Abstract
Protein ubiquitylation is a post-translational modification that controls all aspects of eukaryotic cell functionality, and its defective regulation is manifested in various human diseases. The ubiquitylation process requires a set of enzymes, of which the ubiquitin ligases (E3s) are the substrate recognition components. Modular CULLIN-RING ubiquitin ligases (CRLs) are the most prevalent class of E3s, comprising hundreds of distinct CRL complexes with the potential to recruit as many and even more protein substrates. Best understood at both structural and functional levels are CRL1 or SCF (SKP1/CUL1/F-box protein) complexes, representing the founding member of this class of multimeric E3s. Another CRL subfamily, called CRL3, is composed of the molecular scaffold CULLIN3 and the RING protein RBX1, in combination with one of numerous BTB domain proteins acting as substrate adaptors. Recent work has firmly established CRL3s as major regulators of different cellular and developmental processes as well as stress responses in both metazoans and higher plants. In humans, functional alterations of CRL3s have been associated with various pathologies, including metabolic disorders, muscle, and nerve degeneration, as well as cancer. In this review, we summarize recent discoveries on the function of CRL3s in both metazoans and plants, and discuss their mode of regulation and specificities.
Keywords: BTB domain, Cullin, disease, signalling, ubiquitin
Introduction
Regulation of protein stability by the ubiquitin/proteasome system (UPS) participates in a broad range of physiologically and developmentally controlled processes in all eukaryotes (Ciechanover et al, 2000; Smalle and Vierstra, 2004). A critical step in this pathway involves ubiquitin ligases (also known as E3 enzymes or E3s), which facilitate the transfer of ubiquitin moieties to substrate proteins, as a preparative step for their degradation by the 26S proteasome. Several hundred different E3s have been identified in metazoan and plant genomes, based on specific, commonly shared structural motifs. Among them, CULLIN-RING ubiquitin ligases (CRLs) are the most prevalent class (Petroski and Deshaies, 2005; Hua and Vierstra, 2011). CRLs are multimeric E3s, in which one particular CULLIN protein serves as a molecular scaffold linking up the catalytic module, composed of a RING finger domain protein and a ubiquitin-conjugating (or E2) enzyme, to a specific substrate recognition module, which physically interacts with target proteins. Among the CRL family, the founding member is the SCF (SKP1/CUL1/F-box protein (FBP)) complex (Figure 1A), which employs one of 68 (human) or 700 (Arabidopsis thaliana) FBPs for substrate recognition (Gagne et al, 2002; Jin et al, 2004). Beside CUL1, eukaryotic genomes encode additional cullins (CUL2, CUL3, CUL4, CUL5, and CUL7) (Gieffers et al, 2000; Sarikas et al, 2008) that have likewise been found to form protein complexes with E3 activities, modifying a variety of substrates by using distinct sets of adaptor modules.
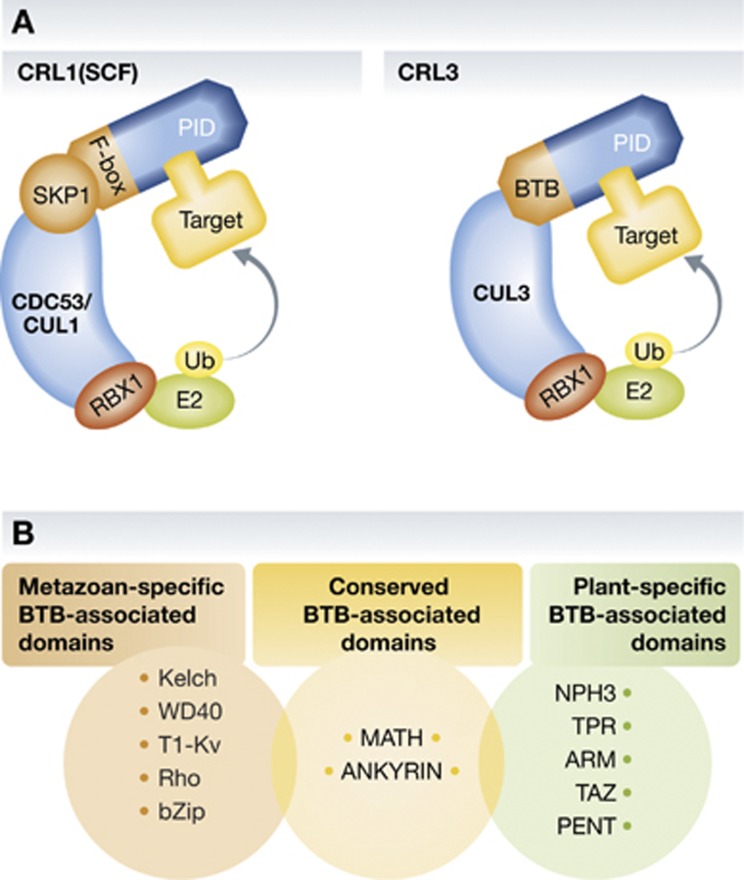
Figure 1.
Structural organization of SCF/CRL1 and the CRL3 complexes. (A) The SCF/CRL1 and the CRL3 complexes share a similar catalytic core module composed of the scaffold proteins CUL1 and CUL3, respectively, and the RING finger protein RBX1 (also known as Hrt1 or ROC1). Single-subunit BTB domain proteins bridge CUL3-RBX1 to substrates, while this function requires an SKP1/FBP heterodimer in SCF/CRL1. Substrate recognition is governed by an independent protein–protein interaction domain (PID) found in most of the FBPs and CUL3-interacting BTB domain proteins. (B) Non-exhaustive list of protein domains is commonly found associated with the BTB domain in CRL3 adaptors. MATH and Ankyrin domains occur in both metazoans and higher plants, while other domains are specific to either kingdom. BTB-KELCH; BTB-WD40; BTB-T1-Kv (voltage-gated potassium channel T1); BTB-Rho (Ras homology); BTB-bZip (basic leucine Zipper); BTB-MATH (Meprin and TRAF homology); BTB-ANKYRIN repeat; BTB-NPH3 (non-phototropic hypocotyl 3); BTB-TPR (Tetratrico Peptide Repeat); BTB-ARM (Armadillo); BTB-TAZ (Transcriptional Adaptor Zinc finger); BTB-PENT (Pentapeptide).
Recent research has firmly established CUL3 as the molecular scaffold of a major class of CRLs controlling different developmental and stress responses (Table I) as well as human pathologies (Table II). CUL3 is a highly conserved CULLIN family member present in the genomes of all eukaryotes. In C. elegans, CUL3 loss-of-function leads to a defect of cytokinesis in single-cell embryos (Kurz et al, 2002), and the deletion of this gene in mouse produces an arrest during early embryogenesis (Singer et al, 1999). In the model plant Arabidopsis thaliana, disruption of the two related CUL3A and CUL3B genes also causes embryo lethality, affecting both embryo pattern formation and endosperm development (Figueroa et al, 2005; Thomann et al, 2005; Gingerich et al, 2007). In contrast to this situation in multicellular organisms, the function of CUL3 orthologues is not essential in either budding or fission yeasts (Geyer et al, 2003; Michel et al, 2003).
Table 1. List of functional CUL3-based ubiquitin ligases and their substrates in different organisms.
BTB-associated domains and substrates supported by strong in vivo evidence are also given.
aSubstrates that may not be degraded and whose ubiquitylation may serve non-proteolytic functions.
bBACURD contains 180 C-terminal residues with no recognizable sequence motif.
Table 2. List of mutations detected in CUL3 and BTB substrate-specific adaptors in patients suffering from indicated diseases.
aDetected mutations, of which predicted effects were not confirmed experimentally.
At the structural level, CUL3 interacts with BTB/POZ (for ‘Bric-a-brac, Tramtrack and Broad Complex/Pox virus and Zinc finger’, hereafter referred to simply as BTB) domain proteins, which function as substrate-specific adaptors (Furukawa et al, 2003; Xu et al, 2003; Pintard et al, 2003b). They bind CUL3 via the BTB domain, and commonly direct substrate specificity through an independent additional protein–protein interaction domain (PID) (Figure 1A), thus uniting the functions of the SKP1/FBP heterodimer in SCF/CRL1 complexes in a single polypeptide. Sequence analyses have so far identified over a dozen different protein domains that are associated, sometimes in combinations, with the BTB domain (Stogios et al, 2005). Some of these are widely distributed throughout eukaryotic genomes (such as the Meprin and TRAF homology (MATH) domain), while others are specific to either metazoans (e.g., the Kelch domain) or plants (e.g., the BTB-non-phototropic hypocotyl 3 (NPH3) domain; Figure 1B). It should be noted that only a subset of all BTB domain proteins actually serve as CRL3 adaptors, and they are set apart from the large fraction of zinc-finger BTB proteins by the presence of an additional paired helical structure (called 3-box motif) positioned C-terminal to the BTB domain, which fulfils an important function in CRL3s assembly analogous to the F-box and SOCS box motifs of other Cullin-based E3s (Zhuang et al, 2009).
