Abstract
The ingestion of dietary protein is of vital importance for the maintenance of fundamental physiological processes. The taste modality umami, with its prototype stimulus, glutamate, is considered to signal the protein content of food. Umami was thought to be mediated by the heterodimeric amino acid receptor, T1R1+T1R3. Based on knockout studies, additional umami receptors are likely to exist. In addition to amino acids, certain peptides can also elicit and enhance umami taste suggesting that protein breakdown products may contribute to umami taste. The recently deorphanized peptone receptor, GPR92 (also named GPR93; LPAR5), is expressed in gastric enteroendocrine cells where it responds to protein hydrolysates. Therefore, it was of immediate interest to investigate if the receptor GPR92 is expressed in gustatory sensory cells. Using immunohistochemical approaches we found that a large population of cells in murine taste buds was labeled with a GPR92-antibody. A molecular phenotyping of GPR92-cells revealed that the vast majority of GPR92-immunoreactive cells express PLCβ2 and can therefore be classified as type II cells. More detailed analyses have shown that GPR92 is expressed in the majority of T1R1-positive taste cells. These results indicate that umami cells may respond not only to amino acids but also to peptides in protein hydrolysates.
Keywords: GPR92, GPR93, LPAR5, gustatory sensory cells, protein breakdown products, receptors, T1R1, taste
Introduction
The ingestion of dietary protein is essential; their structural units, the amino acids, are precursors of many biologically relevant molecules and play a critical role in modulating various physiological processes (Wu 2009; Jahan-Mihan et al. 2011; San Gabriel et al. 2012). Nutrients are first sensed in the oral cavity by gustatory sensory cells organized in taste buds. Protein-rich foods elicit a typical taste perception called umami. Monosodium glutamate (MSG) is found in many protein-containing foods (Maga 1983; Yamaguchi and Ninomiya 2000) and is considered as the prototypic umami taste stimulus (Ikeda 1909, 2002). The heterodimer receptor, T1R1+T1R3, was proposed to mediate umami taste (Nelson et al. 2002; Li et al. 2002). In heterologous expression systems, both the mouse and human T1R1+T1R3 dimer respond to glutamate and several other amino acids, especially in combination with nucleotide monophosphates (Nelson et al. 2002; Li et al. 2002).
However, several recent studies strongly suggest that additional receptor types may also be involved in umami taste transduction. Damak et al. (2003) revealed that T1R3-knockout mice retain significant taste responsiveness to MSG in behavioral experiments and in afferent nerve recordings. This observation was subsequently elaborated: taste buds of mice lacking T1R3 still exhibited significant glutamate-evoked Ca2+ responses, with a similar incidence, but with a decreased amplitude (Maruyama et al. 2006). Finally, single unit recordings on taste afferent neurons also provide strong evidence of umami taste responses that are not dependent on T1R3-containing receptors (Yoshida et al. 2009). In sensory evaluation tests, not only glutamate but also peptides with MW > 1000 elicit and enhance a perception of umami taste (Raksakulthai and Haard 1992; Tamura et al. 1989; Van Den Oord et al. 1997; Schlichtherle-Cerny and Amadò, 2002). Molecular modeling suggests that T1R1+T1R3 binds ligands in a relatively small binding pocket (Zhang et al. 2008). Thus, it seems reasonable that in addition to umami receptors selective for amino acids other receptors responding to protein breakdown products are involved in mediating the umami taste. In this context it is interesting to note that the recently discovered receptor type GPR92 (also named GPR93; LPAR5) is activated by protein-hydrolysates (peptone) (Choi et al. 2007a, b), a mixture of enzymatically derived peptide fragments with MW between 120 and 1200 and free amino acids, that mimics dietary proteins digest in the luminal chyme (Cuber et al. 1990). Therefore, GPR92 is considered as a candidate receptor for sensing protein hydrolysates. This notion is supported by our recent finding that GPR92 is expressed in enteroendocrine cells of the gastric mucosa, G-cells and D-cells, which secret gastrin or somatostatin, respectively, upon stimulation with protein hydrolysates (Haid et al. 2012). Several studies indicate that functional elements of gustatory sensory cells are also expressed in putative chemosensory cells of the gastrointestinal mucosa (for reviews see: Breer et al. 2012; Iwatsuki and Uneyama 2012). Here, we asked the inverse question, whether the gastrointestinal peptone receptor GPR92 is also expressed in amino acid responsive cells of the gustatory system.
Materials and methods
Mice
Analyses were performed with wild type mouse strains C57/BL6J from Charles River (Sulzfeld, Germany). In addition, two previously described transgenic/genetic-targeted mouse lines were used: homozygous PLCβ2-GFP mice which express GFP under the control of the PLCβ2 promotor (Kim et al. 2006), as well as homozygous T1R1-mCherry mice expressing T1R1 promoter-driven mCherry (Voigt et al. 2012). Animals were fed with standard laboratory chow ad libitum and had free access to water. All experiments comply with the Principles of Animal Care, publication no. 85-23, revised 1985, of the National Institutes of Health and with the current laws of Germany.
