Abstract
The bread wheat genome harbors three homoeologs of the barley gene HvAP2, which determines the cleistogamous/non-cleistogamous flowering. The three homoeologs, TaAP2-A, TaAP2-B and TaAP2-D, are derived from the A, B and D genomes. The importance of lodicule swelling in assuring non-cleistogamous flowering in a range of wild and domesticated wheat accessions of varying ploidy level was established. Re-sequencing of wheat AP2 homoeologous genes was carried out to identify natural variation at both the nucleotide and polypeptide level. The sequences of wheat AP2 homoeologs are highly conserved even across different ploidy levels and no functional variants at the key miR172 targeting site were detected. These results indicate that engineering of cleistogamous wheat will require the presence of a functional TaAP2 modification at each of the three homoeologs.
Keywords: Triticum aestivum L., cleistogamy, lodicule, microRNA172
Introduction
Wheat is grown over a larger area than any other crop and, along with maize and rice, provides a large proportion of the human calorific intake. The two major forms of wheat in cultivation are durum, an AABB tetraploid (Triticum durum) and bread, an AABBDD hexaploid (T. aestivum). The diploid progenitor of the A genome is known to be T. urartu (Chapman et al. 1976) and that of the D genome Aegilops tauschii (Kihara 1944, McFadden and Sears 1946). The progenitor of the B genome has yet to be established, but is likely to have been an extant or extinct species belonging to the Sitopsis section of Aegilops (Kilian et al. 2007, Petersen et al. 2006, Riley et al. 1958).
In barley, the major form of cleistogamy is determined by homozygosity for a recessive allele at the cleistogamy1 (cly1) locus on the telomeric region of chromosome 2HL (Kurauchi et al. 1994, Turuspekov et al. 2004). The Cly1 gene encodes an ortholog of Arabidopsis thaliana AP2 and thus was renamed HvAP2 by Nair et al. (2010). The sequence of the recessive HvAP2 allele differs from that of the wild type allele by a single nucleotide change at its miR172 targeting site, with the result that its transcript escapes cleavage. In the presence of sufficient intact HvAP2 transcript, the lodicules fail to develop properly, preventing normal opening of the floret.
Three bread wheat orthologs of HvAP2 have been identified, residing on the chromosomes homeologous to its site in barley (chromosome 2H) (Ning et al. 2013). The three genes, TaAP2-A, TaAP2-B and TaAP2-D, are all abundantly expressed in the wheat flower and particularly in the lodicule and they are all cleaved by miR172. As the role of the lodicule in exposing the stigma and the style at anthesis is the same in wheat as it is in barley, these genes are rational targets for engineering cleistogamy in wheat (Ning et al. 2013). These data proved the high likelihood that wheat three homoeologs are wheat AP2 genes, specifying the lodicule.
Although bread wheat three homoeologous orthologs (TaAP2) of the barley cleistogamy gene cly1 (HvAP2) had been described in a previous work (Ning et al. 2013), it is still a long way before successful application of wheat AP2 homoeolgs in wheat breeding. The point mutations at the miR172 targeting site of cly1 produce the cleistogamous barley. A rational strategy for inducing cleistogamy in wheat would be to identify naturally occurring or induced mutants at the miR172-targeting site for wheat AP2 genes. Here, we report a survey of the natural variation in potential wheat germplasm for wheat AP2 homoeologous sequence across a diverse panel of wild and domesticated wheats, including the (likely) diploid progenitors of the bread wheat A, B and D genomes.
Materials and Methods
Assessment of flowering phenotype
The germplasm panel consisted of 63 accessions, of which 24 were diploids, 23 tetraploids and 16 hexaploids (Table 1). The material was grown in the field at Tsukuba, Japan. Just prior to anthesis, three spikes still attached to the peduncle were detached from each accession and the lemma from the first floret of spikelet in the middle portion of the spike was removed to allow imaging of the lodicules. Lodicule height and depth were obtained from these images using Makijaku v1.1 software (http://cse.naro.affrc.go.jp/iwatah/). Maintaining the spikes in a 100 mg/l solution of 2,4-D for 24 h at room temperature facilitated the assessment of the maximum lodicule width and depth attained.
Table 1.
Source of germplasm utilized
| Species | Type | Genome | Line | Origin or Source | Lodicules before anthesis | Lodicules at anthesis | Haplotype | GenBank Accession No. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||||
| Depth (mm) | Width (mm) | Depth (mm) | Width (mm) | Hap-A | Hap-B(S) | Hap-D | AP2-A | AP2-B(S) | AP2-D | |||||
| T. monococcum L. subsp. aegilopoides (Link) Thell. | Wild | AbAb | KU-101-1 | Collection of College of Agr., Hokkaido Univ., Japan | 0.51 | 0.52 | 0.97 | 0.63 | ND | – | – | ND | – | – |
| T. monococcum L. subsp. aegilopoides (Link) Thell. | Wild | AbAb | KU-101-2 | Balaklava, Crimea, USSR | 0.54 | 0.54 | 1.00 | 0.62 | ND | – | – | ND | – | – |
| T. monococcum L. subsp. aegilopoides (Link) Thell. | Wild | AbAb | KU-103 | Collection of Agr. Exp. Station., Tehran, Iran | 0.49 | 0.49 | 0.83 | 0.61 | ND | – | – | ND | – | – |