It is noteworthy that the number of BTB proteins—and thus potential CRL3s—varies a lot between organisms. The human genome encodes nearly 200 BTB domain proteins, (Stogios et al, 2005), although those lacking the 3-box structures may not be engaged in functional CRL3 complexes, while about 80 BTB proteins have been identified in A. thaliana (Dieterle et al, 2005; Figueroa et al, 2005; Gingerich et al, 2005) and even fewer in D. melanogaster. This contrasts the situation for the SCF/CRL1 complexes, where the large number of FBPs in plants indicates increased versatility (Gagne et al, 2002). However, as will be illustrated below, recent research indicates that this does not mean that CRL3s are of minor importance in plants.
Biological processes involving CRL3s in metazoans
A key regulator of basic cellular functions in metazoans
The ubiquitin/proteasome system is a major regulator of the cell cycle in all eukaryotes, targeting dozens of regulatory proteins for degradation and thus ensuring irreversible cell-cycle stage transitions (Mocciaro and Rape, 2012). While the key E3s for this are the anaphase promoting complex/cyclosome (APC/C) and SCF complexes, more recently CRL3s also entered into the game. In mammalian cells, CRL3s play an essential function in the progression of mitosis and completion of cytokinesis via ubiquitination of Aurora B kinase and thereby preventing chromosomal passenger complex (CPC) accumulation on mitotic chromosomes (Sumara et al, 2007). Aurora B is poly-ubiquitylated on mitotic chromosomes during prometaphase, in a manner dependent on the Kelch-BTB proteins KLHL9 and KLHL13. Rather than triggering its degradation by the proteasome, Aurora B polyubiquitylation during mitosis however seems to serve as a signal for its extraction from chromosomes. During anaphase, another BTB protein, KLHL21, was shown to mono-ubiquitylate Aurora B on microtubules of the spindle midzone (Maerki et al, 2009). Similarly, a CUL3-KLHL22 E3 ligase complex mono-ubiquitylates Polo-like kinase 1 (PLK1) to remove it from kinetochores after chromosomes have achieved bi-orientiation in metaphase (Beck et al, 2013), with this non-degradative PLK1 ubiquitylation being necessary for spindle assembly checkpoint (SAC) silencing and mitotic chromosome segregation. Thus, CUL3 recruits various BTB-containing proteins to target cell-cycle kinases, and possibly also other cell-cycle regulators, at distinct subcellular localizations and at different steps of mitosis. Moreover, the fact that CRL3s not only target substrates for proteasomal degradation, but also reversibly regulate processes by controlling subcellular localization and possibly even the activity and/or interactions of substrates in space and time, significantly expands the versatility and potential roles of CRL3s.
One of the first identified CRL3 substrate adaptors is the well-characterized nematode MATH-BTB protein MEL-26 (reviewed in Pintard et al, 2004). MEL-26 recruits the microtubule-severing katanin protein MEI-1, which is required for meiotic spindle formation but thereafter undergoes rapid CRL3-dependent degradation prior to the onset of mitotic divisions, when its persistence would lead to small and misoriented mitotic spindles. This mechanism appears to be conserved in metazoans, as the mammalian katanin catalytic subunit is also degraded via CRL3-mediated ubiquitylation, involving the Kelch repeat-containing BTB adaptor protein KLHDC5 (Cummings et al, 2009).
In addition to microtubule dynamics, CRL3 regulation also affects the actin cytoskeleton (Chen et al, 2009b). Here, the BTB protein BACURD, which does not contain a known recognizable substrate recognition motif in its C-terminal region, mediates the turnover of the small GTPase RhoA that controls the organization of actin cytoskeleton structure. Failure to degrade RhoA leads to abnormal stress fibres and inhibits the migration capabilities of mammalian cells.
Protein trafficking pathways
CUL3 and its BTB adaptor protein KLHL12 are important regulators of embryonic stem (ES) cell morphology, by affecting the deposition of the extracellular matrix component collagen, which is essential in all metazoans and important for ES cell division (Jin et al, 2012). Similarly to KLHL22, KLHL12 promotes mono-ubiquitylation of its target SEC31, a coat protein of COPII vesicles, and this allows the formation of enlarged COPII vesicle structures required for exocytosis and deposition of rigid, rod-shaped collagen molecules. In addition to secretion, CUL3 has also been implicated in the regulation of late endosome maturation, although the BTB adaptor proteins and their substrates involved in this process remain to be identified (Huotari et al, 2012).
Transcription in developmental signalling
In Drosophila, morphogens such as Hedgehog (Hh) have key roles in developmental processes. A pivotal mediator of Hh signalling, the transcription factor Cubitus Interruptus (Ci), needs to be specifically expressed in and sometimes restricted to specific tissues during development. One way to achieve this is targeted proteolysis, as illustrated by the MATH-BTB protein HIB/SPOP, which is expressed in the Drosophila eye disc posterior to the morphogenic furrow and that promotes Ci degradation to ensure normal eye development (Zhang et al, 2006). This process appears to be conserved in metazoans, as the mammalian SPOP homologue serves as a CRL3 adaptor for degradation of Gli2 and Gli3, two Gli transcription factors homologous to Drosophila Ci (Chen et al, 2009a). Importantly, the work on SPOP CRL3s defines mechanistically how SPOP interacts with its substrates to control transcriptional outputs (Chen et al, 2009a; Zhuang et al, 2009).
In vertebrates, another important signalling protein targeted by a CRL3 complex is Dishevelled (Dsh) (Angers et al, 2006), which constitutes a critical node in cell differentiation/proliferation decisions via the Wnt/β-catenin signalling pathway. Therefore, Dsh protein levels need to be tightly regulated for normal embryonic development, and this is achieved through Wnt signal-dependent Dsh interaction with the BTB-Kelch protein KLHL12, leading to Dsh degradation.
Transcription in stress responses
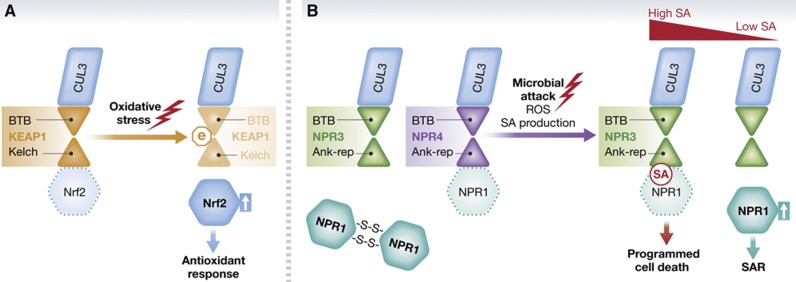
One of the best-understood CRL3 roles in mammalian cells lies in the Keap1-Nrf2 (NF-E2-related factor 2) stress response pathway (for a recent in-depth review, see Taguchi et al, 2011). Nrf2 is a major transcriptional activator that induces expression of numerous protective genes in response to oxidative stress. Under normal growth conditions (i.e., in the absence of cellular stress), the BTB-Kelch substrate adaptor Keap1 triggers Nrf2 ubiquitin-dependent degradation by the proteasome in the cytoplasm (Cullinan et al, 2004; Kobayashi et al, 2004; Zhang et al, 2004). However, several cysteine residues in Keap1 can react with electrophiles produced during stress, which negatively affects CUL3-Keap1 ubiquitin E3 ligase activity (Dinkova-Kostova et al, 2002; Wakabayashi et al, 2004) (Figure 2). Upon oxidative stress, Nrf2 is therefore free to translocate into the nucleus and bind to the anti-oxidant responsive elements (AREs) in the promoter regions of its target genes. Besides Nrf2, Keap1 also recognizes other target proteins, such as the oncogenic kinase IKKβ (Lee et al, 2009) (discussed in more detail below).
Figure 2.
Mode of regulation of CRL3 activity and substrate recognition. (A) Nrf2 is constitutively targeted for Keap1-dependent degradation under normal conditions. In response to oxidative stress, oxidative modifications (denoted as (e), electrophile) on Keap1 impair its activity and result in Nrf2 stabilization. (B) In plant immunity, the transcription coactivator NPR1 is regulated at several levels. In unchallenged cells, NPR1 is predominantly sequestered in the cytoplasm in an oligomeric form through redox-sensitive intermolecular disulphide bonds. Upon pathogen infection, salicylic acid (SA) signals lead to alterations in reduction potential and partially relieves NPR1 to enter the nucleus. High SA concentrations immediately at sites of infection promote its binding to the BTB protein NPR3 and enhance NPR3–NPR1 interaction and subsequent NPR1 degradation, thereby favouring programmed cell death. Lower SA levels in neighbouring cells are insufficient to trigger NPR3-mediated NPR1 ubiquitylation, enabling NPR1 to accumulate and establish systemic acquired resistance (SAR). See text for details.