Isolation of circumvallate, foliate and fungiform taste papillae
Tongues were isolated from freshly decapitated adult C57/BL6J mice and stored in ice-cold Tyrode’s solution (140 mM NaCl, 5 mM KCL, 1mM CaCl2, 1 mM MgCl2, 10 mM HEPES; pH 7.4) for 10 min. Delamination of the epithelia from the tongue muscle layer was carried out as previously described (Striem et al. 1991; Bernhardt et al. 1996; Kretz et al. 1999). After this, circumvallate, foliate and fungiform areas were dissected from the detached lingual epithelium by observation under an inverted microscope (Olympus IX70), transferred into collection tubes and immediately frozen in liquid nitrogen.
RNA isolation and cDNA synthesis
Total RNA was isolated from the dissected circumvallate, foliate and fungiform taste papillae with a NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. To ensure the complete removal of DNA, a DNase digestion (DNase I, Life Technologies, Carlsbad, CA, USA) step was included. Subsequently, 396 ng total RNA was reverse transcribed using oligo(dT) primers and SuperScript III Reverse Transcriptase (RT) (Invitrogen, Carlsbad, CA, USA). RNA integrity of each sample was controlled by the amplification of the housekeeping gene for the ribosomal protein L8 (RpL8) with intron-spanning primers to verify the DNA removal.
Reverse transcriptase polymerase chain reaction (RT-PCR)
RT-PCR amplification was conducted by using normalized cDNA from tissues as described above. PCR amplifications were performed with the following primer combinations: mGPR92 forward, 5′-CCT GGC GGC TGT CGT CTA TT-3′; mGPR92 reverse, 5′-GCC GAA TCC TGG GAG CAG TTG-3′; mRpL8 forward, 5′-GTG CCT ACC ACA AGT ACA AGG C-3′; mRpL8 reverse, 5′-CAG TTT TGG TTC CAC GCA GCC G-3′.
RT-PCR was carried out using High Fidelity PCR Enzyme Mix (Fermentas, St. Leon-Rot, Germany) and a Peltier PTC-200 thermo cycler (MJ Research). For amplification the following PCR cycling profile was used with optimized numbers of amplification cycles. One cycle: 3 min at 94°C; 20 cycles: 30 sec at 94°C, 40 sec at 64°C with −0,5°C per cycle, 40 sec at 72°C; 25 cycles: 30 sec at 94°C, 20 sec at 54°C, 40 sec at 72°C; and one cycle: 1 min at 72°C. PCR products were run on a 1.5 % agarose gel containing EtdBr. Amplification of the housekeeping gene for the ribosomal protein L8 (mRpL8) with intron-spanning primers was used as control to confirm equal quality and quantity of the cDNA preparations. PCR products were subsequently cloned into pGem-T vector (Promega, Madison, Wis., USA) and subjected to sequence analysis in an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, Calif., USA).
Tissue Preparation
For immunohistochemistry tongues were removed and rinsed in 1 x PBS. Afterwards, 1 x PBS (0.85% NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.4) was directly injected under the lingual epithelium followed by a perfusion of 4% ice-cold paraformaldehyde with 0.2% glutardialdehyde (in 150 mM phosphate buffer, pH 7.4). After fixation in the same fixative for 1h to 3h, the tissue was cryoprotected by incubation in 25% sucrose overnight at 4°C. Finally, the tissue was embedded in Tissue Freezing Medium and quickly frozen on dry ice or liquid nitrogen. Cryosections (5 μm) were generated using a CM3050S cryostat (Leica Microsystems) and adhered to Superfrost Plus microscope slides (Menzel Gläser).
Immunohistochemistry
Cryosections were air-dried, rinsed in 1 x PBS for 10 min at room temperature and blocked in 0.3% Triton X-100 in 1 x PBS containing 10% normal goat serum (NGS; Dianova, Hamburg, Germany) for 30 min at room temperature. Primary antibodies were diluted in 0.3% Triton X-100 in 1 x PBS containing 10% NGS. For immunoreactivity to GPR92, a rabbit anti-GPR92 antibody (sc-135237, Santa Cruz Biotechnology, Santa Cruz, Calif., USA) was used in a 1:50 dilution. Blocked sections were incubated with the diluted primary antibody overnight at 4°C. After washing in 1 x PBS, the bound primary antibody was visualized using appropriate secondary antibodies conjugated to Alexa 488 or Alexa 568 (Invitrogen, Karlsruhe, Germany, 1:500) diluted in 1 x PBS with 0.3% Triton X-100 containing 10% NGS for 2 h at room temperature. After three rinses for 5 min in 1 x PBS the sections were counterstained with 4,6- diamidino-2-phenylindole (DAPI; 1 μg/ml, Sigma Aldrich, Schnelldorf, Germany) for 3 min at room temperature, rinsed with bidest. water, and finally mounted in MOWIOL (10% polyvinylalcohol 4–88 (Sigma), 20% glycerol in 1 x PBS). No immunoreactivity could be observed when the primary antibody was omitted.
