SUMMARY
The “CTCF code” hypothesis posits that CTCF pleiotropic functions are driven by recognition of diverse sequences through combinatorial use of its 11 zinc fingers (ZFs). This model, however, is supported by in vitro binding studies of a limited number of sequences. To study CTCF multivalency in vivo, we define ZF binding requirements at ~50,000 genomic sites in primary lymphocytes. We find that CTCF reads sequence diversity through ZF clustering. ZFs 4–7 anchor CTCF to ~80% of targets containing the core motif. Nonconserved flanking sequences are recognized by ZFs 1–2 and ZFs 8–11 clusters, which also stabilize CTCF broadly. Alternatively, ZFs 9–11 associate with a second phylogenetically conserved upstream motif at ~15% of its sites. Individually, ZFs increase overall binding and chromatin residence time. Unexpectedly, we also uncovered a conserved downstream DNA motif that destabilizes CTCF occupancy. Thus, CTCF associates with a wide array of DNA modules via combinatorial clustering of its 11 ZFs.
INTRODUCTION
Chromatin three-dimensional structures have emerged as key drivers of transcription in eukaryotes (Francastel et al., 2000; Misteli, 2007). Local chromatin loops, for instance, facilitate the tethering of promoters with cognate regulatory elements that are often located hundreds of kilobases away (Fraser, 2006). Loops have also been shown to insulate transcription domains from each other to ensure independent function (Felsenfeld et al., 2004) and regulate imprinting of mammalian genes (Murrell et al., 2004).
To date, the best-characterized loop-forming factor in vertebrates is CTCF, an 11 ZF protein initially described as a negative regulator of Myc expression (Lobanenkov et al., 1990). Since its discovery, CTCF’s chromatin structural role has been established within the context of promoter-enhancer interactions, the recruitment of cohesin, X chromosome inactivation, the formation of chromatin barriers against heterochromatin, V(D)J recombination, and insulator function (Bell et al., 1999; Degner et al., 2009; Ebert et al., 2011; Fedoriw et al., 2004; Guo et al., 2011; Ling et al., 2006; Parelho et al., 2008; Wendt et al., 2008; Xu et al., 2007). Most recently, CTCF has been found to modulate messenger RNA (mRNA) splicing by controlling the rate of transcriptional elongation (Shukla et al., 2011). On the basis of the available evidence CTCF is regarded as an essential, pleiotropic genome organizer that links higher-order chromatin structure with complex biological phenomena (Phillips and Corces, 2009). Consistent with this view, CTCF is ubiquitously expressed, and its deletion in the germline is incompatible with cell viability (Heath et al., 2008; Ribeiro de Almeida et al., 2011; Splinter et al., 2006).
As measured by chromatin immunoprecipitation sequencing (ChIP-seq) in more than 20 different cell types, CTCF recognizes ~50,000 uncommonly long and remarkably divergent DNA sequences in humans and mice (Chen et al., 2008; Cuddapah et al., 2009; Kim et al., 2007; Wang et al., 2012; Yamane et al., 2011). Computational and biochemical analyses of these sites uncovered a central ~20 bp core (C) DNA motif critical for CTCF binding (Kim et al., 2007). In some instances, the motif was flanked by additional sequences of unknown function (Boyle et al., 2011; Kim et al., 2007; Rhee and Pugh, 2011; Schmidt et al., 2012; Xie et al., 2007). While a large fraction of binding sites is highly conserved across species (Schmidt et al., 2012), considerable nucleotide variability exists within CTCF core binding motif across the genome (Kim et al., 2007). Furthermore, a substantial number of sites lack the consensus motif altogether (Schmidt et al., 2012).
The association of CTCF with unique DNA sequences is thought to underlie, at least in part, its functional versatility (Filippova, 2008; Ohlsson et al., 2001). However, how CTCF recognizes its vast array of genomic targets is unclear. Under the current model, dubbed the “CTCF code,” CTCF associates with divergent sequences by using different combinations of its 11 ZFs (Ohlsson et al., 2010). The model was derived from in vitro gel shift assays, which showed that deletions or mutations targeting individual or a group of ZFs abrogate CTCF occupancy at a subset of DNA targets (Filippova et al., 1996, 2002; Renda et al., 2007). However, only a limited number of sites were tested by these studies, and the in vivo relevance of the CTCF code remains to be determined. To directly address these questions, we here define the binding behavior of CTCF ZF mutants at ~50,000 genomic targets in primary B lymphocytes.
RESULTS
Genome-wide Binding Profiles of CTCF Zinc Finger Mutants
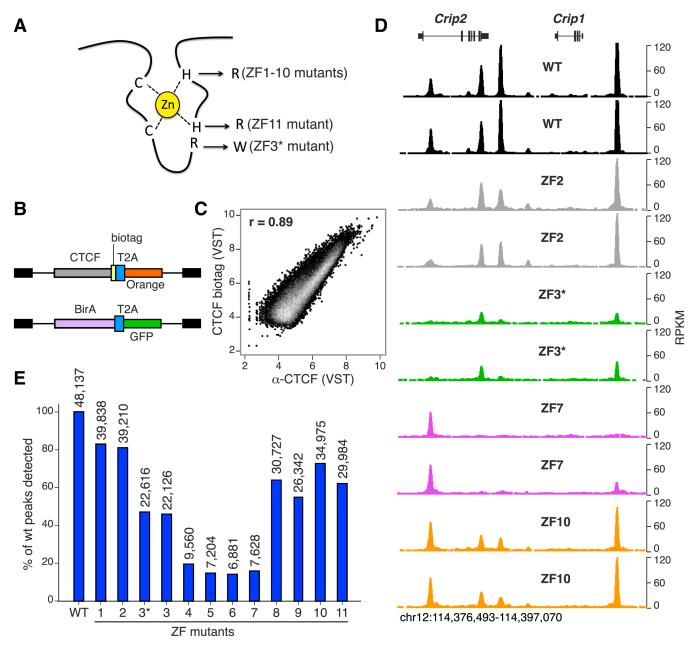
To gain insight into the CTCF code, we disrupted each of CTCF 11 ZFs in retroviral constructs by mutating key histidine residues that coordinate zinc binding (Wolfe et al., 2000). The mutations (H to R substitutions) replaced the first or second histidines within CTCF C2HC (ZF11) or C2H2 (ZFs 1–10) motifs, respectively (Figure 1A). As a control, we also engineered an additional mutation targeting CTCF ZF3 (ZF3*, R to W substitution, Filippova et al., 2002; Figure 1A). The resulting constructs (Figure 1B) were transduced into primary CD43− mouse B cells activated in the presence of lipopolysaccharide and interleukin 4 (LPS + IL-4). To determine genome-wide binding profiles of transduced CTCF, a short biotinylation substrate (biotag, Kim et al., 2009) was fused to CTCF C terminus in all constructs, and B cells were coinfected with retroviruses expressing E. coli biotin ligase BirA (Figure 1B). At 72 hr of culture, doubly infected lymphocytes (GFP+Orange+) were cell sorted, biotinylated proteins were chromatin immunoprecipitated using streptavidin beads, and crosslinked DNA was deep-sequenced. At least three biological replicates were processed for each sample.
Figure 1. Generating a Comprehensive Map of CTCF Multivalency.