Other levels of gene expression control
CRL3s can also control gene expression at other levels than transcriptional activation. In mammalian cells, the CRL3 adaptor SPOP mediates ubiquitylation of the Polycomb group protein BMI1 and the variant histone MacroH2A1 (Hernandez-Munoz et al, 2005), apparently affecting their function in a non-proteolytic fashion. CRL3-SPOP function is required for proper MacroH2A1 localization to the inactive X chromosome, and might thus be actively involved in the epigenetic silencing process that leads to X inactivation.
Further downstream in the process of gene expression, CRL3 was recently implicated in the control of translational homeostasis in mammals (Yanagiya et al, 2012). Here, the BTB adaptor KLHL25 promotes degradation of 4E-BP1, a protein that acts as a repressor of translation initiation. 4E-BP1 is only targeted when it is hypophosphorylated and therefore unable to interact with the mRNA cap-binding protein eIF4E, providing a means to control the levels of translation to maintain cellular homeostasis.
Cell death
In mammalian cells, CUL3 and the adaptor KLHL20 ubiquitylate the death-associated protein kinase (DAPK), an apoptosis mediator involved in interferon (IFN)-induced cell death as well as in response to a variety of other stimuli (Lee et al, 2010). Interestingly, this process is controlled at the level of sequestration of the CRL3 adaptor, whereby IFN induction leads to KLHL20 sequestration in promyelocytic leukaemia (PML) nuclear bodies, thus disrupting its interaction with DAPK and stabilizing the kinase (Lee et al, 2010).
The MATH-BTB protein SPOP is also involved in various apoptotic pathways. In Drosophila, SPOP mediates degradation of the Jun kinase phosphatase Puckered (Puc), which is required for apoptosis depending on the tumour necrosis factor (TNF) Eiger during embryonic segmentation (Liu et al, 2009). Human SPOP is involved in the turnover of the death-associated protein DAXX, an anti-apoptotic regulator (Kwon et al, 2006).
An unexpected mechanism of CUL3 action is exemplified by its role in vertebrate caspase activation (Jin et al, 2009). CUL3-dependent ubiquitylation of caspase-8 does not lead to its degradation, but instead promotes its stabilization and thus apoptosis induction. Moreover, CUL3 directly associates with caspase-8 and may not require a BTB-domain adaptor protein, although the presence of an as-yet unidentified copurified adaptor protein cannot fully be ruled out at this stage. Caspase-8 can also be targeted by an unrelated E3, TRAF2, for polyubiquitylation and proteasomal degradation, in this case to shut off cell-extrinsic apoptosis (Gonzalvez et al, 2012).
CULLIN3-RING ligases in human disease
Given the importance of CRL3s in controlling different cellular and developmental processes, it is perhaps of little surprise that they are also linked to the pathology of various human diseases, including metabolic disorders, muscle and nerve degeneration, but also neoplastic diseases. In this regard, gene dosage alterations and expression regulation of CRL3 complex components appear to be the major underlying pathophysiological mechanisms. In addition, elaborate sequencing approaches and database-mining efforts identified a number of specific mutations in patients suffering from several diseases (Table II). This information helps to understand CRL3 pathways at the molecular level and may in the future even allow their targeting via new therapeutic approaches.
Metabolic diseases
Recent exome sequencing approaches identified numerous recessive and dominant mutations in CUL3 and KLHL3 genes in patients suffering from type II pseudohypoaldosteronism (PHAII) or Gordon’s syndrome, a rare disease featuring hypertension due to misbalance between renal salt reabsorption and electrolyte excretion (Boyden et al, 2012; Louis-Dit-Picard et al, 2012). Previously, mutations in WNK (‘with no lysine’) kinases have been correlated with this pathological condition (Wilson et al, 2003). Interestingly, 9 of 16 dominant mutations were found to cluster within the Kelch propeller of KLHL3 and in the vicinity of the other sites implicated in direct substrate binding, suggesting that KLHL3 mutations may abrogate binding and ubiquitylation of targets normally required for modulation of renal salt K+ and H+ handling in response to physiological challenge (Boyden et al, 2012). This notion gains support from recent studies presenting evidence that WNK kinase isoforms may be the critical CUL3-KLHL3 ubiquitylation targets (Ohta et al, 2013; Shibata et al, 2013; Wakabayashi et al, 2013). Disease-causing mutations in KLHL3 abolish interactions with either CUL3 or WNK kinases, and conversely disease mutations within acidic motifs in WNK1 and WNK4 disrupt interaction with KLHL3 (Ohta et al, 2013; Wakabayashi et al, 2013). The CUL3-RhoBTB1 E3 ligase has also been implicated in hypertension and vascular smooth muscle function, via its regulation of PPARγ and RhoA/Rho-kinase pathways (Pelham et al, 2012), and further support for the importance of CUL3 in blood vessel homeostasis comes from the role of the CUL3–BAZF complex in regulating angiogenesis via Notch signalling (Ohnuki et al, 2012). Finally, CUL3-SPOP controls the stability of the pancreatic duodenal homeobox 1 (Pdx1) transcription factor, and thereby affects pancreatic β cell function in glucose homeostasis (Claiborn et al, 2010). Thus, the CRL3 system emerges as an important regulator of metabolic homeostasis, perhaps by regulating responses to specific stress signals.
Dystrophies
Causative mutations for autosomal dominant Retinitis Pigmentosa (adRP), a heritable form of progressive retinal dystrophy that results in blindness and visual field loss, have been identified in the KLHL7 gene (Kigoshi et al, 2011). While not affecting KLHL7 dimerization, the resulting substitutions of a conserved alanine residue (A153T and A153V) in the KLHL7 BACK domain disrupt interaction with CUL3, consistent with the recently established structural requirement for the BACK domain in CUL3 complex assembly (Canning et al, 2013). As E3 ligase activity was strongly reduced upon mutation of this residue (Kigoshi et al, 2011), adaptor protein interaction with CUL3 but not adaptor protein dimerization status appears to determine CUL3 activity (see below). Similarly, Leucine 95 mutation (L95F) of KLHL9, found in patients suffering from a form of distal myopathy of skeletal muscles, results in reduced interaction with CUL3, although with less pronounced effects (Cirak et al, 2010). In Nemaline myopathy (NEM), one of the most common congenital myopathies, dominant mutations have been identified in the KBTBD13 protein (Sambuughin et al, 2010), later found to be a component of a functional CRL3 complex (Sambuughin et al, 2012). The substitutions R248S, K390N, and R408C are located within the β-sheets of the highly conserved second and fifth Kelch repeats and are predicted to disrupt the molecule’s β-propeller structure (Sambuughin et al, 2010). Another BTB protein, Gigaxonin, is mutated in giant axonal neuropathy (GAN), a severe neuropathy of peripheral nerves and the central nervous system that is characterized by neurofilament accumulation and segmental distension of axons (Bomont et al, 2000). Here, at least one patient mutation, leading to a truncation of the Gigaxonin Kelch propeller, was demonstrated to abolish recruitment of a substrate, the microtubule-binding protein MAP1B (Ding et al, 2002). Further CRL3 links to neurodegeneration come from the BTB adaptor protein KCTD7 (Azizieh et al, 2011), which is found mutated in progressive myoclonic epilepsy (EPM3) (Van Bogaert et al, 2007) as well as in neuronal ceroid lipofuscinosis (NCL), a heritable lysosomal storage disease characterized by childhood-onset neurodegeneration. Here, the identified R184C variant alters KCTD7 subcellular localization patterns and abrogates interaction with CUL3 (Staropoli et al, 2012).
Cancer
CUL3-dependent regulation of Nrf2 pathway in cancer. As discussed earlier, Nrf2 is a master transcriptional activator of genes encoding numerous cytoprotective enzymes and is regulated by CRL3-Keap1. It has been suggested that elevated levels of cytoprotective enzymes provide tumour cells with protective capabilities against environmental stresses, a hallmark of malignancy (Gupta and Massague, 2006). Keap1 has been found to be mutated within its substrate-binding Kelch propeller domain (G364C, G430C) in a lung cancer patient and in lung carcinoma cells, respectively, reducing its affinity to Nrf2 and allowing for high constitutive activation of Nrf2 (Padmanabhan et al, 2006). Likewise, other lung cancer patient mutations within the Kelch domain either abolish interaction with Nrf2 (R415G) (Ohta et al, 2008) or inhibit Keap1-mediated degradation of Nrf2 (G333S, L413R) (Singh et al, 2006). Interestingly, the R272C substitution in the Keap1 BACK domain abolishes E3 activity without affecting binding to CUL3. Since R272 lies in a close proximity to those cysteines (C273 and C288) previously implicated in regulating ubiquitylation of Nrf2, this mutation might lead to an aberrant way of binding to CUL3, and subsequent inhibition of ubiquitylation activity (Ohta et al, 2008). Consistent with the fact that interaction between Nrf2 and the BTB adaptor Keap1 is mandatory for Nrf2 degradation and repression, mutations have been found in lung cancer patients that specifically alter amino acids within the DLG or ETGE substrate recognition motifs of Nrf2 (Shibata et al, 2008). Thus, rapid degradation of Nrf2 by CUL3-Keap1 provides a molecular basis for induction of cytoprotective enzymes both in response to stress and during disease development.