Microscopy and photography
Immunohistochemical staining was documented by using a Zeiss Axiophot microscope (Carl Zeiss MicroImaging, Jena, Germany). Images were captured using a Zeiss Axiocam for transmitted light and a ‘Sensi-Cam’ CCD-camera (PCOimaging, Kelheim, Germany) for fluorescent images. Images were adjusted for contrast in AxioVision LE Rel. 4.3 (Carl Zeiss MicroImaging, Jena, Germany) and arranged in PowerPoint (Microsoft).
Cell quantification
GPR92-immunoreactive cells, T1R1-mCherry-positive cells and PLCβ2-positive cells, respectively, were counted manually on 5 μm sections of circumvallate papillae that were stained with the anti-GPR92 antibody and DAPI to visualize cell nuclei. For the quantitative analyses we counted both the number of GPR92-positive cells and the number of PLCβ2-GPF-positive cells. In addition, the number of cells that were GPR92-positive and also PLCβ2-GFP-positive (double-labeled) was determined. To estimate the proportion (%) of GPR92-positive cells that were also PLCβ2-GFP-positive, we defined the number of GPR92-cells as 100% and calculated the percentage of GPR92-cells that were also PLCβ2-GFP-positive and vice versa. For quantification of co-expression ratios of GPR92 and T1R1-mCherry the calculation was performed as described above. Values are given as mean ± SD.
Results
In order to investigate whether cells of the taste system express the recently described family A receptor GPR92 (also named GPR93; LPAR5), we initially performed RT-PCR experiments using specific primers for the murine GPR92 (mGPR92) on normalized cDNA of circumvallate (CvP), foliate (FoP) and fungiform (FuP) epithelia. Amplicons of the expected size were obtained for tissue samples from all analysed taste tissues (Fig. 1). Subsequent cloning and sequencing of the amplicons revealed that each PCR product corresponded to mGPR92. RT-PCR experiments for the mouse housekeeping gene RpL8 resulted in amplification products of the expected size for tissue samples from all analysed taste tissues confirming equal quality and quantity of the cDNA preparations.
Fig. 1.
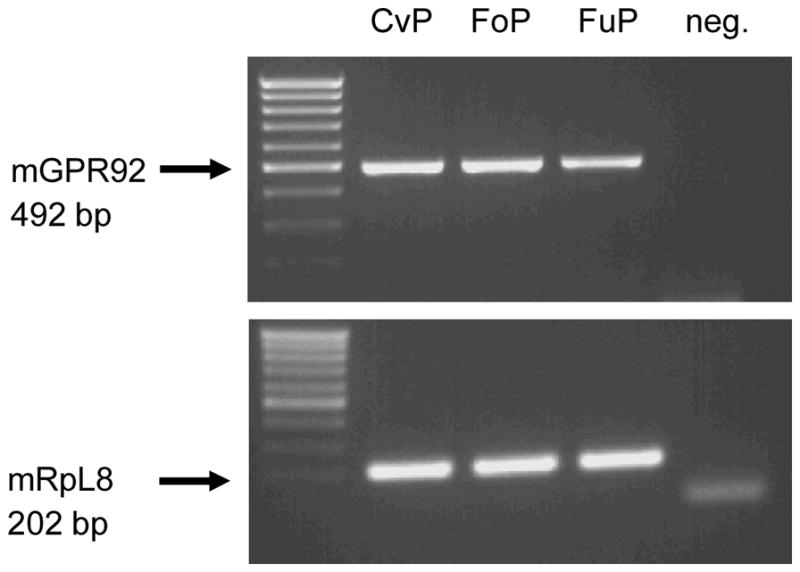
RT-PCR analysis for the peptone receptor GPR92 and ribosomal protein l8 (RpL8) in murine taste papillae. Reverse transcription polymerase chain reaction (RT–PCR) experiments were performed with primer pairs specific for mGPR92 (492 bp) and a control housekeeping mRNA, mRpL8 (202 bp), respectively. Amplicons of the expected size could be observed in all analysed taste papillae types (circumvallate papillae CvP, foliate papillae FoP, fungiform papillae FuP). No bands were observed in negative controls lacking template (neg.)
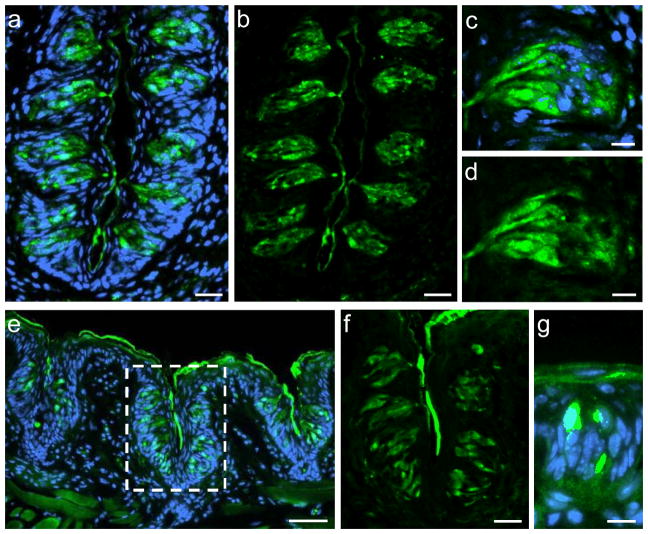
To visualize and identify the cells in taste buds which express GPR92 (GPR93; LPAR5), immunohistochemical experiments were performed on sections through the circumvallate, foliate and fungiform papillae using a GPR92-specific antibody. As depicted in Figure 2, GPR92-immunoreactivity could be visualized in all papillae types. Taste buds of the CvP (Fig. 2a–d) and the FoP (Fig. 2e, f) contained numerous GPR92-positive cells. Higher magnification illustrates the spindle-shaped morphology of GPR92-immunoreactive cells which is reminiscent of the typical taste cell morphology (Fig. 2c, d). In FuP only individual cells were labeled with the GPR92 antibody (Fig. 2g).