(A) Schematic representation of a C2H2 ZF showing the key residues targeted in CTCF ZF mutants. With the exception of ZF3* (R to W), all mutants carry H to R substitutions at either the first (ZF11) or second (ZF1–10) histidines critical for zinc coordination.
(B) Retroviral constructs used to express in activated B cells biotagged CTCF together with Orange fluorescent protein (upper), and the biotinylating enzyme BirA followed by GFP (lower). In both cases, the T2A self-cleaving peptide separates the two proteins upon expression.
(C) Scatterplot comparing ChIP-seq signals from biotagged or endogenous CTCF, immunoprecipitated either with streptavidin beads or an anti-CTCF antibody. Overall correlation between the data sets was calculated via Pearson’s r. ChIP-seq values are represented in variance-stabilizing transformed (VST) format.
(D) CTCF WT or mutant binding profiles at the mouse Crip1/Crip2 locus. Two biological replicates for each sample are shown.
(E) Bar graph representing total ChIP-seq peaks obtained with transduced CTCF WT or ZF mutants (plotted as a percentage of WT). Numbers on top of each bar indicate the absolute number of WT CTCF peaks that passed the SWEMBL peak finder threshold in the different mutants. See also Figures S1, S2, S3, and S4.
To examine the specificity of in vivo CTCF biotinylation, we compared CTCF biotag to endogenous CTCF, immunoprecipitated from uninfected B cells using α-CTCF-specific antibodies (Yamane et al., 2011). We found a high degree of correlation between ectopic and endogenous CTCF (Pearson’s r = 0.89, Figure 1C), comparable to those obtained between biological replicates of wild-type (WT) or ZF mutant samples (Pearson’s r = 0.84–0.97, Figure S1). Further validating the biotag approach, transduced CTCF had no obvious effect on B cell viability, proliferation, or immunoglobulin class switch recombination (μ-γ1) induced by LPS + IL-4 stimulation (Figure S2). We conclude that biotinylated CTCF recapitulates the physiological recruitment of endogenous CTCF and that ectopic expression of CTCF WT or ZF mutants does not interfere with normal activation of primary B cells.
Similar to control samples, biological IP replicates from CTCF mutants were highly correlated (Pearson’s r = 0.71–0.92, Figures S1 and 1D). Notably, however, ZF deletions differentially affected CTCF recruitment at a subset of binding sites, as determined by visual inspection of ChIP-seq libraries using the UCSC genome browser (Figure 1D). This result is in good agreement with previous in vitro binding studies of CTCF mutants at a limited number of sites (Filippova et al., 1996, 2002; Renda et al., 2007). To quantify this phenomenon at a global scale, total peaks from ZF mutant replicates were merged and compared in pairwise fashion to WT controls. Consistent with previous estimates, a total of 48,156 WT CTCF peaks were identified in primary B cells using the SWEMBL peak finder software (Wilder, 2010). By contrast, ZF mutants exhibited in all cases substantially fewer peaks than control, from 39,838 (83% of WT) for ZF1 to 6,881 (14% of WT) for ZF6 (Figure 1E). Mutations affecting “central” ZFs (4, 5, 6, and 7), which have been proposed to mediate CTCF association with the core binding motif (Filippova et al., 1996; Ohlsson et al., 2010; Renda et al., 2007), resulted in the fewest number of peaks, whereas “peripheral” ZF mutants (1–2 and 8–11) were less affected (Figure 1E). Of note, ZF3* (R339W) and ZF3 (H345R) mutants displayed nearly equal number of peaks (22,616 versus 22,126, respectively, Figure 1E), and the two data sets were well correlated (Pearson’s r = 0.93, Figure S3). Thus, we obtained similar phenotypes by disrupting zinc coordination or ZF:DNA interactions for a given ZF. It is important to point out that reduced binding of CTCF mutants was not explained by potential differences in protein stability because transduced cells showed comparable protein levels between WT and ZF mutants (Figure S4). Taken together, the findings demonstrate that disruption of individual ZFs results in distinct and reproducible CTCF binding profiles.
ZF Mutations Differentially Affect CTCF Binding and Nuclear Mobility
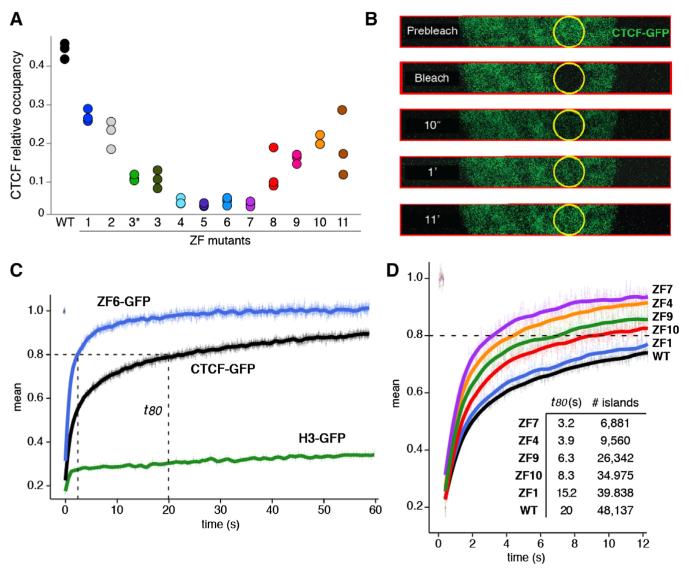
The decreased number of ChIP-seq peaks in mutant samples suggested that individual ZFs directly contribute to CTCF binding. To directly explore this idea, we measured read density at CTCF peaks in the entire data set. The analysis showed an overall decrease of CTCF binding in all mutants relative to control. Consistent with their low number of detected peaks (Figure 1E), the most affected mutants were ZFs 4–7, which exhibited on average a ~5-fold reduction in binding (Figure 2A). Notably, non-core mutants were progressively affected with increasing proximity to the core (Figure 2A), demonstrating that ZFs 3 or 8 contribute more to CTCF binding than ZFs 1 or 11. On the basis of these findings, we conclude that ZF mutations directly impact CTCF binding and that for peripheral ZFs this effect is proportional to their physical distance from the core motif.
Figure 2. ZF Mutations Affect CTCF Binding and Chromatin Residence Time In Vivo.
(A) Relative CTCF occupancy (fraction of reads at binding sites) for the WT or ZF mutant CTCF. Presented are all three biological replicates for each sample.
(B) Nuclear dynamics of the WT or ZF mutant CTCF tagged with green fluorescent protein (GFP) as measured by fluorescent recovery after photobleaching (FRAP). CTCF constructs were transiently expressed in the 3134 mouse cell line. For fast data collection during FRAP, images were collected only in a strip encompassing the circular bleach spot area. Selected time points (t) are shown.
(C) Fluorescence recovery of CTCF-GFP, ZF6-GFP, and histone H3-GFP control following irreversible photobleaching. t80 represents the time (in seconds) when 80% of the original fluorescence at the bleached spot recovers. Data represent the mean values ± SEM, n = 15–30 cells.
(D) Comparison of the FRAP curves obtained with CTCF WT and ZF mutants. The total number of CTCF ChIP-seq peaks (from Figure 1E) and t80 values are provided as a table. Data represent the mean values ± SEM, n = 15–30 cells.