Regulation of stress responses by CUL3 in cancer. Interestingly, other CLR ligases share a similar role in critical stress responses and diseases, for instance, the CUL1-dependent IκB (Amit and Ben-Neriah, 2003) and the CUL2-dependent HIF-1α (Kim and Kaelin, 2004) regulation pathways. It is intriguing that the IκB-IKKβ pathway frequently misregulated in cancer is also a target of the CRL3-Keap1 E3 (Lee et al, 2009; Kim et al, 2010), and that genetic disruptions of the CUL3 complex found in lung cancer patients, at both copy number and gene expression levels, lead to upregulation of IKKβ protein levels and activation of the NF-κB signalling pathway (Thu et al, 2011). Similarly, regulation of CUL3 protein levels has been linked to bladder cancer aggressiveness (Grau et al, 2013) and to liver tumorigenesis (Kossatz et al, 2010). Likewise, CUL3 impinges on hypoxia, an essential feature of the solid tumour environment. Under hypoxic conditions, HIF-1 pathways upregulate the BTB protein KLHL20, and the CUL3-KLHL20 E3-ligase targets the PML protein for degradation to potentiate HIF-1 signalling and disease progression in prostate cancer (Yuan et al, 2011). In this case, CRL3-dependent ubiquitylation requires prior sequential PML modification by CDK1/2 phosphorylation and Pin1-mediated prolyl-isomerization.
Biological roles of CRL3s in plants
Whether the pathways and mechanisms regulated by metazoan CRL3s are similarly under CRL3 control in plants remains unclear at present. For instance, while orchestration of the cell cycle by SCF/CRL1 complexes and the APC/C is clearly conserved in the green plant lineage (Marrocco et al, 2010), possible functions of plant CRL3s in these processes remain unknown. Arguing against this possibility, plants lack homologues of the BTB-Kelch proteins that regulate Aurora B in metazoans. Nevertheless, Arabidopsis possess Aurora kinases with localization patterns reminiscent of metazoan Aurora B and with roles in cell division plane orientation (Van Damme et al, 2011); and lack of CUL3 activity leads to defects in division plane orientation in the embryo (Thomann et al, 2005). Furthermore, RNAi depletion of MAB1, 1 of 25 MATH-BTB proteins in maize that is specifically expressed in germ lineages and the zygote, leads to chromosome segregation defects and short spindles during meiosis (Juranic et al, 2012). This phenotype suggests that MAB1 might function in a similar way as the katanin-degrading nematode MEL-26, although the cellular targets of this maize CRL3-MAB1 remain unknown at this stage. Irrespective of potential CLR3 functions in such basic cellular processes, plant CRL3s have been found to possess important novel functions in various plant signalling pathways and in plant hormone biology.
Regulation of plant hormone biosynthesis and signalling
Ethylene is a gaseous hormone that regulates numerous aspects of plant development, but is also important for plant responses to adverse environments, such as interactions with pathogen. ACS5, a member of the type-2 1-aminocyclo-propane-1-carboxylic acid synthases (ACSs) that catalyse a rate-limiting step in ethylene biosynthesis, was the first identified CRL3 substrate in the plant kingdom (Wang et al, 2004). In Arabidopsis, ACS5 is recognized by the Tetratrico Peptide Repeat (TPR) domain-containing BTB adaptor ETO1, leading to both its inactivation and also its degradation to subsequently repress ethylene production. Mutations of ETO1 or downregulation of CUL3 function results in both ACS5 stabilization and ethylene gas overproduction, affecting plant growth and development (Wang et al, 2004; Thomann et al, 2009). Moreover, two other Arabidopsis BTB-TPR proteins closely related to ETO1, designated as ETO1-like (EOL1) and EOL2, also negatively regulate ethylene synthesis via their ability to target ACS5 and other type-2 ACSs for degradation (Christians et al, 2009). The mechanisms by which ETO1/EOL1/EOL3-CRL3s recognize type-2 ACSs to decrease their stability are still not clearly defined, as initial models of a C-terminal recognition sequence in type-2 ACSs that are negatively regulated by phosphorylation, leading to stabilization of these enzymes (Chae et al, 2003; Wang et al, 2004), was subsequently called into question (Christians et al, 2009; Skottke et al, 2011).
In addition to regulating ethylene biosynthesis, CRL3s are also involved in controlling signalling by another important plant stress hormone, abscisic acid (ABA). The Arabidopsis genome encodes six members of the evolutionarily conserved MATH-BTB protein family (called BPM1-6) (Weber et al, 2005), and all six members were found to interact with a subclade of the class I homeobox-leucine zipper (HD-ZIP) transcription factors (Lechner et al, 2011), including ATHB6, a negative regulator of ABA responses (Himmelbach et al, 2002). CUL3-BPM mediates ubiquitylation and turnover of ATHB6, but this process is slowed down in the presence of ABA, affecting some of the plant responses towards ABA, in particular stomatal closure (Lechner et al, 2011). Note that Arabidopsis MATH-BTB proteins also interact with members of the ETHYLENE RESPONSE FACTOR (ERF)/APETALA2 (AP2) to target their proteasomal degradation, as recently described for WRINKLED1, an ERF/AP2 transcription factor involved in fatty acid metabolism (Chen et al, 2013).
A striking feature of the MATH-BTB class of adaptors is there selective expansion and diversification in some worm and plant species during evolution (Thomas, 2006; Gingerich et al, 2007). For instance, while the Arabidopsis genome encodes only six members of this family, there are 74 predicted MATH-BTB proteins in rice (Gingerich et al, 2007), and it has been speculated that this expansion may reflect positive selection in response to rapid adaption of possible targets such as pathogen proteins (Thomas, 2006; Gingerich et al, 2007). One major route of pathogen entry into plant cells is stomata opening, and not surprisingly pathogens evolved virulence factors to counter stomatal closure and facilitate invasion (Zeng et al, 2010). As noted above, Arabidopsis MATH-BTB mediates ABA-dependent stomatal closure and is therefore strongly expressed in guard cells (Lechner et al, 2011). As many microbial pathogens are known to exploit the ubiquitin pathway to evade host defence mechanisms (Jiang and Chen, 2011), it will be of particular interest to explore possible functions of this class of CRL3 substrate adaptors in innate immunity and bacterial diseases.
Precedent for important plant immunity roles stems from another class of plant BTB proteins, which includes Arabidopsis NPR1, a master regulator of plant immunity that controls the onset of systemic acquired resistance (SAR) to a broad spectrum of pathogens. SAR requires the signalling molecule salicylic acid (SA), a plant hormone synthesised in response to phytopathogen challenge. Upon SA perception, NPR1 moves to the cell nucleus, where it interacts with TGA-bZIP transcription factors to induce defence gene expression. NPR1 combines a BTB domain with an ankyrin repeat motif (Cao et al, 1997). Interestingly, NPR1 in the nucleus is itself degraded by the proteasome in a CUL3-dependent manner, most likely to prevent activation of NPR1 target genes in unchallenged cells (Spoel et al, 2009). On the other hand, NPR1 is phosphorylated in SAR-induced cells, creating an IκB-like phosphodegron motif and also resulting in degradation via the CRL3 pathway to sustain maximum levels of target gene expression.
