Abstract
Advances in chemistry and massively parallel detection underlie DNA sequencing platforms that are poised for application in personalized medicine. In stark contrast, systematic generation of protein-level data lags well-behind genomics in virtually every aspect: depth of coverage, throughput, ease of sample preparation, and experimental time. Here, to bridge this gap, we develop an approach based on simple detergent lysis and single-enzyme digest, extreme, orthogonal separation of peptides, and true nanoflow LC-MS/MS that provides high peak capacity and ionization efficiency. This automated, deep efficient peptide sequencing and quantification (DEEP SEQ) mass spectrometry platform provides genome-scale proteome coverage equivalent to RNA-seq ribosomal profiling and accurate quantification for multiplexed isotope labels. In a model of the embryonic to epiblast transition in murine stem cells, we unambiguously quantify 11,352 gene products that span 70% of Swiss-Prot and capture protein regulation across the full detectable range of high-throughput gene expression and protein translation.
Keywords: Quantitative proteomics, mass spectrometry, LC-MS, MS/MS, gene expression, microarray, protein translation, ribosomal profiling, pluripotency, embryonic stem cells
The field of DNA and RNA sequencing has progressed at a remarkable rate since release of the initial human genome draft sequences1, 2. Genotype-phenotype association studies continue to elucidate large genomic regions that are amplified, deleted, or otherwise altered in the context of human disease, although identification of specific causal gene elements has proven to be a significant challenge3. Despite these and other successes4, primary DNA sequence data does not capture downstream regulatory information for protein translation, degradation, and post-translational modification status5, 6, 7. These data are critical components of studies designed to quantitatively monitor biological response to perturbation or build predictive models of cellular physiology8. As a result, there remains a clear and unmet need for methodologies that can provide systematic and scalable sequence characterization of proteins as an important functional complement to DNA and RNA data.
The wide dynamic range of protein expression and vast array of post-translational modifications, coupled with the lack of an analyte amplification strategy analogous to PCR, presents tremendous challenges for genome-wide protein characterization, particularly for signal transduction and other key regulatory factors that are often present in low abundance. As a result, the majority of shotgun sequencing approaches frequently rely on low throughput pre-fractionation (e.g., prior to LC-MS/MS analysis) of subcellular compartments or intact proteins to improve dynamic range. While these techniques are subject to continued improvements, they have not yet achieved genome-scale protein identification and quantification. Moreover in many cases the labor-intensive nature of pre-fractionation protocols hinders widespread adoption and standardization.
In this work we forgo cellular- or protein-level pre-fractionation altogether and instead rely on direct detergent-based protein solubilization, followed by single-enzyme trypsin digest and extensive, fully automated temporal separation of peptides through multiple physicochemically orthogonal stages: high-pH reversed phase (RP) and strong anion exchange (SAX) dimensions coupled in series and directly with a narrow-bore, extended length (25 μm × 100 cm) low-pH RP analytical column operated in a true nanoflow regime. The latter chromatographic stage provides high peak capacity separation in a third orthogonal dimension, along with increased electrospray ionization efficiency for improved detection. The figures of merit for these individual components combine to yield an automated deep efficient peptide sequencing and quantification (DEEP SEQ) mass spectrometry platform that provides unprecedented separation capacity, rapid sequencing speed, and quantification of proteins across the entire range of mammalian gene expression and protein translation. We utilized multiplexed stable isotope labels in the context of a model designed to profile early perturbations in the naïve, self-renewing ground state of murine embryonic stem cells to quantify 211,535 unique peptide sequences that mapped unambiguously to 11,352 gene products. These results span ∼70% of the highly curated Swiss-Prot database, capture a vast majority of known pluripotent factors, and provide a depth and scale of proteome coverage commensurate with genome-wide analysis of protein translation by RNA-seq ribosomal profiling9.
Results
Peptide quantification by DEEP SEQ mass spectrometry
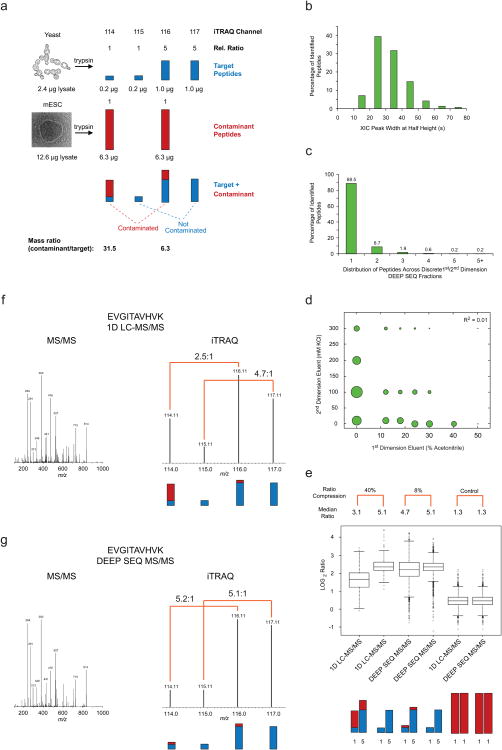
Multiplexed isotope labels have emerged as an enabling technology to increase the throughput of quantitative proteomic studies and provide increased flexibility with respect to experimental variables (time course, dose response, etc.)10. As these reagents have enjoyed wider use, it has become apparent that quantification accuracy degrades significantly as the complexity of target analyte mixtures increases. Simultaneous fragmentation of precursors that overlap in m/z leads to a compression of discrete peptide ratios towards the mean of all measured values11, 12, 13. We reasoned that the extreme temporal separation provided on our protein deep sequencing platform would mitigate deleterious ratio compression effects. To explore this hypothesis we created a mixed-species quantification model12, 13 in which the contaminant peptides were present at a mass ratio of ∼6- and ∼32-fold greater as compared to the target species, respectively (Fig. 1a). We acquired data at a depth of 20 DEEP SEQ fractions (Methods) with an LTQ-Orbitrap XL and used multiplierz14, 15 to compile and analyze extracted ion chromatograms for all detected peptides. We observed an average chromatographic peak width measured at half-height of 27.8 seconds (Fig. 1b), with over 97% of all detected peptides spanning no more than two adjacent fractions (Fig. 1c), yielding an empirical peak capacity16 of ∼1.3E4. Importantly, projection of peptide sequence identifications as a function of first dimension organic and second dimension salt concentrations (Fig. 1d) revealed that peptides were distributed throughout the entire separation space, confirming that our platform provided orthogonal and extensive peptide separation in these experiments. We next plotted iTRAQ ratios for species-specific peptides and compared these to ratios observed in a conventional, single-dimension LC-MS/MS analysis of the same sample mixture. The median ratio for the mixed-species channels (116:114) in the standard LC-MS/MS analysis was compressed by some 40% as compared to the single-species ratios (117:115); however, the equivalent analysis with DEEP SEQ mass spectrometry revealed a relative ratio compression of only 8% (Fig. 1e and Supplementary Data 1). Inspection of individual peptide spectra (Fig. 1f) reveals the marked improvement in quantification data provided by our DEEP SEQ platform.
Figure 1. DEEP SEQ mass spectrometry provides extreme separation and accurate quantification of iTRAQ labeled peptides.
