Summary
Cell surface multi-protein complexes are synthesized in the endoplasmic reticulum (ER) where they undergo co-translational membrane integration and assembly. The quality control mechanisms that oversee these processes remain poorly understood. We show that less hydrophobic transmembrane (TM) regions derived from several single-pass TM proteins can enter the ER lumen completely. Once mislocalized, they are recognized by the Hsp70 chaperone BiP. In a detailed analysis for one of these proteins, the αβT cell receptor (αβTCR), we show that unassembled ER-lumenal subunits are rapidly degraded, whereas specific subunit interactions en route to the native receptor promote membrane integration of the less hydrophobic TM segments, thereby stabilizing the protein. For the TCR α-chain, both complete ER import and subunit assembly depend on the same pivotal residue in its TM region. Thus, membrane integration linked to protein assembly allows cellular quality control of membrane proteins and connects the lumenal ER chaperone machinery to membrane protein biogenesis.
Introduction
In eukaryotic cells, nascent proteins of the secretory pathway enter the endoplasmic reticulum (ER) co-translationally as unfolded polypeptide chains, where they are often glycosylated, form disulfide bonds and oligomerize to ultimately reach their native structure. These events must pass scrutiny by the ER quality control machinery before the protein is allowed to continue along the secretory pathway (Braakman and Bulleid, 2011). Proteins that fail ER quality control standards are retro-translocated to the cytosol and degraded by the proteasome in a process called ER-associated degradation (ERAD) (Vembar and Brodsky, 2008). Lumenal portions of proteins undergo quality control by the chaperone machinery of the ER lumen that is composed of two major branches. The first is centered on the ER-resident Hsp70 chaperone BiP and its co-chaperones (Otero et al., 2010). BiP recognizes exposed hydrophobic peptide stretches as a hallmark of incompletely folded proteins (Blond-Elguindi et al., 1993; Flynn et al., 1991). The second branch relies on the ER-lumenal lectin chaperone system that uses oligosaccharides attached to nascent polypeptide chains as a sensor of their folding status (Hebert and Molinari, 2012).
However, in the case of integral membrane proteins, which comprise roughly one third of the human proteome, we currently do not understand major aspects of their quality control mechanisms. Like their soluble counterparts, they fold, are post-translationally modified and scrutinized before being transported to the cell surface or other intracellular organelles (Houck and Cyr, 2012). Membrane integration generally occurs co-translationally via the Sec61 translocon where hydrophobic sequences stop further transfer into the ER lumen and allow the protein to be integrated into the ER-membrane (Shao and Hegde, 2011). Multipass transmembrane (TM) proteins, however, often possess some TM segments of marginal hydrophobicity (Hessa et al., 2007). In fact, more than 25% of the TM helices in multi-spanning TM proteins of known structure have a predicted unfavorable free energy of membrane integration (Elofsson and von Heijne, 2007; White and von Heijne, 2008). Accordingly, these segments by themselves are not expected to stably integrate into the membrane, and indeed, in some cases they can temporarily enter the ER lumen and must be retrieved and inserted into the membrane post-translocationally (Kanki et al., 2002; Lu et al., 2000; Skach et al., 1994). Of note, TM sequences of low hydrophobicity are often functionally relevant (Illergard et al., 2011) and involved in intra- or intermolecular assembly steps in the lipid bilayer. Thus, the very sequences that are more difficult to be integrated into the ER membrane often determine the function of a TM protein and guide its assembly processes. Accordingly, it is likely that the cell has developed mechanisms of quality control to ensure proper interaction and integration of TM sequences during membrane protein biogenesis. With only a few exceptions (Houck and Cyr, 2012; Lemberg, 2013) little is known if or how intra-membrane assembly steps are linked to mechanisms of cellular quality control in membrane protein biogenesis or if membrane integration itself is scrutinized.
To address this poorly understood and apparently prevalent issue, we chose to examine a number of oligomeric single-pass TM proteins whose subunit interactions are focused on membrane-embedded polar residues. We find that several less hydrophobic TM regions of single-pass integral membrane proteins can enter the ER-lumen completely and engage the ER-chaperone machinery, thus providing a link to the ER quality control system. Building on this finding, we performed a detailed analysis on one of these proteins with less hydrophobic TM sequences, the αβT cell receptor (αβTCR). Rigorous quality control mechanisms act on the αβTCR, as only properly assembled receptors are able to traverse the secretory pathway to the cell surface (Klausner et al., 1990), whereas unassembled receptor chains undergo ERAD (Bonifacino et al., 1989; Huppa and Ploegh, 1997; Lippincott-Schwartz et al., 1988; Yu et al., 1997). Of particular relevance for our study, the ionizable amino acids located in the TM regions of each receptor chain that affect their overall hydrophobicity are critical for both αβTCR assembly as well as degradation if assembly fails (Bonifacino et al., 1990; Cosson et al., 1991; Manolios et al., 1990). Thus, the αβTCR is a well-suited model substrate to delineate the mechanism of how less hydrophobic TM sequences impact on the quality control of integral membrane proteins.
Results
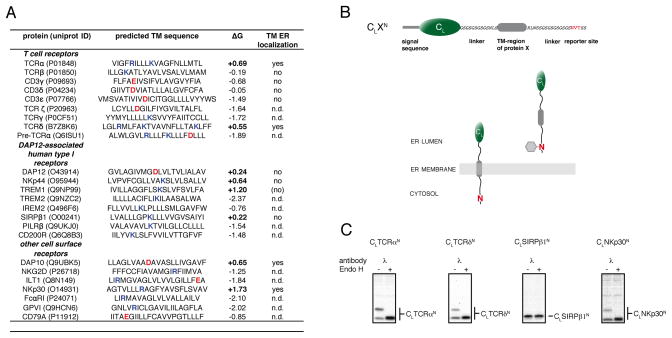
Transmembrane sequences of low hydrophobicity can completely enter the ER lumen
To investigate the role of less hydrophobic TM sequences in protein quality control we examined various immune receptor subunits that possess polar residues in the TM regions that are known to be crucial for correct subunit assembly but also compromise their hydrophobicity (Call and Wucherpfennig, 2007; Feng et al., 2006). Importantly, the selected proteins are all single pass membrane proteins allowing us to dissect membrane integration from subunit assembly for individual TM sequences. We hypothesized that membrane integration itself might be connected to quality control, and thus used a published algorithm (Hessa et al., 2007) to predict the free energy of membrane integration for 24 different receptor-derived TM sequences (Figure 1A). For 8 out of 24 TM sequences, a positive free energy of membrane integration was computed arguing they can not stably integrate into the membrane by themselves. To assess this prediction experimentally, we designed a glycosylation-based reporter system in which the TM sequences of interest were connected to the well-folded, non-glycosylated constant domain of the antibody λ light chain, CL (Hellman et al., 1999). C-terminal of the TM sequence, we engineered an Asn-Val-Thr glycosylation site (Figure 1B). Co-translational membrane integration would localize the reporter sequence to the cytosol, whereas complete transport into the ER lumen would expose the glycosylation site to the ER oligosaccharide transferase machinery allowing its modification (Figure 1B). We detected glycosylation at the C-terminal reporter site for four of the eight TM sequences with a predicted unfavorable free energy of membrane integration (Figures 1C and S1), whereas none of the constructs examined that contained TM sequences with a calculated negative free energy for membrane insertion were glycosylated (Figure 1A). Thus, similar to individual TM segments of multi-pass TM proteins (Kanki et al., 2002; Lu et al., 2000; Ojemalm et al., 2012; Sadlish et al., 2005; Skach et al., 1994), less hydrophobic TM segments of single pass TM proteins can enter the ER lumen completely.
