Abstract
Image registration is typically formulated as an optimization problem with multiple tunable, manually set parameters. We present a principled framework for learning thousands of parameters of registration cost functions, such as a spatially-varying tradeoff between the image dissimilarity and regularization terms. Our approach belongs to the classic machine learning framework of model selection by optimization of cross-validation error. This second layer of optimization of cross-validation error over and above registration selects parameters in the registration cost function that result in good registration as measured by the performance of the specific application in a training data set. Much research effort has been devoted to developing generic registration algorithms, which are then specialized to particular imaging modalities, particular imaging targets and particular postregistration analyses. Our framework allows for a systematic adaptation of generic registration cost functions to specific applications by learning the “free” parameters in the cost functions. Here, we consider the application of localizing underlying cytoarchitecture and functional regions in the cerebral cortex by alignment of cortical folding. Most previous work assumes that perfectly registering the macro-anatomy also perfectly aligns the underlying cortical function even though macro-anatomy does not completely predict brain function. In contrast, we learn 1) optimal weights on different cortical folds or 2) optimal cortical folding template in the generic weighted sum of squared differences dissimilarity measure for the localization task. We demonstrate state-of-the-art localization results in both histological and functional magnetic resonance imaging data sets.
Index Terms: Cross validation error, functional magnetic resonance imaging (fMRI), histology, ill-posed, leave one out error, local maxima, local minima, model selection, objective function, parameter tuning, registration parameters, regularization, space of local optima, tradeoff
I. Introduction
IN medical image analysis, registration is necessary to establish spatial correspondence across two or more images. Traditionally, registration is considered a preprocessing step [Fig. 1(a)]. Images are registered and are then used for other image analysis applications, such as voxel-based morphometry and shape analysis. Here, we argue that the quality of image registration should be evaluated in the context of the application. In particular, we propose a framework for learning the parameters of registration cost functions that are optimal for a specific application. Our framework is therefore equivalent to classic machine learning approaches of model selection by optimization of cross-validation error [33], [43], [58].
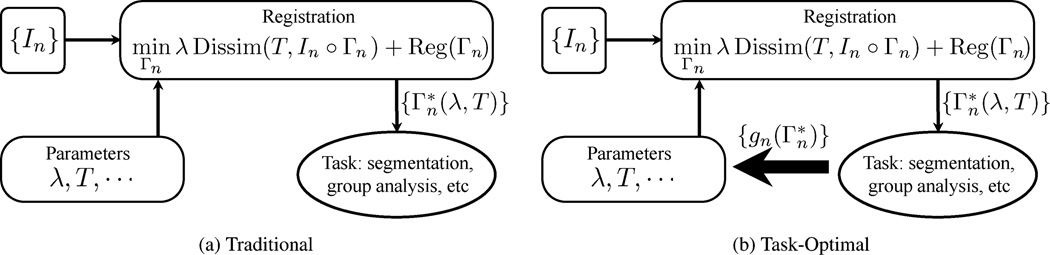
Fig. 1.
Traditional and proposed frameworks for image registration.{In} indicates a collection of images. In image registration, we seek a deformation for each image In. The resulting deformations are then used for other applications, such as segmentation or group analysis. The registration cost function typically contains multiple parameters, such as the tradeoff parameter λ and the template T. Changes in these parameters alter the deformations and thus the outcomes of downstream applications. In our framework (b), we assume a training data set, which allows us to evaluate the quality of the registration as measured by the application performance (or cross-validation error metric) gn for each training subject. This allows us to pick the best parameters that result in good registration as measured by {gn}. Subsequent new subjects are registered using these learned parameters.
A. Motivation
Image registration is typically formulated as an optimization problem with a cost function that comprises an image dissimilarity term and a regularization term [Fig. 1(a)]. The parameters of the cost function are frequently determined manually by inspecting the quality of the image alignment to account for the characteristics (e.g., resolution, modality, signal-to-noise ratio) of the image data. During this process, the final task is rarely considered in a principled fashion. Furthermore, the variability of the results due to these tunable parameters is rarely reported in the literature. Yet, recent work has shown that taking into account the tradeoff between the regularization and similarity measure in registration can significantly improve population analysis [40] and segmentation quality [10], [79].
In addition to improving the performance of applications downstream, taking into account the end-goal of registration could help resolve ambiguities and the ill-posed nature of image registration.
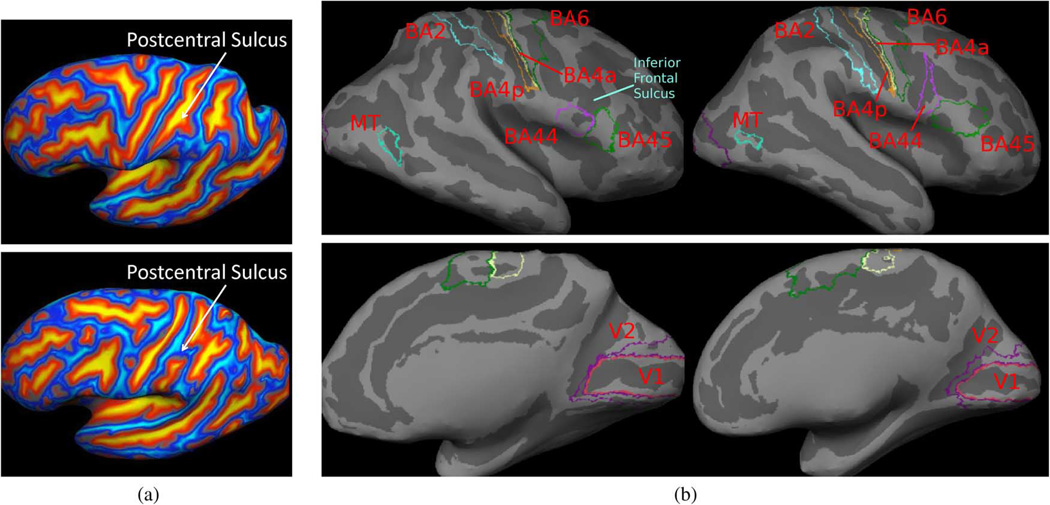
The variability of the folding pattern in the human cerebral cortex is well-documented (see e.g., [45]). Fig. 2(a) shows postcentral sulci of two different subjects. Note the differences in topology between the two sulci. When matching cortical folds, even neuroanatomical experts disagree on whether to join the ends of the broken sulcus or to break up the uninterrupted sulcus.
In population studies of human brain mapping, it is common to align subjects into a single coordinate system by aligning macroanatomy or cortical folding patterns. The pooling of functional data in this common coordinate system boosts the statistical power of group analysis and allows functional findings to be compared across different studies. However, substantial cytoarchitectonic [3], [4], [18] and functional [41], [62]–[64], [77], [78] variability is widely reported. One reason for this variability is certainly misregistration of the highly variable macroanatomy. However, even if we perfectly align the macroanatomy, the underlying function and cellular architecture of the cortex will not be aligned because the cortical folds do not completely predict the underlying brain function [54], [62]. To illustrate this, Fig. 2(b) shows nine Brodmann areas (BAs) projected onto the cortical surfaces of two different subjects, obtained from histology. BAs define cytoarchitectonic parcellation of the cortex closely related to brain function [9]. Here, we see that perfectly aligning the inferior frontal sulcus [Fig. 2(b)] will misalign the superior end of BA44 (Broca’s language area). If our goal is to segment sulci and gyri, perfect alignment of the cortical folding pattern is ideal. However, it is unclear that perfectly aligning cortical folds is optimal for function localization.
Fig. 2.
Examples of ambiguities in image registration, which can potentially be resolved by taking the application at hand into account. (a) Postcentral sulci with different topology. (b) BAs overlaid on cortical surfaces.
In this paper, we propose a task-optimal registration framework that optimizes parameters of any smooth family of registration cost functions on a training set, with the aim of improving the performance of a particular task for a new image [Fig. 1(b)]. The key idea is to introduce a second layer of optimization over and above the usual registration. This second layer of optimization assumes the existence of a smooth cost function or cross-validation error metric [g in Fig. 1(b)] that evaluates the performance of a particular task given the output of the registration step for a training data set. The training data provides additional information not present in a test image, allowing the task-specific cost function to be evaluated during training. For example, if the task is segmentation, we assume the existence of a training data set with ground truth segmentation and a smooth cost function (e.g., Dice overlap measure) that evaluates segmentation accuracy. If the registration cost function employs a single parameter, then the optimal parameter value can be found by exhaustive search [79]. With multiple parameters, exhaustive search is not possible. Here, we establish conditions for which the space of local minima is locally smooth and demonstrate the optimization of thousands of parameters by gradient descent on the space of local minima, selecting registration parameters that result in good registration local minima as measured by the task-specific cost function in the training data set.
We validate our framework on two datasets. The first dataset consists of 10 ex vivo brains with the BAs of each subject obtained via histology [4], [84] and mapped onto the cortical surface representation of each subject obtained via MRI [18]. The second dataset consists of 42 in vivo brains with functional region MT+ (V5) defined using functional magnetic resonance imaging (fMRI). Here, our task is defined to be the localization of BAs and MT+ in the cortical surface representation via the registration of the cortical folding pattern. While it is known that certain cytoarchitectonically or functionally-defined areas, such as V1 or BA28, are spatially consistent with respect to local cortical geometry, other areas, such as BA44, present a challenge for existing localization methods [18], [20].We learn the weights of the weighted sum of squared differences (wSSD) family of registration cost functions and/or estimate an optimal macroanatomical template for localizing the cytoarchitectural and functional regions using only the cortical folding pattern. We demonstrate improvement over existing methods [18].
B. Related Work
An alternative approach to overcome the imperfect correlation between anatomy and function is to directly use the functional data for establishing across-subject functional correspondence [54], [56]. However, these approaches require extra data acquisition (such as fMRI scans) of all future test subjects. In contrast, our method aims to learn the relationship between macro-anatomy and function (or cytoarchitectonics) in a training data set containing information about both macro-anatomy and function (or cytoarchitectonics). We use this information to localize function (or cytoarchitectonics) in future subjects, for which only macro-anatomical information is available.
Our approach belongs to the class of “wrapper methods” for model or feature selection in the machine learning literature [27], [34]. In particular, our model selection criterion of application-specific performance is equivalent to the use of cross-validation error in various model selection algorithms [33], [43], [58]. Unlike feature selection methods that operate in a discrete parameter space, we work in a continuous parameter space. Consequently, standard algorithms in the “wrapper methods” literature do not apply to this problem.
Instead, our resulting optimization procedure borrows heavily from the mathematical field of continuation methods [2]. Continuation methods have been recently introduced to the machine learning community for computing the entire path of solutions of learning problems (e.g., SVM or Lasso) as a function of a single regularization parameter [16], [28], [46]. For example, the cost function in Lasso [67] consists of the tradeoff between a least-squares term and a L1 regularization term. Least-angles regression (LARS) allows one to compute the entire set of solutions of Lasso as a function of the tradeoff parameter [16]. Because we deal with multiple (thousands of) parameters, it is impossible for us to compute the entire solution manifold. Instead, we trace a path within the solution manifold that improves the task-specific cost function. Furthermore, registration is not convex (unlike SVM and Lasso), resulting in several theoretical and practical conundrums that we have to overcome, some of which we leave for future work.
The wSSD similarity measure implicitly assumes an independent Gaussian distribution on the image intensities, where the weights correspond to the precision (reciprocal of the variance) and the template corresponds to the mean of the Gaussian distribution. The weights can be set to a constant value [6], [31] or a spatially-varying variance can be estimated from the intensities of registered images [19]. However, depending on the wSSD regularization tradeoff, the choice of the scale of the variance is still arbitrary [79]. With weaker regularization, the training images will be better aligned, resulting in lower variance estimates.
Recent work in probabilistic template construction resolves this problem by either marginalizing the tradeoff under a Bayesian framework [1] or by estimating the tradeoff with the minimum description length principle [71]. While these methods are optimal for “explaining the images” under the assumed generative models, it is unclear whether the estimated parameters are optimal for application-specific tasks. After all, the parameters for optimal image segmentation might be different from those for optimal group analysis. In contrast, Van Leemput [74] proposes a generative model for image segmentation. The estimated parameters are therefore Bayesian-optimal for segmentation. When considering one global tradeoff parameter, a more direct approach is to employ cross-validation of segmentation accuracy and to perform an exhaustive search over the values of the tradeoff parameter [79]. This is infeasible for multiple parameters.