(a) A mixed-species iTRAQ quantification model consisting of tryptic target peptides from yeast: 0.2 μg (114), 0.2 μg (115), 1.0μg (116), and 1.0μg (117), and contaminant peptides from murine embryonic stem cells (mESC): 6.3 μg (114) and 6.3 μg (116). Contaminant peptides are 6.3× and 31.5× more abundant in the iTRAQ 116 and 114 channels, respectively, as compared to the yeast target peptides. (b) Histogram of extracted ion chromatogram peak width for peptides identified in 20 fraction DEEP SEQ MS/MS analysis of mixed-species model. (c) Analysis of peptide elution profiles demonstrates minimal fraction-to-fraction overlap, with ∼97% of all identified peptides constrained within two adjacent 1st/2nd dimension (high pH RP/SAX) fractions. (d) The number of identified peptides represented as circles of proportional diameter and plotted as a function of 1st and 2nd dimension eluent concentration; low correlation coefficient for least-squares fit of these data (R2 = 0.01) suggests orthogonal fractionation of peptides across high pH reversed phase and strong anion exchange dimensions. (e) Log2 iTRAQ ratios of peptides identified in the mixed-species model displayed in box-plot format for conventional single dimension LC-MS/MS and DEEP SEQ analyses. The data include 272 yeast-specific peptides and 1570 mouse-specific peptides in 1D mode along with 2446 yeast-specific and 20461 mouse-specific peptides identified the DEEP SEQ analysis. Boxes encompass the interquartile range with respective median values indicated with stripes; whiskers represent 1.5× the interquartile range with outliers shown as open circles. Non-transformed, median ratios are listed at the top along with relative ratio compression for contaminated (116:114, blue + red) versus non-contaminated (117:115, blue) iTRAQ ratios. Analysis of mESC contaminant peptides alone (two right-most box plots) provides a positive control for iTRAQ ratios measured in conventional LC-MS/MS and DEEP SEQ analyses, respectively. (f, g) Representative MS/MS spectra for a tryptic peptide (EVGITAVHVK) acquired during (f) typical shotgun LC-MS/MS and (g) DEEP SEQ mass spectrometry analyses. Low-mass m/z region shows iTRAQ signals for each fragment ion spectrum along with measured ratios for contaminated (116:114, blue + red) and non-contaminated (117:115, blue) channels.
Genome-wide protein quantification comparable to ribo-seq
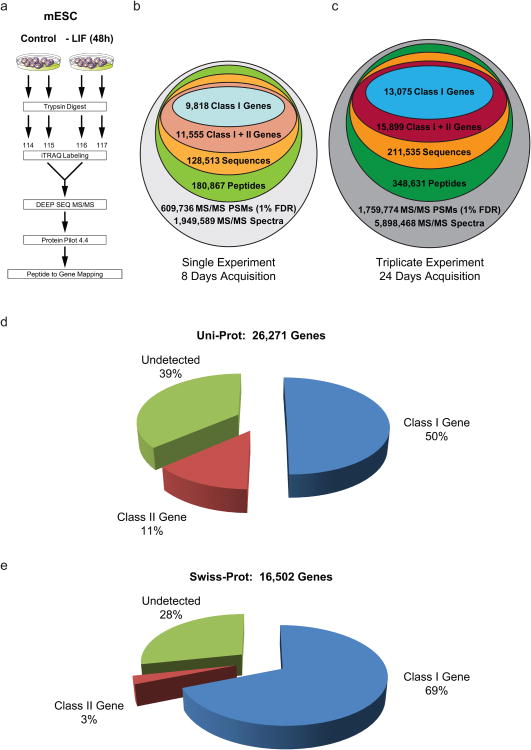
To further explore the performance of our deep protein sequencing platform we sought to quantify changes in mESC protein expression resulting from withdrawal of leukemia inhibitory factor (LIF), a cytokine required to maintain self-renewal in these cells. After iTRAQ labeling we acquired MS/MS data at a depth of 20 DEEP SEQ fractions (Methods) on a Triple TOF 5600 mass spectrometer (Fig. 2a). A single experiment which required approximately 1 day for sample preparation and another 8 days for data acquisition yielded nearly 2 million MS/MS spectra and 0.6 million peptide spectral matches (PSMs) based on a search against the Uniprot database which is composed of Swiss-Prot (16,502 mouse genes) plus Trembl (9,769 mouse genes) and contains 26,271 mouse genes in total. These PSMs corresponded to180,867 peptides (≤1%FDR, including chemical modifications). Following the convention described by Qeli et al.17 we mapped the set of 128,513 non-redundant peptide sequences into the genome as follows: First, 107,722 peptides were uniquely assignable to 9,818 gene IDs; we refer to these as “Class I” peptides and genes, respectively (Fig. 2b). Of the remaining 20,791 peptides, 2,819 sequences mapped to at least two of another 1,737 gene IDs (“Class II” peptides and genes, respectively, Supplementary Data 2); importantly Class II peptides do not map to Class I genes. The remaining 17,972 peptides (“Class III” peptides) map to multiple genes, including one or more Class I genes. Class III peptides are included in the count of total peptide detections but are not otherwise considered for purposes of protein identification. Cumulatively across three biological replicate experiments, comprising 24 days of data acquisition, we obtained ∼5.9 million MS/MS spectra, ∼1.8 million PSMs, and identified 211,535 non-redundant peptide sequences (178,167 Class I, 3,496 Class II, and 29,872 Class III peptides, respectively). This peptide set encompassed 13,075 and 2,824 Class I and II genes, respectively (Fig. 2c), with the former (Class I genes) spanning 50% of UniProt and nearly 70% of the manually curated, non-redundant Swiss-Prot mouse database (Fig. 2d,e).
Figure 2. DEEP SEQ mass spectrometry data spans 70% of protein-coding genes in murine embryonic stem cells.
(a) mESC were cultured in the absence of leukemia inhibitory factor (LIF) for 48 hours. Proteins were solubilized and processed directly (e.g., no sub-cellular or protein-level fractionation) for iTRAQ labeling (two replicates per condition), followed by DEEP SEQ mass spectrometry analysis. (b) A single, 20-fraction DEEP SEQ mass spectrometry analysis provided nearly 2 million MS/MS spectra, ∼610,000 peptide spectral matches (PSMs, <1%FDR), and 180,867 peptides corresponding to 128,513 unique peptide sequences. High-confidence (<1%FDR) peptide sequences were classified based on whether they mapped uniquely to mouse gene I.D.s (Class I peptide, Class I gene) or were shared across two or more mouse genes outside the set of Class I genes (Class II peptides, Class II genes). These data required 8 days of continuous acquisition time and yielded 9,818 Class I gene I.D.s. (blue). (c) Across biological triplicates, DEEP SEQ mass spectrometry analysis identified 13,075 Class I genes derived from ∼5.9 million MS/MS spectra acquired over 24 days. These data spanned (d) 50% of all protein-coding genes in the Uni-Prot database (26,271 entries). (e) The set of Class I peptides mapped unambiguously to nearly 70% of protein-coding genes in the manually annotated, non-redundant Swiss-Prot database (11,352 out of 16,502 total entries).