We reasoned that a reduction in CTCF binding might affect the overall dynamics of CTCF:chromatin interactions. To test this possibility, we expressed CTCF-GFP fusion proteins in mammary 3134 or HeLa cells and carried out fluorescence recovery after photobleaching (FRAP, White and Stelzer, 1999; Figure 2B). The time for complete CTCF recovery was ~11 min (Figure 2C), making it considerably slower than the recoveries of most transcription factors, which exhibit complete recoveries in ~1 min (McNally et al., 2000). On the other hand, CTCF recoveries were still markedly faster than those of core histones (Figure 2C), which require at least several hours for complete recovery. These data therefore suggest that CTCF:chromatin associations are stronger than for most transcription factors, but CTCF still manifests significant exchange with chromatin in living cells. To further test whether these recoveries reflected chromatin interactions, we explored the kinetics of ZF6, which displays seven times fewer peaks than control (Figure 1E). In stark contrast to WT, the initial recovery phase of ZF6 reached 80% 3 s postbleaching, and the overall signal plateaued at ~15 s, or 44 times faster than WT (Figure 2C). These observations indicate that the ZF6 mutation markedly increases nuclear CTCF mobility, changing it to a timescale closer to that seen for most transcription factors. Consistent with these data, all ZF mutants displayed faster FRAP recovery than WT (Figure 2D; data not shown). Importantly, CTCF mobility was proportional to the number of peaks obtained with each particular mutant. For instance, ZF1 was both the least mobile (ts23 80 = 15.2 s, Figure 2D) and the least affected mutant in terms of overall binding (total peaks = 39,838, Figure 1E). On the opposite end of the spectrum, ZF7 recovered to 80% fluorescence in 3.2 s (Figure 2D) and displayed 6,881 peaks across the genome (Figure 1E). Thus, the real-time kinetics of ZF mutants correlates with their genome-wide occupancy as measured by deep sequencing. Based on these data, we conclude that CTCF nuclear dynamics are slower than those of most transcription factors, and that mutations affecting individual ZFs increase CTCF mobility in a manner proportional to their interference with binding.
Clustering of CTCF ZFs
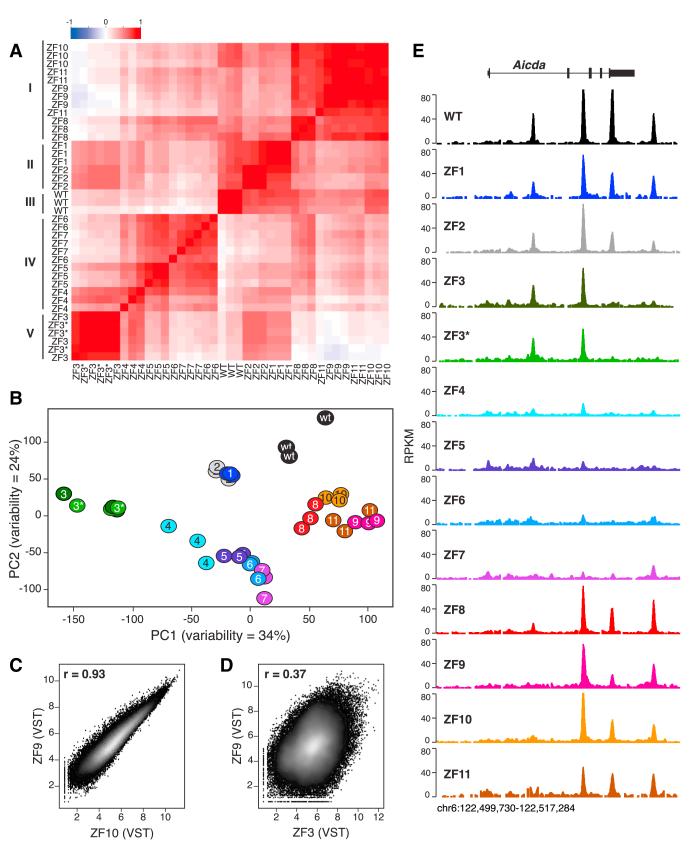
In vitro characterization of CTCF mutants isolated from human tumors (Filippova et al., 1996, 2002) suggests that ZFs can contribute to CTCF binding as independent units. At the same time, cocrystal structures of C2H2 ZF proteins bound to DNA reveal that adjacent ZFs interact cooperatively with DNA bases in an overlapping pattern of contacts (Wolfe et al., 2000). This raises the possibility that contiguous ZFs may cluster into DNA binding subdomains. To directly test this idea, we applied a Pearson’s correlation matrix (Figure 3A) and a principal component analysis (Figure 3B) to variance-stabilized ChIP-seq data. Notably, the two analyses were in good agreement in that they identified five distinct clusters in the data set: (1) ZF8/9/10/11, (2) ZF1/2, (3) WT, (4) ZF4/5/6/7, and (5) ZF3/ZF3* (Figures 3A and 3B). Binding profiles of ZF mutants within a given cluster were highly correlated with an average Pearson’s r of 0.86 (Figure S3). As an example, the correlation between ZF9 and ZF10 profiles (r = 0.93) was comparable to that obtained between biological replicates of the same mutants (compare Figures 3C to S1). Additional examples are provided in Figure S3. Conversely, genome-wide occupancy between members of different clusters was less correlated (Figure 3D), with an average Pearson’s r of 0.65 (p < 0.0001, Mann-Whitney test, Figure S3). Inter- and intracluster correlations are exemplified in Figure 3E for the Aicda locus in mouse chromosome 6. The data are thus consistent with a model where contiguous CTCF ZFs (i.e., 1–2, 4–7, and 8–11) function as discrete DNA binding subdomains.
Figure 3. CTCF ZFs Cluster into DNA Binding Subdomains.
(A) Pearson’s correlation matrix analysis of variance-stabilized CTCF ChIP-seq data at 14,804 sites that showed significant changes in CTCF binding. Five distinct clusters (ZF8–11, ZF1–2, WT, ZF4-7, and ZF3–3*) are highlighted. Scale represents Pearson’s r (from ~1 to 1).
(B) Principal component analysis of CTCF ChIP-seq data sets.
(C) Scatterplot comparison of variance-stabilizing transformed (VST) ChIP-seq data between ZF9 and ZF10 CTCF mutants. Correlation is provided via Pearson’s coefficient r.
(D) Same comparison as (C) between ZF9 and ZF3.
(E) CTCF WT and mutant binding profiles at mouse Aicda locus in chromosome 6. ChIP-seq samples were normalized as RPKM. See also Figure S3.