By learning the weights of the wSSD, we implicitly optimize the tradeoff betweeen the dissimilarity measure and regularization. Furthermore, the tradeoff we learn is spatially varying. Previous work on learning a spatially varying regularization prior suffers from the lack of ground truth (nonlinear) deformations. For example, [10], [25], [35] assume that the deformations obtained from registering a set of training images can be used to estimate a registration regularization to register new images. However, a change in the parameters of the registration cost function used by these methods to register the training images would lead to a different set of training deformations and thus a different prior for registering new images. Furthermore, the methods are inconsistent in the sense that the learned prior applied on the training images will not result in the same training deformations obtained previously.
While there has been efforts in obtaining ground truth human-annotated deformation fields [37], the images considered typically have well-defined correspondences, rather than for example, the brain images of two different subjects. As suggested in the previously presented examples (Fig. 2), the concept of “ground truth deformations” may not always be well-defined, since the optimal registration may be a function of the application at hand. In contrast, image segmentation is generally better defined in the sense that ground truth segmentation is usually known. Our problem therefore differs from recent work on learning segmentation cost functions [42], [70], [83]. In this paper, we avoid the need for ground truth deformations by focusing on the application of registration-based segmentation, where ground truth segmentations are better defined and available. However, our framework is general and can be applied whenever a postregistration application can be well quantified by a smooth application-specific performance cost function.
This paper is organized as follows. In the next section, we introduce the task-optimal registration framework. We specialize the framework to align hidden labels in Section III. We present localization experiments in Section IV and discuss outstanding issues in Section V. This paper extends a previously presented conference article [80] and contains detailed derivations, discussions and experiments that were omitted in the conference version.
We present a framework for learning the parameters of registration cost functions with respect to specific applications. We present an algorithm sufficiently efficient for optimizing thousands of parameters.
We specialize the framework for the alignment of hidden labels, which are not necessarily well-predicted by local image features.
We apply the framework to localizing cytoarchitectural and functional regions using only the cortical folding pattern and demonstrate improvements over existing localization methods [18].
II. Task-Optimal Framework
In this section, we present the task-optimal registration framework for learning the parameters of a registration cost function. Given an image I, let f (w, Γ) denote a smooth registration cost function, with parameters w and a spatial transformation Γ. For example
| (2.1) |
where T is the template image, λ is the tradeoff between the image dissimilarity measure and the regularization on the transformation Γ, I ∘ Γ denotes the deformed and resampled image I. f is therefore also a function of the image I, which we suppress for conciseness. The optimal transformation Γ* minimizes the cost function for a given set of parameters w
| (2.2) |
We emphasize that Γ* is a function of w since a different set of parameters w will result in a different solution to (2.2) and thus will effectively define a different image coordinate system.
The resulting deformation Γ* is used to warp the input image or is itself used for further tasks, such as image segmentation or voxel-based morphometry. We assume that the task performance can be measured by a smooth cost function (or cross-validation error metric) g, so that a smaller value of g (Γ*(w)) corresponds to better task performance. g is typically a function of additional input data associated with a subject (e.g., manual segmentation labels if the task is automatic segmentation), although we suppress this dependency in the notation for conciseness. This auxiliary data is only available in the training set; g cannot be evaluated for the new image.
Given a set of N training subjects, let denote the solution of (2.2) for training subject n for a fixed set of parameters w and gn() denote the task performance for training subject n using the deformation and other information available for the nth training subject. A different set of parameters w would lead to different task performance gn(). We seek the parameters w* that generalize well to a new subject: registration of a new subject with w* yields the transformation Γ*(w*) with a small task-specific cost g(Γ*(w*)). One approach to solve this functional approximation problem [17] is regularized risk minimization. Let Reg(w) denote regularization on w and define
| (2.3) |
Regularization risk minimization seeks
| (2.4) |
The optimization is difficult because while we assume gn to be smooth, the input to gn (·) is itself the local minimum of another nonlinear cost function f. Furthermore, evaluating the cost function G for only one particular set of parameters w requires performing N different registrations!
A. Characterizing the Space of Local Minima
In this section, we provide theoretical characterizations of the optimization problem in (2.4). If Γ*(w) is defined strictly to be a global registration optimum, then Γ*(w) is clearly not a smooth function of w, since a small change in w can result in a big change in the global registration optimum. This definition is also impractical, since the global optimum of a nonlinear optimization problem cannot be generally found in practice. Instead, we relax the definition of Γ*(w) to be a local minimum of the registration cost function for fixed values of w. Here, we derive conditions in which Γ*(w) is locally a smooth function of w, so we can employ gradient descent to optimize (2.4).
Let Γ*(w0) denote a local minimum of the registration cost function for a fixed w = w0. Suppose we perturb w by an infinitestimally small δw, so that Γ*(w0) is no longer the registration local minimum for w = w0 + δw. We consider two representations of this change in local minimum.
Additive deformation models arise when the space of deformations is a vector space, such as the space of displacement fields or positions of B-spline control points. At each iteration of the registration algorithm, deformation updates are added to the current deformation estimates. The additive model is general and applies to many non-convex, smooth optimization problems outside of registration. Most registration algorithms can in fact be modeled with the additive framework.
In some registration algorithms, including that used in this paper, it is more natural to represent deformation changes through composition rather than additions [7], [61], [75]. For example, in the diffeomorphic variants of the demons algorithm [75], [81], [82], the diffeomorphic transformation Γ is represented as a dense displacement field. At each iteration, the transformation update is restricted to be a one parameter subgroup of diffeomorphism parameterized by a stationary velocity field. The diffeomorphic transformation update is then composed with, rather than added to, the current estimate of the transformation, thus ensuring that the resulting transformation is diffeomorphic.
1) Addition Model
Let Γ*(w0 + δw) = Γ*(w0) + δΓ*(w0, δw) denote the new locally optimal deformation for the updated set of parameters w0 + δw. The following proposition characterizes the existence and uniqueness of δΓ*(w0, δw) as δw is varied. In particular, we show that under some mild conditions, δΓ*(w0, δw) is a well-defined smooth function in the neighborhood of (w0, Γ*(w0)). In the remainder, we use and to denote the corresponding partial derivatives.
Proposition 1: If the Hessian1 is positive definite at Γ = Γ*(w0), then there exists an ε > 0, such that for all δw, ‖δw‖ < ε, a unique continuous function δΓ*(w0, δw) exists with δΓ*(w0, 0) = 0. Furthermore, δΓ* has the same order of smoothness as ∂Γ f.
Proof: We define the vector-valued function h(w, Γ) ≜ ∂Γ f (w, Γ). Since Γ*(w0) is a local minimum of f(w0,Γ), we have
| (2.5) |
At (w0, Γ*(w0)), the Hessian matrix is positive definite by the assumption of the proposition and is therefore invertible. By the Implicit Function Theorem [51], there exists an ε > 0, such that for all δw, ‖δw‖ < ε, there is a unique continuous function δΓ*(w0, δw) such that h(w0 + δw, Γ*(w0) + δΓ*(w0, δw)) = 0 and δΓ*(w0, 0) = 0. Furthermore, δΓ*(w0, δw) has the same order of smoothness as h.
Because the Hessian of f is smooth and the eigenvalues of a matrix depend continuously on the matrix [72], there exists a small neighborhood around (w0, Γ*(w0)) in which the eigen-values of are all greater than 0. Since both sufficient conditions for a local minimum are satisfied (zero gradient and positive definite Hessian), Γ*(w0) + δΓ*(w0, dw) is indeed a new local minimum close to Γ*(w0).
Observe that the conditions in Proposition 1 are stronger than those of typical nonlinear optimization problems. In particular, we do not just require the cost functions f and g to be smooth, but also that the Hessian be positive definite at the local minimum. At (w0, Γ*(w0)), by definition, the Hessian is positive semi-definite, so the positive definite condition in Proposition 1 should not be too restrictive. Unfortunately, degeneracies may arise for local minima with a singular Hessian. For example, let Γ be the 1 × 2 vector [a b] and f (Γ, w) = (ab − w)2. Then for any value of w, there is an infinite number of local minima Γ*(w) corresponding to ab = w. Furthermore, the Hessian at any local minimum is singular. In this case, even though f is infinitely differentiable, there is an infinite number of local minima near the current local minimum Γ*(w0), i.e., δΓ*(w0, δw) is not a well-defined function and the gradient is not defined. Consequently, the parameters w of local registration minima whose Hessians are singular are also local minima of the task-optimal optimization (2.4). The proof of Proposition 1 follows the ideas of the Continuation Methods literature [2]. We include the proof here to motivate the more complex composition of deformations model.
2) Composition Model
Let Γ*(w0) be the registration local minimum at w0 and δΓ (υ) denote an update transformation parameterized by υ, so that δΓ (0) corresponds to the identity transform. For example, υ could be a stationary [75], [81], [82], nonstationary [8] velocity field parameterization of diffeomorphism, positions of spline control points [52] or simply displacement fields [59]. In the composition model, Γ*(w0) is a local minimum if and only if there exists an ε > 0, such that f (w0, Γ*(w0)) < f (w0, Γ*(w0) ∘ δΓ (υ)) for all values of ‖υ‖ < ε.
Let Γ*(w0) ∘ δΓ(υ*(w0, δw)) denote the new locally optimal deformation for the new parameters w0 + δw. In general, there might not exist a single update transformation δΓ(υ*(w0, δw)) that leads to a new local minimum under a perturbation of the parameters w, so that there is no correponding version of Proposition 1 for the general composition model. However, in the special case of the composition of diffeomorphisms model [75], [81], [82] employed in this paper, the following proposition characterizes the existence and uniqueness of υ*(w0, δw) as δw is varied.
Proposition 2: If the Hessian is positive definite at υ = 0, then there exists an ε > 0, such that for all δw, ‖δw‖ < ε, a unique continuous function υ*(w0, δw) exists, such that υ*(w0, δw) is the new local minimum for parameters w0 + δw and υ*(w0, 0) = 0. Furthermore, υ*(w0, δw) has the same order of smoothness as f.
Proof: The proof is a more complicated version of Proposition 1 and so we leave the details to Appendix A.
Just like in the case of the additive deformation model, the parameters w of local registration minima that do not satisfy the conditions of Proposition 2 are also local minima of the task-optimal optimization (2.4). In the next section, we derive exact and approximate gradients of G.
B. Optimizing Registration Parameters w
We now discuss the optimization of the regularized task performance G.
1) Addition Model
In the previous section, we showed that at (w0, Γ*(w0)) with a positive definite Hessian, δΓ*(w0, δw) is a smooth well-defined function such that Γ*(w0) + δΓ*(w0, δw) is the new local minimum at w0 + δw. Therefore, we can compute the derivatives of Γ*(w) with respect to w at w0, allowing us to traverse a curve of local optima, finding values of w that improve the task-specific cost function for the training images. We first perform a Taylor expansion of ∂Γ f (w, Γ) at (w0, Γ*(w0))
| (2.6) |
where we dropped the term ∂Γ f (w, Γ)w0, Γ*(w0) = 0. For δΓ = δΓ*(w0,δw), the left-hand side is equal to 0 and we can write
| (2.7) |
Therefore, by taking the limit δw ℩ 0, we get
| (2.8) |
Equation (2.8) tells us the direction of change of the local minimum at (w0, Γ*(w0)). In practice, the matrix inversion in (2.8) is computationally prohibitive for high-dimensional warps Γ. Here, we consider a simplification of (2.8) by setting the Hessian to be the identity
| (2.9) |
Since −∂Γ f is the direction of gradient descent of the cost function (2.2), we can interpret (2.9) as approximating the new local minimum to be in the same direction as the change in the direction of gradient descent as w is perturbed.
Differentiating the cost function in (2.4), using the chain rule, we obtain
| (2.10) |
| (2.11) |
| (2.12) |
We note the subscript n on f indicates the dependency of the registration cost function on the nth training image.