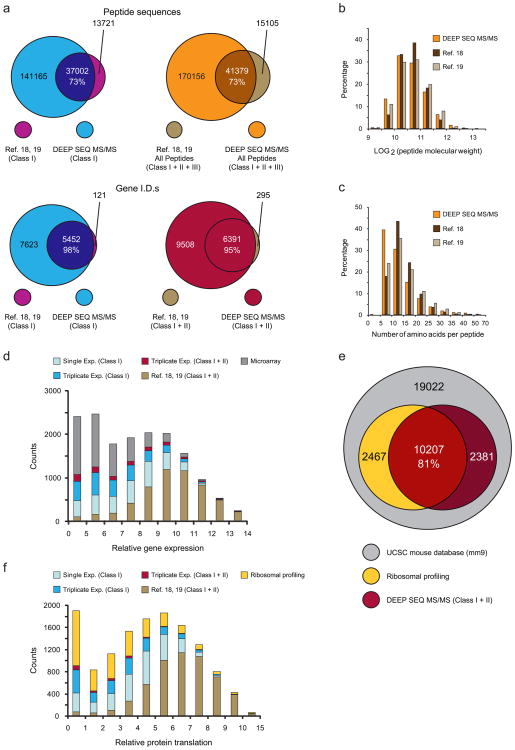
Two recent reports18, 19 that sought to analyze global protein expression in mESC utilized pre-fractionation of sub-cellular compartments, proteins, or peptides (or some combination) prior to LC-MS/MS. We took the union of published data from these studies and found that results from our DEEP SEQ platform encompass ∼73% and ≥95% of the reported peptide and gene IDs, respectively, while adding a significant quantity of new sequence information (Fig. 3a and Supplementary Data 3). Although the fractionation approaches, mass spectrometry instrumentation, and database search engines varied across these three studies we observed very similar physicochemical properties for peptides identified in each data set (Fig. 3b,c). To explore the dynamic range of proteins identified across these data we next overlaid Class I and II genes derived from each peptide set with mRNA expression data for mESC20 (Fig. 3d and Supplementary Data 4). Class I genes identified in our data spanned the full dynamic range of gene expression, with new Class I peptides identified through additional replicate experiments generally mapping to genes expressed at lower levels. Moreover, peptides identified by the two previous studies18, 19 were biased to high-expression genes as compared to those in our data. Strikingly the addition of Class II genes from our DEEP SEQ analysis improved overall proteome coverage only incrementally as compared to the set of Class I genes alone, with a small bias towards low-expression genes. Given the limitations of microarray data as a surrogate for protein expression, we next sought to compare our proteomic data with that from ribosomal profiling by RNA-seq, a technique that monitors protein translation on a genome-wide scale21. In a recent ribosomal profiling study, Ingolia et al.9 measured translation of 12,674 transcripts from 19,022 protein coding genes in mESC. Our set of Class I and II genes spanned an equivalent fraction of the total gene space (63%) and encompassed 81% of all translation events represented in the ribosomal profiling data (Fig 3e and Supplementary Data 5). Again we observed that our deep sequencing data spanned the entire dynamic range of protein translation as represented by ribosomal profiling, including significant coverage (∼42%) of low-frequency translation events (Fig. 3f). As was observed with the comparison to gene expression by microarray, our data exhibited significantly higher coverage of low-expression genes as compared to that from previous studies of the mESC proteome18, 19, while inclusion of DEEP SEQ Class II genes provided only a negligible improvement in proteome coverage. The latter observation is particularly notable given that Class II genes are defined by shared peptides; as a result the data for these gene products is ambiguous with respect to identification and quantification. For these reasons we focused the remainder of our analyses exclusively on Class I genes. Collectively these results suggest that our protein deep sequencing platform provides accurate quantitative data for multiplexed stable isotope reagents while simultaneously maximizing the discovery potential in the analysis of complex mammalian proteomes. In fact, using very stringent criteria for unambiguous protein identification (Class I peptides) our DEEP SEQ mass spectrometry approach provides data that span the full dynamic range of mammalian protein expression.
Figure 3. Protein identification across thefull dynamic range of gene expression and protein translation in mESC.
(a) High-confidence peptides based on the union of published data from two previous mESC proteomic studies18, 19 were mapped to Class I and II gene I.D.s as described (see Methods) and compared with data from this study. High-confidence peptides identified in all three studies exhibited similar physicochemical properties, including (b) molecular weight distribution and (c) number of amino acids per peptide. Comparison of (d) Class I and II gene I.D.s with normalized mRNA levels in mESC demonstrates that DEEP SEQ mass spectrometry data spans the full range of gene expression. (e) The fraction of genes represented in the UCSC mouse database (grey) captured by ribosomal profiling (yellow) and DEEP SEQ mass spectrometry (red). The set of Class I and II genes identified herein encompassed 81% of protein translation events as represented by RNA-seq ribosomal profiling. (f) Overlay of ribosomal profiling data with that from DEEP SEQ mass spectrometry analysis demonstrates that the set of Class I genes spans the full dynamic range of protein translation in mESC.
Functional proteome coverage by DEEP SEQ mass spectrometry
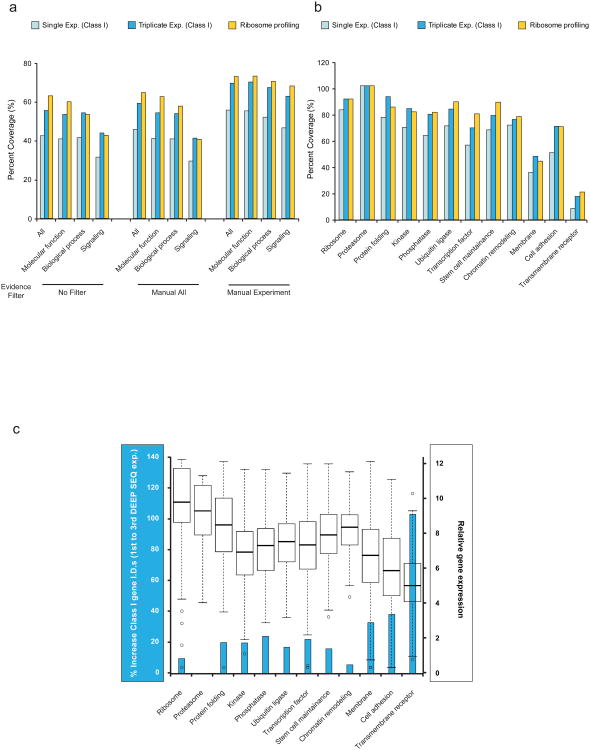
Regulatory proteins such as transcription factors, kinases and other signal transduction factors are often underrepresented in mass spectrometry-based whole-proteome studies. Hence we next sought to evaluate the coverage of functional protein classes provided by our DEEP SEQ platform. We found that the set of Class I peptides mapped unambiguously to ≥40% of genes across Gene Ontology (GO) functional categories, regardless of the evidence filter used (Fig. 4a). We also observed that the coverage varied across categories, from nearly 100% for genes encoding ribosome and proteasome proteins to approximately 20% for transmembrane receptors; the same trend was observed for data derived from ribosomal profiling, suggestive of a general correlation between coverage and protein abundance (Fig 4b). This hypothesis was further corroborated by mapping functional protein classes to gene expression in mESC (Fig. 4c). Consistent with these data we found that replicate experiments tended to augment representation of proteins encoded by genes expressed at low levels. Overall, our set of stringently defined Class I genes spanned 52% of the functional proteome, on par with coverage provided by ribosomal profiling (56% coverage); importantly, our DEEP SEQ mass spectrometry data captured ≥70% of key regulatory protein classes, including kinases, phosphatases, and transcription factors.
Figure 4. DEEP SEQ mass spectrometry provides extensive coverage of the mammalian functional proteome.