| T. monococcum L. subsp. Monococcum | Domesticated | AmAm | KU-104-2 | Japan | 0.43 | 0.59 | 0.81 | 0.63 | ND | – | – | ND | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | KU-199-15 | Baal Bek, Lebanon | ND | ND | ND | ND | A2 | – | – | AB774265 | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | PI428186 | Mardin, Turkey | ND | ND | ND | ND | A8 | – | – | AB774268 | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | PI428230 | Urfa, Turkey | 0.42 | 0.47 | 0.94 | 0.62 | A9 | – | – | AB774269 | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | PI428253 | Arbil, Iraq | 0.38 | 0.48 | 0.88 | 0.73 | A10 | – | – | AB774270 | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | PI428254 | Mus, Turkey | ND | ND | ND | ND | A2 | – | – | AB774267 | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | PI428257 | Armenia | 0.42 | 0.59 | 0.78 | 0.66 | A11 | – | – | AB774271 | – | – |
| T. urartu Tumanian ex Gandilyan | Wild | AA | CItr17664 | Lebanon | 0.42 | 0.56 | 0.66 | 0.60 | A2 | – | – | AB774266 | – | – |
| Ae. speltoides Tausch | Wild | SS | PI170203 | Kirklareli, Turkey | 0.46 | 0.52 | 0.77 | 0.64 | – | ND | – | – | ND | – |
| Ae. speltoides Tausch | Wild | SS | PI499261 | China | 0.42 | 0.49 | 0.78 | 0.64 | – | ND | – | – | ND | – |
| Ae. speltoides Tausch | Wild | SS | PI487231 | Halab, Syria | 0.43 | 0.49 | 0.83 | 0.61 | – | B5 | – | – | AB774246 | – |
| Ae. speltoides Tausch | Wild | SS | PI542238 | Diyarbakir, Turkey | 0.40 | 0.45 | 0.73 | 0.57 | – | B6 | – | – | AB774247 | – |
| Ae. tauschii subsp. tauschii | Wild | DD | AS60 | Middle East | 0.35 | 0.59 | 0.55 | 0.76 | – | – | D2 | – | – | AB774238 |
| Ae. tauschii subsp. tauschii | Wild | DD | AS64 | Canada-2 | 0.32 | 0.58 | 0.55 | 0.75 | – | – | D2 | – | – | AB774239 |
| Ae. tauschii subsp. tauschii | Wild | DD | AS68 | USA | 0.37 | 0.64 | 0.63 | 0.81 | – | – | D2 | – | – | AB774240 |
| Ae. tauschii subsp. tauschii | Wild | DD | AS82 | Xinxiang prefecture, Henan, China | 0.36 | 0.68 | 0.71 | 0.85 | – | – | ND | – | – | ND |
| Ae. tauschii subsp. tauschii (morphological variety ‘typica’) | Wild | DD | KU-20-1 | Derbent, Caucasus, Dagestan, USSR | 0.49 | 0.71 | 1.01 | 0.91 | – | – | D3 | – | – | AB774243 |
| Ae. tauschii subsp. tauschii (morphological variety ‘meyeri’) | Wild | DD | KU-20-10 | 9 km NW of Ramsar (Chalus-Rasht), Iran | 0.27 | 0.65 | 0.69 | 0.84 | – | – | D3 | – | – | AB774244 |
| Ae. tauschii subsp. strangulata (Eig) Tzvelev | Wild | DD | KU-20-9 | 5 km W of Behshahr (Sari-Behshahr), Iran | 0.35 | 0.72 | 0.64 | 0.93 | – | – | D4 | – | – | AB774245 |
| Ae. tauschii subsp. strangulata (Eig) Tzvelev | Wild | DD | AS2386 | Iran | 0.36 | 0.64 | 0.59 | 0.91 | – | – | D3 | – | – | AB774241 |
| Ae. tauschii subsp. strangulata (Eig) Tzvelev | Wild | DD | AS2396 | Israel | 0.34 | 0.50 | 0.62 | 0.77 | – | – | D3 | – | – | AB774242 |
| T. turgidum L. subsp. dicoccoides (Korn. ex Asch. & Graebn.) Thell. | Wild | AABB | KU-108-2 | 20 km NW of Suweida (Cheikh Meskine-Suweida), Syria | 0.45 | 0.57 | 1.05 | 0.76 | A12 | B7 | – | AB774284 | AB774259 | – |
| T. turgidum L. subsp. dicoccoides (Korn. ex Asch. & Graebn.) Thell. | Wild | AABB | KU-8817 | North slope of Jabal Sinjar, N of Kursi, Iraq | 0.64 | 0.84 | 1.13 | 0.99 | A4 | ND | – | AB774283 | ND | – |
| T. turgidum L. subsp. dicoccoides (Korn. ex Asch. & Graebn.) Thell. | Wild | AABB | KU-198 | Collected in Mt. Canaan (Israel) by Dr. Aaronsohn (1906), Israel | 0.53 | 0.73 | 0.94 | 0.90 | A14 | B12 | – | AB774288 | AB774260 | – |
| T. turgidum L. subsp. dicoccum (Schrank ex Schübl.) Thell. | Domesticated | AABB | KU-112 | Peiping, China | 0.62 | 0.81 | 1.33 | 1.00 | A3 | B4 | – | AB774274 | AB774257 | – |
| T. turgidum L. subsp. dicoccum (Schrank ex Schübl.) Thell. | Domesticated | AABB | KU-113 | Collection of Agr. Exp. Sta. of Koonosu, Japan | 0.67 | 0.85 | 1.45 | 1.10 | A3 | B4 | – | AB774275 | AB774258 | – |
| T. turgidum L. subsp. dicoccum (Schrank ex Schübl.) Thell. | Domesticated | AABB | KU-114 | Collection of Agr. Exp. Sta. of Koonosu, Japan | 0.76 | 0.95 | 1.32 | 1.07 | A6 | B1 | – | AB774289 | AB774248 | – |
| T. turgidum L. subsp. durum (Desf.) Husn. | Domesticated | AABB | KU-125 | Collection of College of Agr., Hokkaido Univ., Japan | 0.58 | 0.78 | 1.25 | 1.00 | A5 | B1 | – | AB774285 | AB774251 | – |
| T. turgidum L. subsp. durum (Desf.) Husn. | Domesticated | AABB | KU-135 | Collection of Univ. Wash., Pullman, USA | 0.57 | 0.74 | 1.02 | 0.95 | A5 | B8 | – | AB774286 | AB774262 | – |
| T. turgidum L. subsp. durum (Desf.) Husn. | Domesticated | AABB | KU-146 | Unknown | 0.39 | 0.85 | 0.90 | 1.09 | A3 | B3 | – | AB774279 | AB774255 | – |
| T. turgidum L. subsp. durum (Desf.) Husn. | Domesticated | AABB | KU-185 | Collected in Ethiopia by Dr. Furusato, Ethiopia | 0.57 | 0.74 | 1.03 | 0.90 | A3 | B3 | – | AB774281 | AB774256 | – |
| T. turgidum L. subsp. durum (Desf.) Husn. | Domesticated | AABB | KU-188 | Unknown | 0.44 | 0.70 | 1.08 | 0.93 | A3 | B11 | – | AB774282 | AB774264 | – |
| T. turgidum L. subsp. turanicum (Jakubz.) Á. & D. Löve | Domesticated | AABB | KU-137 | Unknown | 0.37 | 0.63 | 0.90 | 1.03 | A13 | B9 | – | AB774287 | AB774261 | – |