2) Composition Model
In the previous section, we have shown that at (w0, Γ*(w0)), assuming the conditions of Proposition 2 are true, υ*(w0, δw) is a smooth well-defined function such that Γ*(w0) ∘ δΓ (υ*(w0, δw)) is the new local minimum. Therefore, we can compute the derivatives of υ* with respect to w. As before, by performing a Taylor expansion, we obtain
| (2.13) |
| (2.14) |
Appendix B provides the detailed derivations. Differentiating the cost function in (2.4), using the chain rule, we get
| (2.15) |
| (2.16) |
Once again, the subscript n on f indicates the dependency of the registration cost function on the nth training image.
Algorithm 1 summarizes the method for learning the task-optimal registration parameters. Each line search involves evaluating the cost function G multiple times, which in turn requires registering the training subjects, resulting in a computationally intensive process. However, since we are initializing from a local optimum, for a small change in w, each registration converges quickly.
Algorithm 1.
Task-Optimal Registration
| Data: A set of training images {In} | ||
| Result: Parameters w that minimize the regularized task performance G [see (2.4)] | ||
| Initialize w0. | ||
| repeat | ||
| Step | 1. | Given current values of w, estimate , fn (w, Γn), i.e., perform registration of each training subject n. |
| Step | 2. | Given current estimates (w, {Γn (w)}), compute the ∂wG gradient using either
|
| Step | 3. | Perform line search in the direction opposite to ∂wG [47]. |
Since nonlinear registration is dependent on initialization, the current estimates (w, Γ*(w)), which were initialized from previous estimates, might not be achievable when initializing the registration with the identity transform. The corresponding parameters w might therefore not generalize well to a new subject, which are typically initialized with the identity transform. To put this more concretely, suppose our current estimates of w and the registration local minima are (w = 5, Γ* (5) = 2). Next, we perform the gradient decent step and update w accordingly. For argument’s sake, let our new estimates of w and the registration local minima be (w = 5.1, Γ* (5.1) = 1.9). Note that this particular value of Γ* (5.1) = 1.9 is achieved by initializing the registration with Γ = 2. Had we initialized the registration with the identity transform (such as for a new subject), then Γ* (5.1) might instead be equal to 2.1, with possibly poorer application performance than (w = 5, Γ* (5) = 2). To avoid this form of overfitting, after every few iterations, we reregister the training images by initializing with the identity transform, and verify that the value of G is better than the current best value of G computed with initialization from the identity transform.
The astute reader will observe that the preceding discussion on “Addition Model” makes no assumptions specific to the task-optimal registration problem. The framework can therefore also be applied to learn the cost functions in other applications that are formulated as nonlinear optimization problems solved by gradient descent.
III. Learning wSSD for Hidden Label Alignment
We now instantiate the task-optimal registration framework for localizing hidden labels in images. We demonstrate schemes for either 1) learning the weights of the wSSD family of registration cost functions or 2) estimating an optimal template image for localizing these hidden labels. We emphasize that the optimal template is not necessarily the average of the training images, since the goal is not to align image intensities across subjects, but to localize the hidden labels.
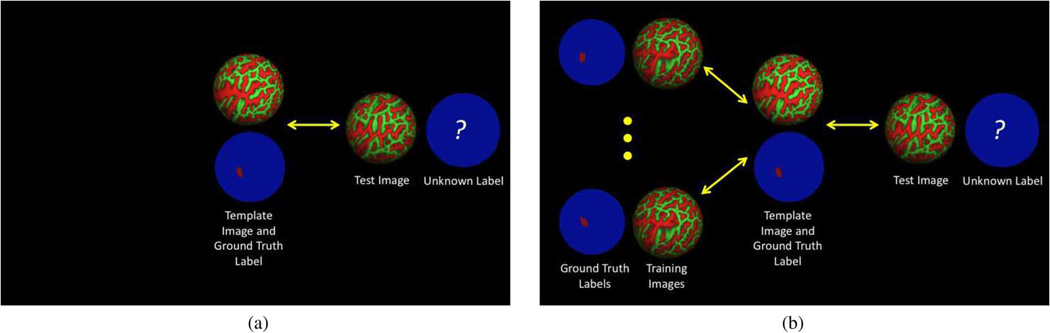
Suppose we have a set of training images {In} with some underlying ground truth structure manually labeled or obtained from another imaging modality (e.g., Brodmann areas from histology mapped onto cortical surface representations). We define our task as localizing the hidden structure in a test image. In the traditional pairwise registration approach [Fig. 3(a)], a single training subject is chosen as the template. After pairwise registration between the template and test images, the ground truth label of the template subject is used to predict that of the test subject. The goal of predicting the hidden structure in the test subject is typically not considered when choosing the training subject or registration algorithm. For hidden labels that are poorly predicted by local image intensity (e.g., BA44 discussed in Section I-A), blind alignment of image intensities lead to poor localization.
Fig. 3.
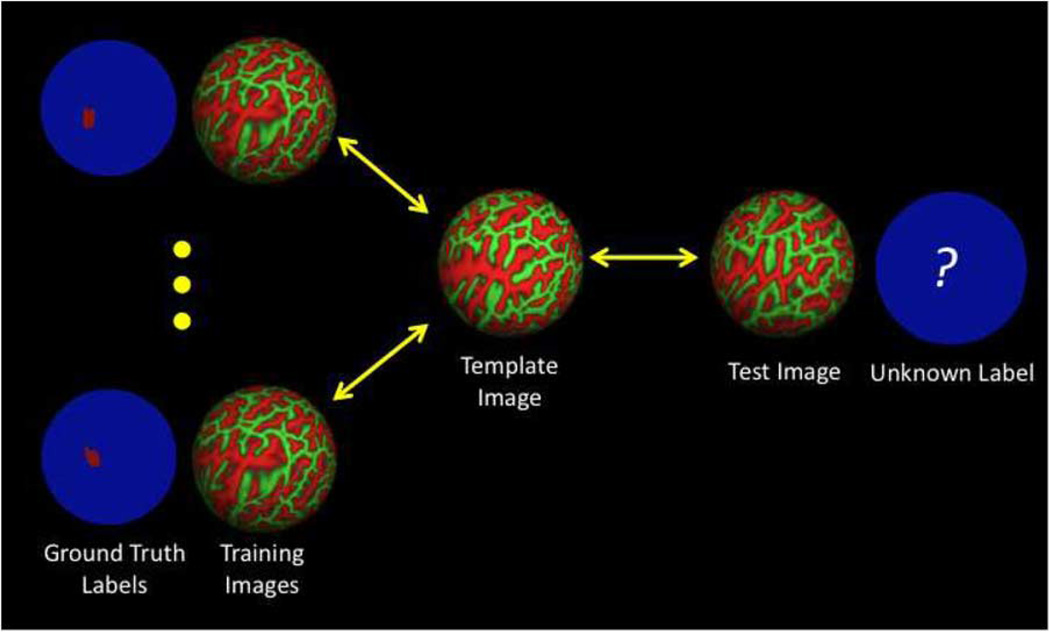
Illustration of the differences between our approach and the pairwise registration approach. In our approach, we use training images and labels to learn an optimal cost function that is optimal for aligning the labels of the training and template subjects. This cost function is then used to register and predict the hidden label in a new subject. (a) Pairwise registration without training using ground truth labels. (b) Task-optimal registration framework.
In contrast, we pick one training subject as the initial template and use the remaining training images and labels [Fig. 3(b)] to learn a registration cost function that is optimal for aligning the labels of the training and template subjects—perfect alignment of the labels lead to perfect prediction of the labels in the training subjects by the template labels. After pairwise registration between the template and test subject using the optimal registration cost function, the ground truth label of the template subject is used to predict that of the test subject.
We limit ourselves to spherical images (i.e., images defined on a unit sphere), although it should be clear that the discussion readily extends to volumetric images. Our motivation for using spherical images comes from the representation of the human cerebral cortex as a closed 2-D mesh in 3-D. There has been much effort focused on registering cortical surfaces in 3-D [14], [15], [24], [30], [65]. Since cortical areas—both structure and function—are arranged in a mosaic across the cortical surface, an alternative approach is to warp the underlying spherical coordinate system [19], [48], [60], [66], [69], [73], [79], [81]. Warping the spherical coordinate system establishes correspondences across the surfaces without actually deforming the surfaces in 3-D. We assume that the meshes have already been spherically parameterized and represented as spherical images: a geometric attribute is associated with each mesh vertex, describing local cortical geometry.
A. Instantiating Registration Cost Function f
To register a given image In to the template image T, we define the following cost function:
where transformation Γn maps a point xi on the sphere S2 to another point Γn (xi) ε S2. The first term corresponds to the wSSD image similiarity. The second term is a percentage metric distortion regularization on the transformation Γn where 𝒩i is a predefined neighborhood around vertex i and dij is the original distance between the neighbors dij = ‖xi − xj‖ [79]. The weights {λi}’s are generalizations of the tradeoff parameter λ, allowing for a spatially-varying tradeoff between the image dissimilarity term and regularization: a higher weight corresponds to placing more emphasis on matching the template image at spatial location xi relative to the regularization. The parameterization of the weights as ensures nonnegative weights.
In this work, we consider either learning the weights or the template T for localizing BA labels or functional labels by aligning cortical folding pattern. Since the weights of the wSSD correspond to the precision of the Gaussian model, by learning the weights of wSSD, we are learning the precision of the Gaussian model and hence the uncertainty of the sulcal geometry. Optimizing leads to placing nonuniform importance on matching different cortical folds with the aim of aligning the underlying cytoarchitectonics or function. For example, suppose there is a sulcus with functional regions that appear on either side of the sulcus depending on the subject. The algorithm may decide to place low weight on the “poorly predictive” sulcus. On the other hand, optimizing T corresponds to learning a cortical folding template that is optimal for localizing the underlying cytoarchitectonics or functional labels of the training subjects. In the case of the previously mentioned “unpredictive sulcus,” the algorithm might learn that the optimal cortical folding template should not contain this sulcus.
We choose to represent the transformation Γn as a composition of diffeomorphic warps {Φk} parameterized by a stationary velocity field, so that Γn = Φ1 ∘ ⋯ ∘ ΦK [75], [81], [82]. We note that our choice of regularization is different from the implicit hierarchical regularization used in Spherical Demons [81] since the Demons regularization is not compatible with our derivations from the previous section. Instead of the efficient 2-Step Spherical Demons algorithm, we will use steepest descent. The resulting registration algorithm is still relatively fast, requiring about 15 min for registering full-resolution meshes with more than 100k vertices, compared with 5 min of computation for Spherical Demons on a Xeon 2.8-GHz single processor machine.
In general, a smooth stationary velocity field υ parameterizes a diffeomorphism Φ via a stationary ODE: ∂tΦ(x,t) = υ⃗(Φ(x, t)) with an initial condition Φ(x,0) = x. The solution at t = 1 is denoted as Φ(x,1) = Φ(x) = exp(υ⃗)(x), where we have dropped the time index. A solution can be computed efficiently using scaling and squaring [5]. This particular choice of representing deformations provides a computationally efficient method of achieving invertible transformations, which is a desirable property in many medical imaging applications. In our case, the velocity field υ⃗ is a tangent vector field on the sphere S2 and not an arbitrary 3-D vector field.
B. Optimizing Registration Cost Function f
To register subject n to the template image T for a fixed set of parameters w, let be the current estimate of . We seek an update transformation exp (υ⃗) parameterized by a stationary velocity field υ⃗
| (3.1) |
Let υ⃗i be the velocity vector tangent to vertex xi, and υ⃗ = {υ⃗i} be the entire velocity field. We adopt the techniques in the Spherical Demons algorithm [81] to differentiate (3.1) with respect to υ⃗, evaluated at υ⃗ = 0. Using the fact that the differential of exp (υ⃗)at υ⃗ = 0 is the identity [44], i.e., [D exp(0)]υ⃗ = υ⃗, we conclude that a change in velocity υ⃗i at vertex xi does not affect exp(υ⃗)(xn) for n ≠ i up to the first order derivatives. Defining to be the 1 × 3 spatial gradient of the warped image In () at xi and to be the 3 × 3 Jacobian matrix of at xi, we get the 1 × 3 derivative
| (3.2) |
We can perform gradient descent of the registration cost function fn using (3.2) to obtain , which can be used to evaluate the regularized task performance G to be described in the next section. We also note that (3.2) instantiates ∂υ⃗ fn within the mixed derivatives term in the task-optimal gradient (2.16) for this application.