(a) Relative coverage provided by DEEP SEQ mass spectrometry and ribosomal profiling as a function of gene-ontology (GO) category and evidence filter. As defined by GO, “Manual All” contains all curator-reviewed experimental and computational annotations while “Manual Experiment” includes only curator-reviewed annotations derived from direct assay (IDA), physical interaction (IPI), mutant phenotype (IMP), genetic interaction (IGI), and expression pattern (IEP) evidence categories. (b) Relative coverage of GO protein families for DEEP SEQ mass spectrometry and ribosomal profiling analyses. Protein membership in each family is derived from the annotations within “Manual Experiment.” (c) Relative mRNA expression level plotted in box-plot format (black and white, y-axis, right) as a function of GO protein families. Boxes encompass the interquartile range with respective median values indicated with stripes; whiskers represent 1.5× the interquartile range with outliers shown as open circles.(blue, y-axis, left) Relative increase in GO protein family representation across bio-triplicate DEEP SEQ mass spectrometry analyses for Class I genes.
Quantification of LIF-dependent functional proteome in mESC
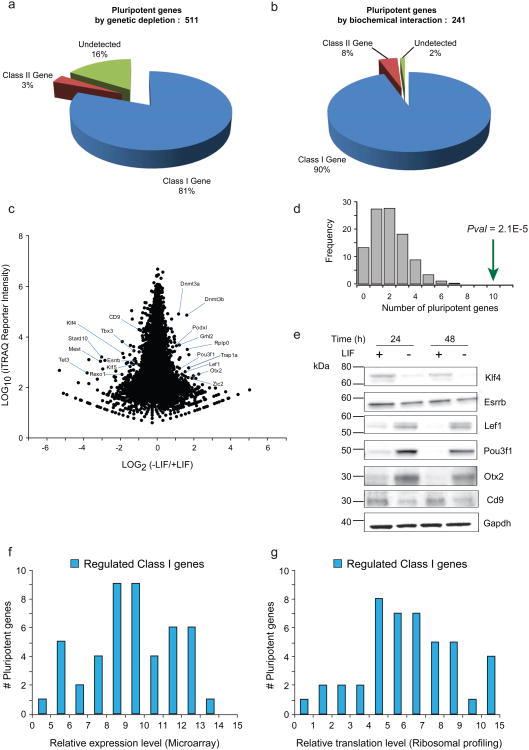
To explore the functional proteome characteristic of mESC, we next compiled two reference sets of pluripotent factors derived from genetic depletion screens22, 23 and proteins biochemically associated with the master regulatory transcription factors Oct4 and Nanog24, 25, 26 (Supplementary Data 6). The set of Class I genes from our biological triplicate analysis encompassed ∼81% (Fig. 5a) and ∼90% (Fig. 5b) of these reference gene sets, respectively, suggesting that our DEEP SEQ mass spectrometry platform can capture regulatory events associated with key mediators of pluripotency in mESC. A plot of quantitative data (Fig. 5c) revealed numerous proteins (Table 1) whose expression level reproducibly increased or decreased in response to LIF withdrawal (Supplementary Data 7). Query of these proteins against the functional and biochemical reference sets demonstrated an enrichment (Fishers exact test, Pval = 2.1E-5) for pluripotent factors (Fig. 5d). Biochemical validation (Fig. 5e) of LIF-mediated expression for a sub-set of proteins confirms the power of DEEP SEQ mass spectrometry to provide accurate quantification for multiplexed isobaric labels in the context of genome-wide proteome profiling.
Figure 5. DEEP SEQ mass spectrometry provides genome-wide proteome quantification in mESC subject to LIF withdrawal.
The set of Class I gene I.D.s from bio-triplicate, 20-fraction DEEP SEQ mass spectrometry experiments encompassed (a) ∼80% of murine pluripotent factors as defined by systematic genetic depletion assays22, 23 and (b) ∼90% of the Nanog and Oct4 transcription factor network as defined through analysis of biochemical interactions24, 25, 26. (c) Scatter plot of iTRAQ log-intensity versus log-ratio for 13,075 class I gene I.D.s identified across bio-triplicate, 20-fraction DEEP SEQ mass spectrometry experiments. Individual iTRAQ signals for each high-confidence Class I peptide were summed across technical and biological replicates; these aggregate ratios were then combined to provide protein-level ratios. (d) Fisher's Exact Test confirms that the set of regulated proteins (Table 1) identified by DEEP SEQ mass spectrometry analysis is enriched (two-sided Pval = 2E-5) for pluripotent factors as defined by systematic loss-of-function and biochemical interaction assays. (e) Selected pluripotent and developmental factors were probed by western blot in mESC at 24 and 48 hrs after withdrawal of LIF. Left-most column indicates molecular weight markers. Overlay of regulated Class I gene products as detected by DEEP SEQ mass spectrometry with (f) relative gene expression as measured by microarray and (g) relative protein translation as measured by RNA-seq ribosomal profiling.
Table 1. mESC proteins whose expression level increases or decreases in response to LIF withdrawal (-LIF, 48hr).
| upregulated | downregulated | |
|---|---|---|
|
| ||
| Def8 | Bpgm | Pygl |
| Dnmt3a | Calb2 | Rexo1 |
| Dnmt3b | Cd9 | Rnf10 |
| Dym | Elovl6 | Rps11 |
| Eif1 | Esrrb | S100a6 |
| Esyt2 | Gjb3 | Slc15a1 |
| Grhl2 | Hacl1 | Slc35b2 |
| Lef1 | Jagn1 | Snx22 |
| Limd2 | Klf4 | Stard10 |
| Lphn1 | Klf5 | Tbx3 |
| Otx2 | Mest | Tcfcp2l1 |
| Pld2 | Padi2 | Tekt3 |
| Podxl | Paf | Tet3 |
| Pou3f1 | Pramef12 | Ubxn1 |
| Rplp0 | Ptrf | Vim |
| Soat1 | Pvrl1 | Zc3hav1 |
| Trap1a | ||
| Zic2 | ||
Discussion
Continued advances in our understanding of genome variation2, gene expression27, 28, and translation9, driven in-part by the advent of next-generation DNA/RNA sequencing technologies3, has effectively re-kindled interest in experiments designed to maximize discovery potential and simultaneously quantify known genomic or other biomolecular events. Establishing a parallel trajectory for this paradigm in proteomics is complicated by the broad range of protein abundance, diverse repertoire and stoichiometry of post-translational modifications, as well as the stochastic nature of discovery-driven or shotgun MS/MS acquisition methods. In fact today, nearly two decades after Marc Wilkens first coined the term, “proteome”29, the dynamic range of protein expression in mammalian systems has represented an insurmountable hurdle for genome-wide proteome quantification. Despite these obstacles, the functional content of protein-level data represents an important complement to genomic-based studies, and hence there remains significant motivation to develop scalable platforms for proteomic analyses.
In order to achieve true, genome-scale sequence coverage along with high-fidelity protein quantification, we sought to significantly improve the analytical figures of merit for each component of our DEEP SEQ mass spectrometry platform. First the use of physicochemically orthogonal high-pH reversed phase and strong anion exchange separation stages16, 30 coupled with a narrow-bore (25 μm I.D.), extended length (100 cm) low-pH analytical column31 provides extreme temporal separation of peptides with an empirical peak capacity of ∼1.3E4. Our experience with online multidimensional separations16, 30 indicates that further optimization of first and second dimension eluent concentrations can yield an improved distribution of peptides across each separation dimension, ultimately providing deeper proteome coverage beyond the ∼70% achieved herein. Second, the final dimension column geometry maintains the integrity of chromatographic separation at ultra-low effluent flow rates (∼5nL/min), thus maximizing electrospray ionization efficiency32, 33. Third, all separation stages in the DEEP SEQ configuration are implemented in microcapillary format and coupled in series, with the final dimension interfaced directly to the mass spectrometer, providing for fully automated operation, along with efficient capture and transfer of peptides across all separation stages.