Despite its BTB domain, however, NPR1 does not directly interact with CUL3; instead, NPR3 and NPR4, two Arabidopsis NPR1 paralogues, are responsible for recruiting it to CUL3 (Fu et al, 2012). Consistently, npr3 npr4 double mutant cells accumulate increased NPR1 protein levels and exhibit enhanced disease resistance, a phenotype opposite to the npr1 mutation. Strikingly, NPR3 and NPR4 not only interact with CUL3 through their BTB domains, but also bind SA, thus representing the long-sought-after receptors for this plant hormone (Fu et al, 2012). This recognition mode resembles that of the plant hormone auxin, whose receptors belong to a six-gene clade of Arabidopsis FBPs including TIR1. Auxin binding to TIR1 in the context of the CRL1–TIR1 E3 complex stabilizes interactions between TIR1 and its Aux/IAA transcription repressor substrates, resulting in their ubiquitin-dependent degradation to trigger auxin transcriptional responses (reviewed in Santner and Estelle, 2009). Indeed, high levels of SA promote NPR3–NPR1 interactions, leading to NPR1 ubiquitylation and degradation at the site of infection; this prevents SAR establishment and leads to effector-triggered cell death of infected cells. However, SA has the opposite effect on NPR4–NPR1 interactions. NPR4 binds NPR1 in the absence of SA and is responsible for constitutive NPR1 degradation in non-infected cells, thus preventing spurious activation of defence gene expression. NPR4’s higher affinity for SA allows it to detect lower SA levels in the cells neighbouring sites of infection, resulting in disruption of NPR4–NPR1 binding, NPR1 stabilization, and establishment of SAR (Figure 2) (Fu et al, 2012).
Light signalling
CRL3s have recently been implicated in phototropism, a process that allows plants to change their growth direction in response to the location of the light source. This involves a large plant-specific protein family called NRL (Pedmale et al, 2010), which usually contain a BTB domain, a central NPH3 domain, and a C-terminal coiled-coil domain. One of them, NPH3, is involved in early phototropism signal transduction downstream of the PHOT1/NPH1 photoreceptor, a light-activated serine/threonine protein kinase (Motchoulski and Liscum, 1999). Arabidopsis NPH3 interacts with CUL3 to ubiquitylate PHOT1 at the plasma membrane in response to blue light (Roberts et al, 2011). An interesting aspect of this process is that PHOT1 poly-ubiquitylation, and thus proteasomal degradation is only stimulated by high-intensity blue light, likely to allow receptor desensitization. On the other hand, low-intensity blue light promotes only PHOT1 mono- and multi-mono-ubiquitylation, which may trigger receptor internalization from the plasma membrane (Roberts et al, 2011). It will now be interesting to address the molecular basis of PHOT1 recognition with respect to light-induced structural changes, as well as the regulation of the CRL3 adaptor NPH3, whose phosphorylation status is itself light dependent (Pedmale and Liscum, 2007). Other members of the NRL family include ROOT PHOTOTROPISM2 (RPT2), which is also involved in phototropic signalling (Sakai et al, 2000), and NPY1/MAB4/ENP (Furutani et al, 2011 and references therein), which regulates Arabidopsis organ formation by controlling transport of the plant hormone auxin. Whether these BTB proteins also function as bona fide CRL3 adaptors remains to be established.
Plant BTB proteins regulate not only blue light responses, but have also been implicated in negatively regulating photomorphogenesis in response to red light (Christians et al, 2012). Here, a pair of so-called LIGHT-RESPONSE BTB proteins (LRB1/2) acts redundantly to limit accumulation of the red light photoreceptors phytochrome B and D, and consequently lrb1 lrb2 double mutant plants are selectively hypersensitive to red light. Both LRB1 and LRB2 interact with CUL3A/B proteins in the nucleus, and may thus target these photoreceptors following red light-induced photoconversion and nuclear import; however, at this stage it remains to be determined whether LRB1/2 are directly involved in phytochrome turnover, for example, via light-induced direct interactions.
Mechanisms of CRL3 regulation
Ubiquitylation and fate of CRL3 substrate proteins can be controlled on various levels, such as substrate recognition, adaptor protein binding, or ligase activity. In the following, we will summarize several key mechanisms of CRL3 regulation.
Regulation by NEDD8 and CAND1
An essential feature for all CULLIN-RING ligases is the dynamic covalent modification of the Cullin scaffold by the ubiquitin-like protein Nedd8/Rub1 (Petroski and Deshaies, 2005; Merlet et al, 2009). Like ubiquitylation, neddylation requires the sequential action of an Nedd8-activating enzyme (NAE), the APPBP1-UBA3 heterodimer, and an NEDD8-conjugating enzyme, UBC12. In addition, DCN1 (defective in cullin neddylation 1) cooperates with the CRL RING finger subunit RBX1 to stimulate the neddylation reaction, thus exerting an E3-like activity for NEDD8 (Scott et al, 2010, 2011). In a reverse reaction, NEDD8 is removed from cullins by the isopeptidase activity of the zinc-dependent metalloenzyme CSN5, a component of the eight-subunit COP9 signalosome (CSN) complex. NEDD8 attachment stimulates processes such as recruitment of ubiquitin-charged E2 enzymes and positioning of the E2 active site for ubiquitin transfer onto substrates (Duda et al, 2008; Saha and Deshaies, 2008). On the other hand, deneddylation promotes dissociation of CRL E3 complexes and Cullin-RING binding to the inhibitory factor CAND1 (cullin-associated and neddylation-disassociated 1), a regulator of CRL complex assembly that binds to un-neddylated cullins (see below) (Liu et al, 2002).
Several lines of evidence suggest that not just cullin neddylation, but dynamic cycles of CRL neddylation and deneddylation are key in CRL activation. CUL3 neddylation was first described in the context of CRL3-MEL26-dependent MEI-1 in C. elegans (Pintard et al, 2003a), with both neddylation and deneddylation being required for MEI-1 degradation. Likewise, while CUL3 neddylation is a prerequisite for Keap1-dependent in vivo ubiquitylation of Nrf2 in mammals, this process is equally affected by siRNA-mediated knockdown of CAND1 (Lo and Hannink, 2006). It is likely the CAND1 regulates CRL3s in a manner similar to its control of SCF/CRL1 complexes, where CAND1 serves as a ‘protein exchange factor’ that allows swapping of FBP substrate adaptors and thereby adjustment of the SCF repertoire to varying substrate demand (Pierce et al, 2013; Wu et al, 2013; Zemla et al, 2013). Therefore, defective Nrf2 degradation upon knockdown of the CAND1 exchange factor may reflect an inability to form additional CRL3-Keap1 modules. Quantitative proteomics results showing as much as 10% of cellular CUL3 bound to CAND1 indicate that this may indeed represent the steady-state levels of CAND1-CUL3 engagement during the course of adaptor assembly (Bennett et al, 2010).
CRL3 dimerization
Most (if not all) of the CRL3s dimerize through the BTB domains of their substrate recognition subunits, which can form tightly intertwined dimers with an extensive hydrophobic interface, as revealed by a crystal structure of the PLZF BTB (Ahmad et al, 1998; Merlet et al, 2009). The best-documented example of a role for such dimerization in substrate recognition comes from biochemical and structural analyses of human Keap1 (McMahon et al, 2006; Taguchi et al, 2011), where two Keap1 molecules in the homodimer bind to a single Nrf2 substrate molecule that employs two distinct binding sites (termed as DLG and ETGE motifs) with differential affinities; these dual interaction sites appear to be important to position Nrf2 in an orientation optimal for its ubiquitylation. Similar features characterize the assembly of CRL3-SPOP and CRL3-HIB complexes with their substrates, with SPOP dimerization allowing engagement of multiple SPOP binding sites in a single substrate (Zhang et al, 2009; Zhuang et al, 2009; Errington et al, 2012). Thus, the use of dimeric substrate recognition modules can increase the avidity for substrates with multiple degrons. Recent structural work on CUL3-KLHL11 modules illustrates how BTB dimerization occurs in the context of a CULLIN3-RING ligase complex, revealing a unique N-terminal extension in CUL3 required for interaction with the adaptor-BTB protein-specific 3-box and also providing a template for modelling quaternary CRL3 assembly such as the one that targets Nrf2 (Canning et al, 2013).
Dimerization of CUL3 E3s may have additional roles beyond substrate recruitment. SPOP-mediated CRL3 dimerization provides a bivalent geometry with dual docking sites for ubiquitin-charged E2 enzyme, thus enhancing both rate and processivity of polyubiquitin chain formation (Zhuang et al, 2009). This notion is supported by functional studies on CRL3-KLHL7, where an adRP disease mutant of KLHL7 is defective for CUL3 binding but still capable of dimerization. Via interaction with wild-type KLHL7, this mutant forms complexes containing dimeric KLHL7 but only a single CUL3 subunit, and such complexes also exhibit reduced ubiquitylation activity (Kigoshi et al, 2011).
In addition to dimerization, higher orders of CRL3 oligomerization have also been observed, such as formation of stable tetramers by the BTB protein KCTD11 (Correale et al, 2011). Moreover, recent structural studies on SPOP complexes also revealed higher-order self-assembly creating oligomeric CRL3-SPOP with enhanced E3 activity (Errington et al, 2012). Here again, E3 oligmerization may increase the catalytic rate of ubiquitylation by locally increasing the concentration of associated E2 enzymes. Interestingly, SPOP can also mediate formation of heterodimeric CRL3 complexes when interacting with its paralogue, SPOP-like (SPOPL). SPOPL has however restricted oligomerization capabilities, and its incorporation therefore limits higher-order SPOP-CRL3 self-assembly and attenuates E3 activity (Errington et al, 2012).