DNA Sequences Flanking the Core Motif Modulate CTCF Binding
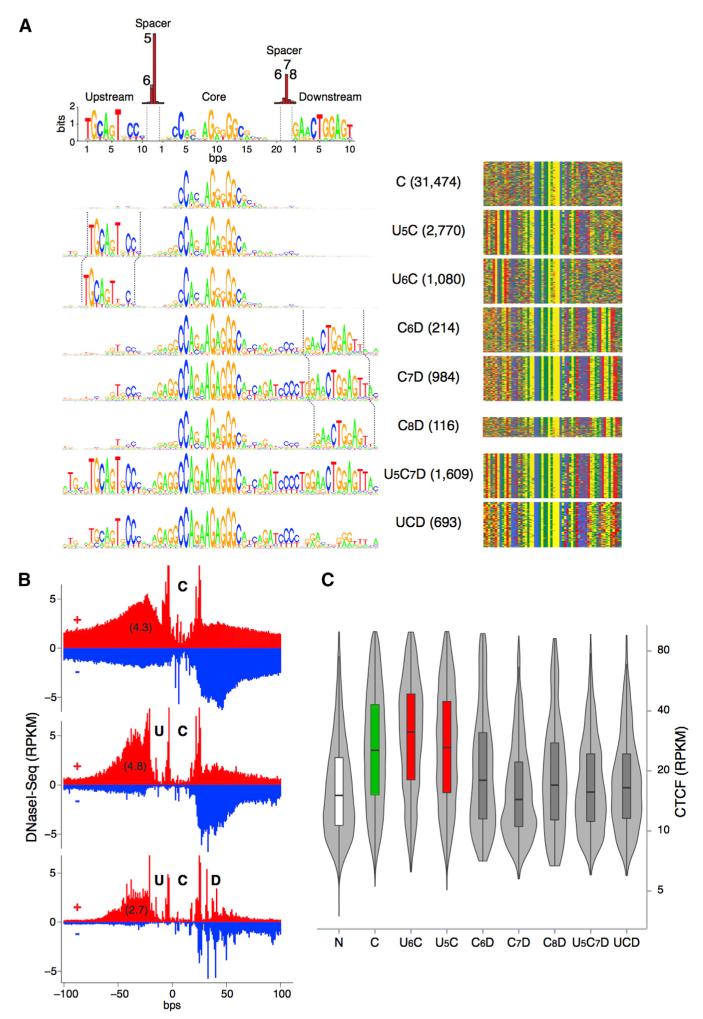
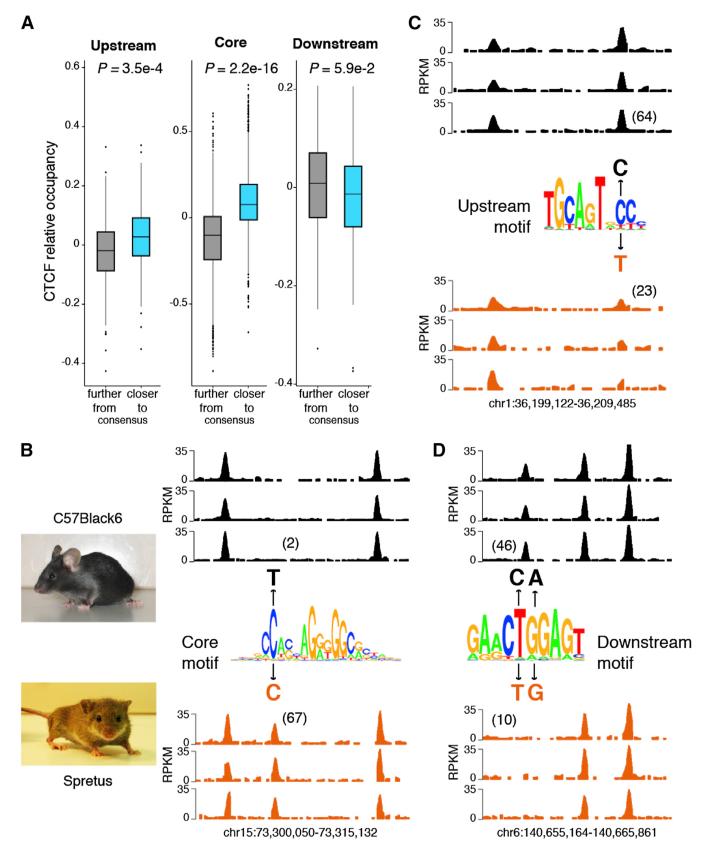
Peripheral ZF clusters might recognize specific DNA binding motifs. To explore this possibility, we revisited CTCF’s DNA recognition sequence by applying MEME motif discovery to our ChIP-seq peaks (Machanick and Bailey, 2011). Consistent with recent genome-wide studies (Boyle et al., 2011; Kim et al., 2007; Rhee and Pugh, 2011; Schmidt et al., 2012), the analysis revealed CTCF’s 20 bp core (C) motif present in 80% (38,940) of all peaks (Figure 4A). Also in agreement with previous work, 13% (6,152) of the sites displayed a 10 bp upstream (U) motif separated from the core sequence by 5 or 6 bp (Figure 4A). Notably, the analysis also uncovered in 8% (3,616) of all peaks a 10 bp motif 6–8 bases downstream (D) of the central core (Figure 4A). In approximately one-third of these sites (1,314), the D motif was associated with the core consensus sequence only, whereas in the 2,302 sites remaining (5% of the total) it was associated both with the core and upstream motifs (Figure 4A). Based on the presence or absence of the three DNA motifs and the spacer sequences separating them, CTCF peaks were classified into eight distinct groups: C, U5C, U6C, C6D, C7D, C8D, U5C7D, and UCD (Figure 4A).
Figure 4. DNA Motifs Associated with CTCF Binding Sites.
(A) Left, analysis of CTCF binding domain using MEME discovery software, which identifies three distinct motifs: upstream, middle core, and a downstream DNA conserved element. Based on the presence or absence of these motifs and the precise base pair distance separating them (top red bars) eight distinct groups are characterized. Absolute number of CTCF peaks for each group are provided in parentheses. Right, color chart representation of 60 bp of DNA sequence comprising the CTCF binding domain centered at the core motif midpoint. Red, green, yellow, and blue represent T, A, G, and C bases, respectively.
(B) Cumulative high-resolution footprinting of C, UC, and UCD CTCF binding sites. Upper (+) and lower (−) strand-specific DNase I-seq signals are represented in red and blue respectively. Cut counts per nucleotide were normalized to a total library size of 1 million reads and multiplied by 1,000 to reflect reads per kilobase per million (RPKM).
(C) Violin plot showing average CTCF signal (RPKM) at the eight CTCF binding groups identified in (A).
To confirm protein interaction at CTCF core and flanking DNA motifs, we applied high-resolution DNase I-seq footprinting (>500 million aligned reads) to the ChIP-seq data as described (Boyle et al., 2011). We found core and upstream motifs to be markedly protected against DNase I digestion and separated by sharp hypersensitive boundaries (Figure 4B). In the presence of the D DNA motif, downstream sequences were characterized by three to four smaller footprints (Figure 4B), indicative of protein binding in vivo. Sites carrying the upstream motif (particularly U6C combinations) displayed on average higher CTCF occupancy than those associated with the core consensus sequence only (Figure 4C). In marked contrast, CTCF binding was consistently reduced in the presence of the downstream motif, irrespective of whether the upstream motif was present or not (Figure 4C). In particular, the average CTCF binding density at D sites were similar to that obtained for sites recruiting CTCF but lacking the C motif (N sites, Figure 4C). Footprinting experiments confirmed these results in that they showed increased (4.8 RPKM) and decreased (2.7 RPKM) DNase I digestion in the presence of upstream and downstream motifs, respectively (Figure 4B).