| T. turgidum L. subsp. carthlicum (Nevski) Á. & D. Löve | Domesticated | AABB | KU-138 | Unknown | 0.56 | 0.70 | 0.91 | 0.89 | A3 | B3 | – | AB774276 | AB774254 | – |
| T. turgidum L. subsp. carthlicum (Nevski) Á. & D. Löve | Domesticated | AABB | KU-187 | Unknown | 0.45 | 0.68 | 1.18 | 0.98 | A3 | B10 | – | AB774277 | AB774263 | – |
| T. turgidum L. subsp. polonicum (L.) Thell. | Domesticated | AABB | KU-141 | Collection of College of Agr., Hokkaido Univ., Japan | 0.61 | 0.70 | 1.10 | 0.93 | A3 | B1 | – | AB774278 | AB774249 | – |
| T. turgidum L. subsp. turgidum | Domesticated | AABB | KU-147 | Collection of College of Agr., Hokkaido Univ., Japan | 0.49 | 0.61 | 1.10 | 0.91 | A3 | B1 | – | AB774280 | AB774250 | – |
| T. turgidum L. subsp. paleocolchicum Á. & D. Löve | Domesticated | AABB | KU-156 | Unknown | 0.49 | 0.75 | 1.24 | 0.89 | A6 | B2 | – | AB774290 | AB774252 | – |
| T. turgidum L. subsp. paleocolchicum Á. & D. Löve | Domesticated | AABB | KU-190-1 | Unknown | 0.62 | 0.76 | 1.50 | 1.05 | A6 | B2 | – | AB774291 | AB774253 | – |
| T. timopheevi (Zhuk.) Zhuk. subsp. armeniacum (Jakubz.) Slageren | Wild | AAGG | KU-1901 | 8 km W of Garni (Erevan-Garni), Armenia, USSR | ND | ND | ND | ND | A15 | – | – | AB774272 | – | – |
| T. timopheevi (Zhuk.) Zhuk. subsp. armeniacum (Jakubz.) Slageren | Wild | AAGG | KU-8735 | SSW of Rowanduz, Iraq | 0.38 | 0.65 | 0.79 | 0.84 | A16 | – | – | AB774273 | – | – |
| T. timopheevi (Zhuk.) Zhuk. subsp. armeniacum (Jakubz.) Slageren | Wild | AAGG | KU-8940 | 39.9 km N from Elazig to Hozat, Turkey | 0.59 | 0.71 | 1.36 | 0.98 | ND | – | – | ND | – | – |
| T. timopheevi (Zhuk.) Zhuk. subsp. timopheevii | Domesticated | AAGG | KU-107-1 | Unknown | 0.53 | 0.77 | 1.38 | 1.05 | ND | – | – | ND | – | – |
| T. timopheevi (Zhuk.) Zhuk. subsp. timopheevii | Domesticated | AAGG | KU-107-4 | Georgia, Collection of All-Union Inst. of Plant Indust., Leningrad, USSR | 0.52 | 0.75 | 1.25 | 1.02 | ND | – | – | ND | – | – |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | KU-163 | Collection of Col Agr. Hokkaido Univ., Japan | 0.44 | 0.76 | 0.92 | 0.92 | A7 | B1 | D3 | AB761172 | AB761176 | AB761191 |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | KU-165 | Correns, Germany | 0.61 | 0.88 | 1.00 | 1.03 | A1 | B1 | D3 | AB761159 | AB761177 | AB761192 |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | KU-265 | Collection of Lab. of Plant Breeding, Facul. of Agr., Kyoto Univ., Japan | 0.51 | 0.80 | 1.00 | 0.99 | A1 | B1 | D3 | AB761160 | AB761179 | AB761193 |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | KU-515 | Tibet, China | 0.39 | 0.75 | 0.68 | 0.90 | A3 | B1 | D3 | AB761163 | AB761180 | AB761194 |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | Fukuho | Japan | ND | ND | ND | ND | A3 | B17 | D3 | AB761164 | AB761188 | AB761189 |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | Norin 61 | Japan | ND | ND | ND | ND | A3 | B1 | D3 | AB761165 | AB761181 | AB761190 |
| T. aestivum L. subsp. aestivum | Domesticated | AABBDD | Chinese Spring | China | 0.52 | 0.86 | 1.38 | 1.17 | A1 | B1 | D1 | AB749311 | AB749312 | AB749313 |
| T. aestivum L. subsp. compactum (Host) Mackey | Domesticated | AABBDD | KU-150 | Collection of College of Agr., Hokkaido Univ., Japan, | 0.51 | 0.74 | 0.91 | 0.93 | A7 | B13 | ND | AB761170 | AB761184 | ND |
| T. aestivum L. subsp. compactum (Host) Mackey | Domesticated | AABBDD | KU-153 | Collection of Univ. Wash., Pullman, USA | 0.46 | 0.75 | 0.96 | 0.94 | A7 | B14 | D3 | AB761171 | AB761185 | AB761195 |
| T. aestivum L. subsp. macha (Dekapr. & A. M. Menabde) Mackey | Domesticated | AABBDD | KU-154 | Unknown | 0.54 | 0.84 | 1.14 | 1.07 | A6 | B1 | D3 | AB761167 | AB761174 | AB761196 |
| T. aestivum L. subsp. macha (Dekapr. & A. M. Menabde) Mackey | Domesticated | AABBDD | KU-193 | Unknown | 0.58 | 0.86 | 0.96 | 0.93 | A6 | B1 | D3 | AB761168 | AB761178 | AB761197 |
| T. aestivum L. subsp. macha (Dekapr. & A. M. Menabde) Mackey | Domesticated | AABBDD | KU-197 | Collection of Ankara Univ. (Agri), Turkey | 0.51 | 0.94 | 1.39 | 1.15 | A6 | B16 | D3 | AB761169 | AB761187 | AB761198 |
| T. aestivum L. subsp. spelta (L.) Thell. | Domesticated | AABBDD | KU-157 | Collection of College of Agr., Hokkaido Univ., Japan | 0.55 | 0.93 | 1.04 | 1.12 | A4 | B1 | D3 | AB761166 | AB761182 | AB761199 |
| T. aestivum L. subsp. sphaerococcum (Percival) Mackey | Domesticated | AABBDD | KU-161 | Unknown | 0.43 | 0.85 | 0.92 | 1.01 | A3 | B2 | D3 | AB761161 | AB761183 | AB761200 |
| T. aestivum L. subsp. sphaerococcum (Percival) Mackey | Domesticated | AABBDD | KU-162-2 | Collection of Islamia College, Pakistan | 0.39 | 0.79 | 0.98 | 0.95 | A3 | B1 | D3 | AB761162 | AB761175 | AB761201 |
| T. aestivum vavilovii Jakubz. | Domesticated | AABBDD | KU-192 | Unknown | 0.27 | 0.76 | 0.95 | 1.19 | A7 | B15 | D5 | AB761173 | AB761186 | AB761202 |
Accessions prefixed with KU provided by the Japanese National BioResource Project (NBRP), those with either PI and CItr by USDA-ARS, those with AS by Sichuan Agricultural University Triticeae Research Institute. ND: not determined. DNA sequences of T. aestivum L. subsp. aestivum were previously published (Ning et al. 2013). Taxonomic classification follows the recommendation of http://www.ars-grin.gov/cgi-bin/npgs/html/index.pl.