Based on a stringent peptide-to-gene I.D. criterion, our DEEP SEQ mass spectrometry analysis of mESC provided quantitative expression data for 11,352 out of 16,502 genes in SwissProt (∼69%, Fig. 2e), a depth of coverage equivalent to a recent study in which RNA-seq ribosomal profiling was used to measure translation of 12,674 out of 19,022 (∼66%) protein coding genes contained in the UCSC mouse database9. Importantly our data provide significant coverage (∼70-85%) for key regulatory protein families, including kinases, ubiquitin ligases, and transcription factors (Fig. 4b). In fact, proteins quantified on our deep sequencing platform span the full dynamic range of corresponding gene expression and protein translation profiles (Fig. 3d-f). Not surprisingly, improvements in proteome coverage across triplicate experiments were most pronounced for low-expression protein families (Fig. 4c).
The absence of protein amplification strategies analogous to PCR, combined with the limited improvements in dynamic range offered by each generation of mass spectrometry hardware, places a disproportionate burden on separation techniques to achieve robust detection of low-abundance proteins. Despite numerous methodological studies, the choice of separation strategies that will provide the best combination of sample yield, experiment time, and ultimately, proteome coverage is unresolved. Our results refute the notion that analysis of tryptic peptides alone is insufficient to characterize proteins across a wide dynamic range of abundance. In practice, DEEP SEQ mass spectrometry achieves significantly higher proteome coverage (Fig. 3a) from only a fraction (<5%) of the input required by techniques that rely on protein- or cellular-level pre-fractionation18, 19. In fact, while chromatographic and sub-cellular fractionation are slow and low-throughput compared to chemical amplification, the time required for our DEEP SEQ analysis (25 days for biological triplicates, including sample preparation) is not dramatically different from other recent attempts at deep protein sequencing by mass spectrometry (for example, 21 days for a study that relied on the use of protein level fractionation and multiple enzymes34), or for that matter genome-wide RNA-seq ribosomal profiling (9-12 days as described in a recent reveiw35). Finally it is worth noting that our DEEP SEQ approach provides the serendipitous benefit of a robust and streamlined sample preparation protocol. Simple detergent solubilization, single enzyme digestion, peptide desalting, and iTRAQ labeling is somewhat reminiscent of the commoditized kits used in conjunction with next-generation DNA/RNA sequencing.
The quality of peptide fractionation provided by our deep protein sequencing platform is also evident in the analysis of a mixed-species iTRAQ quantification model (Fig. 1a). Even with contaminant species present in >30-fold excess relative to the target peptides, our DEEP SEQ mass spectrometry platform provided sufficient separation peak capacity to yield accurate iTRAQ ratios (Fig. 1e). These data are important in light of discrepancies reported between the throughput advantages afforded by multiplexed reagents and the well-documented limitations encountered when using these labels to quantify proteins in complex mixtures11, 12, 13, 36. In fact, results from three of these studies12, 13, 36 demonstrated unequivocally that two dimensions of peptide fractionation are insufficient to abrogate suppression of multiplexed ratios for analysis of samples intended to represent complex proteomes. Importantly, DEEP SEQ mass spectrometry provides a platform to reconcile these juxtaposed observations and fully leverage the advantages of isobaric reagents in studies designed to quantify proteome response to perturbation on a genome-wide scale. Moreover, recent reports37, 38 suggest the possibility of significant increases in the degree of multiplexing for these stable isotope reagents. These advances, along with improvements in mass spectrometry instrumentation39, 40 in addition to new chromatographic41, 42, 43 and electrophoretic44, 45, 46 separation platforms, will yield a concomitant increase in the throughput of DEEP SEQ mass spectrometry analysis.
Finally, our data provide insight into the transition of mESC from a naïve ground state, similar to that observed in the embryo inner cell mass prior to implantation, to a post-implantation epiblast-like stage (mEpiSC) that is poised for directed differentiation47. LIF is a critical cytokine that supports self-renewal in mESC through the Jak-Stat and Pi3k-Akt signaling axes, while opposing pathways, including Wnt-Gsk3β and Mek-Erk, mediate transcription programs that degrade pluripotent potential48. How these and other exogenous stimuli coordinately influence the core pluripotent genes (Oct4 and Sox2) and other peripheral factors to enforce the transcriptional ground state or mediate lineage commitment is not yet fully resolved.
Strikingly we detect regulated protein expression for downstream targets linked to each of these discrete pathways (Klf4, Klf549, Lef150, Esrrb, Tcfcp2l151 and Tbx348). Proteins having roles in other developmental contexts (Otx252, Pou3f1/Oct653, and CD954, 55) along with epigenetic factors (Dnmt3a/b) that are critical for high-fidelity gene expression56, were also regulated in a LIF-dependent manner. In addition to these studies that targeted specific genes or pathways, our DEEP SEQ analysis also quantified the majority of pluripotent factors previously defined by high-throughput functional genetic22, 23 and biochemical interaction24, 25, 26 assays (Fig. 5a,b). Indeed, we reproducibly observed regulated expression for 50 proteins (Table 1) and found that these were enriched for pluripotent and developmental genes (Fig. 5d). Importantly, this set of putative LIF-dependent protein targets spans the full dynamic range of gene expression and protein translation in mESC (Fig. 5f,g). Thus, though results presented herein represent a single time point in the LIF-mediated transition between naïve and primed epiblast states, the depth and scale of these data provide compelling evidence that our DEEP SEQ mass spectrometry platform can capture the vast majority of the mESC functional proteome in the context of more complex experiments57, 58 designed to decipher individual contributions of the above pathways to self-renewal and lineage commitment. More generally our DEEP SEQ mass spectrometry platform represents a powerful and scalable approach for genome-wide profiling of protein expression and post-translational modification status in mammalian systems.
Methods
Cell culture
Yeast cells were grown and processed under conditions similar to those described previously30. S. cerevisiae, strain S288C, BY4741 (ATCC 201388, MATa his3Δ1 leu2Δ0met15Δ0 ura3Δ0) was grown in yeast extract peptone dextrose (YEPD) liquid medium to log phase, OD600 ≈0.7 at 30°C. Cells were lysed by the addition of boiling SDS (50mM Tris-HCl, pH 7.5, 7.5% SDS, 5% glycerol, 50mM DTT, 5mM EDTA), followed by centrifugation, with the supernatant stored at −80 °C.
Mouse embryonic stem cell (mESC) line J1 was generously provided by Dr. Stuart Orkin (Dana-Farber Cancer Institute and Children's Hospital, Boston, MA). Initially, 10cm Nunclon tissue culture dishes (ThermoFisher Scientific, Waltham, MA) were coated with 0.1% Gelatin (EMD Millipore, Billerica, MA) at room temperature for 30 minutes. Gelatin was aspirated and J1 mESC were plated at a density of ∼6 × 104/cm2 in Dulbecco's modified Eagle's medium with high glucose supplemented with L-glutamine (Gibco Life Technologies, Carlsbad, CA), 15% embryonic stem cell validated FCS (Stem Cell Technologies, Vancouver BC, Canada), 2-mercaptoethanol, nucleosides (EMD Millipore, Billerica, MA), nonessential amino acids (Gibco Life Technologies, Carlsbad, CA), and 10 ng/mL murine LIF6 (EMD Millipore, Billerica, MA). Perturbation experiments were performed by establishing J1 mESC under the above conditions and then removing LIF6 for 48 hours. At the time of harvest, plates were washed with cold PBS to remove serum proteins and adherent cells were lysed by the addition of a boiling SDS (50mM Tris-HCl, pH 7.5, 7.5% SDS, 5% glycerol, 50mM DTT, 5mM EDTA). Lysed cells were centrifuged and the supernatant extract was stored at −80 °C.