C. Instantiating Regularized Task Performance G
We represent the hidden labels in the training subjects as signed distance transforms on the sphere {Ln} [36]. We consider a pairwise approach, where we assume that the template image T has a corresponding labels with distance transform LT and set the task-specific cost function to be
| (3.3) |
A low value of gn indicates good alignment of the hidden label maps between the template and subject n, suggesting good prediction of the hidden label.
We experimented with a prior that encourages spatially constant weights and template, but did not find that the regularization lead to improvements in the localization results. In particular, we considered the following smoothness regularization on the registration parameters depending on whether we are optimizing for the weights λi or the template T:
| (3.4) |
| (3.5) |
A possible reason for this lack of improvement is that the reregistration after every few line searches already helps to regularize against bad parameter values. Another possible reason is that the above regularization assumes a smooth variation in the relationship between structure and function, which may not be true in reality. Unfortunately, the relationship between macro-anatomical structure and function is poorly understood, making it difficult to design a more useful regularization that could potentially improve the results. In the experiments that follow, we will discard the regularization term of the registration parameters (i.e., set Reg(w) = 0). We also note that Reg(w) is typically set to 0 in machine learning approaches of model selection by optimization of cross-validation error [33], [43], [58].
D. Optimizing Task Performance G
To optimize the task performance G over the set of parameters w, we have to instantiate the task-optimal gradient specified in (2.16). We first compute the derivative of the task-specific cost function with respect to the optimal update υ⃗*. Once again, we represent υ⃗* as the collection , where is a velocity vector at vertex xi. Defining to be the 1 × 3 spatial gradient of the warped distance transform of the nth subject Ln at xi, we get the 1 × 3 derivative
| (3.6) |
Weight Update: To update the weights {λj} of the wSSD, we compute the derivative of the registration local minimum update υ⃗* with respect to the weights. Using the approximation in (2.14), we obtain the 3 × 1 derivative of the velocity update υ⃗ with respect to the weights of the wSSD cost function
| (3.7) |
| (3.8) |
| (3.9) |
| (3.10) |
Here δ (k, i) if k = i and is zero otherwise. Since (3.10) is in the same direction as the first term of the gradient descent direction of registration [negative of (3.2)], increasing will improve the intensity matching of vertex xk of the template. Substituting (3.10) and (3.6) into (2.16) provides the gradient for updating the weights of the wSSD cost function.
Template Update: To update the template image T used for registration, we compute the 3 × 1 derivative of the velocity update υ⃗ with respect to the template T
| (3.11) |
| (3.12) |
| (3.13) |
| (3.14) |
Since (3.14) is in the same direction as the first term of the gradient descent direction of registration [negative of (3.2)], when T (xk) is larger than In , increasing the value T (xk) of will warp vertex xk of the template further along the direction of increasing intensity in the subject image. Conversely, if T (xk) is smaller than In , decreasing the value of T (xk) will warp vertex xk of the template further along the direction of decreasing intensity in the subject image. Substituting (3.14) and (3.6) into (2.16) provides the gradient for updating the template used for registration. Note that the template subject’s hidden labels are considered fixed in template space and are not modified during training.
We can in principle optimize both the weights {λi} and the template T. However, in practice, we find that this does not lead to better localization, possibly because of too many degrees-of-freedom, suggesting the need to design better regularization of the parameters. A second reason might come from the fact that we are only using an approximate gradient rather than the true gradient for gradient descent. Previous work [82] has shown that while using an approximate gradient can lead to reasonable solutions, using the exact gradient can lead to substantially better local minima. Computing the exact gradient is a challenge in our framework. We leave exploration of efficient means of computing better approximations of the gradient to future work.
IV. Experiments
We now present experiments on localizing BAs and fMRI-defined MT+ (V5) using macro-anatomical cortical folding in two different data sets. For both experiments, we compare the framework with using uniform weights [31] and FreeSurfer [19].
A. BA Localization
We consider the problem of localizing BAs in the surface representations of the cortex using only cortical folding patterns. In this study, ten human brains were analyzed histologically postmortem using the techniques described in [57] and [84]. The histological sections were aligned to postmortem MR with nonlinear warps to build a 3-D histological volume. These volumes were segmented to separate white matter from other tissue classes, and the segmentation was used to generate topologically correct and geometrically accurate surface representations of the cerebral cortex using a freely available suite of tools [21]. Six manually labeled BA maps (V1, V2, BA2, BA44, BA45, MT) were sampled onto the surface representations of each hemisphere, and errors in this sampling were manually corrected (e.g., when a label was erroneously assigned to both banks of a sulcus). A morphological close was then performed on each label to remove small holes. Finally, the left and right hemispheres of each subject were mapped onto a spherical coordinate system [19]. The BAs on the resulting cortical representations for two subjects are shown in Fig. 2(b). We do not consider BA4a, BA4p, and BA6 in this paper because they were not histologically mapped by the experts in two of the ten subjects in this particular data set (even though they exist in all human brains).
As illustrated in Fig. 2(c) and discussed in multiple studies [3], [4], [18], we note that V1, V2, and BA2 are well-predicted by local cortical geometry, while BA44, BA45, and MT are not. For all the BAs however, a spherical morph of cortical folding was shown to improve their localization compared with only Talairach or nonlinear spatial normalization in the Euclidean 3-D space [18]. Even though each subject has multiple BAs, we focus on each structure independently. This allows for an easier interpretation of the estimated parameters, such as the optimal template example we provide in Section IV-A3. A clear future direction is to learn a registration cost function that is jointly optimal for localizing multiple cytoarchitectural or functional areas.
We compare the following algorithms.
-
Task-Optimal. We perform leave-two-out cross-validation to predict BA location. For each test subject, we use one of the remaining nine subjects as the template subject and the remaining eight subjects for training. When learning the weights of the wSSD, the weights {λj} are globally initialized to 1 and the template image T is fixed to the geometry of the template subject. When learning the cortical folding template T, the template image is initialized to that of the template subject and the weights {λj} are globally set to 1.
Once the weights or template are learned, we use them to register the test subject and predict the BA of the test subject by transferring the BA label from the template to the subject. We compute the symmetric mean Hausdorff distance between the boundary of the true BA and the predicted BA on the cortical surface of the test subject—smaller Hausdorff distance corresponds to better localization [13]. The symmetric mean Hausdorff distance between two curves is defined as follows. For each boundary point of the first curve, the shortest distance to the second curve is computed and averaged. We repeat by computing and averaging the shortest distance from each point of the second curve to the first curve. The symmetric mean Hausdorff distance is obtained by averaging the two values. We consider all 90 possibilities of selecting the test subject and template, resulting in a total of 90 trials and 90 mean Hausdorff distances for each BA and for each hemisphere.
Uniform-Weights. We repeat the process for the uniform-weight method that fixes the template T to the geometry of the template subject, and sets all the weights {λj} to a global fixed value λ without training. We explore 14 different values of global weight λ, chosen such that the deformations range from rigid to flexible warps. For each BA and each hemisphere, we pick the best value of λ leading to the lowest mean Hausdorff distances. Because there is no cross-validation in selecting the weights, the uniform-weight method is using an unrealistic oracle-based version of the strategy proposed in [79].
FreeSurfer. Finally, we use FreeSurfer [19] to register the 10 ex vivo subjects to the FreeSurfer Buckner40 atlas, constructed from the MRI of 40 in vivo subjects [21]. Once registered into this in vivo atlas space, for the same 90 pairs of subjects, we can use the BAs of one ex vivo subject to predict another ex vivo subject. We note that FreeSurfer also uses the wSSD cost function, but a more sophisticated regularization that penalizes both metric and areal distortion. For a particular tradeoff between the similarity measure and regularization, the Buckner40 template consists of the empirical mean and variance of the 40 in vivo subjects registered to template space. We use the reported FreeSurfer tradeoff parameters that were used to produce prior state-of-the-art BA alignment [18].
We note that both the task-optimal and uniform-weights methods use a pairwise registration framework, while FreeSurfer uses an atlas-based registration framework. Under the atlas-based framework, all the ex vivo subjects are registered to an atlas (Fig. 4). To use the BA of a training subject to predict a test subject, we have to compose the deformations of the training subject to the atlas with the inverse deformation of the test subject to the atlas. Despite this additional source of error from composing two warps, it has been shown that with carefully constructed atlases, using the atlas-based strategy leads to better registration because of the removal of template bias in the pairwise registration framework [6], [23], [26], [31], [32], [39], [79].
Fig. 4.
FreeSurfer’s atlas-based registration approach. Training and test subjects are registered to an atlas. The BA of a training subject can then be used to predict that of the test subject.
We run the task-optimal and uniform-weights methods on a low-resolution subdivided icosahedron mesh containing 2562 vertices, whereas FreeSurfer results were computed on high-resolution meshes of more than 100k vertices. In our implementation, training on eight subjects takes on average 4 h on a standard PC (AMD Opteron, 2GHz, 4GB RAM). Despite the use of the low-resolution mesh, we achieve state-of-the-art localization accuracy. We also emphasize that while training is computationally intensive, registration of a new subject only requires one minute of processing time since we are working with low-resolution meshes.
1) Quantitative Results
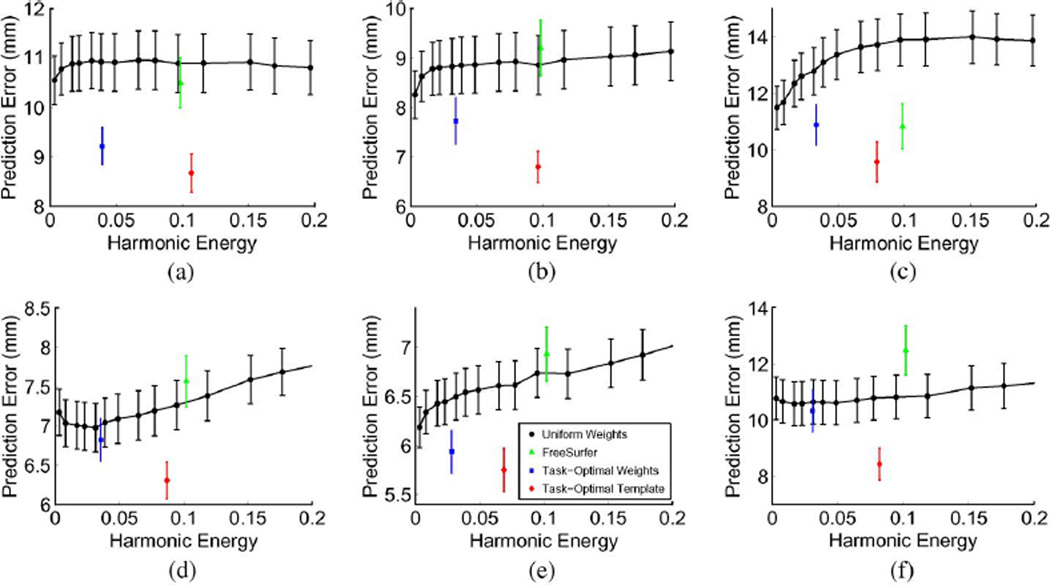
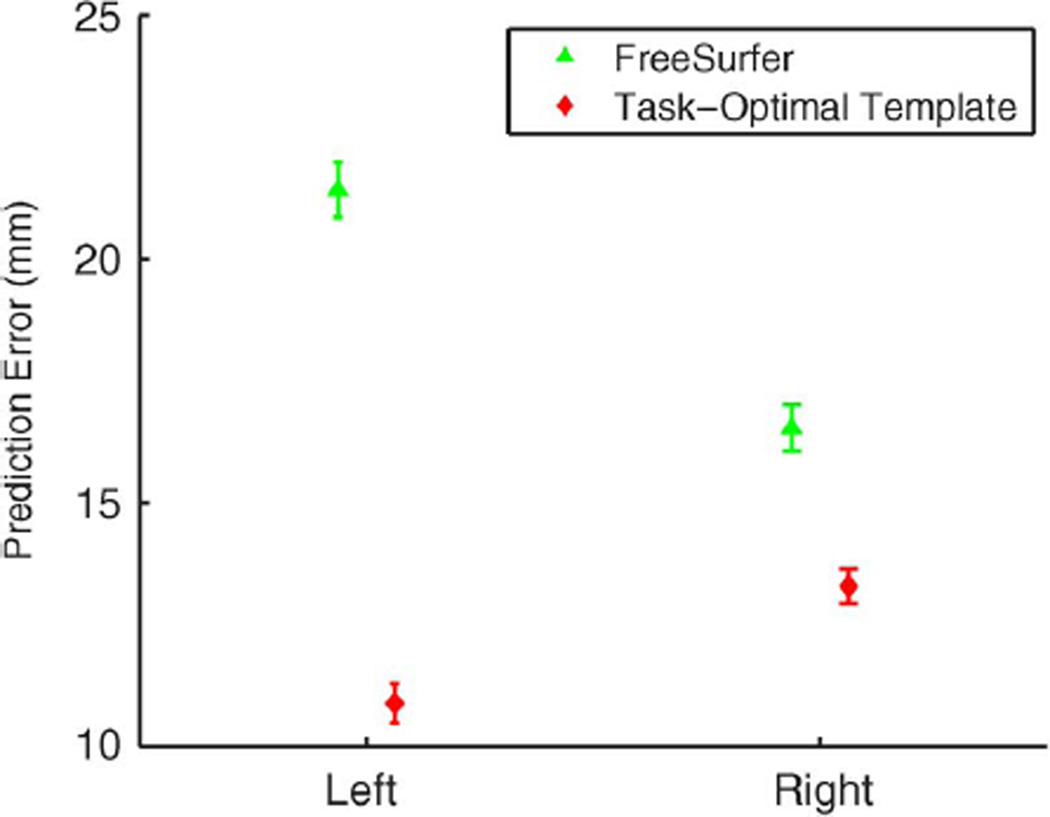
Fig. 5 displays the mean and standard errors from the 90 trials of leave-two-out. On average, task-optimal template performs the best, followed by task-optimal weights. Permutation tests show that task-optimal template outperforms FreeSurfer in five of the six areas, while task-optimal weights outperforms FreeSurfer in four of the six areas after corrections for multiple comparisons (see Fig. 5 for more details). For the Broca’s areas (BA44 and BA45) and MT, this is not surprising. Since local geometry poorly predicts these regions, by taking into account the final goal of aligning BAs instead of blindly aligning the cortical folds, our method achieves better BA localization. FreeSurfer and the uniform-weights method have similar performance because a better alignment of the cortical folds on a finer resolution mesh does not necessary improve the alignment of these areas.