DNA amplification and sequencing
Genomic DNA was extracted from young, freshly harvested leaves according to Komatsuda et al. (1998). We used A, B and D genome ortholog-specific PCR primers designed in our previous work (Ning et al. 2013). The three TaAP2 homoeologous sequences were amplified from their start to their stop codon using primers detailed in Table 2. Each 50 μl PCR targeting TaAP2-A fragment 1 contained 0.2 × GC Buffer II; amplification reactions targeting TaAP2-A fragment 2, TaAP2-B and TaAP2-D were based on 1 × Ex Taq polymerase buffer. All reactions contained 0.25U Ex Taq polymerase (Takara, Tokyo, Japan), 0.6 μM of each primer, 0.2 mM dNTP, 2.0 or 2.5 mM MgCl2, 8 or 12% v/v dimethyl sulphoxide and 40 ng genomic DNA. The PCR regime comprised a denaturation step (94°C/5 min), followed by 30 cycles of 94°C/1 min, 57 or 65°C (primer-dependent)/1 min, 72°C/2 min and a final extension step (72°C/10 min). The resulting amplicons were electrophoresed through 1% agarose (Iwai Kagaku, Tokyo, Japan) in 0.5 × TBE buffer and were visualized using EtBr staining to check that amplification had been achieved. The amplicons were purified using a QIAquick PCR purification kit (QIAGEN, Germantown, MD, USA) in preparation for their cycle sequencing using a Big Dye Terminator kit (Applied Biosystem, Foster, CA, USA). The sequencing reactions were purified by Agencourt CleanSEQ (Beckman, Beverly, MA, USA) and analyzed using an ABI PRISM 3130 genetic analyzer (Applied Biosystem).
Table 2.
PCR primer sequences used for the amplification of wheat AP2 homoeologs
| Target | Primer name | Sequence of primer (5′-3′) | Primer name | Sequence of primer (5′-3′) | PCR mixture | Tm (°C) | size (kb) | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Buffer | MgCl2 (mM) | DMSO (%) | |||||||
| AP2-A_fragment 1 | U1005A23 | GCAGACCAGAGAGAGGCTAGAGG | 2223L20 | CTGCAAGGCCAATTACAGGT | 0.2 × GC Buffer II | 2.5 | 8 | 57 | 1.2 |
| AP2-A_fragment 2 | F695 | TGCGGCAAGCAGGTCTATCTG | A3794L19 | CCCATGCTCCTCCGTGATC | 1 × Ex Taq polymerase buffer | 2.0 | 8 | 65 | 2.0 |
| AP2-B | F-est2 | AGAGCAGGGCAGAGGGAGGCGTAGGG | R-est1543 | GCTGGCTGCTCTCGACGGATGGT | 1 × Ex Taq polymerase buffer | 2.0 | 8 | 65 | 2.8 |
| AP2-D_fragment 1 | 55U24 | GCAAGCAGGGAGGGGAGCTAGCCA | R1690 | GGCTCGAACTCCTCGGCG | 1 × Ex Taq polymerase buffer | 2.5 | 12 | 65 | 1.8 |
| AP2-D_fragment 2 | F695 | TGCGGCAAGCAGGTCTATCTG | 3897L20 | TGGAGCTGGTCTTGATGGTC | 1 × Ex Taq polymerase buffer | 2.5 | 8 | 65 | 2.0 |
Sequence alignment and phylogenetic analysis
Multiple sequence alignments at both the nucleotide and predicted polypeptide level were performed using DNAMAN v6.0 software (Lynnon Biosoft, Quebec, Canada). Phylogenetic trees were constructed based on the neighbor-joining method, using MEGA v5 software (Tamura et al. 2011). Bootstrap analysis was based on 1,000 replicates.