Sample preparation for DEEP SEQ analysis
Proteins were precipitated by adding six volumes of cold (−20°C) acetone and resolubilized in digestion buffer containing 8 M urea and 0.1 M NH4HCO3. Total protein levels were measured by BCA. Dithiothreitol (DTT) was added to a final concentration of 10 mM and incubated for 30 minutes at 60 °C, followed by addition of methyl methanethiosulfonate (MMTS) (ThermoFisher Scientific, Waltham, MA) to 20 mM. After 30 min. incubation in the dark at room temperature, excess MMTS was quenched by addition of DTT to a final concentration of 20 mM. Reduced and alkylated proteins were diluted in 0.1M ammonium bicarbonate, followed by addition of trypsin, with overnight digestion 37 °C and end-over-end rotation. Digested sample solutions were loaded onto SepPak C18 reverse phase cartridges (Waters Corp., Milford, MA) to remove urea and other salts. Eluted peptides (45% acetonitrile in water with 0.1% trifluoroacetic acid) were lyophilized by vacuum centrifugation.
Peptides were labeled with 4-plex iTRAQ reagents (AB Sciex, Framingham, MA). Two aliquots of 0.2 μg yeast peptides were labeled with 114 and 115. Separately, two other aliquots of 1.0 μg yeast peptides were labeled with 116 and 117. In addition, two aliquots of 6.3 μg peptides from mESC were labeled with 114 and 116. For the mESC perturbation experiment, peptides from the cells incubated without LIF were labeled as technical replicates with iTRAQ116 and 117, while peptides from the cells incubated with LIF were labeled as technical replicates with iTRAQ 114 and 115. For each reaction, peptides were resuspended in 500mM triethylammonium bicarbonate and mixed with the appropriate iTRAQ reagent in ethanol. Labeling was allowed to proceed at room temperature for one hour; samples were then combined and dried by vacuum centrifugation. Fresh aliquots of mESC were processed as described above to provide biological triplicates.
DEEP SEQ multi-dimension separation
Three dimension peptide separation was performed on a modified Waters (Milford, MA) NanoAcquity UHPLC system with binary and isocratic pumps, along with an autosampler and additional 6-port, 2-position valve (Valco, Austin, TX). The 1st dimension reversed phase (RP) column consisted of a 200 μm I.D. capillary packed with 20 cm of 5 μm dia. XBridge C18 resin (Waters Corp., Milford, MA). An anion exchange column (200 μm I.D. × 20 cm) was packed with 5μm dia. SAX resin (Sepax Technologies, Neward, DE) and connected to the outlet of the 1st dimension RP column. The 3rd dimension consisted of reversed phase pre- (100 μm I.D. ×4 cm of 10μm dia. POROS 10R2 resin) and analytical (25μm I.D. ×100 cm of 5 μm dia. Monitor C18 [Column Engineering, Ontario, CA], with integrated 1 μm dia. emitter tip) columns configured in a vented geometry59, 60. The autosampler picked up and delivered either peptide samples or 1st (acetonitrile in 20mM ammonium formate, pH 10) and 2nd (KCl in 20 mM ammonium formate, pH 10) dimension eluents at 2μL/min. through the sample loop. Injection of each 1st or 2nd dimension eluent constitutes a “DEEP SEQ fraction” (Supplementary Data 8). The binary pump delivered 0.1% formic acid at 8μL/min. to dilute the organic content and acidify the 1st/2nddimension effluent prior to the 3rd dimension pre-column, or provided for gradient elution (2-50% B in 580 minutes, A = 0.1% formic acid, B = acetonitrile with 0.1% formic acid) of peptides from the 3rd dimension reversed phase columns for LC-MS/MS analysis at a flow rate of ∼5nL/min. A Digital PicoView electrospray source platform (New Objective, Woburn, MA) was used on both the Orbitrap XL and 5600 Triple TOF mass spectrometers to automatically position the emitter tip at the source inlet during LC-MS/MS acquisition or beneath a gravity-driven drip station during injection of peptide samples or 1st/2nd dimension eluents.
DEEP SEQ sample capacity and experiment time
Based on our published and unpublished studies, we estimate that the total loading capacity of our first dimension RP column (200 μm I.D. × 20 cm) is currently ∼100 μg. The DEEP SEQ platform is easily tailored to a variety of sample types. Generally, first and second dimension eluent concentrations in a range of 7 to 55% acetonitrile and 10 to 300 mM KCl represent the useful boundaries for peptide elution. Importantly, our experience to date suggests that these conditions are robust with respect to biological input, obviating the need to run repeated pilot experiments for every sample. Total time of analysis is another important consideration when using multidimensional fractionation techniques. For example, DEEP SEQ experiments performed at a depth of 20 fractions required 8 days for data acquisition. System reliability is particularly important given that a single, DEEP SEQ mass spectrometry analysis will typically require several days of continuous instrument time. Importantly, all data presented herein were acquired using a single 25μm I.D. ×100 cm resolving column, demonstrating the robustness of our platform.
DEEP SEQ mass spectrometry data acquisition parameters
The LTQ-Orbitrap XL mass spectrometer (Thermo, Waltham, MA) was operated in data dependent mode, such that the top 10 most abundant precursors in each MS scan were subjected to MS/MS in both CAD and HCD mode (CAD in the linear trap, normalized collision energy = 35%, precursor isolation width = 1.9 Da, intensity threshold for precursor selection = 20,000 HCD in the orbitrap, normalized collision energy = 47%, precursor isolation width = 1.0 Da, intensity threshold for precursor selection = 20,000). Dynamic exclusion was enabled with a repeat count of 1 and exclusion duration set to 20 seconds. Electrospray voltage was 2.2 kV. We enabled the Lock Mass feature and selected m/z = 445.120025 ([Si(CH3)2O]6) as the internal calibrant.
The 5600 Triple TOF (AB Sciex, Framingham, MA) was operated in information dependent mode (IDA), with the top 50 precursors (charge state +2 to +5, >100 counts) in each MS scan (800 ms, scan range 350-1500 m/z) subjected to MS/MS (minimum time 140 ms, scan range 100-1400m/z). A dynamic exclusion window of 20 s was used with unit resolution for precursor isolation. Electrospray voltage was 2.2 kV.