Fig. 5.
Mean Hausdorff distances over an entire range of harmonic energy for BA44, BA45, and MT. First row corresponds to left hemisphere. Second row corresponds to right hemipshere. * indicates that task-optimal template is statistically significantly better than FreeSurfer. † indicates that task-optimal weights is statistically significantly better than FreeSurfer. Statistical threshold is set at 0.05, FDR corrected with respect to the 24 statistical tests performed in this section. FreeSurfer is not statistically better than either of the task-optimal methods in any of the Brodmann areas. (a) Left BA44 *, †. (b) Left BA45 *, †. (c) Left MT. (d) Right BA44 *. (e) Right BA45 *, †. (f) Right MT*, †.
Since local cortical geometry is predictive of V1, V2, and BA2, we expect the advantages of our framework to vanish. Surprisingly, as shown in Fig. 6, task-optimal template again achieve significant improvement in BAs alignment over the uniform-weights method and FreeSurfer. Task-optimal weights is also significantly better than the uniform-weights method, but only slightly better than FreeSurfer. Permutation tests show that task-optimal template outperforms FreeSurfer in five of the six areas, while task-optimal weights is outperforms FreeSurfer in three of the six areas after corrections for multiple comparisons (see Fig. 6 for more details). This suggests that even when local geometry is predictive of the hidden labels and anatomy-based registration achieves reasonable localization of the labels, tuning the registration cost function can further improve the task performance. We also note that in this case, FreeSurfer performs better than the uniform-weights method on average. Since local cortical folds are predictive of these areas, aligning cortical folds on a higher resolution mesh yields more precise alignment of the cortical geometry and of the BAs.
Fig. 6.
Mean Hausdorff distances over an entire range of harmonic energy for V1, V2, and BA2. First row corresponds to left hemisphere. Second row corresponds to right hemisphere. * indicates that task-optimal template is statistically significantly better than FreeSurfer. † indicates that task-optimal weights is statistically significantly better than FreeSurfer. Statistical threshold is set at 0.05, FDR corrected with respect to the 24 statistical tests performed in this section. FreeSurfer is not statistically better than either of the task-optimal methods in any of the Brodmann areas. (a) Left V1 *. (b) Left V2 *, †. (c) Left BA2 *, †. (d) Right V1 *. (e) Right V2 *, †. (f) Right BA2.
We note that the FreeSurfer Buckner40 atlas utilizes 40 in vivo subjects consisting of 21 males and 19 females of a wide-range of age. Of these, 30 are healthy subjects whose ages range from 19 to 87. 10 of the subjects are Alzheimer’s patients with age ranging from 71 to 86. The average age of the group is 56 (see [12] for more details). The T1-weighted scans were acquired on a 1.5T Vision system (Siemens, Erlangen Germany), with the following protocol: two sagittal acquisitions, FOV = 224, matrix = 256 × 256, resolution = 1 × 1 × 1.25 mm, TR = 9.7 ms, TE = 4 ms, Flip angle = 10°, TI = 20 ms and TD = 200 ms. Two acquisitions were averaged together to increase the contrast-to-noise ratio. The histological data set includes five male and five female subjects, with age ranging from 37 to 85 years old. The subjects had no previous history of neurologic or psychiatric diseases (see [4] for more details). The T1-weighted scans of the subjects were obtained on a 1.5T system (Siemens, Erlangen, Germany) with the following protocol: flip angle 40°, TR = 4 ms, TE = 5 ms and resolution = 1 × 1 × 1.17 mm. While there are demographic and scanning differences between the in vivo and ex vivo data sets, the performance differences between FreeSurfer and the task-optimal framework cannot be solely attributed to this difference. In particular, we have shown in previous work that FreeSurfer’s results are worse when we use an ex vivo atlas for registering ex vivo subjects ([81,Table III]). Furthermore, FreeSurfer’s results are comparable with that of the uniform-weights baseline algorithm, as well as previously published results [18], where we have checked for gross anatomical misregistration. We emphasize that since the goal is to optimize Brodmann area localization, the learning algorithm might take into account the idiosyncrasies of the registration algorithm in addition to the relationship between macro-anatomy and cytoarchitecture. Consequently, it is possible that the performance differences are partly a result of our algorithm learning a registration cost function with better local minima, thus avoiding possible misregistration of anatomy.
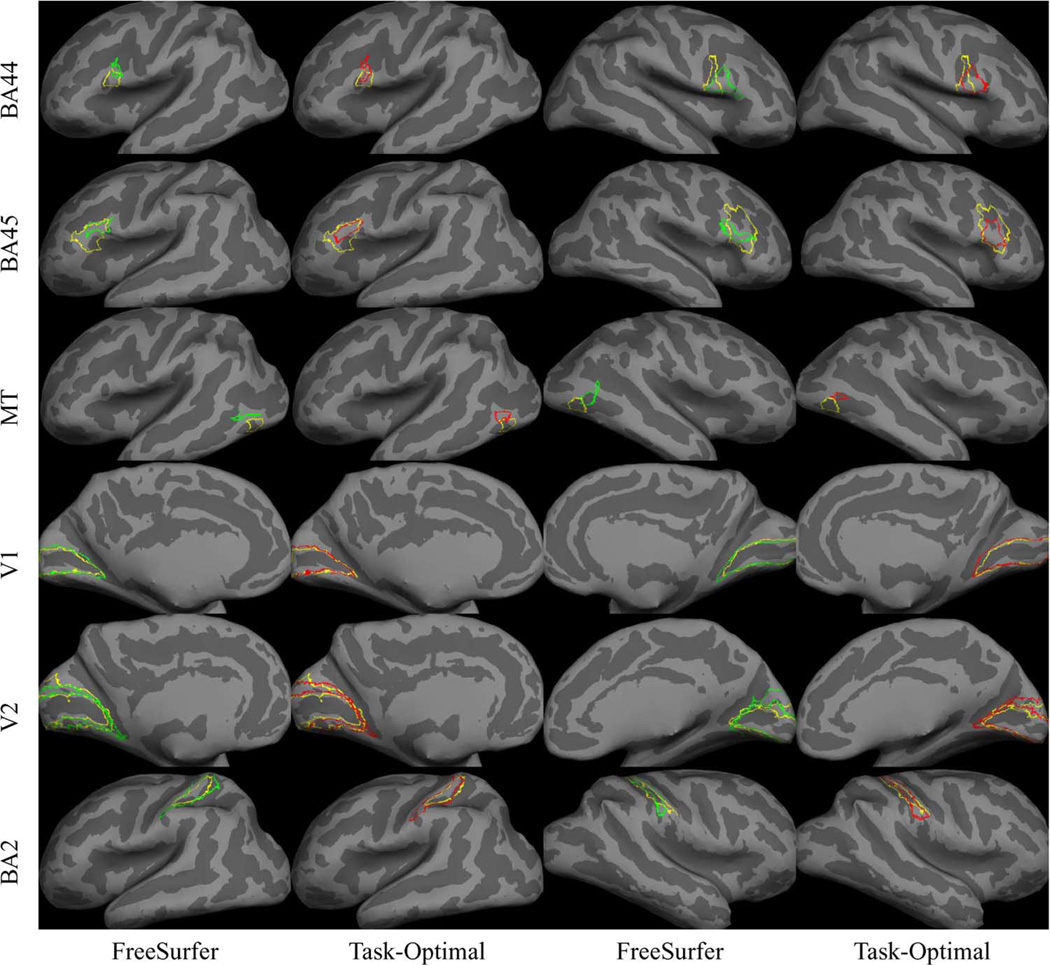
2) Qualitative Results
Fig. 7 illustrates representative localization of the BAs for FreeSurfer and task-optimal template. We note that the task-optimal boundaries (red) tend to be in better visual agreement with the ground truth (yellow) boundaries, such as the right hemisphere BA44 and BA45.
Fig. 7.
Representative BA localization in 90 trials of leave-two-out for FreeSurfer and task-optimal template. Yellow indicates ground truth boundary. Green indicates FreeSurfer prediction. Red indicates Task-Optimal prediction. The representative samples were selected by finding subjects whose localization errors are close to the mean localization errors for each BA. Furthermore, for a given BA, the same subject was selected for both methods to simplify the comparison.
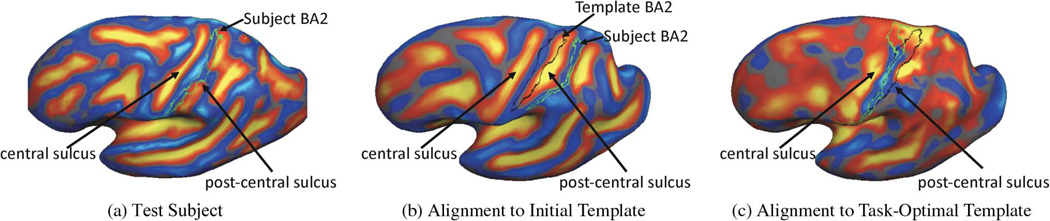
3) Interpreting the Template
Fig. 8 illustrates an example of learning a task-optimal template for localizing BA2. Fig. 8(a) shows the cortical geometry of a test subject together with its BA2. In this subject, the central sulcus is more prominent than the postcentral sulcus. Fig. 8(b) shows the initial cortical geometry of a template subject with its corresponding BA2 in black outline. In this particular subject, the postcentral sulcus is more prominent than the central sulcus. Consequently, in the uniform-weights method, the central sulcus of the test subject is incorrectly mapped to the postcentral sulcus of the template, so that BA2 is misregistered. Fig. 8(b) also shows the BA2 of the test subject (green) overlaid on the cortical geometry of the template subject after registration to the initial template geometry. During task-optimal training, our method interrupts the geometry of the postcentral sulcus in the template because the uninterrupted postcentral sulcus in the template is inconsistent with localizing BA2 in the training subjects. The final template is shown in Fig. 8(c). We see that the BA2 of the subject (green) and the task-optimal template (black) are well-aligned, although there still exists localization error in the superior end of BA2.
Fig. 8.