The above findings argue that DNA motifs flanking the core sequence modulate CTCF binding by enhancing (U motif) or reducing (D motif) its affinity for DNA. To directly test this idea, we explored whether nucleotide changes at the consensus sequence of flanking motifs impact CTCF occupancy in vivo. To this end, we compared CTCF binding in activated B cells from Mus musculus (C57BL/6) and Mus Spretus (Spretus), which differ from each other at millions of loci (Keane et al., 2011). We identified a total of 5,192 single nucleotide variants (SNVs) that fall within one of three DNA motifs at CTCF targets. Consistent with previous findings (Maurano et al., 2012), the vast majority of SNVs (4,661, or 89%) mapped to the C motif, whereas 310 and 221 overlapped with U and D motifs, respectively. To simplify the analysis, indels, structural variants, or SNVs affecting more than one motif per CTCF binding site were not considered. The potential effects of SNVs on CTCF recruitment were examined by comparing CTCF occupancy to the motif position weight matrix (PWM) score. PWM scores can be used to calculate the contribution of each nucleotide to the protein-DNA interaction energy at a given site (Wasserman and Sandelin, 2004). For the core motif, we found a positive correlation between these parameters in that the most energetically favorable sites displayed higher CTCF occupancy than sites where SNVs decreased the overall PWM score (p < e-15, Figure 5A, center plot). For instance, a C to T variant in chromosome 15 of C57BL/6, which falls on a high information position within the core motif, abolishes CTCF occupancy in that strain relative to Spretus (2 RPKM in C57BL/6 versus 67 RPKM in Spretus, Figure 5B). A similar correlation was observed between CTCF occupancy and PWM scores calculated for the upstream motif (p = 3.5e-4, Figure 5A, left plot). As an example, Figure 5C shows that a C to T substitution at position 8 within the U motif results in a ~3-fold reduction (64 versus 23 average RPKM) in CTCF binding ~in Spretus vis-à-vis C57BL/6. In contrast, the D motif showed an inverse relationship, in that CTCF binding was generally reduced the closer the motif sequence was to the consensus, although this tendency did not reach significance likely due to fewer SNVs targeting the D motif (p = 5.9e-2, Figure 5A, right graph). As an example, Figure 5D shows no detectable CTCF at a site carrying an optimal D motif in Spretus, whereas CTCF is present in C57BL/6 B cells where the motif is mutated away from the consensus at positions 5 and 6. Additional examples for all three motifs are provided in Figure S5. The results are thus consistent with the notion that SNVs affect CTCF occupancy by modulating CTCF-DNA interactions. Furthermore, the findings support the proposal that the upstream and downstream motifs up- and downmodulate CTCF binding in vivo.
Figure 5. DNA Motifs Flanking the Core Sequence Modulate CTCF Occupancy.
(A) Spretus CTCF binding sites carrying a single SNP were classified based on whether the nucleotide variation decreased (gray box) or increased (blue box) the motif score relative to C57Black7 (i.e., whether the sequence approached or moved away from the consensus). Only binding sites carrying a single SNP at either U (310 sites), C (4,631), or D (220) motifs were considered. P values were calculated using a two-sided Wilcoxon rank sum test.
(B–D) Examples of C, U, and D sites where SNPs differentially affect CTCF binding in C57BL/6 or Spretus mouse strains. Sequence logos are as described in Figure 4A. Numbers in parentheses represent the RPKM average value at the given CTCF binding site for the three biological replicates.
See also Figure S5.
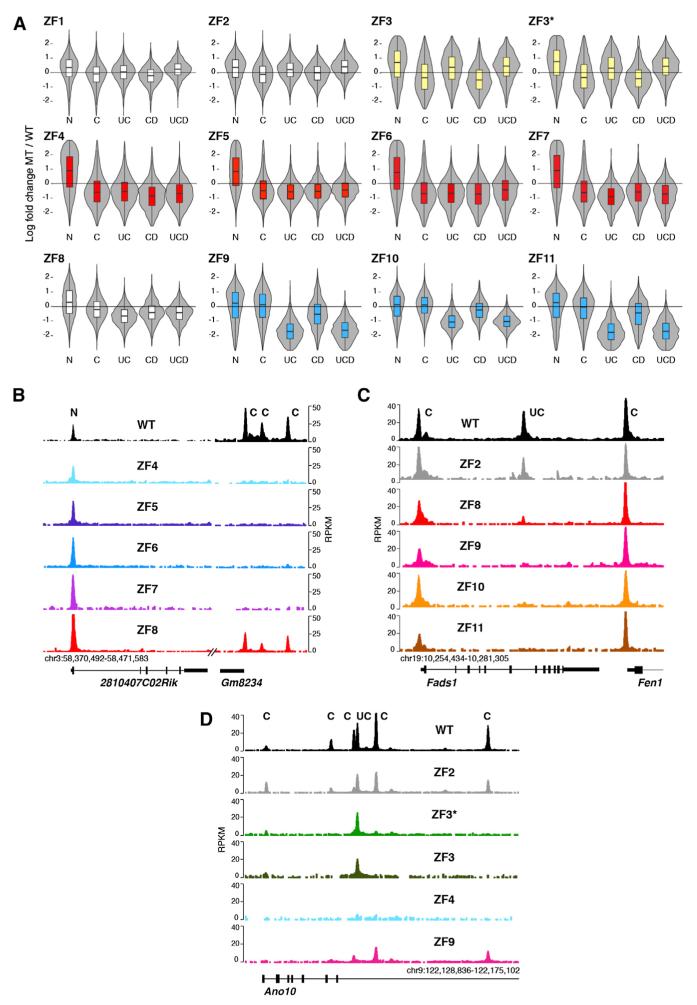
Recognition of CTCF Binding Motifs by ZF Clusters
We next sought to address two related questions: whether CTCF associates with C, U, or D motifs in vivo and whether these associations are mediated by specific ZF clusters. To this end, we sorted CTCF ChIP-seq peaks as N, C, UC, DC, and UCD based on the classifications shown in Figure 4. For each group, we calculated the ZF mutant to WT density ratio, and the data were plotted as moderated log ratios, where 0 represents no relative change. The analysis revealed three important features of CTCF multivalency in vivo. First, it provided direct evidence that ZFs 4–7 as a group recognize the core DNA binding motif, as all four mutants displayed impaired CTCF binding to sites carrying the C motif irrespective of the presence or absence of peripheral motifs (Figure 6A, highlighted in red). Notably, binding of ZF4–7 mutants to genomic sites lacking the core motif (N sites) was less affected (Figure 6A). These features are represented in Figure 6B, which provides examples of N (binding) and C (no binding) CTCF sites at the Gm8234 locus in mouse chromosome 3. For additional examples, see Figure S6A. The findings are thus consistent with the notion that the ZF4–7 cluster is required to recognize the C motif but dispensable for CTCF deposition at sites lacking the motif.
Figure 6. CTCF Uses Different ZF Clusters to Recognize U and C DNA Motifs.
(A) Violin plots showing effects of ZF mutations on CTCF binding sites based on N, C, UC, and UCD classifications described in Figure 4. Data are graphed as the log fold change of mutant to WT ratio. Data were adjusted for global decreases in CTCF binding. Three distinct clusters were highlighted either in yellow (ZF3/3*), red (ZF4–7), or blue (ZF9–11).
(B) Gm8234 mouse locus showing lack of core ZF mutant occupancy at C sites but normal binding to N sites.
(C) Fads1/Fen1 mouse locus depicts defective binding of ZF9–11 mutants to U-containing sites while displaying WT recruitment to sites lacking the motif. ChIP-seq values are plotted as RPKM.
(D) Ano10 locus showing lack of ZF3/3* recruitment to C sites but normal occupancy at binding sites associated with the upstream (U) motif. See also Figure S6.
