Abstract
Sleep is important for maintenance of normal physiology in animals. In mammals, neuropeptide Y (NPY), a homolog of Drosophila neuropeptide F (NPF), is involved in sleep regulation, with different effects in human and rat. However, the function of NPF on sleep in Drosophila melanogaster has not yet been described. In this study, we investigated the effects of NPF and its receptor-neuropeptide F receptor (NPFR1) on Drosophila sleep. Male flies over-expressing NPF or NPFR1 exhibited increased sleep during the nighttime. Further analysis demonstrated that sleep episode duration during nighttime was greatly increased and sleep latency was significantly reduced, indicating that NPF and NPFR1 promote sleep quality, and their action on sleep is not because of an impact of the NPF signal system on development. Moreover, the homeostatic regulation of flies after sleep deprivation was disrupted by altered NPF signaling, since sleep deprivation decreased transcription of NPF in control flies, and there were less sleep loss during sleep deprivation and less sleep gain after sleep deprivation in flies overexpressing NPF and NPFR1 than in control flies, suggesting that NPF system auto-regulation plays an important role in sleep homeostasis. However, these effects did not occur in females, suggesting a sex-dependent regulatory function in sleep for NPF and NPFR1. NPF in D1 brain neurons showed male-specific expression, providing the cellular locus for male-specific regulation of sleep by NPF and NPFR1. This study brings a new understanding into sleep studies of a sexually dimorphic regulatory mode in female and male flies.
Introduction
Sleep, consisting of a period of sustained quiescence that is associated with an increased arousal threshold, is a common phenomenon that widely exists in animals from vertebrates to invertebrates [1]. Flies share with mammals a similar sleep regulatory mechanism that involves two relatively independent processes: the circadian system that is responsible for consolidating sleep during the night and the homeostatic system that is responsible for wakefulness and sleep drive. Longer waking periods lead to longer and more intense sleep periods [2]. Sleep regulation in Drosophila melanogaster is anatomically located in the mushroom body, known to be involved in learning and memory [3], while approximately 150 clock neurons in the central nervous system are involved in setting circadian rhythms. These clock neurons are divided into three lateral neuron groups - dorsolateral neurons (LN ds), PDF-positive ventrolateral neurons (LN Vs) and the fifth small ventrolateral neuron (5th small LNv) - and three dorsal neuron groups - dorsal neurons 1, 2, and 3 (DN1, DN2, DN3). The LN vs in the circadian neuronal system contribute to sleep regulation by promoting wakefulness controlled by the GABA receptor and PER protein [4,5,6]. Dorsal neurons function in modulating sleep suppression during starvation [7].
Neuropeptide Y (NPY), a 36-amino-acid peptide from the pancreatic polypeptide (PP) family, is one of the more abundant peptides in the central and peripheral nervous system in mammals [8]. NPY is involved in several physiological functions, such as food intake, hormonal release, circadian rhythms, cardiovascular disease, thermoregulation, stress response and anxiety [9,10,11,12]. In humans, NPY enhances sleep by prolonging the sleep period and reducing sleep latency and wakefulness [13]. In contrast, NPY in rats increases wakefulness and decreases sleep [14]. These findings indicate that NPY regulates sleep differently in different animal species, probably due to their diurnal or nocturnal difference. NPY acts through its trans-membrane receptor (a G-protein-coupled receptor) and through Gi/o signaling pathways (mediated by α or βγ subunits) to inhibit cAMP formation and intracellular Ca2+ mobilization, and to modulate Ca2+ and K+ channels [15].
In invertebrates, neuropeptide F (NPF) and its receptor (NPFR1 is the only identified functional receptor for NPF in D. melanogaster) show conserved structure and function with NPY and NPY receptors in mammals [16]. NPF in D. melanogaster has been characterized for involvement in diverse behavioral responses, such as food intake, hypermobility, cooperative burrowing, alcohol sensitivity and locomotor rhythm [17,18,19,20]. However, regulation of sleep by NPF in invertebrates has not yet been characterized. In this study, we explored NPF function in sleep by over-expressing npf and npfr1 in brain-specific neurons.
Materials and Methods
Fly strains
The npf-gal4, npfr1-gal4, UAS-npf, UAS-npf dsRNA, UAS-npfr1, UAS-npfr1 dsRNA, and UAS-mCD8-GFP strains have been previously described [18,21,22]. The tubP-GAL80[ts] (stock NO. 7018) was purchased from the Bloomington Drosophila Stock Center (Indiana University). The clk 8.0-Gal4 flies were from the laboratory of Dr. Paul E. Hardin (Texas A&M University, Texas, U.S.). Flies were reared on a standard cornmeal–yeast–agar medium at 25°C and 65% relative humidity in a 12 hr L:12 hr D cycle.