Template estimation in the task-optimal framework improves localization of BA2. (a) Cortical geometry of test subject with corresponding BA2 (in green). (b) Initial cortical geometry of template subject with corresponding BA2 (in black). In (b), we also show the BA2 of the test subject (in green) after registration to the intial template. (c) Final cortical geometry of template subject after task-optimal training. BA2 of the test subject (in green) after registration to the task-optimal template demonstrates significantly better alignment with the BA2 of the template subject.
In the next section, we turn our attention to a fMRI data set. Since the task-optimal template performed better than the task-optimal weights, we will focus on the comparison between the task-optimal template and FreeSurfer.
B. fMRI-MT+ Localization
We now consider the application of localizing fMRI-defined functional areas in the cortex using only cortical folding patterns. Here, we focus on the so-called MT+ area localized in 42 in vivo subjects using fMRI. The MT+ area defined functionally is thought to include primarily the cytoarchitectonically-defined MT and a small part of the medial superior temporal (MST) area (hence the name MT+). The imaging paradigm involved subjects viewing an alternating 16 s blocks of moving and stationary concentric circles. The structural scans were processed using the FreeSurfer pipeline [21], resulting in spherically parameterized cortical surfaces [11], [19]. The functional data were analyzed using the general linear model [22]. The resulting activation maps were thresholded by drawing the activation boundary centered around the vertex with maximal activation. The threshold was varied across subjects in order to maintain a relatively fixed ROI area of about 120 mm2 (±5%) as suggested in [68]. The subjects consist of 10 females and 32 males, with age ranging from 21 to 58 years old. 23 of the 42 subjects are clinically diagnosed with schizophrenia, while the other 19 subjects are healthy controls. Imaging took place on a 3T MR scanner (Siemens Trio) with echoplanar (EP) imaging capability. Subjects underwent two conventional high-resolution 3-D structural scans, constituting a spoiled GRASS (SPGR) sequence (128 sagittal slices, 1.33 mm thickness, TR = 2530 ms, TE = 3.39 ms, Flip angle = 7°, voxel size = 1.3 × 1 × 1.3 mm). Each functional run lasted 224 s during which T2*-weighted echoplanar (EP) images were acquired (33 × 3-mm-thick slices, 3 × 3 × 3 mm voxel size) using a gradient echo (GR) sequence (TR = 2000 ms; TE = 30 ms; Flip angle = 90°). To maximize training data, no distinction is made between the healthy controls and schizophrenia patients.
1) Ex Vivo MT Prediction of In Vivo MT+
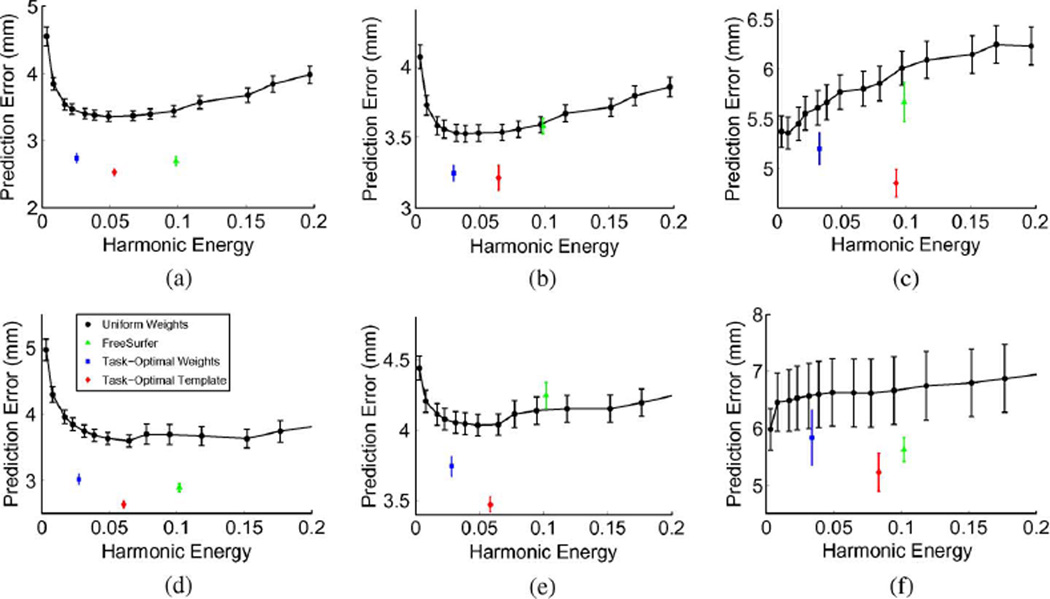
In this experiment, we use each of the 10 ex vivo subjects as a template and the remaining nine subjects for training a task-optimal template for localizing MT. We then register each task-optimal template to each of the 42 in vivo subjects and use the template subject’s MT to predict that of the test subjects’ MT+. The results are 420 Hausdorff distances for each hemisphere. For FreeSurfer, we align the 42 in vivo subjects to the Buckner40 atlas. Once registered in this space, we can use MT of the ex vivo subjects to predict MT+ of the in vivo subjects.
Fig. 9 reports the mean and standard errors of the Hausdorff distances for both methods on both hemispheres. Once again, we find that the task-optimal template significantly outperforms the FreeSurfer template (p < 10−5 for both hemispheres). We note that the errors in the in vivo subjects (Fig. 9) are significantly worse than those in the ex vivo subjects (Fig. 5). This is not surprising since functionally defined MT+ is slightly different from cytoarchitectonically defined MT. Furthermore, the ex vivo surfaces tend to be noisier and less smooth than those acquired from in vivo subjects [81]. Since our framework attempts to leverage domain specific knowledge about MT from the ex vivo data, one would expect these mismatches between the data sets to be highly deterimental to our framework. Instead, FreeSurfer appears to suffer more than our framework.
Fig. 9.
Mean Hausdorff distances using ex vivo MT to predict MT+ in in vivo scans. Permutation testing shows that the differences between FreeSurfer and task-optimal template are statistically significant (p < 10−5).
2) In Vivo MT Prediction of In Vivo MT+
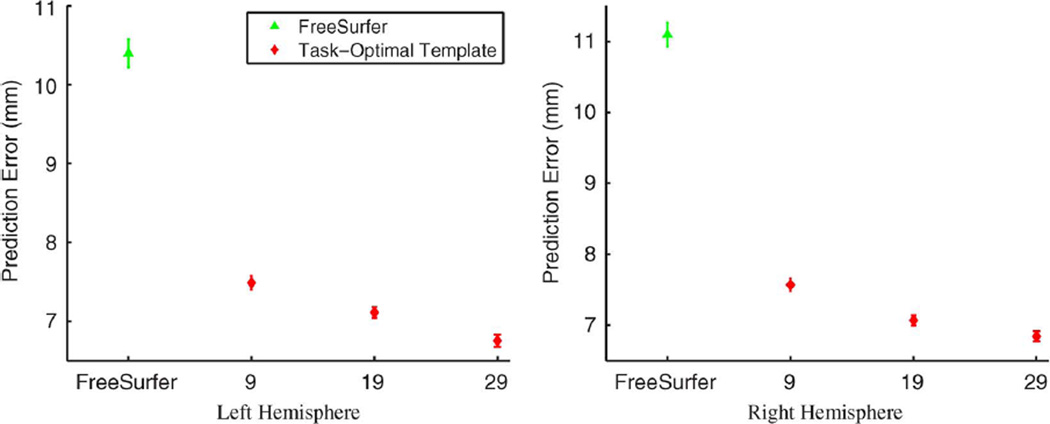
To understand the effects of the training set size on localization accuracy, we perform cross-validation within the fMRI data set. For each randomly selected template subject, we consider 9, 19, or 29 training subjects. The resulting task-optimal template is used to register and localize MT+ in the remaining 32, 22, or 12 test subjects, respectively. The cross-validation trials were repeated 100, 200, and 300 times, respectively, resulting in a total of 3200, 4400, and 3600 Hausdorff distances. This constitutes thousands of hours of computation time. For FreeSurfer, we perform a pairwise prediction of MT+ among the in vivo subjects after registration to the Buckner40 atlas, resulting in 1722 Hausdorff distances per hemisphere.
Fig. 10 reports the mean and standard errors of the Hausdorff distances for FreeSurfer and task-optimal template on both hemispheres. We see that the FreeSurfer alignment errors are now commensurate with the ex vivo results (Fig. 5). However, the task-optimal template still outperforms FreeSurfer (p < 10−5 for all cases).We also note that the accuracy of MT+ localization improves with the size of the training set. The resulting localization error with a training set of 29 subjects is less than 7 mm for both hemispheres. For all training set sizes, the localization errors are also better than the ex vivo MT experiment (Fig. 5).
Fig. 10.
Plot of mean hausdorff errors for MT+ from cross-validation of the fMRI data set using either FreeSurfer or in vivo trained task-optimal template. For the task-optimal framework, we tried different number of training subjects. Test errors decrease as we go from 9 to 19 to 29 training subjects. Once again, permutation testing shows that the differences between FreeSurfer and task-optimal template are statistically significant (p < 10−5).
V. Discussion and Future Work
The experiments in the previous section demonstrate the feasibility of learning registration cost functions with thousands of degrees-of-freedom from training data. We find that the learned registration cost functions generalize well to unseen test subjects of the same (Sections IV-A and IV-B2), as well as different imaging modality (Section IV-B1). The almost linear improvement with increasing training subjects in the fMRI-defined MT+ experiment (Fig. 10) suggests that further improvements can be achieved (in particular in the histological data set) with a larger training set. Unfortunately, histological data over a whole human hemisphere is difficult to obtain, while fMRI localization experiments tend to focus on single functional areas. Therefore, a future direction of research is to combine histological and functional information obtained from different subjects and imaging modalities during training.
Since our measure of localization accuracy uses the mean Hausdorff distance, ideally we should incorporate it into our task-specific objective function instead of the SSD on the distance transform representing the BA. Unfortunately, the resulting derivative is difficult to compute. Furthermore, the gradient will be zero everywhere except at the BA boundaries, causing the optimization to proceed slowly. On the other hand, it is unclear how aligning the distance transform values far from the boundary helps to align the boundary. Since distance transform values far away from the boundary are larger, they can dominate the task-specific objective function g. Consequently, we utilize the distance transform over the entire surface to compute the gradient, but only consider the distance transform within the boundary of the template BA when evaluating the task performance criterion g.
The idea of using multiple atlases for segmentation has gained recent popularity [29], [49], [50], [53], [55], [76]. While we have focused on building a single optimal template, our method can complement the multiatlas approach. For example, one could simply fuse the results of multiple individually-optimal templates for image segmentation. A more ambitious task would be to optimize for multiple jointly-optimal templates for segmentation.
In this work, we select one of the training subjects as the template subject and use the remaining subjects for training. The task-specific cost function g evaluates the localization of the hidden labels via the template subject. During training (either for learning the weights or template in the registration cost function), the Brodmann areas of the template subject are held constant. Because the fixed Brodmann areas are specific to the template subject, the geometry of the template subject should in fact be the best and most natural initialization. It does not make sense to use the geometry of another subject (or average geometry of the training subjects) as initialization for the template subject’s Brodmann areas, especially since the geometry of this other subject (or average geometry) is not registered to the geometry of the template subject. However, the use of a single subject’s Brodmann (or functional) area can bias the learning process. An alternative groupwise approach modifies the task-specific cost function g to minimize the variance of the distance transforms across training subjects after registration. In this case, both the template geometry and Brodmann (functional) area are estimated from all the training subjects and dynamically updated at each iteration of the algorithm. The average geometry of the training subjects provided a reasonable template initialization. However, our initial experiments in the ex vivo data set do not suggest an improvement in task performance over the pairwise formulation in this paper.
While this paper focuses mostly on localization of hidden labels, different instantiations of the task-specific cost function can lead to other applications. For example, in group analysis, the task-specific cost function could maximize differences between diseased and control groups, while minimizing intra-group differences, similar to a recent idea proposed for discriminative Procrustes alignment [38].
VI. Conclusion
In this paper, we present a framework for optimizing the parameters of any smooth family of registration cost functions, such as the image dissimilarity-regularization tradeoff, with respect to a specific task. The only requirement is that the task performance can be evaluated by a smooth cost function on an available training data set. We demonstrate state-of-the-art localization of Brodmann areas and fMRI-defined functional regions by optimizing the weights of the wSSD image-similarity measure and estimating an optimal cortical folding template. We believe this work presents an important step towards the automatic selection of parameters in image registration. The generality of the framework also suggests potential applications to other problems in science and engineering formulated as optimization problems.