A second key finding was that mutations targeting ZFs 9, 10, or 11 preferentially affect CTCF recruitment to the 6,152 genomic sites carrying the upstream consensus motif (Figure 6A, highlighted in blue). Unexpectedly, this effect was not obvious for ZF8 mutants (Figure 6A), which, as previously shown, display binding profiles analogous to those of ZF9–11 mutants when all 48,137 CTCF sites are considered (Figure 3). The Fads1-Fen1 locus provides a good example of these profiles by showing normal CTCF recruitment to C sites at Fads1 and Fen1 promoters but defective association of ZF9–11 mutants with the UC site within Fads1 intron 6 (Figure 6C). In like manner, the previously characterized CTCF site downstream of Myc’s P2 promoter (Filippova et al., 1996) did not recruit ZF9–11 mutants (Figure S6B), consistent with the observation that these fingers are required for CTCF binding to this site in gel shift assays (Filippova et al., 1996). The data thus indicate that CTCF interacts with the upstream DNA motif via ZFs 9, 10, and 11 but with little or no contribution from ZF8. This view is consistent with the prediction that a polydactyl protein would require only three ZFs to associate with a 10 bp DNA binding sequence such as the U motif (Persikov and Singh, 2011; Wolfe et al., 2000).
Finally, the analysis showed that, analogous to ZF4–7 mutants, ZF3 and ZF3* exhibit lower occupancy for CTCF sites carrying the core consensus sequence (Figure 6A). Notable exceptions, however, were sites associated with the U motif, whose presence appears to compensate for the loss of ZF3 (Figure 6A). The Ano10 and Slc38a10 loci are illustrative of this behavior (Figures 6D and S6C). We conclude that ZF3 is not required for CTCF binding in vivo in the presence of the U motif, but becomes essential in its absence.
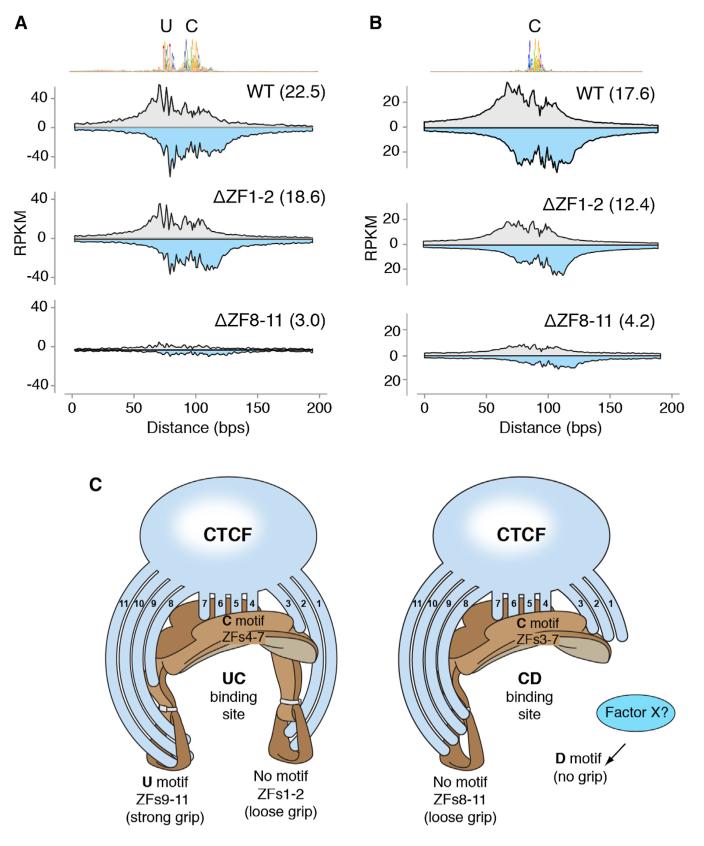
Peripheral ZFs Provide Binding Stability in the Absence of Flanking DNA Motifs
The above results agree with the proposed inverted orientation of CTCF on its binding site (Renda et al., 2007), where CTCF is expected to interact with 5′ most sequences (e.g., U motif) via fingers downstream of ZF7. On the other hand, the analysis provided no obvious link between ZFs 1 and 2 and the downstream DNA motif (Figure 6A). Also, as discussed above, there is little or no ZF8 contribution to U motif binding, even though at the vast majority of CTCF target sites ZF8 recapitulates the binding pattern of ZF9–11 mutants (Figure 4). We thus entertained the possibility that ZF1–2 and ZF8–11 clusters might associate with nonconserved core flanking sequences. To directly address this question, we generated CTCF mutants carrying deletions (Δ) in ZFs 1–2 and 8–11 and determined their binding profiles via ChIP-exo. This technique increases the spatial resolution and quantitative accuracy of ChIP-seq by incorporating an exonuclease step that reduces extraneous DNA contamination (Rhee and Pugh, 2011). In agreement with previous findings (Rhee and Pugh, 2011), WT CTCF displayed multiple exonuclease-derived borders, coincident with the location of the upstream and central core motifs (Figure 7A). We found that while the ΔZF1–2 mutant recapitulates WT profiles, binding at UC sites was slightly reduced relative to control (22.5 versus 18.6 average RPKM, Figure 7A), indicating that these ZFs contribute to CTCF binding in the absence of defined DNA motifs downstream of C. As expected, we detected little or no CTCF binding in DZF8–11 mutants when the upstream domain was present (3.0 RPKM, Figure 7A). At sites carrying only the C motif, CTCF recruitment was also affected in ΔZF1–2 and most markedly in DZF8–11 mutants (Figure 7B). These findings are thus consistent with the proposition that both ZF1–2 and ZF8–11 clusters help stabilize CTCF occupancy in the apparent absence of DNA binding motifs flanking the C domain. The fact the CTCF binding is reduced in ZF8 relative to WT indicates that ZF8 on its own stabilizes CTCF to C sites (Figure S6D).
Figure 7. Contribution of ZF Clusters in the Absence of Flanking Motifs.
(A) ChIP-exo raw sequencing tags distributed around 3,850 UC CTCF targets centered by the core motif midpoint. Gray and light blue indicate forward and reverse strand tags respectively. Samples were WT or CTCF carrying deletions (Δ) in ZFs 1–2 or 8–11. Values were normalized as RPKM and numbers in parenthesis represent the average of total tags per group per genomic site.
(B) Same as (A) but for sites carrying only the consensus core motif.
(C) “Saddle” model of CTCF multivalency. Left schematics, CTCF associates strongly to UC sites by interacting with the consensus core motif (represented by the seat of the saddle) via ZFs 4–7. The upstream motif (left stirrup) is recognized by the ZF9–11 cluster, which stabilizes CTCF overall binding (strong grip). To a lesser extent, ZFs 1–2 contribute to binding by associating with DNA sequences lacking a consensus motif (loose grip) downstream of the core. In the presence of the D motif, such as at CD sites (right schematics) either the ZF1–2 cluster loses affinity for DNA or an unknown factor × outcompetes it for binding (no grip). In the absence of U, ZF8 clusters with ZFs 9–11 and stabilizes CTCF binding probably by associating with random DNA sequences 5′ of the core motif (loose grip). Finally, the contribution of ZF3 to CTCF binding becomes essential at sites lacking U sequences. Figure design by Ethan Tyler, from the NIH Office of Medical Arts.