Subcellular sequestration of CRL3s
CRL3 substrate adaptors can become sequestered in specific subcellular compartments, separate from their substrates, to additionally control CRL3 ubiquitylation activity. As discussed earlier, the CRL3-KLHL20 substrate DAPK is stabilized upon KLHL20 sequestration in PML nuclear bodies in IFN-α-treated cells, forming the basis for IFN sensitivity of tumour cells (Lee et al, 2010). In a different example, CUL3 restriction to the cytoplasm stabilizes Steroid Receptor Coactivator-3 (SRC-3), a coactivator of nuclear receptors, in the nucleus under basal conditions. Retinoic acid treatment, however, leads to CUL3 translocation in the nucleus, allowing ubiquitylation and degradation of its substrate SRC-3 (Ferry et al, 2011).
Post-translational modification of substrates and adaptors
E3 recognition sequences in substrates, called ‘degrons’ (Varshavsky, 1991), frequently undergo post-translational modification that affects their recognition by substrate binding domains. This has been found particularly common for phosphorylation, which often leads to formation of a phosphorylation-dependent recognition sequence of ‘phospho-degron’. For instance, signal-dependent SRC-3 ubiquitylation by CRL3-SPOP depends on a phospho-degron (Ferry et al, 2011; Li et al, 2011). Similarly, NPR1 phosphorylation in the presence of SA stimulates its CUL3-dependent turnover in Arabidopsis (Spoel et al, 2009). In other cases, however, phosphorylation inhibits rather than promotes substrates recruitment by CRL3s. For example, phosphorylation of the conserved threonine residue within the Nrf2 ETGE degron (see above) has the capability of disrupting its interaction with Keap1 and stabilising Nrf2 in cells (Lo et al, 2006). Similarly, substrate peptides containing phospho-serine or phospho-threonine substitutions in their SPOP binding element abolish in vitro binding to SPOP (Zhuang et al, 2009), although the occurrence and significance of phosphorylation-modulated recognition in vivo remains to be established.
Like target proteins, substrate adaptor proteins also are subjected to regulation by post-translational modifications. The best example is again Nrf2 regulation by CUL3-Keap1, where oxidative stress and generation of electrophiles result in modification of thiol groups in Keap1 cysteine residues, and hence attenuation of E3 activity (see Figure 2A and earlier discussion) (Cullinan et al, 2004; Kobayashi et al, 2004; Zhang et al, 2004); although it is still under debate how exactly Keap1 post-translational modification affects Nrf2 ubiquitylation at the molecular level (Taguchi et al, 2011). Finally, albeit not constituting a bona fide covalent post-translational modification, the earlier discussed SA binding-modulated interactions of plant BTB proteins NPR3 and NPR4 with their target NPR1 provide an additional example of CRL3 stress sensor abilities regulated at the substrate adaptor level (Fu et al, 2012).
Perspectives
While CRL3s are not unique in the way they are regulated or the way they target a broad range of regulatory proteins for ubiquitylation, there is still much that remains to be discovered to fully appreciate the nature of this class of E3 enzymes. One challenge is to better understand degradative versus non-degradative CRL3 roles at the mechanistic levels. While most of the BTB-containing adaptor proteins described in the current literature are linked to ubiquitin-dependent substrate proteolysis, several examples have highlighted non-proteolytic CRL3 functions. In human cells, CRL3-SPOP promotes MacroH2A mono-ubiquitylation and subsequent relocalization to the X chromosome, an important step for stable X chromosome inactivation (Hernandez-Munoz et al, 2005). However, as described above, the same E3 complex is also involved in ubiquitin-dependent proteolysis of other substrates like Gli2/3 (Chen et al, 2009a) or SRC-3 (Li et al, 2011), most likely via K48-linked polyubiquitin chains. Similarly, Aurora B and caspase-8 are regulated by non-proteolytic ubiquitylation and Sec31A and PLK1 are mono-ubiquitylated by CRL3s in human cells (Jin et al, 2009; Maerki et al, 2009; Beck et al, 2013). Another interesting example is the light intensity-dependent differential regulation of poly- versus mono-/multi-mono-ubiquitylation of PHOT1 by CRL3-NPH3 in plants (Roberts et al, 2011). How CRL3 E3 ligases can switch from poly-ubiquitin chain formation to mono-ubiquitylation, sometimes on the same substrate, remains to be understood. In this respect, it has to be mentioned that E2 enzymes also have major influence on the outcome of ubiquitylation and ubiquitin chain specificity. However, since the cognate E2s for CRL3s in vivo remain to be decisively clarified, many of the studies reported so far have relied on utilization of the promiscuous E2 enzyme UbcH5, which may have affected the respective conclusions from CRL3 in vitro ubiquitylation. Nevertheless, the findings on non-proteolytic ubiquitylation by CRL3s certainly expand the possible roles of CRL3s and suggest that additional cellular signalling functions of CULLIN3 ligases remain to be discovered. Future studies will also be needed to discover downstream effectors and to understand how CRL3s regulate protein localization and function in the above examples.
Another still puzzling issue concerns the real repertoire of cellular substrates for each CRL3. The degron motifs recognized by E3s are often short, poorly conserved peptide sequences within a target protein. For example, the substrate adaptor SPOP binds a five-residue peptide (called SPOP binding consensus, SBC) including a stretch of serine/threonine residues, which is present in several of its substrates, such as Puc, MacroH2A, Ci, and Daxx (Zhuang et al, 2009). However, these sequences are not sufficiently informative to easily identify them at the proteome level, rendering bioinformatics-based CRL3 substrate prediction uncertain, if not outright impossible. Therefore, various strategies to establish novel high-throughput approaches for identifying E3 substrates have been proposed (Yen and Elledge, 2008; Merbl and Kirschner, 2009). Given that substrate phosphorylation may not be a general prerequisite for CRL3 substrate recognition, in vitro ubiquitylation performed with commercially spotted protein arrays may offer a convenient approach for such studies in this case, as illustrated by the successful identification of several potential CUL3-KLHL22 substrates, many of which are involved in cell-cycle functions (Beck et al, 2013).
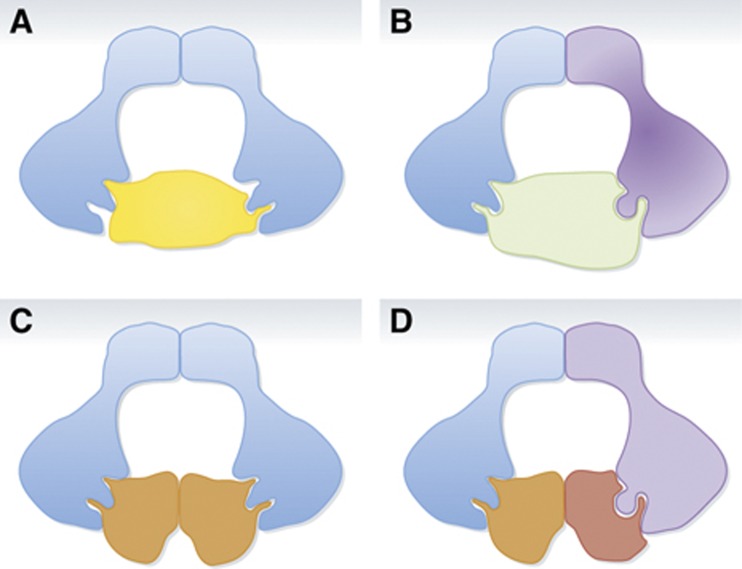
Finally, an intriguing question concerns the structural complexity of CRL3s. As indicated above, a hallmark of CRL3s is their capacity to dimerize through their substrate receptors. In particular, homodimerization is well established for a number of these complexes, but heterodimerization through related MATH-BTB subunits has also been reported (Errington et al, 2012). Moreover, we have recently obtained evidence for in vivo heterodimerization among all six MATH-BTB proteins in Arabidopsis (our unpublished results). Therefore, one may speculate about the actual heterodimerization potential within larger, expanded classes of CRL3 substrate adaptors in some metazoan and plant genomes, such as the predicted 52 human BTB-Kelch proteins, the 74 members of the rice MATH-BTB family, or the ∼30 NPH3-BTB proteins in Arabidopsis (Thomas, 2006; Gingerich et al, 2007; Canning et al, 2013). Furthermore, some substrates themselves are also known to dimerize, such as dimerization of the transcription factor AtHB6 with its paralogues AtHB5 and AtHB16 (Lechner et al, 2011). It is therefore tempting to speculate that through combinatorial heterodimerization at the levels of both adaptors and their substrates, CRL3s may be able to dramatically diversify their substrate repertoire and to adapt their recognition potential in many ways (Figure 3).