DEEP SEQ mass spectrometry data processing
Our API-based multiplierz software framework was used to extract and format MS/MS data from the Orbitrap XL for subsequent search against the Uniprot mouse database (downloaded on 11/02/2011) and a S. cerevisiae database (downloaded from http://www.yeastgenome.org/01/06/2010). MS/MS data was searched using Protein Pilot V4.4 (AB Sciex, Framingham, MA) with the following parameters: instrument =“Orbi/FT MS (sub-ppm), LTQ MS/MS” for data acquired with the Orbitrap XL, or “5600 TripleTOF” for data acquired with the 5600 Triple TOF mass spectrometer. A fixed modification of +42 corresponding to MMTS of cysteine was also included. The sample type was set to “iTRAQ 4-plex (peptides labeling)”. All peptide spectral matches (PSM) from biotriplicate acquisitions were combined for the FDR assessment. Only those peptides with scores at or above a PSM FDR threshold of 1% were further considered. All peptides were mapped back to the genome without consideration of splice isoforms from the same gene. Following the procedure of Qeli et al.17, peptides passing the 1% FDR filter were classified as either “unique” or “shared” based on whether they could be assigned to only one or more than one gene, respectively. Class I genes were defined as those identified only by uniquely assignable peptides. Class II genes were identified based on shared peptides that could not be assigned to any Class I gene. The results reported herein are based on Class I and Class II genes; no Class III genes were included in our analyses. Multiplierz scripts were used to systematically extract XICs and calculate corresponding peak widths for all identified peptides. To estimate the orthogonality of peptide fractionation, the number of unique peptides identified in each third dimension LC-MS/MS run was represented as a circle of proportional diameter and projected onto a 2D plot at the corresponding 1st dimension acetonitrile (x-axis) and 2nd dimension salt (y-axis) concentrations used in the experiment. Linear regression was performed using the function stats. linregress from Numpy version 1.4.1, with each unique peptide sequence used as a separate data point.
In the mixed species quantification experiments the iTRAQ116/iTRAQ114 and iTRAQ117/iTRAQ115 ratios were first normalized based on all spectra identified (final normalization factor was 1.14). We used R scripts to create box-plots for all iTRAQ ratios. For the quantitative analyses of mESC, we summed iTRAQ channels in each biological condition (±LIF) for Class I peptides to generate a final ratio for each Class I gene. Across three biological triplicate experiments a protein was considered to be regulated in expression (Table 1) if the following criteria were met in two out of the three experiments: (i) the iTRAQ intensities exceeded 200 counts and (ii) the Log2 iTRAQ ratio was ≥1 or ≤ −1. Enrichment of pluripotent factors within the set of genes whose expression was reproducibly regulated across biotriplicate experiments was estimated using a Fisher's exact test. A null distribution and two-sided p-value were calculated based on code freely available at: (http://research.microsoft.com/en-us/um/redmond/projects/MSCompBio).
Comparisons to mESC microarray and ribosomal profiling data
Comparison of DEEP SEQ mass spectrometry data to that from ribosome profiling for mESC was performed as follows: The UCSC mouse genome database (mm9) (http://hgdownload.soe.ucsc.edu/goldenPath/mm9/database/) was downloaded on 10/02/2012. We aligned the peptides identified in triplicate DEEP SEQ mass spectrometry analyses against sequences in the UCSC database. A similar alignment was performed for mESC ribosomal profiling data previously reported9. In 1,034 cases we were unable to reconcile gene names from the UCSC and UniProt databases; these genes were removed from further consideration. Average gene expression levels across three wild type mESC lines (B21, J1 and R1) was calculated using published datasets (accession codes GSM338369, GSM338371, GSM338373)20. We used the R/Bioconductor software environment to re-normalize all microarray data based on the multi-array averaging method (RMA) and to re-map all probe sequences to Entrez Gene IDs using a custom Chip Definition File (CDF) from the Michigan Microarray Lab (version 13). The average mRNA expression level (in Log2 space) between the three arrays was calculated and the Entrez Gene IDs were converted to UniProt gene symbols. UniProt entries mapping to more than one Entrez Gene ID were excluded. The final list contained 15,705 entries of (average Log2 expression value) | (UniProt gene symbol) pairs.
Gene ontology and reference gene sets for pluripotency
Positive reference sets (PRS) of pluripotent genes were created from RNAi screening data downloaded from two previous studies22, 23. Similarly, pluripotency reference genes based on biochemical interactors of Nanog and Pou5f1 (Oct4) were created from three previous studies24, 25, 26.
The Gene Ontology (GO) database was downloaded on 03/15/2012. The GO subcategories were filtered based on different identifiers: Molecular function (GO:0003674), Biological process (GO:0008150), Signaling (GO:0023052), Ribosome (GO:0005840), Proteasome (GO:0000502), Protein folding (GO:0006457), Kinase (combination of GO:0004672 and GO:0016301), Phosphatase (combination of GO:0016791 and GO:0004721), Ubiquitin ligase (GO:0004842), Transcription factor (combination of GO:0006351 and GO:0008134), Stem cell maintenance (GO:0019827), Chromatin remodeling (GO:0006338), Membrane (GO:0016020), Cell adhesion (GO:0007155) and Transmembrane receptor (GO:0004888).
Western Blotting
Protein lysates from ∼3 × 105 murine ESC (about 30μg total protein) incubated with or without LIF were separated on 4-12% Bis-Tris gels (Life Science) under reducing conditions and transferred onto polyvinylidene difluoride membranes. The blot was stained with antibodies against Lef1 (Cell Signaling Technology, 1:1000), Pou3f1 (Abcam, 1:1000), Esrrb (Cell Signaling Technology, 1:1000), Otx2 (Abcam, 1:1000), Klf4 (Cell Signaling Technology, 1:1000), Cd9 (Abcam, 1:1000), and Gapdh (Cell Signaling Technology, 1:6000). The bands were visualized by enhanced chemiluminescence (Thermo, SuperSignal West Femto Chemiluminescent Substrate).
Supplementary Material
Acknowledgments
Generous support for this work was provided (to J.A.M) by the Dana-Farber Cancer Institute and the National Institutes of Health (NINDS, P01NS047572). Y.L. acknowledges support from a National Institutes of Health training grant in transfusion medicine (T32HL66987-11). C.J.L. acknowledges support from the National Blood Foundation and the Department of Pathology at the Brigham and Women's Hospital. The authors thank Dr. Stuart Orkin (Dana-Farber Cancer Institute and Children's Hospital, Boston, MA) for kindly providing the gel-adapted J1 murine embryonic stem cells used in this study. The authors thank Dr. Nicholas Ingolia (Carnegie Institution for Science, Baltimore, MD) for helpful discussions related to mESC ribosomal profiling data. In addition the authors thank Christie Hunter and Sean Seymour of AB Sciex for support and technical assistance with the 5600 Triple TOF mass spectrometer and Protein Pilot search software.
Footnotes
Author contributions: F.Z. designed the DEEP SEQ platform, planned and executed the experiments, organized and analyzed gene- and protein-level data, prepared figures, and wrote the manuscript. Y.L. planned and performed time-course experiments in murine embryonic stem cells. W.J. contributed reagents and performed biochemical assays. S.B.F. and G.A. contributed to data analysis. C.J.L. provided intellectual contributions. J.A.M. conceived of the study, directed the research, and wrote the manuscript.
Competing financial interest: The authors declare that they have no competing financial interests.