Acknowledgment
The authors would like to thank P. Parillo for discussion on the optimization aspects of this paper. The authors would also like to thank C. Brun, S. Durrleman, T. Fletcher, and W. Mio for helpful feedback on this work.
This work was supported in part by the NAMIC (NIH NIBIB NAMIC U54-EB005149), in part by the NAC (NIH NCRR NAC P41-RR13218), in part by the mBIRN (NIH NCRR mBIRN U24-RR021382), in part by the NIH NINDS R01-NS051826 Grant, in part by the NSF CAREER 0642971 Grant, in part by the National Institute on Aging (AG02238), in part by the NCRR (P41-RR14075, R01 RR16594-01A1), in part by the NIBIB (R01 EB001550, R01EB006758), in part by the NINDS (R01 NS052585-01), and in part by the MIND Institute. Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation. The work of B. T. Thomas Yeo was supported by the A*STAR, Singapore.
Appendix A
Proof of Proposition 2
In this appendix, we prove Proposition 2: If the Hessian is positive definite at υ = 0, then there exists an ε > 0, such that for all δw, ‖δw‖ < ε, a unique continuous function υ*(w0, δw) exists, such that υ*(w0, δw) is the new local minimum for parameters w0 + δw and υ*(w0, 0) = 0. Furthermore, υ*(w0, δw) has the same order of smoothness as f.
In the next section, we first prove that the Hessian is equal to the mix-derivatives matrix under the composition of diffeomorphisms model [75], [81], [82]. We then complete the proof of Proposition 2.
A. Proof of the Equivalence Between the Hessian and Mix-Derivatives Matrix for the Composition of Diffeomorphisms Model
We will only provide the proof for when the image is defined in ℝ3 so as not to obscur the main ideas behind the proof. To extend the proof to a manifold (e.g., S2), one simply need to extend the notations and bookkeeping by the local parameterizing the velocity fields υ1 and υ2 using coordinate charts. The same proof follows.
Let us define some notations. Suppose the image and there are M voxels. Let x⃗ be the ℝ3M rasterized coordinates of the M voxels. For conciseness, we define for the fixed parameters w0
| (A.1) |
Therefore, p is a function from ℝ3M to ℝ. Under the composition of diffeomorphisms model, δΓ (υ) is the diffeomorphism parameterized by the stationary velocity field υ defined on the M voxels, so that δΓ (υ)(·) is a function from ℝ3M to ℝ. To make the dependence of δΓ (υ) on υ explicit, we define
| (A.2) |
and so ϒ is a function from ℝ3M × ℝ3M to ℝ3M. In other words, we can rewrite
| (A.3) |
and
| (A.4) |
Now that we have gotten the notations out of the way, we will now show that
| (A.5) |
Hessian: We first compute the 1× 3M Jacobian via the chain rule
| (A.6) |
From the above equation, we can equivalently write down the jth component of the 1 × 3M Jacobian
| (A.7) |
where ϒn and denote the nth and j th components of ϒ and υ1, respectively. Now, we compute the (i, j) th component of the 3M × 3M Hessian using the product rule
| (A.8) |
| (A.9) |
| (A.10) |
Because ∂υ1 ϒ|υ1=0 is the identity matrix and the 1 × 3M Jacobian ∂υ1 p(ϒ(υ1, x⃗))|υ1=0 = (∂ϒp)(∂υ1 ϒ)|υ1=0 = 0 (because derivative is zero at local minimum), we get ∂ϒp |υ1=0 = 0, and so the second term in (A.10) is zero.
To simplify the first term of (A.10), we once again use the fact that ∂υ1 ϒ|υ1=0 is the identity matrix, and so the summand is zero unless k = i and n = j. Consequently, (A.10) simplifies to
| (A.11) |
or equivalently
| (A.12) |
Mix-Derivatives Matrix: We first compute the 1 × 3M Jacobian via the chain rule
| (A.13) |
| (A.14) |
From the above equation, we can equivalently write down the jth component of the 1 × 3M Jacobian
| (A.15) |
Now, we compute the (i, j)th component of the 3M × 3M mix-derivatives matrix using the product rule
| (A.16) |
| (A.17) |
Like before, we have ∂ϒp|υ1=υ2=0 = 0, and so the second term is zero. Because ∂υ1 ϒ|υ1=0 is the identity, is zero unless k = i. Since , is also equal to zero unless n = j. Therefore, we get
| (A.18) |
or equivalently
| (A.19) |
B. Completing the Proof of Proposition 2
We now complete the proof of Proposition2. Let h(w, υ1) ≜ ∂υ2 f (w, Γ*(w0) ∘ δΓ (υ1) ∘ δΓ (υ2))|υ2=0. Since δΓ(0) = Id, we have
| (A.20) |
| (A.21) |
| (A.22) |
where the last equality comes from the definition of Γ*(w0) being a local minimum for the composition model.
Since the mix-derivatives matrix ∂υ1 h(w, υ1)|υ1=0 is invertible by the positive-definite assumption of this proposition, by the Implicit Function Theorem, there exists an ε > 0, such that for all δw, ‖δw‖ < ε, there is a unique continuous function υ* (w0, δw), such that h(w0 + δw, υ* (w0, δw)) = 0 and υ* (w0, 0) = 0. Furthermore, υ* (w0, δw) has the same order of smoothness as f.
Let . Then k (w0, 0) is positive definite at υ1 = 0 by the assumption of the proposition. By the smoothness of derivatives and continuity of eigenvalues, there exists a small neighborhood around (w0, υ1 = 0) in which the eigenvalues of k (w, υ1) are all greater than zero. Therefore, Γ*(w0) ∘ δΓ (υ*(w0, δw)) does indeed define a new local minimum close to Γ*(w0).
Appendix B
Computing the Derivative∂w υ*
To compute ∂w υ*, we perform a Taylor expansion
| (B.1) |
| (B.2) |
and rearranging the terms for υ1 = υ*, we get
| (B.3) |
Footnotes
Here, we assume that the transformation Γ is finite dimensional, such as the parameters of affine transformations, positions of spline control points or dense displacement fields defined on the voxels or vertices of the image domain.
Contributor Information
B. T. Thomas Yeo, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA (ythomas@csail.mit.edu).
Mert R. Sabuncu, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA 02129 USA (msabuncu@csail.mit.edu).
Tom Vercauteren, Mauna Kea Technologies, 75010 Paris, France (tom.vercauteren@maunakeatech.com).
Daphne J. Holt, Massachusetts General Hospital Psychiatry Department, Harvard Medical School, Charlestown, MA 02139 USA (dholt@partners.org)
Katrin Amunts, Department of Psychiatry and Psychotherapy, RWTH Aachen University and the Institute of Neuroscience and Medicine, Research Center Jülich, 52425 Jülich, Germany (kamunts@ukaachen.de).
Karl Zilles, Institute of Neuroscience and Medicine, Research Center Jülich and the C.&O. Vogt-Institute for Brain Research, University of Düsseldorf, 52425 Jülich, Germany (k.zilles@fz-juelich.de).
Polina Golland, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA (polina@csail.mit.edu).
Bruce Fischl, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA; Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA 02129 USA; Department of Radiology, Harvard Medical School and the Divison of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA (fischl@nmr.mgh.harvard.edu).
References
- 1.Allassonniere S, Amit Y, Trouvé A. Toward a coherent statistical framework for dense deformable template estimation. J. R. Stat. Soc., Series B. 2007;vol. 69(no. 1):3–29. [Google Scholar]
- 2.Allgower E, Georg K. Introduction to Numerical Continuation Methods. Philadelphia, PA: SIAM; 2003. [Google Scholar]
- 3.Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. Brodmann’s areas 17 and 18 brought into stereotaxic space—Where and how variable? NeuroImage. 2000;vol. 11:66–84. doi: 10.1006/nimg.1999.0516. [DOI] [PubMed] [Google Scholar]
- 4.Amunts K, Schleicher A, Burgel U, Mohlberg H, Uylings H, Zilles K. Broca’s region revisited: Cytoarchitecture and intersubject variability. J. Comparative Neurol. 1999;vol. 412(no. 2):319–341. doi: 10.1002/(sici)1096-9861(19990920)412:2<319::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Arsigny V, Commowick O, Pennec X, Ayache N. A log-euclidean framework for statistics on diffeomorphisms. Proc. Int. Conf. Med. Image Computing Computer Assist. Intervent. (MICCAI); LNCS; 2006. pp. 924–931. [DOI] [PubMed] [Google Scholar]
- 6.Avants B, Gee J. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage. 2004;vol. 23:139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Baker S, Matthews I. Lucas-Kanade 20 years on: A unifying framework. Int. J. Comput. Vis. 2004;vol. 56(no. 3):221–255. [Google Scholar]
- 8.Beg M, Miller M, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vis. 2005;vol. 61(no. 2):139–157. [Google Scholar]
- 9.Brodmann K. Vergleichende Lokalisationslehre der Groβhirnrinde in Ihren Prinzipien Dargestellt auf Grund des Zellenbaues. 1909 [Google Scholar]
- 10.Commowick O, Stefanescu R, Fillard P, Arsigny V, Ayache N, Pennec X, Malandain G. Incorporating statistical measures of anatomical variability in atlas-to-subject registration for conformal brain radiotherapy. Proc. Int. Conf. Med. Image Computing and Computer Assist. Intervent. (MICCAI); LNCS; 2005. pp. 927–934. [DOI] [PubMed] [Google Scholar]
- 11.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis I: Segmentation and surface reconstruction. NeuroImage. 1999;vol. 9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 12.Desikan R, Segonne F, Fischl B, Quinn B, Dickerson B, Blacker D, Buckner R, Dale A, Maguire R, Hyman B, Albert M, Killiany R. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;vol. 31(no. 3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Dubuisson M, Jain A. A modified Hausdorff distance for object matching. Proc. 12th IAPR Int. Conf. Pattern Recognit. 1994;vol. 1:566–568. [Google Scholar]
- 14.Durrleman S, Pennec X, Trouvé, P. A, Ayache N. Inferring brain variability from diffeomorphic deformations of currents: An integrative approach. Med. Image Anal. 2008;vol. 12(no. 5):626–637. doi: 10.1016/j.media.2008.06.010. PMID: 18658005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckstein I, Joshi A, Kuo CJ, Leahy R, Desbrun M. Generalized surface flows for deformable registration and cortical matching. Proc. Int. Conf. Med. Image Computing Computer Assist. Intervent. (MICCAI); LNCS; 2007. pp. 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann. Stat. 2004:407–451. [Google Scholar]
- 17.Evgeniou T, Pontil M, Poggio T. Advances in Computational Mathematics. Cambridge, MA: MIT Press; 2000. Regularization networks and support vector machines; pp. 1–50. [Google Scholar]
- 18.Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BT, Mohlberg H, Amunts K, Zilles K. Cortical folding patterns and predicting cytoarchictecture. Cerebral Cortex. 2008;vol. 18(no. 8):1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischl B, Sereno M, Tootell R, Dale A. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapp. 1999;vol. 8(no. 4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischl B, Stevens A, Rajendran N, Yeo BT, Greve D, Van Leemput K, Polimeni J, Kakunoori S, Buckner R, Pacheco J, Salat D, Melcher J, Frosch M, Hyman B, Grant PE, Rosen BR, van der Kouwe A, Wiggins G, Wald L, Augustinack J. Predicting the location of entorhinal cortex from MRI. Neuroimage. 2009;vol. 47(no. 1):8–17. doi: 10.1016/j.neuroimage.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiki Freesurfer. [Online]. Available: http://surfer.nmr.mgh.har-vard.edu/fswiki/freesurferwiki/ [Google Scholar]
- 22.Friston K, Holmes A, Worsley K, Poline J-P, Frith C, Frackowiak R. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapp. 1995;vol. 2(no. 4):189–210. [Google Scholar]
- 23.Geng X, Christensen G, Gu H, Ross T, Yang Y. Implicit reference-based group-wise image registration and its application to structural and functional MRI. NeuroImage. 2009;vol. 47(no. 4):1341–1351. doi: 10.1016/j.neuroimage.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng X, Kumar D, Christensen G. Transitive inverse-consistent manifold registration. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2005. pp. 468–479. [DOI] [PubMed] [Google Scholar]