Results
Flower opening and lodicule swelling in wheats of varying ploidy level
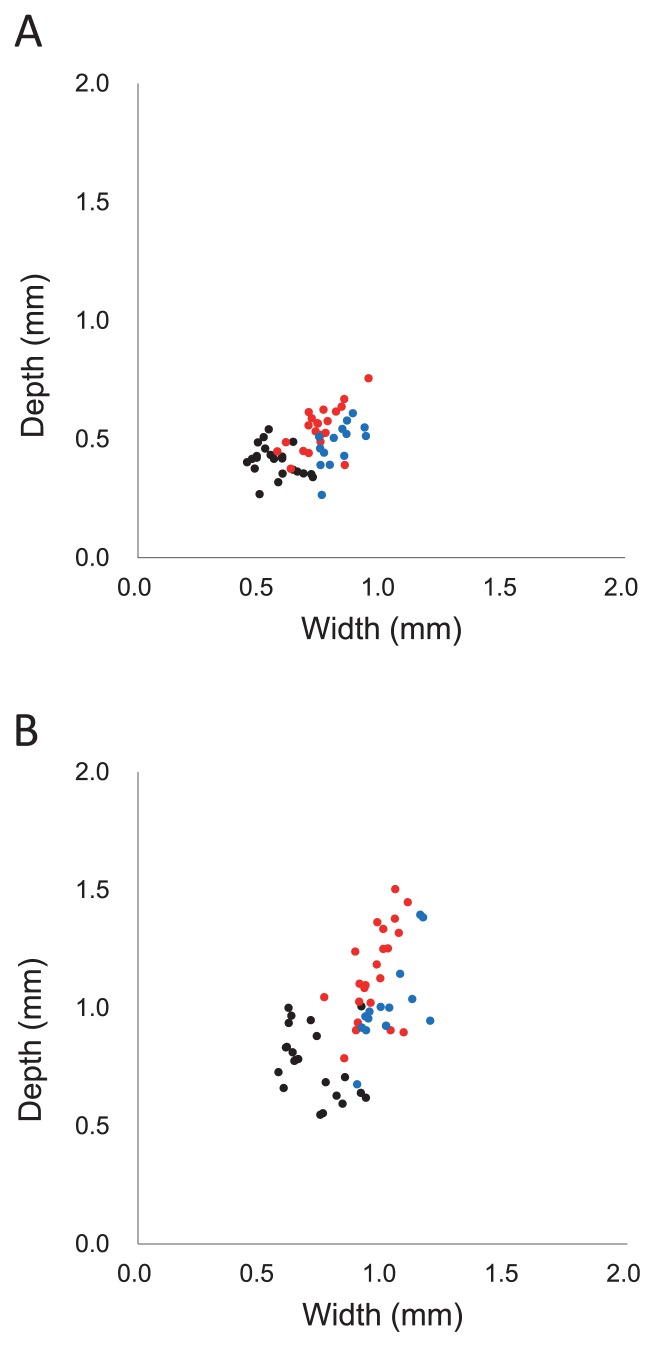
Anthesis was reached over the period early May to mid June, although six of the 63 accessions did not reach flowering. All plants formed normal lodicules and prior to anthesis, all florets were enclosed tightly by the palea and lemma. At anthesis, anther extrusion was driven by the swelling of the lodicules (Fig. 1). The range in lodicule width and depth displayed by the accessions prior to and at anthesis is given in Table 1. Their width prior to anthesis ranged from 0.40–0.72 mm among the diploid accessions, 0.57–0.95 mm among the tetraploids and 0.74–0.94 mm among the hexaploids; while at anthesis, the respective ranges were 0.57–0.93 mm, 0.76–1.10 mm and 0.90–1.17 mm. Similarly lodicule depth varied from 0.27–0.54 mm, 0.38–0.76 mm and 0.27–0.61 mm prior to anthesis and 0.55–1.01 mm, 0.79–1.45 mm and 0.91–1.39 mm at anthesis (Fig. 2). Overall, lodicule depth at anthesis was about double that prior to anthesis. Accession to accession variation in lodicule size was considerable (the largest was more than twice the size of the smallest) but continuous. In barley, variation in lodicule size was discontinuous, representing cleistogmous and non-cleistogmous phenotypes (Nair et al. 2010). Therefore the results in wheat were unlike in barley.
Fig. 1.
Variation in lodicule size across wheat ploidy levels. A: T. urartu (AA) PI428230, B: Ae. speltoides (SS) PI487231, C: Ae. tauschii (DD) KU-20-1, D: T. durum (AABB) KU-125, E: bread wheat (AABBDD) cv. ‘Chinese Spring’. Lodicule width and depth indicated by arrows. Left panel: prior to anthesis, right panel: at anthesis. Bar: 1 mm.
Fig. 2.
Variation in lodicule size displayed by 57 accessions of diploid (black), tetraploid (red) and hexaploid wheat (blue). Lodicule size was measured (A) prior to and (B) at anthesis.
TaAP2: nucleotide sequences
The full set of wheat AP2 homoeologous sequences has been deposited in GenBank as accession AB761159 to AB761202, AB774238 to AB774291 (Table 1). All three homoeologs comprised ten exons, with a 21nt miR172 targeting site sited in the tenth exon.
A sample of 43 AP2 homoeologous sequences of the A genome, obtained from seven T. urartu, 18 T. turgidum (AABB tetraploid), two T. timopheevi (AAGG tetraploid) and 16 T. aestivum accessions, revealed 16 haplotypes (Fig. 3). The most common haplotype (Hap-A3) was present in 14 accessions (nine T. turgidum and five T. aestivum accessions) and bread wheat cv. Shinchunaga (Ning et al. 2013), followed by Hap-A6 (six accessions: three T. turgidum and three T. aestivum) and Hap-A7 (four T. aestivum accessions). Both Hap-A1 and Hap-A2 were present in three accessions (the former all T. aestivum and the latter all T. urartu). Hap-A4 and Hap-5 each were represented by two accessions (the former comprising one T. turgidum and one T. aestivum accession and the latter two T. turgidum accessions). The remaining nine haplotypes were unique to a single accession: Hap-A8 to -A11 were specific to T. urartu, Hap-A12 to -A14 to T. turgidum and A15 to -A16 to T. timopheevi. Exon variation involved four single nucleotide substitutions and four indels; five of these eight polymorphisms induced an altered peptide sequence; the intron variation involved 20 single nucleotide substitutions and three indels (Fig. 3).
Fig. 3.