Figure 1. TM sequences of low hydrophobicity can enter the ER lumen.
(A) Analysis of TM sequences of human single pass cell surface receptor subunits whose assembly is focused on intramembrane polar residues. TM sequence assignments were taken from the uniprot database (www.uniprot.org). ΔG values were calculated according to (Hessa et al., 2007) (http://dgpred.cbr.su.se/). Positive ΔG values predict an unfavorable free energy of membrane integration (bold). Basic residues are highlighted in blue, acidic residues in red. TM regions that were tested experimentally for lumenal localization are indicated as yes/no, and those not examined are described as not determined (n.d.). The bracketed no for TREM1 indicates a very weak band detected for the C-terminally glycosylated species. Data for TCRβ and CD3γ, δ and ε are derived from Figure 2.
(B) Top: schematic outline of the reporter construct used to monitor ER-import of TM sequences. It is composed of the non-glycosylated immunoglobulin λ light chain CL domain connected by flexible linkers and short amino acid stretches derived form the TCR α-chain to the TM sequence of interest followed by a C-terminal glycosylation site (NVT, marked in red). Bottom: A cartoon showing that membrane integration of the reporter construct would not allow its C-terminal NVT site to become glycosylated, whereas complete ER import would expose it to the ER glycosylation machinery allowing it to become glycosylated (shown as grey hexagon).
(C) COS-1 cells transfected with the indicated constructs were metabolically labeled and cell lysates were immunoprecipitated with anti-mouse λ antiserum. Precipitated material was split and treated with or without EndoH prior to SDS-PAGE analysis. See also Figure S1.
Unassembled authentic TCR α-chains can fully enter the ER lumen
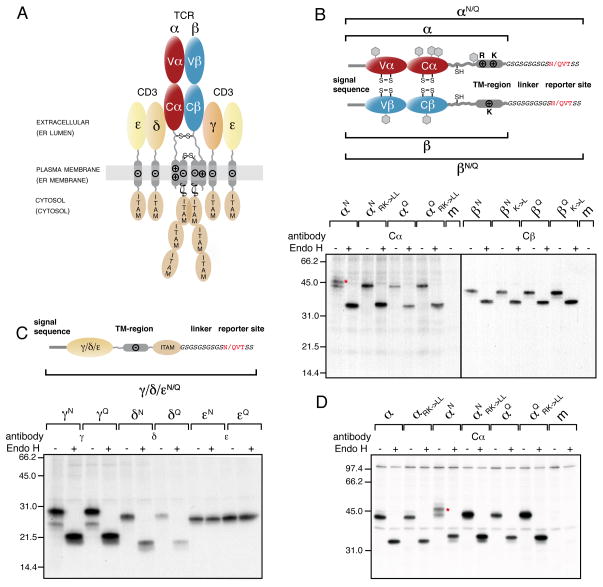
This finding provides a possible hint towards mechanisms of quality control for integral membrane proteins that act on the TM segment itself. Therefore, to investigate this in more detail and deduce a molecular mechanism for a possible TM segment-focused quality control of integral membrane proteins, we chose the αβTCR (Figure 2A) whose assembly and degradation have been linked to the very residues that limit the hydrophobicity of its TM regions (Bonifacino et al., 1990; Call et al., 2002; Cosson et al., 1991; Fayadat and Kopito, 2003; Manolios et al., 1990; Shin et al., 1993). First, we determined if full-length unassembled αβTCR chains, all of which are single pass TM proteins with hydrophilic residues in their TM regions (Figures 1A and 2A), could also enter the ER-lumen. We engineered C-terminal tags that possessed either NVT (glycosylation consensus sites) or QVT (control sites that can not be glycosylated) motifs to the α- and β-chain (designated as αN or αQ and βN or βQ, respectively, Figure 2B) and each of the CD3 co-receptor chains (designated as γN or γQ, δN or δQ and εN or εQ, respectively, Figure 2C) of the human A6 αβTCR (Utz et al., 1996). To dissect early steps in the TCR biosynthesis process we performed metabolic labeling experiments in COS-1 cells (Hall et al., 1991). We observed two species for αN, separated by the size of a single glycan moiety (Figure 2B). Enzymatic deglycosylation by EndoH revealed that the two species originate from different extents of glycosylation of the α-chain (Figure 2B). Inspection of the α-chain construct with the QVT motif at the C-terminus (αQ) revealed a single band that migrated with the faster mobility band observed for αN (Figure 2B). Thus, the additional band corresponds to glycosylation of the C-terminal reporter site and indicates that an isolated full-length TCR α-chain can enter the ER lumen completely, including its designate TM region.
Figure 2. The TCR α-chain TM region is mislocalized into the ER lumen.
(A) The αβTCR consists of the clonotypic α- and β-chains, made up of one variable (Vα and Vβ, respectively) and one constant domain (Cα and Cβ, respectively) each and the co-receptor chains, CD3γ, δ and ε, as well as ζ. The receptor has three topological layers whose final disposition in the cell as well as their localization during the initial biosynthesis steps (shown in brackets) are indicated. Transmembrane basic (+) and acidic (−) residues and interchain disulfide bonds are shown. Immunoreceptor tyrosine-based activation motifs (ITAMs) initiate downstream signaling events upon TCR-MHC binding.
(B) Top: Outline and nomenclature of glycosylation reporter constructs to monitor ER-membrane integration of the TCR α- and β-chain. The engineered C-terminal glycosylation sites (NVT) and respective control sites (QVT) are marked in red. Predicted endogenous glycosylation sites are shown as grey hexagons. Bottom: Glycosylation analysis of radiolabeled TCR α- and β-chain reporter constructs in COS-1 cells. Mutations of the TM basic residues against Leu are indicated (RK->LL for the α-chain or K->L for the β-chain, respectively). Lysates from metabolically labeled cells were immunoprecipitated with antibodies specific for the human Cα or Cβ domains and treated with or without EndoH prior to SDS-PAGE analysis. The red asterisk marks the α-chain glycosylated at its C-terminal reporter site.
(C) Top: Schematics and nomenclature of analogous reporter constructs to monitor ER-membrane integration of the CD3γ, δ and ε chain. Bottom: Glycosylation analysis of radiolabeled CD3γ, δ and ε chain reporter constructs in COS-1 cells were performed as in (B) using the indicated chain-specific antibodies.
(D) Glycosylation analysis of radiolabeled TCR α-chain constructs in Jurkat J.RT-T3.1 cells. Nomenclature of the constructs is as in (B). The red asterisk marks the α-chain glycosylated at its C-terminal reporter site. Where indicated, radiolabeled proteins were deglycosylated with Endo H after immunoprecipitation with anti-Cα antibodies (m: mock transfection). See also Figure S2.
If the two TM basic residues (Arg and Lys, Figure 2B) of the α-chain TM region were replaced by Leu (αNRK->LL), only a single band was observed, which migrated identically to αQ (Figure 2B). Thus, full entry of the TCR α-chain TM region into the ER lumen is dependent on the presence of these basic residues. In contrast to the α-chain, neither the glycosylation site-tagged β-chain (Figure 2B) nor any of the CD3 chains (Figure 2C) were further modified at C-terminal reporter sites and thus appear to be properly integrated into the ER membrane in agreement with their predicted negative free energy of membrane integration (Figure 1A).