Fig. 2.
Visualization of GPR92 in murine taste papillae. Immunolabeling of cross sections through the murine circumvallate papillae (a–d), the foliate papillae (e–f) or the fungiform papillae (g).
(a, b) Numerous GPR92-positive cells are located within taste buds of the circumvallate papilla.
(c, d) Magnification of a circumvallate taste bud. GPR92-positive cells show the typical, spindle-shaped morphology of taste cells.
(e) GPR92-immunoreactive cells in taste buds of foliate papillae.
(f) Magnification of the dotted area in (e). GPR92-immunoreactive cells in foliate taste buds.
(g) GPR92-positive cell in a fungiform papilla.
Sections are counterstained with DAPI (blue). Scale bars: a, b, f = 20 μm; c, d, g = 10 μm; e = 50 μm
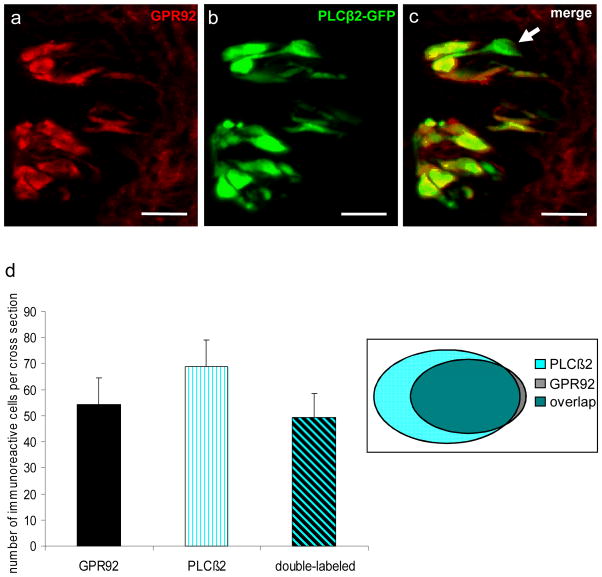
Mammalian taste buds contain three different types of cells classified on the basis of their morphological, molecular and functional properties as types I, II and III (Yee et al. 2001; Chaudhari and Roper, 2010). One of these categories, type II receptor cells, is characterized by the expression of G protein-coupled receptors (GPCRs; T1Rs, T2Rs) and the downstream effector enzyme phospholipase C beta 2 (PLCβ2) (Rössler et al. 1998; Clapp et al. 2004; DeFazio et al. 2006). Therefore, type II taste cells can directly respond to sweet, bitter and umami tastants (Clapp et al. 2004; Tomchik et al. 2007). To assess whether the G protein-coupled receptor GPR92 (GPR93; LPAR5) is expressed in this taste cell type we analysed PLCβ2-GFP transgenic mice (Kim et al. 2006). A typical section through the CvP of a PLCβ2-GFP transgenic mouse is shown in Figure 3. PLCβ2-positive cells can be visualized by their intrinsic GFP-fluorescence (Fig. 3a). Higher magnification revealed that GFP-positive cells exhibit the typical spindle-shaped morphology of taste cells and are exclusively located within taste buds (Fig. 3b, c). Employing a GPR92-specific antibody on cross sections through the CvP of a PLCβ2-GFP transgenic mouse revealed that the vast majority of GPR92-immunoreactive cells showed intrinsic GFP-fluorescence (Fig. 4a–c) and can therefore be classified as type II receptor cells. To quantify the co-expression of GPR92 and PLCβ2, we counted the number of GPR92-positive cells, GFP-positive cells and double-labeled cells on cross sections through the CvP of PLCβ2-GFP transgenic mice immunostained with the GPR92 antibody (Fig. 4d). The analyses revealed that 91.4 % of GPR92-immunoreactive cells were GFP-positive (54.2 ± 10.5 GPR92-positive cells; 49.5 ± 9.2 double-labeled cells) and that 72.0% of GFP-positive cells showed GPR92-immunoreactivity (68.7 ± 10.3 GFP-positive cells; 49.5 ± 9.2 double-labeled cells).
Fig. 3.