References
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. Genomics: ENCODE explained. Nature. 2012;489(7414):52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo JM, Buck MJ. Key principles and clinical applications of “next-generation” DNA sequencing. Cancer Prev Res (Phila) 2012;5(7):887–900. doi: 10.1158/1940-6207.CAPR-11-0432. [DOI] [PubMed] [Google Scholar]
- 4.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450(7172):991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 7.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 8.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans C, Noirel J, Ow SY, Salim M, Pereira-Medrano AG, Couto N, et al. An insight into iTRAQ: where do we stand now? Anal Bioanal Chem. 2012;404(4):1011–1027. doi: 10.1007/s00216-012-5918-6. [DOI] [PubMed] [Google Scholar]
- 11.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. iTRAQ underestimation in simple and complex mixtures:“the good, the bad and the ugly”. J Proteome Res. 2009;8(11):5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 12.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8(11):937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, et al. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat Methods. 2011;8(11):933–935. doi: 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Askenazi M, Parikh JR, Marto JA. mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat Methods. 2009;6(4):240–242. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parikh JR, Askenazi M, Ficarro SB, Cashorali T, Webber JT, Blank NC, et al. multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics. 2009;10:364. doi: 10.1186/1471-2105-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, Garg B, Adelmant GO, Webber JT, et al. Online Nanoflow Multi-dimensional Fractionation Strategies for High Efficiency Phosphopeptide Analysis. Mol Cell Proteomics. 2011;10:M111.011064. doi: 10.1074/mcp.O111.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qeli E, Ahrens CH. PeptideClassifier for protein inference and targeted quantitative proteomics. Nat Biotechnol. 2010;28(7):647–650. doi: 10.1038/nbt0710-647. [DOI] [PubMed] [Google Scholar]
- 18.Graumann J, Hubner NC, Kim JB, Ko K, Moser M, Kumar C, et al. Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7(4):672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Li QR, Xing XB, Chen TT, Li RX, Dai J, Sheng QH, et al. Large scale phosphoproteome profiles comprehensive features of mouse embryonic stem cells. Mol Cell Proteomics. 2011;10(4):M110 001750. doi: 10.1074/mcp.M110.001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirt SE, Adler AS, Gebala V, Weimann JM, Schaffer BE, Saddic LA, et al. G1 arrest and differentiation can occur independently of Rb family function. The Journal of cell biology. 2010;191(4):809–825. doi: 10.1083/jcb.201003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 23.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23(7):837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 25.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell stem cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell stem cell. 2010;6(4):382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 29.Wilkins MR, Sanchez JC, Gooley AA, Appel RD, HumpherySmith I, Hochstrasser DF, et al. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnology and Genetic Engineering Reviews. 1996;13:19–50. doi: 10.1080/02648725.1996.10647923. [DOI] [PubMed] [Google Scholar]
- 30.Zhou F, Sikorski TW, Ficarro SB, Webber JT, Marto JA. Online Nanoflow Reversed Phase-Strong Anion Exchange-Reversed Phase Liquid Chromatography-Tandem Mass Spectrometry Platform for Efficient and In-Depth Proteome Sequence Analysis of Complex Organisms. Anal Chem. 2011;83(18):6996–7005. doi: 10.1021/ac200639v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou F, Lu Y, Ficarro SB, Webber JT, Marto JA. Nanoflow low pressure high peak capacity single dimension LC-MS/MS platform for high-throughput, in-depth analysis of mammalian proteomes. Anal Chem. 2012;84(11):5133–5139. doi: 10.1021/ac2031404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ficarro SB, Zhang Y, Lu Y, Moghimi AR, Askenazi M, Hyatt E, et al. Improved Electrospray Ionization Efficiency Compensates for Diminished Chromatographic Resolution and Enables Proteomics Analysis of Tyrosine Signaling in Embryonic Stem Cells. Anal Chem. 2009;81(9):3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 33.Shen YF, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74(16):4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 34.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7(1):548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mertins P, Udeshi ND, Clauser KR, Mani D, Patel J, Ong Se, et al. iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol Cell Proteomics. 2012;11(6) doi: 10.1074/mcp.M111.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dephoure N, Gygi SP. Hyperplexing: a method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Science signaling. 2012;5(217):rs2. doi: 10.1126/scisignal.2002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Everley RA, Kunz RC, McAllister FE, Gygi SP. Increasing Throughput in Targeted Proteomics Assays: 54-Plex Quantitation in a Single Mass Spectrometry Run. Anal Chem. 2013;85(11):5340–5346. doi: 10.1021/ac400845e. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Zhu G, Dovichi NJ. Comparison of the LTQ-Orbitrap Velos and the Q-Exactive for proteomic analysis of 1-1000 ng RAW 264.7 cell lysate digests. Rapid Commun Mass Spectrom. 2013;27(1):157–162. doi: 10.1002/rcm.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding C, Jiang J, Wei J, Liu W, Zhang W, Liu M, et al. A Fast Workflow for Identification and Quantification of Proteomes. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.O112.025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Jong EP, Griffin TJ. Online nanoscale ERLIC-MS outperforms RPLC-MS for shotgun proteomics in complex mixtures. J Proteome Res. 2012;11(10):5059–5064. doi: 10.1021/pr300638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Q, Gu Y, Wu SL, Rejtar T, Karger BL. Two-dimensional strong cation exchange/porous layer open tubular/mass spectrometry for ultratrace proteomic analysis using a 10 microm id poly(styrene- divinylbenzene) porous layer open tubular column with an on-line triphasic trapping column. Electrophoresis. 2008;29(8):1604–1611. doi: 10.1002/elps.200700741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritorto MS, Cook K, Tyagi K, Pedrioli PG, Trost M. Hydrophilic Strong Anion Exchange (hSAX) Chromatography for Highly Orthogonal Peptide Separation of Complex Proteomes. J Proteome Res. 2013;12(6):2449–2457. doi: 10.1021/pr301011r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu G, Sun L, Yan X, Dovichi NJ. Single-shot proteomics using capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry with production of more than 1250 Escherichia coli peptide identifications in a 50 min separation. Anal Chem. 2013;85(5):2569–2573. doi: 10.1021/ac303750g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heemskerk AA, Busnel JM, Schoenmaker B, Derks RJ, Klychnikov O, Hensbergen PJ, et al. Ultra-low flow electrospray ionization-mass spectrometry for improved ionization efficiency in phosphoproteomics. Anal Chem. 2012;84(10):4552–4559. doi: 10.1021/ac300641x. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Fonslow BR, Wong CC, Nakorchevsky A, Yates JR., 3rd Improving the comprehensiveness and sensitivity of sheathless capillary electrophoresis-tandem mass spectrometry for proteomic analysis. Anal Chem. 2012;84(20):8505–8513. doi: 10.1021/ac301091m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols J, Smith A. Naive and primed pluripotent states. Cell stem cell. 2009;4(6):487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 48.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460(7251):118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 49.Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell stem cell. 2009;5(6):597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Huang C, Qin D. Role of Lef1 in sustaining self-renewal in mouse embryonic stem cells. J Genet Genomics. 2010;37(7):441–449. doi: 10.1016/S1673-8527(09)60063-1. [DOI] [PubMed] [Google Scholar]
- 51.Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, et al. Esrrb Is a Pivotal Target of the Gsk3/Tcf3 Axis Regulating Embryonic Stem Cell Self-Renewal. Cell stem cell. 2012;11(4):491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9(21):2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 53.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, et al. The POU factor Oct-6 is required for the progression of Schwann cell differentiation in peripheral nerves. Science. 1996;273(5274):507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 54.Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287(5451):319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- 55.Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287(5451):321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- 56.Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27(24):8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13(7):838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Heeft E, ten Hove GJ, Herberts CA, Meiring HD, van Els CA, de Jong AP. A microcapillary column switching HPLC-electrospray ionization MS system for the direct identification of peptides presented by major histocompatibility complex class I molecules. Anal Chem. 1998;70(18):3742–3751. doi: 10.1021/ac9801014. [DOI] [PubMed] [Google Scholar]
- 60.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74(13):3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.