- 25.Glocker B, Komodakis N, Navab N, Tziritas G, Paragios N. Dense registration with deformation priors. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2009. pp. 540–551. [DOI] [PubMed] [Google Scholar]
- 26.Guimond A, Meunier J, Thirion J-P. Average brain models: A convergence study. Comput. Vis. Image Understand. 2000;vol. 77(no. 2):192–210. [Google Scholar]
- 27.Guyon I, Elisseeff A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003;vol. 3:1157–1182. [Google Scholar]
- 28.Hastie T, Rosset S, Tibshirani R, Zhu J. The entire regularization path for the support vector machine. J. Mach. Learn. Res. 2004;vol. 5:1391–1415. [Google Scholar]
- 29.Heckemann R, Hajnal J, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 2006;vol. 33(no. 1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 30.Jaume S, Ferrant M, Warfield S, Macq B. Multiresolution parameterization of meshes for improved surface-based registration. Proc. SPIE Med. Imag. 2001;vol. 4322:633–642. [Google Scholar]
- 31.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;vol. 23:151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 32.Klein A, Ghosh SS, Avants B, Yeo BT, Fischl B, Ardekani B, Gee JC, Mann JJ, Parsey RV. Evaluation of volume-based and surface-based brain image registration methods. Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Int. Joint Conf. Artif. Intell. 1995;vol. 14:1137–1145. [Google Scholar]
- 34.Kohavi R, John G. Wrappers for feature subset selection. Artif. Intell. 1997;vol. 97(no. 1–2):273–324. [Google Scholar]
- 35.Lee D, Hofmann M, Steinke F, Altun Y, Cahill N, Schlkopf B. Learning the similarity measure for multi-modal 3-D image registration; Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit; 2009. Jun. pp. 186–193. [Google Scholar]
- 36.Leventon M, Grimson W, Faugeras O. Statistical shape influence in geodesic active contours; Proc. Int. Conf. Comput. Vis. Pattern Recognit; 2000. pp. 1316–1323. [Google Scholar]
- 37.Liu C, Freeman WT, Adelson EH, Weiss Y. Human-assisted motion annotation; Proc. Int. Conf. Comput. Vis. Pattern Recognit; 2008. pp. 1–8. [Google Scholar]
- 38.Loog M, de Bruijne M. Discriminative shape alignment. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2009. pp. 459–466. [DOI] [PubMed] [Google Scholar]
- 39.Lyttelton O, Boucher M, Robbins S, Evans A. An unbiased iterative group registration template for cortical surface analysis. NeuroImage. 2007;vol. 34(no. 4):1535–1544. doi: 10.1016/j.neuroimage.2006.10.041. [DOI] [PubMed] [Google Scholar]
- 40.Makrogiannis S, Verma R, Karacali B, Davatzikos C. A joint transformation and residual image descriptor for morphometric image analysis using an equivalence class formulation; Proc. Workshop Math. Methods Biomed. Image Anal., Int. Conf. Comput. Vis. Pattern Recognit; New York. 2006. [Google Scholar]
- 41.McGonigle D, Howseman A, Athwal B, Friston K, Frackowiak R, Holmes A. Variability in fMRI: An examination of intersession differences. NeuroImage. 2000;vol. 11(no. 6):708–734. doi: 10.1006/nimg.2000.0562. [DOI] [PubMed] [Google Scholar]
- 42.McIntosh C, Hamarneh G. Is a single energy functional sufficient? Adaptive energy functionals and automatic initialization. Proc. Int. Conf. Med. Image Computing Computer Assisted Intervent. (MICCAI); LNCS; 2007. pp. 503–510. [DOI] [PubMed] [Google Scholar]
- 43.Moore A, Lee M. Efficient algorithms for minimizing cross validation error; Proc. 11th Int. Conf. Mach. Learn; 1994. pp. 190–198. [Google Scholar]
- 44.Olver P. Applications of Lie Groups to Differential Equations. 2nd ed. New York: Springer-Verlag; 1993. [Google Scholar]
- 45.Ono M, Kubick S, Abernathey C. Atlas of the Cerebral Sulci. 1st ed. Germany: Georg Thieme Verlag; 1990. [Google Scholar]
- 46.Park M, Hastie T. l1 -regularization path algorithm for generalized linear models. J. R. Stat. Soc., Series B. 2007;vol. 69:659–677. [Google Scholar]
- 47.Press W, Teukolsky S, Vetterling W, Flannery B. Numerical Recipes in C: The Art of Scientific Computing. 2nd ed. Cambridge, UK: Cambridge Univ. Press; 1992. [Google Scholar]
- 48.Qiu A, Miller M. Cortical hemisphere registration via large deformation diffeomorphic metric curve mapping. Proc. Int. Conf. Medical Image Computing Computer Assisted Intervent. (MICCAI); LNCS; 2007. pp. 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohfling T, Brandt R, Menzel R, Maurer C., Jr Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage. 2004;vol. 21(no. 4):1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Rohfling T, Russakoff D, Maurer C. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans. Med. Imag. 2004 Aug.vol. 23(no. 8):983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- 51.Rudin W. Principles of Mathematical Analysis. New York: Mc-Graw-Hill; 1976. [Google Scholar]
- 52.Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Non-rigid registration using free-form deformations: Application to breast MR images. IEEE Trans. Med. Imag. 1999 Aug.vol. 18(no. 8):712–720. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 53.Sabuncu MR, Balci S, Shenton M, Golland P. Image-driven population analysis through mixture-modeling. IEEE Trans. Med. Imag. 2009 Sep.vol. 28(no. 9):1473–1487. doi: 10.1109/TMI.2009.2017942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabuncu MR, Singer B, Conroy B, Bryan R, Ramadge P, Haxby J. Function-based inter-subject alignment of the cortical anatomy. Cerebral Cortex. 2010;vol. 20(no. 1):130–140. doi: 10.1093/cercor/bhp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabuncu MR, Yeo BT, Van Leemput K, Fischl B, Golland P. Supervised nonparameteric image parcellation. Proc. Int. Con. Med. Image Computing and Computer Assisted Intervent. (MICCAI); LNCS; 2009. pp. 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saxe R, Brett M, Kanwisher N. Divide and conquer: A defense of functional localizers. NeuroImage. 2006;vol. 30(no. 4):1088–1096. doi: 10.1016/j.neuroimage.2005.12.062. [DOI] [PubMed] [Google Scholar]
- 57.Schormann T, Zilles K. Three-dimensional linear and non-linear transformations: An integration of light microscopical and MRI data. Human Brain Mapp. 1998;vol. 6:339–347. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<339::AID-HBM3>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao J. Linear model selection by cross-validation. J. Am. Stat. Assoc. 1993:486–494. [Google Scholar]
- 59.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imag. 2002 Nov.vol. 21(no. 11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 60.Shi Y, Morra J, Thompson P, Toga A. Inverse-consistent surface mapping with laplace-beltrami eigen-features. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2009. pp. 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shum H-Y, Szeliski R. Construction of panoramic image mosaics with global and local alignment. Int. J. Comput. Vis. 2000;vol. 16(no. 1):63–84. [Google Scholar]
- 62.Thirion B, Flandin G, Pinel P, Roche A, Ciuciu P, Poline J-B. Dealing with the shortcomings of spatial normalization: Multi-subject parcellation of fMRI datasets. Human Brain Mapp. 2006;vol. 27:678–693. doi: 10.1002/hbm.20210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thirion B, Pinel P, Mériaux S, Roche A, Dehaene S, Poline J-B. Analysis of a large fMRI cohort: Statistical and methodological issues for group analyses. NeuroImage. 2007;vol. 35:105–120. doi: 10.1016/j.neuroimage.2006.11.054. [DOI] [PubMed] [Google Scholar]
- 64.Thirion B, Pinel P, Tucholka A, Roche A, Ciuciu P, Mangin J-F, Poline J. Structural analysis of fMRI data revisited: Improving the sensitivity and reliability of fMRI group studies. IEEE Trans. Med. Imag. 2007 Sep.vol. 26(no. 9):1256–1269. doi: 10.1109/TMI.2007.903226. [DOI] [PubMed] [Google Scholar]
- 65.Thompson P, Toga A. A surface-based technique for warping 3-dimensional images of the brain. IEEE Trans. Med. Imag. 1996 Aug.vol. 15(no. 4):1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- 66.Thompson P, Woods R, Mega M, Toga A. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Human Brain Mapp. 2000;vol. 9(no. 2):81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Series B (Methodological) 1996:267–288. [Google Scholar]
- 68.Tootell R, Taylor J. Anatomical evidence for MT and additional cortical visual areas in humans. Cerebral Cortex. 1995;vol. 5:39–55. doi: 10.1093/cercor/5.1.39. [DOI] [PubMed] [Google Scholar]
- 69.Tosun D, Prince J. Cortical surface alignment using geometry driven multispectral optical flow. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2005. pp. 480–492. [DOI] [PubMed] [Google Scholar]
- 70.Tu Z, Narr K, Dollar P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans. Med. Imag. 2008 Apr.vol. 27(no. 4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Twining C, Cootes T, Marsland S, Petrovic V, Schestowitz R, Taylor C. A unified information-theoretic approach to groupwise non-rigid registration and model building. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2005. pp. 1611–3349. [DOI] [PubMed] [Google Scholar]
- 72.Tyrtyshnikov E. A Brief Introduction to Numerical Analysis. Boston, MA: Birkhäuser; 1997. [Google Scholar]
- 73.Van Essen D, Drury H, Joshi S, Miller M. Functional and structural mapping of human cerebral cortex: Solutions are in the surfaces. Proc. Nat. Acad. Sci. 1996;vol. 95(no. 3):788–795. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Leemput K. Encoding probabilistic brain atlases using bayesian inference. IEEE Trans. Med. Imag. 2009 Jun.vol. 28(no. 6):822–837. doi: 10.1109/TMI.2008.2010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage. 2009;vol. 45(no. 1):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 76.Warfield SK, Zou KH, Wells WM. Simultaneous Truth and Performance Level Estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imag. 2004 Jul.vol. 23(no. 7):903. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White T, O’Leary D, Magnotta V, Arndt S, Flaum M, Andreasen N. Anatomic and functional variability: The effects of filter size in group fMRI data analysis. NeuroImage. 2001;vol. 13(no. 4):577–588. doi: 10.1006/nimg.2000.0716. [DOI] [PubMed] [Google Scholar]
- 78.Xiong J, Rao S, Jerabek P, Zamarripa F, Woldorff M, Lancaster J, Fox P. Intersubject variability in cortical activations during a complex language task. NeuroImage. 2000;vol. 12(no. 3):326–339. doi: 10.1006/nimg.2000.0621. [DOI] [PubMed] [Google Scholar]
- 79.Yeo BT, Sabuncu M, Desikan R, Fischl B, Golland P. Effects of registration regularization and atlas sharpness on segmentation accuracy. Med. Image Anal. 2008;vol. 12(no. 5):603–615. doi: 10.1016/j.media.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeo BT, Sabuncu M, Golland P, Fischl B. Task-optimal registration cost functions. Proc. Int. Conf. Med. Image Computing and Computer Assist. Intervent. (MICCAI); LNCS; 2009. pp. 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeo BT, Sabuncu MR, Vercauteren T, Ayache N, Fischl B, Golland P. Spherical demons: Fast diffeomorphic landmark-free surface registration. IEEE Trans. Med. Imag. 2010 Mar.vol. 29(no. 3):650–668. doi: 10.1109/TMI.2009.2030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yeo BT, Vercauteren T, Fillard P, Peyrat J-M, Pennec X, Golland P, Ayache N, Clatz O. DT-REFinD: Diffusion tensor registration with exact finite-strain differential. IEEE Trans. Med. Imag. 2009 Dec.vol. 28(no. 12):1914–1928. doi: 10.1109/TMI.2009.2025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou SK, Comaniciu D. Shape regression machine. Proc. Int. Conf. Inf. Process. Med. Imag; LNCS; 2007. pp. 13–25. [DOI] [PubMed] [Google Scholar]
- 84.Zilles K, Schleicher A, Palomero-Gallagher N, Amunts K. Quantitative Analysis of Cyto- and Receptor Architecture of the Human Brain. New York: Elsevier; 2002. [Google Scholar]