Figure 3.
Speculative models on substrate accommodation by BTB protein dimerization. The model in (A) corresponds to the well-described binding of the Nrf2 transcription factor via two different binding sites to the Kelch domains of the Keap1 homodimer (McMahon et al, 2006). Models (B–D) represent speculative variations involving substrate recognition by BTB protein heterodimers (B) or cases considering both BTB protein and substrate homo- and hetero-dimerization (C, D).
Glossary.
- ABA
abscisic acid
- adRP
autosomal dominant Retinitis Pigmentosa
- BTB/POZ
Bric-a-brac, Tramtrack and Broad Complex/Pox virus and Zinc finger
- CAND1
Cullin-Associated and Neddylation-Disassociated 1
- Ci
Cubitus Interruptus
- CPC
Chromosomal Passenger Complex
- CRL
CULLIN-RING Ubiquitin Ligase
- DCN1
Defective in Cullin Neddylation 1
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin ligase
- FBP
F-box protein
- IFN
Interferon
- MATH
Meprin and TRAF homology
- NAE
Nedd8-Activating Enzyme
- NPH3
Nonphototropic Hypocotyl 3
- Nrf2
NF-E2-related factor 2
- PID
protein interaction domain
- PLK1
Polo-Like Kinase 1
- SA
Salicylic Acid
- SAC
Spindle Assembly Checkpoint
- SAR
Systemic Acquired Resistance
- SCF
SKP1/CUL1/F-box protein
- TPR
Tetratrico Peptide Repeat
- UPS
Ubiquitin/Proteasome System
Acknowledgments
We wish to thank the members of our laboratories for helpful suggestions on the manuscript. The work in Genschik’s laboratory is supported by CNRS and Labex NetRNA (ANR-10-LABX-36) and IS is supported by the IGBMC, ATIP-AVENIR program from CNRS and INSERM, Sanofi-Aventis and Fondation ARC pour la recherche sur le cancer.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahmad KF, Engel CK, Prive GG (1998) Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci USA 95: 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit S, Ben-Neriah Y (2003) NF-kappaB activation in cancer: a challenge for ubiquitination- and proteasome-based therapeutic approach. Semin Cancer Biol 13: 15–28 [DOI] [PubMed] [Google Scholar]
- Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT (2006) The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol 8: 348–357 [DOI] [PubMed] [Google Scholar]
- Azizieh R, Orduz D, Van Bogaert P, Bouschet T, Rodriguez W, Schiffmann SN, Pirson I, Abramowicz MJ (2011) Progressive myoclonic epilepsy-associated gene KCTD7 is a regulator of potassium conductance in neurons. Mol Neurobiol 44: 111–121 [DOI] [PubMed] [Google Scholar]
- Beck J, Maerki S, Posch M, Metzger T, Persaud A, Scheel H, Hofmann K, Rotin D, Pedrioli P, Swedlow JR, Peter M, Sumara I (2013) Ubiquitylation-dependent localization of PLK1 in mitosis. Nat Cell Biol 15: 430–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW (2010) Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143: 951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M (2000) The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 26: 370–374 [DOI] [PubMed] [Google Scholar]
- Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Valimaki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR et al. (2012) Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce MW, Boronenkov IV, Anderson RA (2008) Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem 283: 8678–8686 [DOI] [PubMed] [Google Scholar]
- Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Marsden BD, Gileadi O, Knapp S, von Delft F, Bullock AN (2013) Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem 288: 7803–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT (2009a) Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev 23: 1910–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F (2009b) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35: 841–855 [DOI] [PubMed] [Google Scholar]
- Chen L, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H (2013) Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell (advance online publication, 21 June 2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD (2012) The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol 160: 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22: 442–451 [DOI] [PubMed] [Google Scholar]
- Cirak S, von Deimling F, Sachdev S, Errington WJ, Herrmann R, Bonnemann C, Brockmann K, Hinderlich S, Lindner TH, Steinbrecher A, Hoffmann K, Prive GG, Hannink M, Nurnberg P, Voit T (2010) Kelch-like homologue 9 mutation is associated with an early onset autosomal dominant distal myopathy. Brain 133(Pt 7): 2123–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborn KC, Sachdeva MM, Cannon CE, Groff DN, Singer JD, Stoffers DA (2010) Pcif1 modulates Pdx1 protein stability and pancreatic beta cell function and survival in mice. J Clin Invest 120: 3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correale S, Pirone L, Di Marcotullio L, De Smaele E, Greco A, Mazza D, Moretti M, Alterio V, Vitagliano L, Di Gaetano S, Gulino A, Pedone EM (2011) Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie 93: 715–724 [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CM, Bentley CA, Perdue SA, Baas PW, Singer JD (2009) The Cul3/Klhdc5 E3 ligase regulates p60/katanin and is required for normal mitosis in mammalian cells. J Biol Chem 284: 11663–11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Thomann A, Renou JP, Parmentier Y, Cognat V, Lemonnier G, Muller R, Shen WH, Kretsch T, Genschik P (2005) Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J 41: 386–399 [DOI] [PubMed] [Google Scholar]
- Ding J, Liu JJ, Kowal AS, Nardine T, Bhattacharya P, Lee A, Yang Y (2002) Microtubule-associated protein 1B: a neuronal binding partner for gigaxonin. J Cell Biol 158: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P (2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA 99: 11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington WJ, Khan MQ, Bueler SA, Rubinstein JL, Chakrabartty A, Prive GG (2012) Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure 20: 1141–1153 [DOI] [PubMed] [Google Scholar]
- Ferry C, Gaouar S, Fischer B, Boeglin M, Paul N, Samarut E, Piskunov A, Pankotai-Bodo G, Brino L, Rochette-Egly C (2011) Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proc Natl Acad Sci USA 108: 20603–20608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, Deng XW (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, Dong X (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y (2003) Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 5: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Furutani M, Sakamoto N, Yoshida S, Kajiwara T, Robert HS, Friml J, Tasaka M (2011) Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 138: 2069–2078 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA (2003) BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 12: 783–790 [DOI] [PubMed] [Google Scholar]
- Gieffers C, Schleiffer A, Peters JM (2000) Cullins and cell cycle control. Protoplasma 211: 20–28 [Google Scholar]
- Gingerich DJ, Gagne JM, Salter DW, Hellmann H, Estelle M, Ma L, Vierstra RD (2005) Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J Biol Chem 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Gingerich DJ, Hanada K, Shiu SH, Vierstra RD (2007) Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19: 2329–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S, Pham VC, Stephan JP, Lill J, Ashkenazi A (2012) TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell 48: 888–899 [DOI] [PubMed] [Google Scholar]
- Grau L, Luque-Garcia JL, Gonzalez-Peramato P, Theodorescu D, Palou J, Fernandez-Gomez JM, Sanchez-Carbayo M (2013) A quantitative proteomic analysis uncovers the relevance of CUL3 in bladder cancer aggressiveness. PLoS One 8: e53328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J (2006) Cancer metastasis: building a framework. Cell 127: 679–695 [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA 102: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Huotari J, Meyer-Schaller N, Hubner M, Stauffer S, Katheder N, Horvath P, Mancini R, Helenius A, Peter M (2012) Cullin-3 regulates late endosome maturation. Proc Natl Acad Sci USA 109: 823–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Chen ZJ (2011) The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol 12: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18: 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M (2012) Ubiquitin-dependent regulation of COPII coat size and function. Nature 482: 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A (2009) Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137: 721–735 [DOI] [PubMed] [Google Scholar]
- Juranic M, Srilunchang KO, Krohn NG, Leljak-Levanic D, Sprunck S, Dresselhaus T (2012) Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24: 4974–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigoshi Y, Tsuruta F, Chiba T (2011) Ubiquitin ligase activity of Cul3-KLHL7 protein is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J Biol Chem 286: 33613–33621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI (2010) Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal 22: 1645–1654 [DOI] [PubMed] [Google Scholar]
- Kim WY, Kaelin WG (2004) Role of VHL gene mutation in human cancer. J Clin Oncol 22: 4991–5004 [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossatz U, Breuhahn K, Wolf B, Hardtke-Wolenski M, Wilkens L, Steinemann D, Singer S, Brass F, Kubicka S, Schlegelberger B, Schirmacher P, Manns MP, Singer JD, Malek NP (2010) The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. J Clin Invest 120: 3820–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Pintard L, Willis JH, Hamill DR, Gonczy P, Peter M, Bowerman B (2002) Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295: 1294–1298 [DOI] [PubMed] [Google Scholar]
- Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol JH, Baek SH, Chiba T, Tanaka K, Bang OS, Joe CO, Chung CH (2006) BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem 281: 12664–12672 [DOI] [PubMed] [Google Scholar]
- Lechner E, Leonhardt N, Eisler H, Parmentier Y, Alioua M, Jacquet H, Leung J, Genschik P (2011) MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev Cell 21: 1116–1128 [DOI] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, Li CW, Ding Q, Liao TL, Lai CC, Lin AC, Chang YH, Tsai SF, Li LY, Hung MC (2009) KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 36: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH (2010) The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J 29: 1748–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, O'Malley BW (2011) Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 30: 4350–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10: 1511–1518 [DOI] [PubMed] [Google Scholar]