As glycosylation of sites near the C-terminus can be inefficient, we used subcellular fractionation and sodium carbonate extraction experiments to confirm that our glycosylation reporter system adequately monitored membrane integration. Indeed, we found that constructs that did not become C-terminally glycosylated could only be extracted with detergent in keeping with stable membrane integration (Figure S2A). Conversely, α-chain constructs for which we had observed C-terminal glycosylation could be partially extracted with sodium bicarbonate or urea, indicative of improper membrane integration (Figure S2A). Importantly, the wild-type α-chain behaved like the α-chain with the reporter glycosylation site (Figure S2A). We obtained identical results for another human anti-hemagglutin TCR (HA TCR) α-chain with a different ER-import sequence and Vα domain (Hewitt et al., 1992), revealing that these regions do not influence mislocalization of the α-chain into the ER lumen (Figures S2B and S2C).
To assess if TCR α-chains can also completely enter the ER-lumen in the context of authentic T cells, we used the Jurkat cell line J.RT-T3.1. It is deficient in the expression of the α-chain (Saito et al., 1987), allowing analysis of our reporter constructs. In this cell line αN restored surface expression of the αβTCR to the same extent as the wild type α-chain (data not shown), arguing that the reporter site does not interfere with receptor assembly and transport. Importantly, similar to COS-1 cells, we observed glycosylation of both the A6 (Figure 2D) and HA TCR (Figure S2C) α-chains at the C-terminal reporter site, but not for the αNRK->LL or αQ constructs (Figures 2D and S2C). Thus, even though TCRβ and all co-receptor chains are present in the J.RT-T3.1 line (Saito et al., 1987) and expression levels of the transfected TCR α-chain and the co-receptor chains were comparable (Figure S2D), the TCR α-chain can also fully enter the ER in the authentic environment of T cells.
The ER-resident Hsp70 chaperone BiP binds to ER-lumenal TM regions
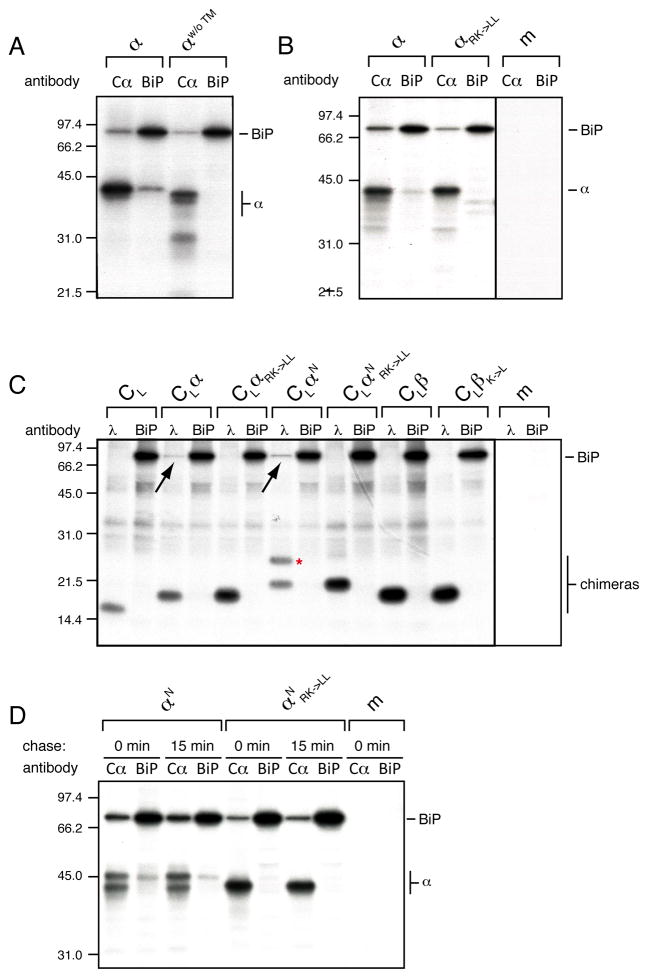
ER-localized TM sequences provide a possible point of action for the lumenal ER quality control system that has not been investigated yet. Exposed hydrophobic peptide stretches are the typical signature of Hsp70 binding sites, so we asked if the ER-resident Hsp70 chaperone BiP could recognize lumenally disposed TM regions. We co-expressed the various α-chain constructs with hamster BiP (Lee et al., 1999) and analyzed their interactions. In agreement with previous work, the full-length α-chain co-immunoprecipitated with BiP (Figure 3A), which had been presumed to occur via interactions with either the Vα or Cα domains that are in the ER lumen during TCR biosynthesis (Suzuki et al., 1991) (Figure 2A). However, when an α-chain construct without its TM region was similarly analyzed, we could no longer co-immunoprecipitate the α-chain with BiP (Figure 3A). Thus, the presence of the TM region appeared necessary to allow co-immunoprecipitation of the α-chain with BiP by our antibody. Reciprocally, BiP was co-immunoprecipitated with the α-chain even in the absence of its TM region arguing that additional binding sites for BiP exist in the lumenal domains (Figure 3A). Our finding led us to speculate that BiP might directly recognize the TCR α-chain TM region. In agreement with this idea, when we used the membrane-integrated αRK–>LL (Figure 2B), we no longer co-immunoprecipitated the α-chain with BiP and the amount of BiP co-immunoprecipitating with αRK–>LL was decreased (Figure 3B). Conversely, the β-chain co-immunoprecipitated with BiP independent of the presence of its TM region (Figure S3A) or of its single TM Lys residue (Figure S3B) via the domains that are normally exposed to the ER lumen.
Figure 3. BiP binds the TM region of ER-lumenal α-chains.
(A) Metabolically labeled lysates from COS-1 cells transfected with the full-length α-chain construct (α) or one devoid of its TM region (αw/o ™) and BiP were split and immunoprecipitated with either an anti-Cα antibody or anti-BiP antiserum and analyzed by SDS-PAGE.
(B) COS cells co-transfected with BiP and either the wild type α-chain construct (α) or one in which the TM basic residues were mutated to leucine (αRK->LL) were analyzed as in (A).
(C) Interaction of radiolabeled BiP with CL-based reporter constructs (see Figure 1). Metabolically labeled lysates of cells co-transfected with BiP and one of the indicated constructs were divided and immunoprecipitated with either anti-mouse λ antiserum or anti-BiP antiserum. The construct glycosylated at its C-terminal reporter site is marked with a red asterisk. Co-immunoprecipitating BiP is indicated with a black arrow.
(D) Analysis of the interaction of αN and αNRK->LL with BiP. Cells co-transfected with the indicated constructs and BiP were subjected only to a 30 min pulse (0 min chase) or chased for an additional 15 min prior to cell lysis and immunoprecipitation with the indicated antibodies (m: mock transfection). See also Figure S3.
To further test if BiP was binding to the TCR α-chain TM region when it was mislocalized to the ER lumen, we generated fusion proteins of the CL domain, which is not a BiP substrate (Hellman et al., 1999), tagged to different α-chain TM region variants and tested for their interaction with BiP. We found that if the CL domain was fused to the TM region of the TCR α-chain (CLα), BiP co-immunoprecipitated with the chimeric protein (Figure 3C), but not vice versa arguing that regions N-terminal of the TM region influence the ability of our antibody to co-immunoprecipitate substrates with BiP. Interaction with BiP was abolished if the basic residues in the α-chain TM region were replaced with Leu (CLαRK->LL, Figure 3C), corroborating the notion that BiP directly binds to an ER-lumenal α-chain TM region. CLαN with the C-terminal glycosylation reporter site (Figure 2B) recapitulated the behavior of the authentic α-chain, as it populated two glycoforms if the TM basic residues were present and only a single glycoform if the basic amino acids in the TM region were replaced with Leu (CLαN RK->LL). Only CLαN but not CLαN RK->LL co-immunoprecipitated BiP (Figure 3C). Thus, independent of the protein context, the α-chain TM region defines localization and is directly bound by BiP once ER-lumenal. In agreement with this idea, no interaction of BiP with fusion protein containing any of the TCR β-chain TM region variants was observed (Figure 3C).