Haplotype variation in wheat AP2 homoeolog of A genome in a sample of diploid, tetraploid and hexaploid wheats. Variation from the bread wheat cv. Chinese Spring (CS) is indicated. Hap-A1: represented by accessions CS, KU-165 and -265; Hap-A2 by KU-199-15, Cltr17664 and PI428254; Hap-A3 by KU-112, -113, -138, -141, -146, -147, -161, -162-2, -515, -187, -185, -188, cv. Fukuho, cv. Norin 61 and cv. Shinchunaga (Ning et al. 2013); Hap-A4 by KU-8817 and -157; Hap-A5 by KU-125 and -135; Hap-A6 by KU-114, -154, -156, -190-1, -193 and -197; Hap-A7 by KU-150, -153, -163 and -192; Hap-A8 by PI428186; Hap-A9 by PI428230; Hap-A10 by PI428253; Hap-A11 by PI428257; Hap-A12 by KU-108-2; Hap-A13 by KU-137; Hap-A14 by KU-198; Hap-A15 by KU-1901; Hap-A16 by KU-8735. Genomic sequences between start and stop codon were aligned. Exonic polymorphisms indicated in bold and those generating a changed peptide by asterisks.
AP2 homoeologous sequences of B and S genome were recovered from two accessions of Ae. speltoides (SS), 17 of T. turgidum (eight subspecies) and 16 of T. aestivum. A total of 17 haplotypes was recognized (Fig. 4). The most common haplotype (Hap-B1) was present in four T. turgidum and nine T. aestivum accessions, and bread wheat cv. Shinchunaga (Ning et al. 2013), Hap-B2 in two T. turgidum and two T. aestivum accessions, Hap-B3 in three T. turgidum accessions and Hap-B4 in two T. turgidum accessions. The remaining haplotypes were unique to a single accession: Hap-B5 and -B6 to Ae. speltoides, -B7 to -B12 to T. turgidum and -B13 to -17 to T. aestivum. The variation in intron sequence involved 65 single nucleotide substitutions and 25 indels and that in the exon sequence 34 single nucleotide substitutions and 25 indels. Of the latter, 24 induced an altered peptide sequence (Fig. 4).
Fig. 4.
Haplotype variation in wheat AP2 homoeolog of B(S) genome in a sample of diploid, tetraploid and hexaploid wheats. Variation from the bread wheat cv. Chinese Spring (CS) is indicated. Hap-B1: KU-114, -125, -141, -147, -154, -162-2, -163, -165, -193, -265, -515, CS, cv. Norin 61 and cv. Shinchunaga (Ning et al. 2013); Hap-B2: KU-156, -157, -161, -190-1; Hap-B3: KU-138, -146 and -185; Hap-B4: KU-112, -113; Hap-B5: PI487231; Hap-B6: PI542238; Hap-B7: KU-108-2; Hap-B8: KU-135; Hap-B9: KU-137; Hap-B10: KU-187; Hap-B11: KU-188; Hap-B12: KU-198; Hap-B13: KU-150; Hap-B14: KU-153; Hap-B15: KU-192; Hap-B16: KU-197; Hap-B17: cv. Fukuho. Genomic sequences between start and stop codon were aligned. Exonic polymorphisms indicated in bold and those generating a changed peptide by asterisks.
The AP2 homoeologous sequences of D genome were obtained from eight Ae. tauschii and 15 T. aestivum accessions. The most common of the five haplotypes recognized was Hap-D3, which was present in four Ae. tauschii and 13 of the 15 T. aestivum accessions and bread wheat cv. Shinchunaga (Ning et al. 2013). Hap-D2 was represented in three Ae. tauschii accessions, while Hap-D1 and -D5 each had one T. aestivum accession and Hap-D4 one Ae. tauschii accession. There were 16 single nucleotide substitutions and five indels in the intronic sequence and nine single nucleotide substitutions and three indels in the exonic sequence; five of the latter induced an altered peptide sequence (Fig. 5).
Fig. 5.

Haplotype variation in wheat AP2 homoeolog of D genome in a sample of diploid, tetraploid and hexaploid wheats. Variation from the bread wheat cv. Chinese Spring (CS) is indicated. Hap-D1: CS; Hap-D2: AS60, AS64 and AS68; Hap-D3: KU-20-1, -20-10, AS2386, AS2396 and all the hexaploid wheat accessions including cv. Shinchunaga (Ning et al. 2013) except KU-192; Hap-D4: KU-20-9; Hap-D5: KU-192. Genomic sequences between start and stop codon were aligned. Exonic polymorphisms indicated in bold and those generating a changed peptide by asterisks.
With respect to the miR172 targeting sites of wheat AP2 homoeologs, the second nucleotide was uniformly a cytosine in B(S) genome and a thymine in A and D genomes (Fig. 8).
Fig. 8.

Sequence variation in the miR172 targeting site of barley HvAP2 and wheat AP2 homoeologs.
Wheat AP2 homoeologs: peptide sequences
At the polypeptide sequence level, the 16 haplotypes from AP2 homoeologous sequences of A genome collapsed into seven distinct polypeptides: the products translated from Hap-A1 and -A7 were indistinguishable from one another, as were those from -A2, -A8, -A9, -A10 and -A11, those from -A3, -A6 and -A13, those from -A15 and -A16 and those from -A5 and -A14. The gene products of Hap-A4 and -A12 were different from the other five, as well as from one another. For B(S) genome, the 17 haplotypes collapsed into ten distinct polypeptides: the products translated from Hap-B1, -B4 and -B17 were indistinguishable from one another, as were those from -B2 and -B13 and those from -B3, -B8, -B10, -B11 and -B15. The remaining haplotypes (Hap-B5, -B6, -B7, -B9, -B12, -B14 and -B16) each produced a unique polypeptide. For D genome, the five haplotypes collapsed into four distinct polypeptides: the products translated from Hap-D3 and -D5 were indistinguishable from one another, while the others each produced a unique polypeptide. A comparison of the three AP2 homoeologous protein sequences is shown in Fig. 6. Their structural organization was identical and they shared several conserved sequence features, namely the central core of the AP2 polypeptide (residues 118–276; boxed in Fig. 6), including AP2 domains R1 and R2 and the α-helix, the highly basic ten residue domain thought to be a nuclear localization signal (residues 108–117, boxed in Fig. 6), the AASSFGF box (residues 460–477, boxed in Fig. 6) and the three motifs (residues 1–5, 50–56 and 289–294, boxed in Fig. 6) (Jofuku et al. 1994, Tang et al. 2007). The only variant detected involved the homoeologous sequence of A genome present in T. timopheevi KU-1901 and KU-8735 and the homoeologous sequence of B genome in KU-197, both of which had a single residue substitution at, respectively, positions 271 and 274 (Fig. 6).