- Liu J, Ghanim M, Xue L, Brown CD, Iossifov I, Angeletti C, Hua S, Negre N, Ludwig M, Stricker T, Al-Ahmadie HA, Tretiakova M, Camp RL, Perera-Alberto M, Rimm DL, Xu T, Rzhetsky A, White KP (2009) Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science 323: 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Hannink M (2006) CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol 26: 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M (2006) Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J 25: 3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis-Dit-Picard H, Barc J, Trujillano D, Miserey-Lenkei S, Bouatia-Naji N, Pylypenko O, Beaurain G, Bonnefond A, Sand O, Simian C, Vidal-Petiot E, Soukaseum C, Mandet C, Broux F, Chabre O, Delahousse M, Esnault V, Fiquet B, Houillier P, Bagnis CI et al. (2012) KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 44: 456–460 S451–453 [DOI] [PubMed] [Google Scholar]
- Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, Sumara I, Peter M (2009) The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol 187: 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K, Bergdoll M, Achard P, Criqui MC, Genschik P (2010) Selective proteolysis sets the tempo of the cell cycle. Curr Opin Plant Biol 13: 631–639 [DOI] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD (2006) Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a ‘tethering’ mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem 281: 24756–24768 [DOI] [PubMed] [Google Scholar]
- Merbl Y, Kirschner MW (2009) Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci USA 106: 2543–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlet J, Burger J, Gomes JE, Pintard L (2009) Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66: 1924–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel JJ, McCarville JF, Xiong Y (2003) A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J Biol Chem 278: 22828–22837 [DOI] [PubMed] [Google Scholar]
- Mocciaro A, Rape M (2012) Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. J Cell Sci 125(Pt 2): 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E (1999) Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Nam S, Min K, Hwang H, Lee HO, Lee JH, Yoon J, Lee H, Park S, Lee J (2009) Control of rapsyn stability by the CUL-3-containing E3 ligase complex. J Biol Chem 284: 8195–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki H, Inoue H, Takemori N, Nakayama H, Sakaue T, Fukuda S, Miwa D, Nishiwaki E, Hatano M, Tokuhisa T, Endo Y, Nose M, Higashiyama S (2012) BAZF, a novel component of cullin3-based E3 ligase complex, mediates VEGFR and Notch cross-signaling in angiogenesis. Blood 119: 2688–2698 [DOI] [PubMed] [Google Scholar]
- Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T (2013) The CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 451: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S (2008) Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 68: 1303–1309 [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21: 689–700 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Celaya RB, Liscum E (2010) Phototropism: mechanism and outcomes. Arabidopsis Book 8: e0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E (2007) Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J Biol Chem 282: 19992–20001 [DOI] [PubMed] [Google Scholar]
- Pelham CJ, Ketsawatsomkron P, Groh S, Grobe JL, de Lange WJ, Ibeawuchi SR, Keen HL, Weatherford ET, Faraci FM, Sigmund CD (2012) Cullin-3 regulates vascular smooth muscle function and arterial blood pressure via PPARgamma and RhoA/Rho-kinase. Cell Metab 16: 462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan SO, Deshaies RJ (2013) Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B (2003a) Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol 13: 911–921 [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M (2004) Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J 23: 1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, Peter M (2003b) The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316 [DOI] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, Zheng N, Hannink M, Genschik P, Liscum E (2011) Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3(NPH3). Plant Cell 23: 3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K (2000) RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuughin N, Swietnicki W, Techtmann S, Matrosova V, Wallace T, Goldfarb L, Maynard E (2012) KBTBD13 interacts with Cullin 3 to form a functional ubiquitin ligase. Biochem Biophys Res Commun 421: 743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuughin N, Yau KS, Olive M, Duff RM, Bayarsaikhan M, Lu S, Gonzalez-Mera L, Sivadorai P, Nowak KJ, Ravenscroft G, Mastaglia FL, North KN, Ilkovski B, Kremer H, Lammens M, van Engelen BG, Fabian V, Lamont P, Davis MR, Laing NG et al. (2010) Dominant mutations in KBTBD13, a member of the BTB/Kelch family, cause nemaline myopathy with cores. Am J Hum Genet 87: 842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Sarikas A, Xu X, Field LJ, Pan ZQ (2008) The cullin7 E3 ubiquitin ligase: a novel player in growth control. Cell Cycle 7: 3154–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Rongo C (2006) KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell 17: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA (2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA (2010) A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell 39: 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S (2008) Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA 105: 13568–13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP (2013) Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci USA 110: 7838–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JD, Gurian-West M, Clurman B, Roberts JM (1999) Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 13: 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S (2006) Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A (2011) Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet 7: e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sobieszczuk DF, Poliakov A, Xu Q, Wilkinson DG (2010) A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev 24: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staropoli JF, Karaa A, Lim ET, Kirby A, Elbalalesy N, Romansky SG, Leydiker KB, Coppel SH, Barone R, Xin W, MacDonald ME, Abdenur JE, Daly MJ, Sims KB, Cotman SL (2012) A homozygous mutation in KCTD7 links neuronal ceroid lipofuscinosis to the ubiquitin-proteasome system. Am J Hum Genet 91: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG (2005) Sequence and structural analysis of BTB domain proteins. Genome Biol 6: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, Peter M (2007) A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell 12: 887–900 [DOI] [PubMed] [Google Scholar]
- Taguchi K, Motohashi H, Yamamoto M (2011) Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123–140 [DOI] [PubMed] [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U, Genschik P (2005) Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J 43: 437–448 [DOI] [PubMed] [Google Scholar]
- Thomann A, Lechner E, Hansen M, Dumbliauskas E, Parmentier Y, Kieber J, Scheres B, Genschik P (2009) Arabidopsis CULLIN3 genes regulate primary root growth and patterning by ethylene-dependent and -independent mechanisms. PLoS Genet 5: e1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH (2006) Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res 16: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu KL, Pikor LA, Chari R, Wilson IM, Macaulay CE, English JC, Tsao MS, Gazdar AF, Lam S, Lam WL, Lockwood WW (2011) Genetic disruption of KEAP1/CUL3 E3 ubiquitin ligase complex components is a key mechanism of NF-kappaB pathway activation in lung cancer. J Thorac Oncol 6: 1521–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bogaert P, Azizieh R, Desir J, Aeby A, De Meirleir L, Laes JF, Christiaens F, Abramowicz MJ (2007) Mutation of a potassium channel-related gene in progressive myoclonic epilepsy. Ann Neurol 61: 579–586 [DOI] [PubMed] [Google Scholar]
- Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E (2011) Arabidopsis alpha Aurora kinases function in formative cell division plane orientation. Plant Cell 23: 4013–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (1991) Naming a targeting signal. Cell 64: 13–15 [DOI] [PubMed] [Google Scholar]
- Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, Chiga M, Kikuchi E, Nomura N, Mori Y, Matsuo H, Murata T, Nomura S, Asano T, Kawaguchi H, Nonoyama S, Rai T, Sasaki S, Uchida S (2013) Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep 3: 858–868 [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA 101: 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Weber H, Bernhardt A, Dieterle M, Hano P, Mutlu A, Estelle M, Genschik P, Hellmann H (2005) Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol 137: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson FH, Kahle KT, Sabath E, Lalioti MD, Rapson AK, Hoover RS, Hebert SC, Gamba G, Lifton RP (2003) Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci USA 100: 680–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA (2013) CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun 4: 1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW (2003) BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321 [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N (2012) Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell 46: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322: 923–929 [DOI] [PubMed] [Google Scholar]
- Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, Tsai CH, Chen HY, Chiang CT, Lai CK, Lu LT, Chen CH, Gu DL, Pu YS, Jou YS, Lu KP, Hsiao PW, Shih HM, Chen RH (2011) A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 20: 214–228 [DOI] [PubMed] [Google Scholar]
- Zemla A, Thomas Y, Kedziora S, Knebel A, Wood NT, Rabut G, Kurz T (2013) CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun 4: 1641. [DOI] [PubMed] [Google Scholar]
- Zeng W, Melotto M, He SY (2010) Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr Opin Biotechnol 21: 599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B, Jiang J (2009) Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci USA 106: 21191–21196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J (2006) A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell 10: 719–729 [DOI] [PubMed] [Google Scholar]
- Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA (2009) Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell 36: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]