DISCUSSION
CTCF has been described as a multivalent protein on the basis that it can bind diverse DNA sequences presumably by using different combinations of ZFs (Ohlsson et al., 2010). This model, however, relies on in vitro binding studies of a limited number of genomic sites, including CTCF targets at the myc promoter (Filippova et al., 1996), the Igf2/H19 imprinted locus (Bell and Felsenfeld, 2000; Renda et al., 2007), the human APP promoter (Quitschke et al., 2000), and the β-globin insulator (Filippova et al., 2002). By expressing ZF mutants in primary lymphocytes, our studies now reveal the ZF requirements for CTCF recruitment to ~50,000 targets. This high-resolution multivalency map conceptually redefines the CTCF code hypothesis by showing that CTCF associates with its diverse array of sequences via ZF clustering. Rather than using arbitrary ZF combinations, the data are consistent with a model where CTCF functionally groups contiguous ZFs into distinct binding subdomains, including ZFs 1–2, ZFs 3–7, ZFs 4–7, ZFs 8–11, and ZFs 9–11. As discussed in detail below, which ZF clusters are important for binding a given site depends on the DNA modules present.
Similar to other cell types (Kim et al., 2007), about 80% of CTCF genomic targets identified in mouse B cells carry the consensus core motif. In gel shift assays, the presence of this motif is sufficient to promote CTCF binding to DNA probes (Holohan et al., 2007; Kim et al., 2007; Renda et al., 2007; Rhee and Pugh, 2011; Schmidt et al., 2012; Xie et al., 2007). In vivo, we have found that recognition of this motif requires ZFs 4-7. The functional clustering of these ZFs is most clearly illustrated by the fact that mutations targeting any one of them preferentially affect CTCF recruitment to C sites, whereas binding to N sites lacking the consensus sequence is less affected. Crystallographic studies of other C2H2 “polydactyl” proteins provide a rationale to CTCF ZF clustering in that adjacent ZFs are predicted to recognize four base pair binding domains that overlap by one nucleotide (Persikov and Singh, 2011; Wolfe et al., 2000). Under this model, CTCF is expected to contact key nucleotides at core or flanking DNA motifs with more than one ZF. Although direct proof of this idea awaits crystallographic characterization of the CTCF-DNA interface, it agrees well with the high degree of correlation obtained between binding profiles of contiguous ZF mutants.
How CTCF associates with domains lacking the core motif, however, is unclear. One possibility is that CTCF recognizes sequences at such sites that only remotely resemble the C motif and that thus fall below the detection limit of the motif discovery algorithm (Machanick and Bailey, 2011). Alternatively, CTCF may associate with N sites indirectly by interacting with prebound factors, perhaps via CTCF N- or C-terminal domains (Ohlsson et al., 2010). We favor this hypothesis based on the fact that mutations targeting core ZFs have little or no effect on CTCF recruitment to N sites. In addition, the hypothesis fits well with the proposed tethering role of CTCF in the establishment of protein-protein interactions and nuclear architecture in general (Handoko et al., 2011; Phillips and Corces, 2009). One caveat of our analysis is that it cannot distinguish direct from indirect CTCF associations; thus, additional techniques will need to be applied to fully answer this question.
In addition to core ZFs, we have shown that peripheral ZFs clearly modulate CTCF binding in vivo. Mutations disrupting zinc coordination at ZFs 1–3 and 8–11 decrease both CTCF overall chromatin residence time and the total number of ChIP-seq peaks. In addition, we have found that the precise contribution of peripheral ZFs to CTCF occupancy wanes proportionally to the distance that separates them from the core motif (Figure 2A). At least for ZFs 3 and 8, this phenomenon might be attributed to partial recognition of core nucleotides. This would be consistent with the predicted model of DNA binding by “polydactyl” proteins as alluded above. At the same time, the finding underscores the central role of the core motif in securing CTCF to DNA and suggests that peripheral ZFs play a rather stabilizing role. Figure 7C illustrates these functions by likening CTCF binding sites to a saddle, where the saddle seat represents the core motif and the stirrups, which provide overall balance, symbolizing flanking DNA sequences (Figure 7C, left schematics).
Similar to core ZFs, peripheral ZFs associate with flanking DNA as functional clusters. The most notable example being ZFs 9–11, which recognize a phylogenetically conserved DNA motif located 5–6 bp upstream of the core sequence (Boyle et al., 2011; Rhee and Pugh, 2011; Schmidt et al., 2012). Although only present at a fraction of CTFC target sites (~15%), this element is associated with a well-defined DNase I footprint and enhances CTCF binding. We provide direct proof of this by showing that SNVs decreasing the PWM score of the U motif downmodulate CTCF binding in Spretus B cells relative to C57BL/6. In the absence of a recognizable consensus sequence upstream of C, our results indicate CTCF still associates with DNA via ZFs 8–11 (Figure 7C, right schematics). This binding is likely weak considering that protection from DNase I attack is not complete at upstream sequences in core-only sites (Figure 4B upper graph). Even so, ChIP-exo analysis clearly indicates that the contribution of ZF8–11 to CTCF binding is substantial. A similar argument can be made for the ZF1–2 cluster, which is expected to interact with DNA sequences 3′ of the core motif (Figure 7C, right schematics). Finally, the role of ZF3 is intriguing. On the one hand, ZF3 recapitulates the binding spectrum of core ZF mutants at C sites. On the other hand, in the presence of the U motif ZF3 contribution to CTCF binding seems redundant. Considering the proposed geometry of ZF-DNA interface discussed above, ZF3 would be expected to contact one or a few key residues at the 3′ end of the core motif. It is important to point out that this contact is likely to occur independently of the presence or absence of U (Figure 7C).
CTCF binding profiles between different tissues exhibit substantial concordance (Cuddapah et al., 2009; Kim et al., 2007). For instance, up to 70% of binding sites are common between any two given cell types (Wang et al., 2012). Where variability has been described, it appears to result from differential DNA methylation, particularly at two key CpGs within CTCF core binding motif (Wang et al., 2012). DNA methylation, however, cannot account for tissue-specific variability, as marked changes in CTCF deposition have been described at sites where methylation profiles are constant during development. At least some of these changes can be explained by neighboring DNA binding factors that may directly modulate CTCF affinity for chromatin, or maintain CTCF binding motifs in an unmethylated state during ontogeny (Weth and Renkawitz, 2011). Several DNA binding proteins have been proposed to modulate CTCF recruitment in vivo, including YY1, SMADs, TAF3, Oct4, VEZF1, and cohesin (Donohoe et al., 2007, 2009; Liu et al., 2011; Parelho et al., 2008; Rubio et al., 2008; Wendt et al., 2008). By associating with flanking sequences, these factors might help stabilize or even destabilize CTCF affinity for chromatin. Destabilization might be the predominant outcome when neighboring DNA elements directly overlap with the CTCF footprint. In this context, our studies have uncovered a conserved downstream DNA motif (6–8 bp from the core) that negatively impacts CTCF recruitment. Supporting this claim, our studies show that CTCF recruitment diminishes the closer D is to the consensus. In addition, the PWM score of C is higher in the presence of D (Figure S5D), suggesting that there is evolutionary pressure for the C motif to approach the consensus when CTCF binding sites include D. Presumably, this feature might in part compensate for the inhibitory activity of the D sequence itself or a putative factor recruited therein. The prospect that this motif truly recruits a CTCF-competing factor(s) is intriguing, as it would provide a means to regulate CTCF activity in a cell-type-specific manner (i.e., by controlling expression of the competitor[s]).