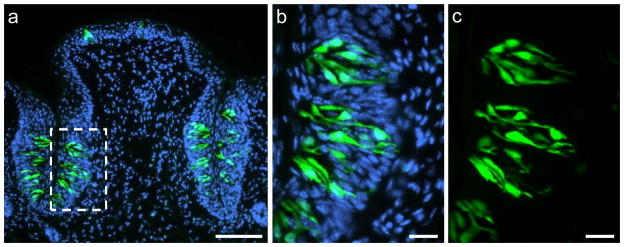
Intrinsic GFP-fluorescence in the circumvallate papilla of a PLCβ2 transgenic mouse.
(a) Overview of a cross section through the circumvallate papilla. GFP-positive cells (green) are located within taste buds.
(b, c) Magnification of the dotted area in (a). GFP-positive cells show the typical, spindle-shaped morphology of taste cells.
Sections are counterstained with DAPI (blue). Scale bars: a = 100 μm; b, c = 20 μm
Fig. 4.
GPR92 is expressed in type II taste cells.
Immunohistochemistry employing an GPR92 antibody on cross sections through the circumvallate papilla of a PLCβ2 transgenic mouse.
(a) GPR92-immunoreactive cells in circumvallate taste buds.
(b) Numerous GFP-positive cells in the same circumvallate taste buds.
(c) The overlay of (a) and (b) clearly reveals that the vast majority of GPR92-immunoreactive cells is also GFP-positive. However, a subset of GFP-expressing cells shows no labeling for GPR92 (arrowhead).
(d) Quantitative analyses of co-expression patterns. GPR92-positive cells, PLCβ2-GFP-positive cells and double-labeled cells (GPR92-positive and PLCβ2-GFP-positive) in the circumvallate papilla were counted on cross sections. The analyses revealed that 91.4% of GPR92-immunoreactive cells are GFP-positive. In contrast, 72.0% of GFP-expressing cells show GPR92-immunoreactivity. Twenty sections from the circumvallate papilla of one mouse were analyzed. Data are expressed as mean numbers ± SD. Sections are counterstained with DAPI (blue). Scale bars: a–c = 20 μm
The finding that a relatively high proportion of PLCβ2-positive type II cells in the CvP express the receptor GPR92 was an unexpected result; especially in the light of three other subsets of type II cells depending on the receptor types T1R1, T1R2 or T2R. Interestingly, a relatively high number of labeled cells was also described for the T1R1 receptor in circumvallate papillae of mice (Kim et al. 2003; Kusakabe et al. 2005). This raised the question whether the amino acid receptor T1R1 and the peptone receptor GPR92 may be co-expressed in the same cells. Unfortunately, this question could not be assessed by double-immunohistochemistry because available antibodies to the two proteins are both generated in the same host species. Nevertheless, we conducted immunohistochemical staining on consecutive sections of the CvP. These experiments preliminarily suggested that GPR92 and T1R1 may indeed be co-expressed in taste cells of the CvP (data not shown).
In order to verify this notion, we made use of T1R1-mCherry gene-targeted mice in which T1R1-expressing taste cells can be identified by their intrinsic mCherry-fluorescence (Voigt et al. 2012). Cross sections of the CvP from T1R1-mCherry mice were immunostained with the GPR92-specific antibody. Numerous GPR92- and mCherry-positive cells were visible in taste buds of the CvP (Fig. 5a–c). Higher magnification revealed that the vast majority of GPR92-immunoreactive cells showed mCherry-fluorescence (Fig. 5d–f). To quantify the co-expression of GPR92 and mCherry, we counted the number of GPR92-positive cells, mCherry-positive cells and double-labeled cells on cross sections through the CvP of T1R1-mCherry gene-targeted mice immunostained with the GPR92 antibody. The results are summarized in Figure 5d. We found that 94.5% of GPR92-immunoreactive cells were mCherry-positive (54.4 ± 8.3 GPR92-positive cells; 51.4 ± 8.5 double-labeled cells) and that 97.9% of mCherry-labeled cells express GPR92 (52.5 ± 8.3 mCherry-positive cells; 51.4 ± 8.5 double-labeled cells). Thus, the peptone-receptor GPR92 (GPR93; LPAR5) is co-expressed in T1R1-positive taste cells suggesting that these cells may respond to both amino acids and peptides from protein hydrolysates to generate umami taste.
Fig. 5.
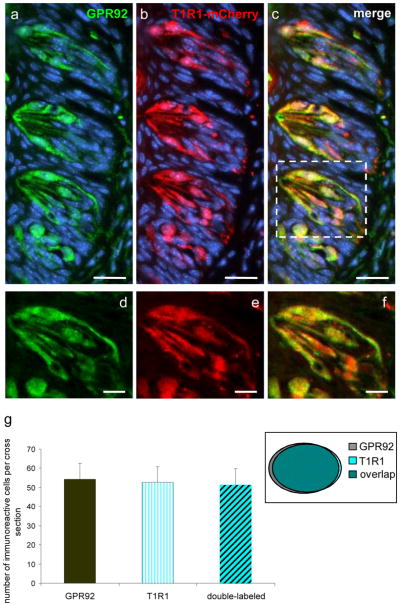
GPR92 is expressed in T1R1-expressing taste cells of the circumvallate papilla. Immunohistochemistry on cross sections through the circumvallate papilla of a T1R1-mCherry gene-targeted mouse.