Fig. 6.
Alignment of polypeptide sequences of wheat AP2 homoeologs. Accessions within each of the haplotypes given in the legends to Figs. 3, 4, 5. The various key features of AP2 proteins are shown boxed, and the α-helix present in the core region of each AP2 domain shown delimited by arrows.
Wheat AP2 homoeologs phylogeny
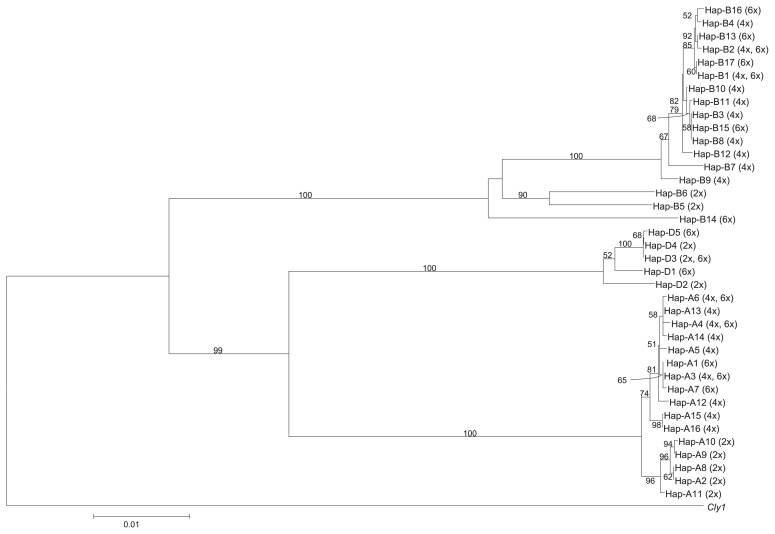
The phylogeny of the wheat AP2 homoeologs based on full-length nucleotide sequences is shown as Fig. 7. Each of the three homoeologs formed a distinct clade. The homoeologs from A and D genome clades appeared to be more closely related to one another than to the homoeologs from B(S) genome clade. Within the homoeolog from A genome clade, two sub-groups were recognizable: one comprised the T. urartu sequences Hap-A2, -A8, -A9, -A10 and -A11 and the other the A genome sequences present in T. timopheevi, T. turgidum and T. aestivum. For homoeolog from B(S) genome, all but three of the haplotypes clustered into one large clade; Hap-B14 (present in T. aestivum subsp. compactum) was an outlier, as were the two haplotypes Hap-B5 and -B6, both present in Ae. speltoides. The phylogeny of the sequences of homoeolog from D genome was less easy to interpret, because only five haplotypes were represented. However, the sequences Hap-D3 (Aegilops tauschii ssp. tauschii morphological varieties ‘typica’: KU-20-1 and morphological varieties ‘meyeri’ KU-20-10, two lines of ssp. strangulata and most of wheats), Hap-D4 (ssp. strangulate KU-20-9 ) and -D5 (T. aestivum vavilovii KU-192) formed a clear subgroup, which was not closely related to either the -D1 (Chinese Spring) or the -D2 (Aegilops tauschii subsp. tauschii: AS60, AS64 and AS68) sequence.
Fig. 7.
Phylogeny based on the full length genomic sequence using the neighbor-joining method. The sequence of barley Cly1 (HvAP2) used as the outgroup. Bootstrap values (%) based on 1,000 replicates.
Discussion
Lodicule swelling at anthesis is ubiquitous in wheat and its near relatives at all ploidy levels; the mechanical pressure which this process generates is sufficient to prise apart the lemma and the palea and allow the stamens to be extruded. The identical mechanism is used by non-cleistogamous barley cultivars (Nair et al. 2010). All three homeologous versions of TaAP2 were structured into ten exons, and their sequences were highly homologous with one another. The sequence variation between them was concentrated more in the intronic than in the exonic DNA (Figs. 3, 4, 5). The sequence of the miR172 targeting site was very highly conserved throughout, with the only variants detected among the 101 re-sequenced wheat AP2 homoeologous genes being at the second nucleotide (Fig. 8). The miR172 targeting site sequences matched those present in the TaAP2 homoeologs of the bread wheat cv. Shinchunaga, which are all successfully cleaved by miR172 (Ning et al. 2013), while the T/C polymorphism (which discriminated the AP2-B version from that of both AP2-A and -D) in barley is known to have no effect on cleistogamy (Nair et al. 2010). At the polypetide level, all of the AP2 homoeologous products shared the same structural organization and retained the key features of the protein, namely the AP2 domain, the putative nuclear localizing signal, the AASSFGF box and the three motifs (Jofuku et al. 1994, Tang et al. 2007). The only departures from this conservation related to AP2-A homoeolog in T. timopheevi accessions KU-1901 and KU-8735 and AP2-B homoeolog in accession KU-197, but in both of these accessions, the lodicules became swollen at anthesis, meaning that their TaAP2 polymorphism was non-functional in terms of cleistogamy – presumably because it had no effect on TaAP2 transcript abundance. Thus, the indication is that all three AP2 homoeologous genes are regulated by miR172 cleavage, in the same way as occurs in barley, and that as a result, non-cleistogamy in wheat is determined by the successful cleavage of AP2 homoeologous mRNA (Nair et al. 2010, Ning et al. 2013).