To further verify our assumption that BiP directly binds to the α-chain TM region we examined which glycoform of αN BiP bound. If indeed the α-chain can only be co-immunoprecipitated with BiP via its TM region, we would expect only the completely ER-lumenal species of αN (i.e., only the species glycosylated at its reporter site) to co-immunoprecipitate with BiP, which is exactly what we observed (Figure 3D). As we obtained this result after a 30 min labeling pulse either without any subsequent chase period or after a short 15 min chase (Figure 3D), entry of at least a portion of the α-chain TM region into the ER is a reasonably fast process. Furthermore, these data suggest that the less glycosylated species has not fully entered the ER as opposed to glycosylation at the reporter site being inefficient, since BiP does not co-immunoprecipitate this species. Taken together our data demonstrate that the ER-lumenal chaperone BiP directly binds to the putative TM region of the TCR α-chain once it enters the ER lumen. To establish if the binding of BiP to lumenally exposed TM segments was a more general phenomenon, we examined BiP interactions with the reporter constructs that we had tested for their ability to enter the ER lumen (Figure 1). BiP co-immunoprecipitated with all the chimeric proteins that were C-terminally glycosylated, indicative of their entry into the ER-lumen (Figures 1A and S3C), providing a more general link between unstable membrane integration of TM segments and the lumenal ER quality control machinery.
Mislocalization underlies rapid degradation of the α-chain
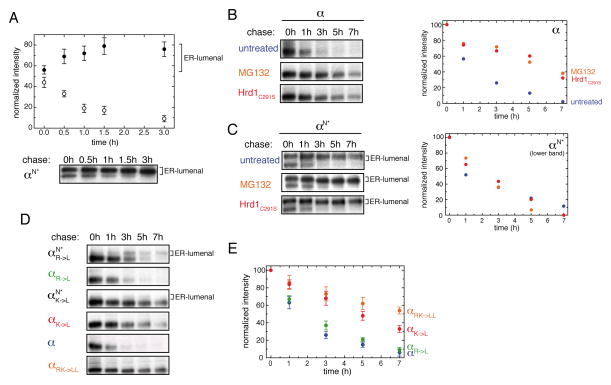
In order for quality control to rely on ER-import of the TM segments of unassembled α-chains, all unassembled α-chains would need to ultimately pass entirely into the ER lumen, a behavior that has not been previously described. To examine this possibility, pulse-chase experiments were performed. As αN became additionally glycosylated at a cryptic glycosylation site over time, complicating quantifications, we used a construct devoid of this cryptic glycosylation site, denoted as αN* (for details see Figure S4A). Immediately after pulse-labeling, a little over half the α-chain was glycosylated at the engineered C-terminal site (Figure 4A). Over time, the less glycosylated species disappeared while glycosylation at the reporter site increased until almost the entire pool of the α-chain ultimately became modified (Figure 4A). The increase in the amount of C-terminally glycosylated α-chain was paralleled by more α-chain co-immunoprecipitating with BiP over time (Figure S4B). These data argue that some unassembled α-chains enter the ER initially while the rest continue to do so post-translationally.
Figure 4. Degradation of the TCR α-chain correlates with localization of its TM region into the ER lumen.
(A) Quantification of the two glycospecies for αN*. Cells expressing the αN* construct were pulse-labeled and chased for the indicated times before immunoprecipitation and SDS-PAGE analyses (Bottom). Signal averages for the ER-lumenal, C-terminally glycosylated species are shown as closed circles, data for the species not C-terminally glycosylated are indicated with open circles (n=4±SD) (Top). Note that in this and most subsequent experiments a construct designated as αN* was used instead of αN (for details see Figure S4A).
(B) Inhibition of α-chain degradation was carried out by addition of MG132 (orange) or co-expression of Hrd1C291S (red) as indicated and compared to the degradation of the α-chain in the absence of any treatment (blue). Quantifications are shown on the right.
(C) Disappearance of the lower, not C-terminally glycosylated band for αN* was measured and quantified under the same conditions as in (B). Quantifications are shown on the right.
(D) Pulse-chase experiments were performed for different α-chain constructs containing either a single, none, or both TM basic residues replaced by Leu as indicated. For mutants with single TM basic residue replaced, ER-lumenal localization was additionally assessed by the αN* reporter construct. Species glycosylated at their C-terminal reporter site are indicated with “ER-lumenal” on the side.
(E) Quantifications (n=3±SD) of the degradation kinetics for the wt α-chain (blue), αRK->LL (orange), αR->L (green) and αK->L (red). See also Figure S4.
To further verify continued post-translational entry of the α-chain TM region as opposed to the less glycosylated species being rapidly degraded, we blocked degradation of the α-chain with the proteasome inhibitor MG132 (Huppa and Ploegh, 1997; Yu et al., 1997) or a dominant negative mutant of the E3 ubiquitin ligase Hrd1 (Hrd1C291S) (Kikkert et al., 2004; Nadav et al., 2003). Both, MG132 and Hrd1C291S expression stabilized the wild type α-chain against degradation (Figure 4B). When our glycosylation reporter construct was similarly examined, we continued to observe the disappearance of the α-chain species that was not glycosylated at its C-terminal reporter site (Figure 4C). These data support the idea that a precursor:product relationship exists between the C-terminally unglycosylated and glycosylated forms of this α-chain construct. The fact that the additional C-terminal glycosylation of the α-chain increased its stability (Figures S4A and C) allowed us to perform this type of analysis without interfering with the ERAD machinery. When we performed similar experiments on the chimeric proteins examined in Figure 1 we also observed further post-translational ER import for several of the TM sequences that entered the ER lumen initially, which was most striking for the construct comprising the NKP30 TM region (Figure S4D).
To more fully define the relationship between assembly and full egress into the ER-coupled quality control, we generated mutants of the α-chain with each of the basic residues in the TM region singly exchanged against Leu, as their respective roles in assembly have been previously determined (Call et al., 2002). Constructs were made in the presence of the glycosylation reporter site (to monitor import) or in its absence (to monitor degradation kinetics). The TM Arg single mutant of the reporter construct (αN*R->L) still entered the ER lumen (Figure 4D), arguing that the central lysine alone was sufficient to prevent stable ER membrane integration. Of note, it did so primarily post-translationally and thus was initially apparently recognized as a stop-transfer sequence. In agreement with it being able to enter the ER lumen, the αR->L construct was degraded only slightly slower than the wild-type α-chain (Figures 4D an E). In contrast, when the TM Lys was mutated to Leu (αN*K->L), the resulting protein did not detectably enter the ER lumen and the αK->L construct was significantly stabilized (Figures 4D and E). In agreement with this connection between mislocalization and rapid turnover, changing the single TM Lys residue in the β-chain to Leu had no effect on either localization (Figure 2B) or degradation (Figure S4E). Taken together, we observed a strong correlation between the ability of the α-chain to completely enter the ER lumen and its rapid degradation. These findings thus provide a link between localization of the α-chain TM region and degradation of the protein, explaining the destabilizing role of unpaired basic residues in the α-chain TM region.