(a) The GPR92 antibody labels numerous cells in taste buds of the circumvallate papilla.
(b) T1R1-expressing taste cells visualized by their intrinsic mCherry-fluorescence.
(c) Overlay of (a) and (b); the vast majority of GPR92-immunoreactive cells shows also mCherry-fluorescence.
(d, e, f) Magnification of the dotted area in (c). GPR92 is co-expressed in mCherry-labeled cells.
(g) Quantitative analyses of co-expression patterns. GPR92-positive cells, mCherry-positive cells and double-labeled cells (GPR92-positive and T1R1-mCherry-positive) were counted on cross sections through the circumvallate papillae of T1R1-mCherry gene-targeted mice. About 94.5% of GPR92-labeled cells show mCherry-fluorescence and 97.9% of T1R1-mCherry expressing cells are GPR92-immunoreactive. Thirty-six sections from circumvallate papillae of two mice were analyzed. Data are expressed as mean numbers ± SD. Sections are counterstained with DAPI (blue). Scale bars: a–c = 20 μm; d–f = 10 μm
Discussion
An adequate nutrient supply is of vital importance to maintain fundamental physiological processes. The ingestion of dietary protein is an essential element of nutritionally balanced diets. Besides their role as energy source, their building blocks, the amino acids, are precursors of many biologically relevant molecules. Moreover, dietary proteins play a critical role in modulating a variety of physiological processes including the regulation of food intake and gastrointestinal functions (Jahan-Mihan et al. 2011; San Gabriel et al. 2012). The first site for detecting dietary protein is the oral cavity which is endowed with specialized gustatory sensory cells. Previous studies have shown that a typical taste perception, the umami taste, is a signal for the ingestion of dietary protein. Many protein-rich foods, especially cured meats, aged cheeses, germinated grains and cooked foods contain partially hydrolyzed proteins and certain peptides in protein hydrolysates have been reported to elicit umami taste (Van den Oord and Wasenaar 2007; Schlichtherle-Cerny and Amadò 2002). Therefore, we considered that in addition to receptors responsive to amino acids, other taste receptors may exist for detecting peptides indicative of high-protein foods, and may contribute to perceived “protein” taste. Our findings demonstrating that the peptone receptor GPR92 is expressed in gustatory sensory cells, particularly those that are likely umami-sensing cells support this notion. The GPR92 receptor type was shown to be responsive to protein hydrolysates (Choi et al. 2007a, b), a mixture of enzymatically derived peptide fragments with different molecular weights (Cuber et al. 1990). This characteristic ligand spectrum makes the GPR92 receptor a suitable candidate to render gustatory sensory cells responsive to protein breakdown products. In this context, it is interesting to note that previous studies have shown that not only glutamate but also peptides with MW > 1000 provoke and enhance umami taste perception (Raksakulthai and Haard 1992). The view that the receptor type GPR92 may contribute to the responsiveness of gustatory sensory cells to protein fragments is supported by our recent finding that GPR92 is expressed in specific enteroendocrine cells of the gastric mucosa, the G-cells and D-cells (Haid et al. 2012). The secretion of their peptide hormones, gastrin and somatostatin, respectively, depends on the amount of proteins and their breakdown products in the luminal content of the stomach (Saffouri et al. 1984). Moreover, the observation that gastrointestinal cells and gustatory sensory cells express the same receptor type is in line with several recent studies demonstrating that gustatory and gastrointestinal cells share the expression of gustatory signaling elements and receptors (for reviews see: Breer et al. 2012; Iwatsuki and Uneyama 2012).
Recent research unraveling the basis of gustatory sensation has revealed that each taste modality is mediated by subsets of gustatory sensory cells which are endowed with a distinct molecular recognition and transduction machinery dedicated to the detection of the relevant chemical compounds (Zhang et al. 2003; Clapp et al. 2004; Chaudhari and Roper, 2010). For sweet and umami taste the specificity of the corresponding cells was proposed to be mediated by a single dimeric receptor, T1R1+T1R3 for umami and T1R2+T1R3 for sweet (Nelson et al. 2001, 2002; Li et al. 2002). Our findings that T1R1-expressing cells also express the peptone receptor GPR92 suggests that umami-sensing cells are endowed with at least two different umami receptors. Further, another candidate umami receptor, mGluR4, was also shown to be co-expressed in many PLCβ2-expressing cells in CvP, FuP and palatal taste buds (Chaudhari et al. 2009). The existence of multiple umami receptors in individual cells may render such gustatory cells more broadly tuned sensors for the detection of different protein-related ligands. Chaudhari et al. (2009) have speculated that similar to the chemically diverse bitter substances, the chemical diversity of umami tastants may require more broadly tuned sensors than the spectrum of T1R1+T1R3; one way to accomplish this requirement is the endowment of taste cells with multiple receptor types. In fact, due to the importance of dietary proteins, omnivorous animals may utilize diverse receptors to identify free amino acids, nucleotides, peptides and possibly other likely indicators of protein. The co-expression of the amino acid receptor T1R1+T1R3 together with the peptone receptor GPR92 in umami cells may make these cells more effective for monitoring the protein content of ingested food.