In summary, our studies support a model where the extent of CTCF occupancy depends on intrinsic ZF clusters that recognize specific DNA modules and extrinsic factors that either stabilize or destabilize binding. This strategy likely underlies how CTCF executes diverse functions in different contexts and cell types.
EXPERIMENTAL PROCEDURES
Fluorescence Recovery after Photobleaching
3134 cells were transiently transfected by electroporation with GFP-tagged mouse CTCF (or mutants ZF1–11) and grown overnight in coverglass chambers (Lab-Tec) at a density of 2 × 105 in phenol red-free DMEM containing 10% fetal bovine serum (HyClone). Fluorescence recovery after photobleaching (FRAP) experiments were carried out on a Zeiss 510 confocal microscope with a 100×/1.3 numerical aperture oil immersion objective, and the cells were kept at 37°C using an air stream stage incubator (Nevtek, Williamsville, VA). Bleaching was performed with a circular spot using the 488 and 514 nm lines from a 45 mW argon laser operating at 96% laser power. A single iteration was used for the bleach pulse, and fluorescence recovery was monitored at low laser intensity (0.5% for a 45 mW laser) at 58.6 ms intervals. To determine the complete recovery of the WT CTCF-GFP, the FRAP measurements were extended to over 11 min and for the last 10 min the fluorescence recovery was monitored at 559 ms intervals. Data from at least three independent experiments were collected and used to generate corresponding average FRAP curves (±SEM). Curves were normalized as previously described (Stavreva and McNally, 2004).
B Cell Activation, Transduction, and Sorting
B lymphocytes were isolated from spleens of wild-type C57BL6 male mice by immunomagnetic depletion using CD43 MACS beads (Miltenyi Biotec). Purified cells were cultured at 0.3 × 106 cells per ml in B cell media (Advanced RPMI 1640, 10% FCS, 1 × antibiotic-antimycotic, 1% glutamine, 50 μM 2-β-mercaptoethanol, and 10 mM HEPES). Cells were preactivated overnight in the presence of 0.5 mg/ml of aCD180 (RP105) antibody (RP/14, BD PharMingen). At 0, 8, and 24 hr, cells were transduced with Vector1 (pMy-CTCFbiotag-T2A-mOrange) and Vector2 (pMy-BirA-T2A-eGFP) by centrifugation for 90 s at 2,500 rpm, at 32°C. B cell media was supplemented with 50 mg/ml of LPS (Escherichia coli 0111:B4; Sigma-Aldrich), 2.5 ng/ml of IL-4 (Invitrogen), and 0.5 mg/ml of aCD180. At 32 hr, cells were diluted to 0.1 3 106 cells per ml. Seventy-two hours after first infection, B cells were harvested and GFP/mOrange double positives were cell sorted using a BD FACSAria III (Becton Dickinson). The percentage of double-positive cells was 30%–40%. All animal experiments were performed according to the National Institutes of Health guidelines for laboratory animals and were approved by the Scientific Committee of the NIAMS Animal Facilities.
ChIP-Seq
Sorted cells (10–20 × 106 cells) were crosslinked for 10 s at 37°C with 1% (v/v) formaldehyde, followed by quenching with 0.125 M glycine (final concentration). Crosslinked cell samples were then sonicated with a Covaris sonicator to obtain DNA fragments 200–300 bp in length. Biotinylated samples were incubated with 40 μl of Dynabeads M-280 Streptavidin Beads (Invitrogen) or 5 μg of anti-CTCF antibody (07-729, Millipore) overnight at 4°C in RIPA buffer (10 mM Tris [pH 7.6], 1 mM EDTA, 0.1% [w/v] SDS, 0.1% [w/v] sodium deoxycholate and 1% [v/v] Triton X-100). Beads were washed twice with Wash buffer 1 (2% [v/v] SDS), once with Wash buffer 2 (0.1% [v/v] deoxycholate, 1% [v/v], once with Wash buffer 3 (250 mM LiCl, 0.5% [v/v] NP-40, 0.5% [v/v] deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.1]), and then twice with TE buffer (10 mM Tris-HCl [pH 7.5] and 1 mM EDTA). ChIP DNA was then extracted for 4 hr at 65°C in Tris-EDTA buffer with 0.3% (w/v) SDS and proteinase K (1 mg/ml). Samples were processed for microsequencing and run on a Genome Analyzer IIx or HiSeq2000 analyzer as previously described (Yamane et al., 2013).
For further details on the materials and methods used in this study, please refer to the Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Gutierrez from the NIAMS genomics facility for technical assistance and Ethan Tyler for designing Figure 7C. This work was supported in part by the Intramural Research Program of NIAMS and NCI, NIH. The study made use of the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov) and the resources of NCI’s High-Throughput Imaging Facility. L.V. was supported in part by an American-Italian Cancer Foundation postdoctoral research fellowship.
Footnotes
ACCESSION NUMBERS
Deep-sequencing data are available at the NCBI SRA database under accession number GSE33819.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.024.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
REFERENCES
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J. Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Burgess-Beusse B, Farrell C, Gaszner M, Ghirlando R, Huang S, Jin C, Litt M, Magdinier F, Mutskov V, et al. Chromatin boundaries and chromatin domains. Cold Spring Harb. Symp. Quant. Biol. 2004;69:245–250. doi: 10.1101/sqb.2004.69.245. [DOI] [PubMed] [Google Scholar]
- Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, Loukinov DI, Pugacheva EM, Klenova EM, Grundy PE, et al. Tumorassociated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- Francastel C, Schübeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat. Rev. Mol. Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- Fraser P. Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat. Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Wang H, Kutyavin T, Stamatoyannopoulos JA. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 2012;8:e1002599. doi: 10.1371/journal.pgen.1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parentspecific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Singh M. An expanded binding model for Cys2His2 zinc finger protein-DNA interfaces. Phys. Biol. 2011;8:035010. doi: 10.1088/1478-3975/8/3/035010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke WW, Taheny MJ, Fochtmann LJ, Vostrov AA. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 2000;28:3370–3378. doi: 10.1093/nar/28.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Stadhouders R, de Bruijn MJ, Bergen IM, Thongjuea S, Lenhard B, van Ijcken W, Grosveld F, Galjart N, Soler E, Hendriks RW. The DNA-binding protein CTCF limits proximal Vk recombination and restricts k enhancer interactions to the immunoglobulin k light chain locus. Immunity. 2011;35:501–513. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, McNally JG. Fluorescence recovery after photobleaching (FRAP) methods for visualizing protein dynamics in living mammalian cell nuclei. Methods Enzymol. 2004;375:443–455. doi: 10.1016/s0076-6879(03)75027-7. [DOI] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Weth O, Renkawitz R. CTCF function is modulated by neigh-boring DNA binding factors. Biochem. Cell Biol. 2011;89:459–468. doi: 10.1139/o11-033. [DOI] [PubMed] [Google Scholar]
- White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Wilder S. SWEMBL: a generic peak-calling program. 2010 http://www.ebi.ac.uk/~swilder/SWEMBL/
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat. Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC, Casellas R. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.