Interaction with CD3δ and ε retains the TCR α-chain in the membrane, stabilizing it against degradation
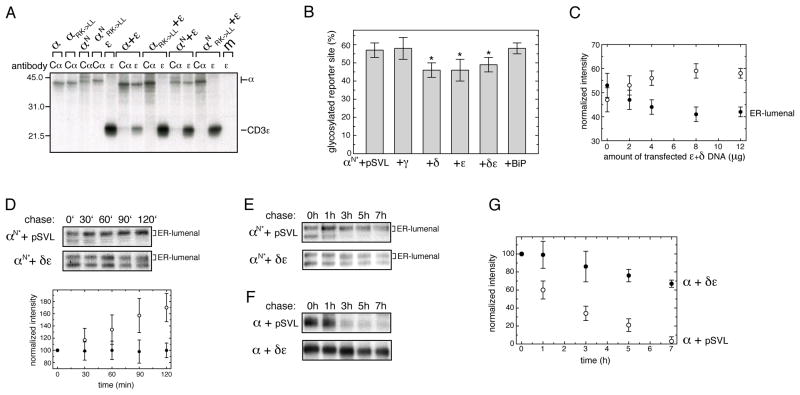
For quality control of unassembled α-chains to rely on mislocalization to the ER lumen, correct assembly must prevent this from occurring. Elements of the CD3 co-receptor are the best candidates for retention of the α-chain in the ER membrane, as they possess acidic residues in their TM regions (Figure 2A) and assemble with the TCR α chain prior to the β- and ζ-chains during TCR biosynthesis (Kearse et al., 1995). Therefore, we assessed the impact of different CD3 chains on the localization of the α chain. In agreement with previous studies (Call et al., 2002; Manolios et al., 1990), no significant co-immunoprecipitation between CD3γ and the TCR α-chain was observed (Figure S5A). CD3δ and ε, however, strongly interacted with the α-chain (Figures 5A and S5B). These interactions are dependent on the acidic residues in the TM regions of the two CD3 chains and the lysine residue in the α-chain TM segment (Call et al., 2002). As expected, the interactions between these proteins were abolished by replacing the α-chain TM basic residues (Figures 5A and S5B). Most importantly, only the αN species that was not glycosylated at its C-terminal reporter site could be co-immunoprecipitated with CD3δ and ε (Figures 5A and S5B). Accordingly, we next asked if this interaction could inhibit TCR α-chain mislocalization and thus influence the amount of α-chain that fully enters the ER. Indeed, less of the C-terminal reporter site was glycosylated on the α-chain if either CD3δ or ε were co-expressed individually or together to allow formation of the naturally occurring CD3δε dimer (Manolios et al., 1991) (Figure 5B). In keeping with the absence of association of CD3γ with the α-chain, there was no reduction of C-terminal glycosylation of αN* upon CD3γ co-expression (Figure 5B) nor did BiP over-expression change the amount of reporter-site glycosylation (Figure 5B). Thus, BiP does not seem to play an active role in α-chain localization but rather binds its TM region once it becomes ER-localized. In agreement with the low efficiency of αβTCR complex assembly (Minami et al., 1987), we only found a modest reduction of TCR α-chain entering the ER lumen upon CD3δ or ε co-expression. By increasing the amounts of CD3δ and ε DNA co-transfected to a six-fold excess over that of the α-chain DNA, we further increased α-chain integration to ~60% (Figure 5C). Importantly, even though CD3δ and ε could not prevent the entire α-chain pool from entering the ER, together they completely inhibited further import of the TCR α-chain into the ER lumen over time (Figures 5D and E) and stabilized the TCR α-chain against degradation (Figures 5F and G). The fact that both CD3δ and ε were needed for efficient retention and stabilization (Figures S5C–F) might be partially attributable to the intrinsic instability of CD3δ when expressed alone (Wileman et al., 1993) which is stabilized upon co-expression of CD3ε (data not shown). However, CD3ε is stable by itself, it could not retain the α-chain in the membrane or stabilize it against degradation as efficiently as CD3δ and ε together (Figures 5D–G and S5C–F). Taken together, we observed a very good correlation between the retention of the α-chain reporter construct in the membrane and stabilization of the wt α-chain against degradation when the CD3 co-receptor subunits were co-expressed in the various combinations.
Figure 5. CD3δ and ε retain the α-chain in the ER membrane and stabilize it against degradation.
(A) The indicated α-chain constructs were co-expressed with the CD3ε chain and radiolabeled lysates were immunoprecipitated with antibodies to Cα or CD3ε. Mutations of the TM basic residues in the α-chain to Leu are indicated (RK->LL) (m: mock transfection).
(B) Quantification of the reporter site glycosylation species for αN* as a percent of the total amount of protein immunoprecipitated was performed by phosphorimager analysis. Data that were statistically significantly different from the αN* construct in the presence of empty pSVL vector are marked with an asterisk (n=7±SD, Student’s t test, p<0.005).
(C) The amount of C-terminal glycosylation for αN* in the presence of increasing amounts of CD3δ and ε was assessed. Two microgram of αN* were co-transfected with the indicated amounts of δ+ε DNA (equal amounts of both CD3 constructs were used). Closed circles denote the relative amount of αN* chains that are C-terminally glycosylated and completely ER-lumenal, and open circles the relative amount of αN* that is not C-terminally glycosylated (n=3±SD).
(D) Pulse-chase experiments were conducted over an interval of 2 h on cells co-expressing either αN* together with empty pSVL vector or with both CDδ and ε. The upper autoradiograph shows data for αN* in the presence of empty pSVL vector, the lower autoradiograph in the presence of CDδ and ε. Quantifications of the upper band, corresponding to the ER-lumenal species, are shown below the autoradiographs. Open circles are derived from αN*+pSVL and closed circles from αN*+CDδ and ε (n=6±SD). The signal present at t=0′ is arbitrarily set to 100 in both cases.
(E) Pulse-chase experiments to follow the ER import of αN* over time in the presence of either empty pSVL vector or a combination of CD3δ and ε. C-terminally glycosylated, ER-lumenal species are indicated.
(F) Pulse-chase experiments conducted on the wt α-chain expressed alone or in the presence of either empty pSVL vector or CD3δ and ε to assess its stability.
(G) Quantifications of the data shown in (F) (n=3±SD). Open circles show the degradation of α+pSVL, and closed circles indicate that of α when co-expressed with CDδ and ε. See also Figure S5.