Acknowledgments
We would like to thank Kerstin Bach for excellent technical assistance and Prof. Meyerhof for providing the T1R1-mCherry mice. This work was supported by the Deutsche Forschungsgemeinschaft, BR 712/25-1.
Abbreviations
- DAPI
4′,6-Diamidino-2-phenylindole
- GPCR
G-protein coupled receptor
- GPR92
G-protein coupled receptor 92 (GPR93; LPAR5)
- mGluR4
metabotropic glutamate receptor 4
- T1R
taste receptor type 1
- T2R
taste receptor type 2
- T1R1
taste receptor type 1, member 1
- T1R3
taste receptor type 1, member 3
References
- Bernhardt SJ, Naim M, Zehavi U, Lindemann B. Changes in IP3 and cytosolic Ca2+ in response to sugars and non-sugar sweeteners in transduction of sweet taste in the rat. J Physiol. 1996;490 ( Pt 2):325–36. doi: 10.1113/jphysiol.1996.sp021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breer H, Eberle J, Frick C, Haid D, Widmayer P. Gastrointestinal chemosensation: chemosensory cells in the alimentary tract. Histochem Cell Biol. 2012;138(1):13–24. doi: 10.1007/s00418-012-0954-z. [DOI] [PubMed] [Google Scholar]
- Chaudhari N, Pereira E, Roper SD. Taste receptors for umami: the case for multiple receptors. Am J Clin Nutr. 2009;90(3):738S–742S. doi: 10.3945/ajcn.2009.27462H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N, Roper SD. The cell biology of taste. J Cell Biol. 2010;190(3):285–96. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Lee M, Shiu AL, Yo SJ, Aponte GW. Identification of a protein hydrolysate responsive G protein-coupled receptor in enterocytes. Am J Physiol Gastrointest Liver Physiol. 2007a;292(1):G98–G112. doi: 10.1152/ajpgi.00295.2006. [DOI] [PubMed] [Google Scholar]
- Choi S, Lee M, Shiu AL, Yo SJ, Halldén G, Aponte GW. GPR93 activation by protein hydrolysate induces CCK transcription and secretion in STC-1 cells. Am J Physiol Gastrointest Liver Physiol. 2007b;292(5):G1366–75. doi: 10.1152/ajpgi.00516.2006. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. Morphologic characterization of rat taste receptor cells that express components of the phospholipase C signaling pathway. J Comp Neurol. 2004;468(3):311–21. doi: 10.1002/cne.10963. [DOI] [PubMed] [Google Scholar]
- Cuber JC, Bernard G, Fushiki T, Bernard C, Yamanishi R, Sugimoto E, Chayvialle JA. Luminal CCK-releasing factors in the isolated vascularly perfused rat duodenojejunum. Am J Physiol. 1990;259(2 Pt 1):G191–7. doi: 10.1152/ajpgi.1990.259.2.G191. [DOI] [PubMed] [Google Scholar]
- Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Varadarajan V, Zou S, Jiang P, Ninomiya Y, Margolskee RF. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science. 2003;301(5634):850–3. doi: 10.1126/science.1087155. [DOI] [PubMed] [Google Scholar]
- DeFazio RA, Dvoryanchikov G, Maruyama Y, Kim JW, Pereira E, Roper SD, Chaudhari N. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J Neurosci. 2006;26(15):3971–80. doi: 10.1523/JNEUROSCI.0515-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid D, Jordan-Biegger C, Widmayer P, Breer H. Receptors responsive to protein breakdown products in g-cells and d-cells of mouse, swine and human. Front Physiol. 2012;3:65. doi: 10.3389/fphys.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K. New seasonings. J Tokyo Chem Soc. 1909;30:820–836. (in Japanese) [Google Scholar]
- Ikeda K. New seasonings. Chem Senses. 2002;27(9):847–9. doi: 10.1093/chemse/27.9.847. [DOI] [PubMed] [Google Scholar]
- Iwatsuki K, Uneyama H. Sense of taste in the gastrointestinal tract. J Pharmacol Sci. 2012;118(2):123–8. doi: 10.1254/jphs.11r08cp. [DOI] [PubMed] [Google Scholar]
- Jahan-Mihan A, Luhovyy BL, El Khoury D, Anderson GH. Dietary proteins as determinants of metabolic and physiologic functions of the gastrointestinal tract. Nutrients. 2011;3(5):574–603. doi: 10.3390/nu3050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Kusakabe Y, Miura H, Shindo Y, Ninomiya Y, Hino A. Regional expression patterns of taste receptors and gustducin in the mouse tongue. Biochem Biophys Res Commun. 2003;312(2):500–6. doi: 10.1016/j.bbrc.2003.10.137. [DOI] [PubMed] [Google Scholar]
- Kim JW, Roberts C, Maruyama Y, Berg S, Roper S, Chaudhari N. Faithful expression of GFP from the PLCbeta2 promoter in a functional class of taste receptor cells. Chem Senses. 2006;31(3):213–9. doi: 10.1093/chemse/bjj021. [DOI] [PubMed] [Google Scholar]