Although the germplasm studied here was extremely diverse, covering a range of ploidy levels and ranging from cultivated varieties to wild relatives, the plants were uniformly non-cleistogamous, and their AP2 homoeologous sequences were highly conserved. No functional mutations at the miR172 targeting site were identified. Because of failure of amplification, the AP2 homoeologous sequences of individual materials were not determined in Table 1, possibly by the variations in primer binding sites. Non-cleistogamy is the norm in barley as well, but two naturally occurring cleistogamous types carrying distinct alleles at HvAP2 have been identified (Nair et al. 2010). As both of these alleles are recessive to the wild type non-cleistogamous allele, the implication is that in polyploid wheat, functional wheat AP2 homoeologous mutants are unlikely to display a cleistogamous phenotype, because their effect will be hidden by the presence of a wild type allele at the other homeolocus(loci). Engineering a cleistogamous hexaploid wheat will require the presence of a functional wheat AP2 homoeologous mutant at each of the three homoeoloci. Given the absence of any such alleles among the 101 re-sequenced wheat AP2 homoeologous genes, the likelihood of finding a naturally occurring mutant appears to be rather low. Therefore, the TILLING approach (Henikoff et al. 2004) could provide an attractive platform for detecting allelic variants. Alternatively, site-specific nucleases which have been designed by fusing the DNA cleavage domain of FokI and a custom-designed DNA binding domain, such as the C2H2 zinc-finger motif for zinc finger nucleases (ZFNs) (Urnov et al. 2010) and the truncated transcription activator-like effector (TALE) domain for TALE nucleases (Miller et al. 2011) could lend itself readily to engineering the miR172 targeting site in wheat.
The D genome donor Ae. tauschii has been taxonomically divided on the basis of its morphology into four types, of which three (‘typica’, ‘meyeri’ and ‘anathera’) have been grouped together to form subsp. tauschii, while strangulata forms its own subspecies. It was indicated that the donor of the bread wheat D genome belonged to subsp. strangulata (Dvorak et al. 1998, Jaaska 1980, Xiang et al. 2009). The two strangulata accessions and the single accessions of typica and meyeri all shared the same AP2 homoeologous sequence, which was also present in all bar two of the T. aestivum accessions. The two subsp. strangulata lines and the ‘typica’ and ‘meyeri’ representatives proved to be phylogenetically close to T. aestivum but distinct from the other subsp. tauschii accessions, providing evidence to support the hypothesis that ‘typica’ and ‘meyeri’ are equidistant from subsp. strangulata, although both belong to the strangulata genepool (Dvorak et al. 1998).
Acknowledgements
We thank the Japanese National BioResource Project (NBRP), the USDA-ARS and the Sichuan Agricultural University Triticeae Research Institute (Chengdu, China) for the gift of germplasm used in this study. We thank T. Kawahara, Y. Nagamura, J. Song, G. Chen, C. Liu, C. Li, H. Sassa and S. Kikuchi for their help and advice. This research was funded by the Japanese Ministry of Agriculture, Forestry and Fisheries (Genomics for Agricultural Innovation grants no. TRG1004) to T.K. S.N. appreciates the award of a Japanese Government (Monbukagakusho: MEXT) scholarship.
Literature Cited
- Chapman, V., Miller, T.E. and Riley, R (1976) Equivalence of the A genome of bread wheat and that of Triticum urartu. Genet. Res. 27: 69–76 [Google Scholar]
- Dvorak, J., Luo, M.-C., Yang, Z.-L. and Zhang, H.-B. (1998) The structure of Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97: 657–670 [Google Scholar]
- Henikoff, S., Till, B.J. and Comai, L (2004) TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135: 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaska, V. (1980) Electrophoretic survey of seedling esterases in wheats in relation to their phylogeny. Theor. Appl. Genet. 56: 273–284 [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G., Montagu, M.V. and Okamuro, J.K. (1994) Control of Arabidopsis flower and seed developmentby the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, H. (1944) Discovery of the DD-analyser, one of the an ancestors of Triticum vulgare. Agric. Hortic. 19: 889–890 [Google Scholar]
- Kilian, B., Özkan, H., Deusch, O., Effgen, S., Brandolini, A., Kohl, J., Martin, W. and Salamini, F. (2007) Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol. Biol. Evol. 24: 217–227 [DOI] [PubMed] [Google Scholar]
- Komatsuda, T., Nakamura, I., Takaiwa, F. and Oka, S. (1998) Development of STS markers closely linked to the vrs1 locus in barley, Hordeum vulgare. Genome 41: 680–685 [Google Scholar]
- Kurauchi, N., Makino, T and Hirose, S (1994) Inheritance of cleistogamy-chasmogamy in barley. Barley Genet. Newsl. 23: 19 [Google Scholar]
- McFadden, E.S. and Sears, E.R. (1946) The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–89 [DOI] [PubMed] [Google Scholar]
- Miller, J.C., Tan, S, Qiao, G, Barlow, K.A., Wang, J, Xia, D.F., Meng, X, Paschon, D.E., Leung, E and Hinkley, S.J.et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat. Biot. 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Nair, S.K., Wang, N., Turuspekov, Y., Pourkheirandish, M., Sinsuwongwat, S., Chen, G., Sameri, M., Tagiri, A., Honda, I. and Watanabe, Y.et al. (2010) Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. USA 107: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, S., Wang, N, Sakuma, S, Pourkheirandish, M, Wu, J, Matsumoto, T, Koba, T and Komatsuda, T (2013) Structure, transcription and post-transcriptional regulation of the bread wheat orthologs of the barley cleistogamy gene Cly1. Theor. Appl. Genet. 126: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Petersen, G., Seberg, O, Yde, M and Berthelsen, K (2006) Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39: 70–82 [DOI] [PubMed] [Google Scholar]
- Riley, R., Unrau, J and Chapman, V (1958) Evidence on the origin of the B genome of wheat. J. Hered. 49: 91–98 [Google Scholar]
- Tamura, K., Peterson, D, Peterson, N, Stecher, G, Nei, M and Kumar, S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, M., Li, G. and Chen, M. (2007) The phylogeny and expression pattern of APETALA2-like genes in rice. J. Genet. Genomics 34: 930–938 [DOI] [PubMed] [Google Scholar]
- Turuspekov, Y., Mano, Y, Honda, I, Kawada, N, Watanabe, Y and Komatsuda, T (2004) Identification and mapping of cleistogamy genes in barley. Theor. Appl. Genet. 109: 480–487 [DOI] [PubMed] [Google Scholar]
- Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. and Gregory, P.D. (2010) Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11: 636–646 [DOI] [PubMed] [Google Scholar]
- Xiang, Z-G., Zhang, L-Q., Ning, S-Z., Zheng, Y-L. and Liu, D-C. (2009) Evaluation of Aegilops tauschii for heading date and gene location in a re-synthesized hexaploid wheat. Agricultural Sciences in China 8: 1–7 [Google Scholar]