Discussion
A new mechanism underlying quality control of integral membrane protein complexes
Assembly within the membrane is a crucial step in the biosynthesis of many multimeric integral membrane protein complexes. However, it has been unclear how the cell scrutinizes the outcome of this process. We find that the cell can infer the assembly status from the localization of the TM segment as several less hydrophobic TM sequences derived from multimeric TM protein complexes can enter the ER lumen completely, where they are recognized as substrates of the chaperone BiP, which in turn can initiate degradation of the unassembled subunit. Indeed, most TM segments are likely to contain Hsp70 binding sequences (Blond-Elguindi et al., 1993; Flynn et al., 1991), and BiP is readily available at the translocon (Brodsky and Schekman, 1993; Hamman et al., 1998), suggesting a general link between proper integration of TM segments and the ER lumenal chaperone machinery. When applied to the αβTCR, these concepts solve the longstanding puzzle of how ionizable residues in the TCR α-chain TM region can both direct subunit assembly, and if this does not occur, promote degradation. A possible role for membrane integration in this process has long been debated, but due to conflicting results obtained with chimeric constructs (Bonifacino et al., 1991; Fayadat and Kopito, 2003; Ishikura et al., 2010; Shin et al., 1993; Soetandyo et al., 2010) has remained controversial. We find the TM region of an authentic α-chain indeed fully enters the ER and contains the signal for degradation by exposing unresolvable chaperone binding sites in its TM region (Figure 6). Our finding that membrane integration initially fails for ~50% of the TCR α-chain with the remaining pool entering the ER lumen over time may provide an explanation for the different results obtained with the various αTM:chimeric proteins that have been observed. Based on our finding of BiP binding to the α-chain TM region, the reported ubiquitination of both the C-terminus (Ishikura et al., 2010) and TM region (Anania et al., 2013) of the α-chain suggests retrotranslocation is initiated from the C-terminus of the α-chain. This might further reconcile conflicting results on the impact of failure to integrate into the membrane on degradation, as the constructs used varied in the inclusion of the more recently identified ubiqitination sites (Anania et al., 2013; Bonifacino et al., 1991; Ishikura et al., 2010; Shin et al., 1993).
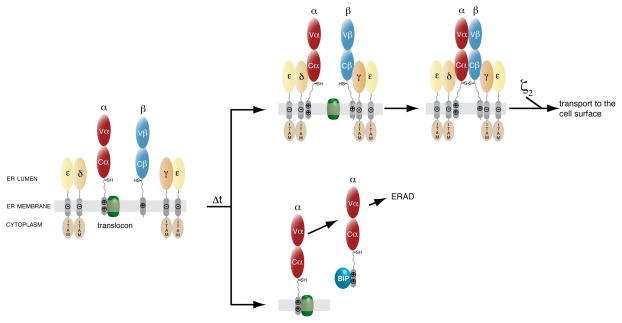
Figure 6. Quality control of the αβTCR by assembly-dependent membrane integration.
Model for the assembly-dependent quality control mechanism in αβTCR biosynthesis. All chains except for the α-chain are properly integrated in the ER membrane co-translationally. Heterodimers between CD3γ and ε or CD3δ and ε form soon after their biosynthesis. Two pathways can be taken by the TCR α-chain. Either, the CD3δε heterodimer interacts with the α-chain and allows it to stably integrate into the ER membrane. Subsequently, the CD3δε-α and the CD3γε-β hetero-trimers can move in the membrane, assemble covalently, pair with the ζ dimer and be transported to the cell surface. Alternatively, newly synthesized α-chains that fail to assemble with the CD3δε heterodimer completely enter the ER lumen over time and their TM domains are recognized by the ER chaperone BiP allowing them to be targeted for ERAD. Some of the TCR α-chain its TM region is initially missed as a stop-transfer sequence and it directly enters the ER lumen to be degraded.
We find that membrane integration of the TCR α-chain is promoted if a complex is formed with the CD3δε dimer, a process occurring early in αβTCR biogenesis (Kearse et al., 1995). Increasing levels of CD3δε can further reduce the amount of α-chain entering the ER-lumen (Figure 5C), however at an elevated biosynthetic cost and a certain percentage of the α-chain is initially missed as a stop-transfer sequence and will be lost for further assembly steps. Even in T cells most TCR α-chains are ultimately degraded (Minami et al., 1987). Evolution has likely tolerated this trade-off to increase fidelity of TCR assembly. Indeed, only small quantities of the αβTCR are needed at the cell surface for proper functioning in the immune system, but quality control of this protein complex must be very reliable to ensure adequate immune responses (Guy et al., 2013). The correlation between CD3 subunits being able to retain the α-chain in the membrane and to stabilize it against degradation argues that membrane-integration underlies the previously described stabilizing effect of CD3δ and ε on the TCR α-chain (Bonifacino et al., 1990; Bonifacino et al., 1989; Lippincott-Schwartz et al., 1988; Manolios et al., 1991) (Figure 6). We find that within the TM region of the α-chain the central Lys residue is much more destabilizing for membrane integration than the more N-terminal Arg residue. Of note, the same central Lys residue in the α-chain is responsible for the interaction with the acidic TM residues in CD3δ and ε (Call et al., 2002). Thus, the very residue that defines localization of the α-chain is the one that interacts with CD3δ and ε to stably integrate the α-chain in the membrane. Ideally, CD3-mediated membrane-retention of the TCR α-chain should report on the completeness of the assembly status. Indeed, we find that both, CD3δ and ε together, are needed to retain the α-chain for more prolonged times in the membrane and stabilize it against degradation, further explaining previous data that a three-helical interaction motif between TCRα and CD3δε is the most stable (Call et al., 2002). Interaction with the ζ chain occurs via interaction with the α-chain TM Arg residue (Call et al., 2002), which we found had no significant effect on localization.
The translocon in the assembly process of integral membrane proteins
A significant part of the α-chain TM region is localized to the ER lumen in both COS-1 and Jurkat cells almost immediately after biosynthesis, arguing that its TM region is rather poorly recognized by the translocon as a stop-transfer sequence. The completely ER-lumenal pool continues to increase post-translationally, a characteristic that we also observed for a variety of other TM segments of low hydrophobicity. Thus, while less hydrophobic TM regions can be initially missed as a stop-transfer sequence, a pool exists in a state that allows them to more slowly enter into the ER lumen in the absence of subunit assembly. The slow ER entry of the TCR α-chain TM region, as judged by its kinetics of post-translational glycosylation, suggests there might be a kinetic window to allow TM segments to assemble correctly, and perhaps more lumenal regions to fold, before mislocalization of their TM segments occur. Of note, post-translational ER entry seemed to be faster for constructs that contained the well-folded CL domain instead of the authentic Vα and Cα domain (Figures 4A and S4D), suggesting that the domains that have already entered the ER lumen might influence the rate of post-translational ER entry. We cannot completely rule out that our C-terminal glycosylation reporter underestimates the amount of TCR α-chain that initially enters the ER lumen or the kinetics of ER entry as C-terminal glycosylation can be inefficient and slow (Ruiz-Canada et al., 2009). However, C-terminal glycosylation of NXT sites (our reporter comprises an NVT site) can be efficient and fast (Shrimal et al., 2013). Furthermore, the inability of the underglycosylated form of our reporter construct to co-immunoprecipitate with BiP (Figures 3D and S4B) and the fact that CD3δε completely inhibit any further C-terminal glycosylation of the α-chain (Figure 5D) together argue for our C-terminal glycosylation site being a benign reporter for ER-import.
Our finding that the central Lys residue is more detrimental to membrane integration of the α-chain than the less-centrally localized Arg is in agreement with data on model TM helices (Hessa et al., 2007), but we also observed some interesting deviations between the predicted free energy of membrane integration and our model proteins being able to become C-terminally glycosylated (Figure 1A). In particular co- and post-translational entry of the ER lumen might rely on somewhat different rules for TM sequence hydrophobicity as exemplified by our construct with its TM region Arg residue replaced by Leu that apparently entered the ER-lumen exclusively post-translationally.