- Kretz O, Barbry P, Bock R, Lindemann B. Differential expression of RNA and protein of the three pore-forming subunits of the amiloride-sensitive epithelial sodium channel in taste buds of the rat. J Histochem Cytochem. 1999;47(1):51–64. doi: 10.1177/002215549904700106. [DOI] [PubMed] [Google Scholar]
- Kusakabe Y, Kim MR, Miura H, Shindo Y, Ninomiya Y, Hino A. Regional expression patterns of T1r family in the mouse tongue. Chem Senses. 2005;30(Suppl 1):i23–4. doi: 10.1093/chemse/bjh094. [DOI] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci U S A. 2002;99(7):4692–6. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga JA. Flavor potentiators. Crit Rev Food Sci Nutr. 1983;18(3):231–312. doi: 10.1080/10408398309527364. [DOI] [PubMed] [Google Scholar]
- Maruyama Y, Pereira E, Margolskee RF, Chaudhari N, Roper SD. Umami responses in mouse taste cells indicate more than one receptor. J Neurosci. 2006;26(8):2227–34. doi: 10.1523/JNEUROSCI.4329-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106(3):381–90. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature. 2002;14;416(6877):199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Raksakulthai N, Haard NF. Correlation between the concentration of peptides and amino acids and the flavour of fish sauce. ASEAN Food Journal. 1992;7(2):86–90. [Google Scholar]
- Rössler P, Kroner C, Freitag J, Noè J, Breer H. Identification of a phospholipase C beta subtype in rat taste cells. Eur J Cell Biol. 1998;77(3):253–61. doi: 10.1016/s0171-9335(98)80114-3. [DOI] [PubMed] [Google Scholar]
- Saffouri B, DuVal JW, Makhlouf GM. Stimulation of gastrin secretion in vitro by intraluminal chemicals: regulation by intramural cholinergic and noncholinergic neurons. Gastroenterology. 1984;87:557–561. [PubMed] [Google Scholar]
- San Gabriel A, Uneyama H. Amino acid sensing in the gastrointestinal tract. Amino Acids. 2012 doi: 10.1007/s00726-012-1371-2. [DOI] [PubMed] [Google Scholar]
- Schlichtherle-Cerny H, Amadò R. Analysis of taste-active compounds in an enzymatic hydrolysate of deamidated wheat gluten. J Agric Food Chem. 2002;50(6):1515–22. doi: 10.1021/jf010989o. [DOI] [PubMed] [Google Scholar]
- Striem BJ, Naim M, Lindemann B. Generation of Cyclic AMP in Taste Buds of the Rat Circumvallate Papilla in Response to Sucrose. Cell Physiol Biochem. 1991;1:46–54. [Google Scholar]
- Tamura M, Nakatsuka T, Tada M, Kawasaki Y, Kikuchi E, Okai A. The relationship between taste and primary structure of “delicious peptide” (Lys-Gly-Asp-Glu-Glu-Ser-Leu-Ala) from beef soup. Agric Biol Chem. 1989;53:319–325. [Google Scholar]
- Tomchik SM, Berg S, Kim JW, Chaudhari N, Roper SD. Breadth of tuning and taste coding in mammalian taste buds. J Neurosci. 2007;27(40):10840–8. doi: 10.1523/JNEUROSCI.1863-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Oord AHA, Van Wassenaar PD. Umami peptides: assessment of their alleged taste properties. Eur Food Res Technol. 1997;205:25–130. [Google Scholar]
- Voigt A, Hübner S, Lossow K, Hermans-Borgmeyer I, Boehm U, Meyerhof W. Genetic Labeling of Tas1r1 and Tas2r131 Taste Receptor Cells in Mice. Chem Senses. 2012;37(9):897–911. doi: 10.1093/chemse/bjs082. [DOI] [PubMed] [Google Scholar]
- Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37(1):1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Ninomiya K. Umami and food palatability. J Nutr. 2000;130(4S Suppl):921S–6S. doi: 10.1093/jn/130.4.921S. [DOI] [PubMed] [Google Scholar]
- Yee CL, Yang R, Böttger B, Finger TE, Kinnamon JC. “Type III” cells of rat taste buds: immunohistochemical and ultrastructural studies of neuron-specific enolase, protein gene product 9.5, and serotonin. J Comp Neurol. 2001;440(1):97–108. doi: 10.1002/cne.1372. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Yasumatsu K, Shirosaki S, Jyotaki M, Horio N, Murata Y, Shigemura N, Nakashima K, Ninomiya Y. Multiple receptor systems for umami taste in mice. Ann N Y Acad Sci. 2009;1170:51–4. doi: 10.1111/j.1749-6632.2009.03902.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112(3):293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- Zhang F, Klebansky B, Fine RM, Xu H, Pronin A, Liu H, Tachdjian C, Li X. Molecular mechanism for the umami taste synergism. Proc Natl Acad Sci U S A. 2008;105 (52):20930–4. doi: 10.1073/pnas.0810174106. [DOI] [PMC free article] [PubMed] [Google Scholar]