Hsieh FK, Studitsky VM (2010) RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci USA 107: 11325–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, Li W, Gozani O (2011) NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 44: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T (2002) The many faces of histone lysine methylation. Curr Opin Cell Biol 14: 286–298 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD (2010) Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA 107: 14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR 3rd, Workman JL (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP (2002) Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277: 49383–49388 [DOI] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G (2006) PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24: 309–316 [DOI] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH (2010) Crosstalk between mRNA 3' end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, Martinez-Martinez Y, Chesi M, Bergsagel PL, Zhou MM, Waxman S, Leibovitch BA, Walsh MJ, Licht JD (2008) The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 111: 3145–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K (2008) The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem 283: 9040–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K (2011) Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB (2008) Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A (2010) Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y (2009) A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460: 287–291 [DOI] [PubMed] [Google Scholar]
- Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K (2013) BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol 33: 2497–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA (2010) c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM (2011) The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31: 2641–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, Almouzni G (2011) Dynamics of histone h3 deposition in vivo reveal a nucleosome gap-filling mechanism for h3.3 to maintain chromatin integrity. Mol Cell 44: 928–941 [DOI] [PubMed] [Google Scholar]
- Roeder RG (2005) Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett 579: 909–915 [DOI] [PubMed] [Google Scholar]
- Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K, Kato S (2010) A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev 24: 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K (2005) Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO et al. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT (1998) WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 7: 1071–1082 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL (2011) Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80: 473–499 [DOI] [PubMed] [Google Scholar]
- Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, Milburn PJ, Parish CR, Tremethick DJ, Rao S (2009) Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol 29: 1972–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S (2010) Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11: 264–275 [DOI] [PubMed] [Google Scholar]
- Tamura T, Smith M, Kanno T, Dasenbrock H, Nishiyama A, Ozato K (2009) Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem 284: 12217–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S (2007) Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397: 161–168 [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL (2012) Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489: 452–455 [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M (2010) Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol 17: 617–619 [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C, Bell O, Schubeler D (2005) Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev 19: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545 [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.