It is noteworthy that the TM regions of our model proteins continued to enter the ER-lumen post-translationally, suggesting their membrane passage might be gated, if so, likely via Sec61. Indeed, the translocon has been shown to remain associated with more hydrophilic TM sequences for prolonged times due to either specific interactions with components of the translocon (Cross and High, 2009; Do et al., 1996; Heinrich et al., 2000) or to its inherent ability to provide retention space for more than one TM segment in the process of biosynthesis and assembly (Beckmann et al., 2001; Hanein et al., 1996). In agreement with this idea, we find that a significant part of the α-chain can be extracted from the membrane by the chemical denaturant urea that unfolds proteins and disrupts protein-protein interactions (Figure S2A). We were unable, however, to detect any direct interactions between the TCR α-chain and either Sec61α, Sec61β or TRAM (data not shown). This point thus warrants further study, in particular since a TM segment that remained translocon-associated would impose certain requirements on the assembly process with other TM sequences when they are dispersed over a number of different polypeptide chains. If subunit assembly required the various chains to enter through the same translocon, it would raise very interesting questions about how translation and assembly could possibly be coordinated (Gilmore and Mandon, 2012) and which role the translocon plays in this process (Skach, 2009). It is noteworthy is this context that we found CD3δ and ε together being able to completely inhibit any further ER entry of the α-chain TM region arguing that assembly between CD3δε and the α-chain is rapid and thus would not lead to prolonged occupation of translocons by unassembled α-chains.
Implications for the quality control and evolution of integral membrane proteins
Diverting polar residues in TM segments over different polypeptide chains within a complex can reduce the efficiency of membrane integration of unassembled subunits, while at the same time allowing more rigorous quality control and providing greater evolutionary flexibility in reshuffling different subunits in comparison to multipass TM proteins. However, unlike the group of single pass proteins examined in this study the TM segments of single pass TM proteins are generally close to ideal TM helices (Hessa et al., 2007). In contrast, this is often not the case for multipass TM proteins where a significant amount of TM helices are predicted to be unable to insert into the membrane by themselves (Elofsson and von Heijne, 2007; Hessa et al., 2007; White and von Heijne, 2008), and in some cases have either been directly shown or predicted to enter the ER lumen transiently (Hessa et al., 2007; Kanki et al., 2002; Kauko et al., 2010; Lu et al., 2000; Meindl-Beinker et al., 2006; Ojemalm et al., 2012; Ota et al., 1998; Sadlish et al., 2005; Skach et al., 1994). Thus, based on our findings it is reasonable to suggest that one way of monitoring their failure to integrate would be their exposure to the chaperone machinery, which could initiate their degradation (Buck and Skach, 2005). Conversely, correct intrachain assembly would promote membrane integration of these TM segments and thus shield them from chaperones. Conceptually, this is very similar to the transient exposure of hydrophobic residues to the solvent in the folding process of soluble proteins.
In addition to the principles we have uncovered in this study, a complimentary mechanism has been identified for the quality control of integral membrane proteins that also relies on the recognition of unpaired hydrophilic residues but in this case they are detected in the membrane environment itself (Lemberg, 2013). Recognition of these residues in the membrane leads to the unassembled protein’s ubiquitination, rendering it an ERAD substrate, which can include cleavage of the hydrophilic TM segment (Erez and Bibi, 2009; Fleig et al., 2012; Loureiro et al., 2006; Reggiori and Pelham, 2002; Sato et al., 2009). Of note, efficient detection and cleavage of the pre-TCRα, which is predicted to be membrane integrated (Figure 1A), occurs by this mechanism, while cleavage of the TCR α-chain TM region, which we show relies on a localization-coupled quality control mechanism is very inefficient (Fleig et al., 2012).
Finally it should be noted that the mechanism we have unveiled in this study might explain the molecular basis for certain diseases where polar residues in TM regions arise due to genetic mutations (Partridge et al., 2004) but also has the potential to significantly extend the known repertoire of TM protein functions (Illergard et al., 2011). It allows the cell to utilize TM segments of largely different composition and hydrophobicity, even those that would be unlikely to serve as membrane anchors by themselves, without sacrificing fidelity in TM protein folding and assembly.
Experimental Procedures
Construct design
Individual constructs were amplified from synthetic TCR genes. Chimeric constructs were generated using a previously described λ light chain CL domain construct (Hellman et al., 1999) fused to the different TM regions. Mutants were generated by site directed mutagenesis. A hamster BiP construct was used (Lee et al., 1999). Details can be found in the Supplemental Experimental Procedures.
Cell transfections
COS-1 cells were transfected using GeneCellin (BioCellChallenge, Toulon, France) according to the manufacturer’s protocol.
Jurkat J.RT-T3.1 cells (Saito et al., 1987) were transfected via electroporation according to the manufacturer’s protocol. Details can be found in the Supplemental Experimental Procedures.
Metabolic labeling, pulse-chase experiments and immunoprecipitations
For metabolic labeling, cells were starved for 30 min in media without Met and Cys and subsequently labeled for 30 min with EasyTag™ EXPRESS35S Protein Labeling Mix (Perkin Elmer, Waltham, MA). For pulse-chase experiments cells were chased in complete medium supplemented with additional 2 mM of cold Met and Cys after the labeling pulse. If present, MG132 (Sigma-Aldrich, St. Louis, MO) was added 2.5 h before the pulse, during the starving period and during the subsequent pulse and chase. Cells were lysed in appropriate buffers for each experiment. Details can be found in the Supplemental Experimental Procedures. Immunoprecipitations were performed with commercially available antibodies or the previously described antiserum against BiP (Hendershot et al., 1995). EndoH deglycosylation experiments were performed according to the manufacturer’s protocol. Samples were run on 10% or 14% SDS-PAGE gels, gels were dried and either used in autoradiography or phosphorimager analysis. Further details can be found in the Supplemental Experimental Procedures.
Subcellular fractionation and western blot
Subcellular fractionation was essentially performed as published (Lai et al., 2012). Details can be found in the Supplemental Experimental Procedures.
Statistical analysis
Results are shown as means ±one standard deviation. P values associated with all comparisons were derived from paired two-tailed Student’s t-tests.
Supplementary Material
Highlights.
Transmembrane (TM) segments of low hydrophobicity can enter the ER lumen completely
The Hsp70 chaperone BiP binds ER-lumenal TM sequences
ER-lumenal TM proteins are recognized by ER quality control and rapidly degraded
Assembly retains less hydrophobic TM segments in the membrane, inhibiting degradation
Acknowledgments
We are grateful to Julia Behnke and Drs. Johannes Buchner, Joseph Opferman, Joel Otero, Janet Partridge and Brenda Schulman for helpful comments on the manuscript. We thank Art Weiss, UCSF for kindly sharing J.RT-T3.1 cells and Yuval Reiss, Proteologics/Israel for sharing the Hrd1C291S plasmid.
MJF acknowledges funding by the German Academy of Sciences Leopoldina, grant number LPDS 2009-32 and by the Paul Barrett-endowed fellowship of St. Jude. We are grateful to the Hartwell center of St. Jude for DNA sequencing. This work was funded by NIH grants R03 AI097733 and R01 GM54068, and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
Footnotes
MJF and LMH designed the study. MJF performed experiments. MJF and LMH analyzed data and wrote the manuscript.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anania VG, Bustos DJ, Lill JR, Kirkpatrick DS, Coscoy L. A Novel Peptide-Based SILAC Method to Identify the Posttranslational Modifications Provides Evidence for Unconventional Ubiquitination in the ER-Associated Degradation Pathway. Int J Proteomics. 2013;2013:857918. doi: 10.1155/2013/857918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Shah N, Klausner RD. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Suzuki CK, Lippincott-Schwartz J, Weissman AM, Klausner RD. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J Cell Biol. 1989;109:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Skach WR. Differential stability of biogenesis intermediates reveals a common pathway for aquaporin-1 topological maturation. J Biol Chem. 2005;280:261–269. doi: 10.1074/jbc.M409920200. [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- Cross BC, High S. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J Cell Sci. 2009;122:1768–1777. doi: 10.1242/jcs.046094. [DOI] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- Erez E, Bibi E. Cleavage of a multispanning membrane protein by an intramembrane serine protease. Biochemistry. 2009;48:12314–12322. doi: 10.1021/bi901648g. [DOI] [PubMed] [Google Scholar]
- Fayadat L, Kopito RR. Recognition of a single transmembrane degron by sequential quality control checkpoints. Mol Biol Cell. 2003;14:1268–1278. doi: 10.1091/mbc.E02-06-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006;4:e142. doi: 10.1371/journal.pbio.0040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell. 2012;47:558–569. doi: 10.1016/j.molcel.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Mandon EC. Understanding integration of alpha-helical membrane proteins: the next steps. Trends Biochem Sci. 2012;37:303–308. doi: 10.1016/j.tibs.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CS, Vignali KM, Temirov J, Bettini ML, Overacre AE, Smeltzer M, Zhang H, Huppa JB, Tsai YH, Lobry C, et al. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat Immunol. 2013;14:262–270. doi: 10.1038/ni.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Berkhout B, Alarcon B, Sancho J, Wileman T, Terhorst C. Requirements for cell surface expression of the human TCR/CD3 complex in non-T cells. Int Immunol. 1991;3:359–368. doi: 10.1093/intimm/3.4.359. [DOI] [PubMed] [Google Scholar]
- Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. Flagging and docking: dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem Sci. 2012;37:404–410. doi: 10.1016/j.tibs.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hellman R, Vanhove M, Lejeune A, Stevens FJ, Hendershot LM. The in vivo association of BiP with newly synthesized proteins is dependent on the rate and stability of folding and not simply on the presence of sequences that can bind to BiP. J Cell Biol. 1999;144:21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM, Wei JY, Gaut JR, Lawson B, Freiden PJ, Murti KG. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hewitt CR, Lamb JR, Hayball J, Hill M, Owen MJ, O’Hehir RE. Major histocompatibility complex independent clonal T cell anergy by direct interaction of Staphylococcus aureus enterotoxin B with the T cell antigen receptor. J Exp Med. 1992;175:1493–1499. doi: 10.1084/jem.175.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck SA, Cyr DM. Mechanisms for quality control of misfolded transmembrane proteins. Biochim Biophys Acta. 2012;1818:1108–1114. doi: 10.1016/j.bbamem.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- Illergard K, Kauko A, Elofsson A. Why are polar residues within the membrane core evolutionary conserved? Proteins. 2011;79:79–91. doi: 10.1002/prot.22859. [DOI] [PubMed] [Google Scholar]
- Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem. 2010;285:23916–23924. doi: 10.1074/jbc.M110.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Sakaguchi M, Kitamura A, Sato T, Mihara K, Hamasaki N. The tenth membrane region of band 3 is initially exposed to the luminal side of the endoplasmic reticulum and then integrated into a partially folded band 3 intermediate. Biochemistry. 2002;41:13973–13981. doi: 10.1021/bi026619q. [DOI] [PubMed] [Google Scholar]
- Kauko A, Hedin LE, Thebaud E, Cristobal S, Elofsson A, von Heijne G. Repositioning of transmembrane alpha-helices during membrane protein folding. J Mol Biol. 2010;397:190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- Kearse KP, Roberts JL, Singer A. TCR alpha-CD3 delta epsilon association is the initial step in alpha beta dimer formation in murine T cells and is limiting in immature CD4+ CD8+ thymocytes. Immunity. 1995;2:391–399. doi: 10.1016/1074-7613(95)90147-7. [DOI] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Lippincott-Schwartz J, Bonifacino JS. The T cell antigen receptor: insights into organelle biology. Annu Rev Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- Lai CW, Otero JH, Hendershot LM, Snapp E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem. 2012;287:7969–7978. doi: 10.1074/jbc.M111.311290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg MK. Sampling the membrane: function of rhomboid-family proteins. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Bonifacino JS, Yuan LC, Klausner RD. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 2006;441:894–897. doi: 10.1038/nature04830. [DOI] [PubMed] [Google Scholar]
- Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolios N, Bonifacino JS, Klausner RD. Transmembrane helical interactions and the assembly of the T cell receptor complex. Science. 1990;249:274–277. doi: 10.1126/science.2142801. [DOI] [PubMed] [Google Scholar]
- Manolios N, Letourneur F, Bonifacino JS, Klausner RD. Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO J. 1991;10:1643–1651. doi: 10.1002/j.1460-2075.1991.tb07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl-Beinker NM, Lundin C, Nilsson I, White SH, von Heijne G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006;7:1111–1116. doi: 10.1038/sj.embor.7400818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Weissman AM, Samelson LE, Klausner RD. Building a multichain receptor: synthesis, degradation, and assembly of the T-cell antigen receptor. Proc Natl Acad Sci U S A. 1987;84:2688–2692. doi: 10.1073/pnas.84.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun. 2003;303:91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- Ojemalm K, Halling KK, Nilsson I, von Heijne G. Orientational preferences of neighboring helices can drive ER insertion of a marginally hydrophobic transmembrane helix. Mol Cell. 2012;45:529–540. doi: 10.1016/j.molcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K, Sakaguchi M, von Heijne G, Hamasaki N, Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol Cell. 1998;2:495–503. doi: 10.1016/s1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- Otero JH, Lizak B, Hendershot LM. Life and death of a BiP substrate. Semin Cell Dev Biol. 2010;21:472–478. doi: 10.1016/j.semcdb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge AW, Therien AG, Deber CM. Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins. 2004;54:648–656. doi: 10.1002/prot.10611. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Saito T, Weiss A, Gunter KC, Shevach EM, Germain RN. Cell surface T3 expression requires the presence of both alpha- and beta-chains of the T cell receptor. J Immunol. 1987;139:625–628. [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Lee S, Strominger JL. Translocation of TCR alpha chains into the lumen of the endoplasmic reticulum and their degradation. Science. 1993;259:1901–1904. doi: 10.1126/science.8456316. [DOI] [PubMed] [Google Scholar]
- Shrimal S, Trueman SF, Gilmore R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J Cell Biol. 2013;201:81–95. doi: 10.1083/jcb.201301031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR, Shi LB, Calayag MC, Frigeri A, Lingappa VR, Verkman AS. Biogenesis and transmembrane topology of the CHIP28 water channel at the endoplasmic reticulum. J Cell Biol. 1994;125:803–815. doi: 10.1083/jcb.125.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetandyo N, Wang Q, Ye Y, Li L. Role of intramembrane charged residues in the quality control of unassembled T-cell receptor alpha-chains at the endoplasmic reticulum. J Cell Sci. 2010;123:1031–1038. doi: 10.1242/jcs.059758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki CK, Bonifacino JS, Lin AY, Davis MM, Klausner RD. Regulating the retention of T-cell receptor alpha chain variants within the endoplasmic reticulum: Ca(2+)-dependent association with BiP. J Cell Biol. 1991;114:189–205. doi: 10.1083/jcb.114.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz U, Banks D, Jacobson S, Biddison WE. Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: evidence for oligoclonal expansion. J Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- Wileman T, Kane LP, Young J, Carson GR, Terhorst C. Associations between subunit ectodomains promote T cell antigen receptor assembly and protect against degradation in the ER. J Cell Biol. 1993;122:67–78. doi: 10.1083/jcb.122.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.