Sleep analysis and statistics
Flies were placed in 65 mm × 5 mm glass tubes containing standard fly food at one end. They were acclimated in behavior tubes for at least 24 hr at 25°C in LD conditions, and then data were collected in LD for 4 days with the DAM System (Trikinetics, Waltham, MA) in 1-min bins. Sleep parameters were measured with Pysolo software (obtained from the website: http://www.pysolo.net) using averages over 4 days of LD [23]. Sleep deprivation was performed using a mechanical method to keep flies awake for 12 hours during the night after 3 days of baseline measurement [24]. Statistical analysis was performed in a SPSS program (SPSS, Chicago, IL, USA). Significance levels were determined by independent t-test, and * indicates p < 0.05, ** indicates p < 0.01, and ***indicates p < 0.001.
Mean sleep episode duration and sleep bout number are important for analysis of sleep quality [25]. Sleep latency - the time for flies to fall asleep once the lights are turned off or on during LD cycles - reflects the sleep pressure, which influences sleep-initiation neuronal firing [26]. Here, sleep latency was measured and analyzed from the time of lights off or lights on to the onset of the first sleep episode as previously reported [26].
Immunohistochemistry
NPF-expressing cells were identified through immunofluorescence methods in female and male brains as described previously [20]. The brain samples were viewed with Nikon ECLIPSE TE2000-E and Nikon D-ECLIPSE (Nikon, Tokyo, Japan) confocal microscopes. Confocal images were obtained at an optical section thickness of 1–2 µm, and NPF immunofluorescence was quantified by ImageJ (http://rsb.info.nih.gov/ij/index.html) as previously described [20].
Quantitative real-time PCR for measurements of npf expression level
Npf expression level was analyzed as described in a previous report [20]. Npf amount was determined by the mean of three independent replicates. Significant differences were determined as mentioned above.
Results
The efficiency of Gal4-UAS transgenic system for npf and npfr1 expression
The npf-gal4, UAS-npf, npfr1-gal4, UAS-npfr1, UAS-npf dsRNA i and UAS-NPFR dsRNAi lines have been widely used for regulation of npf and npfr1 expression [18,27]. Our experiments further demonstrated a one and a half - two-fold increase at the transcript level in npf and npfr1 over-expression lines, and a 50% decrease in npf and npfr1 knockdown (dsRNAs) lines (Figure S1).
Sleep regulation by NPF and NPFR1 in D. melanogaster
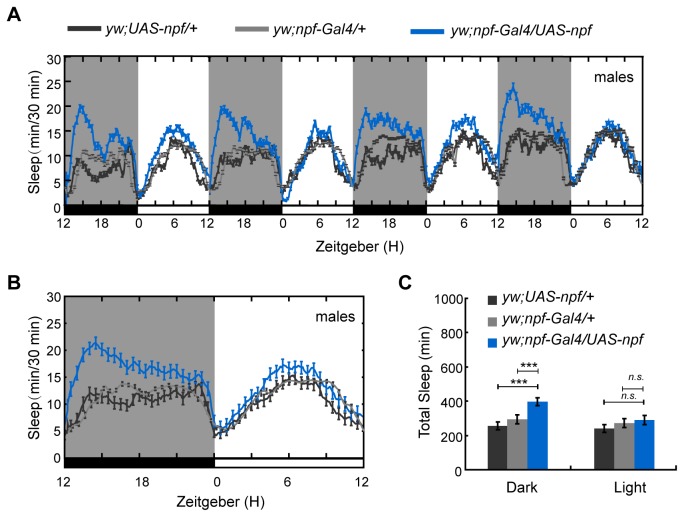
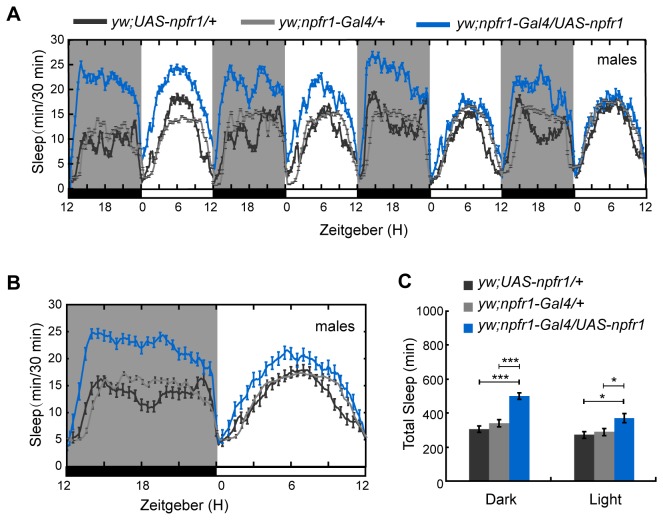
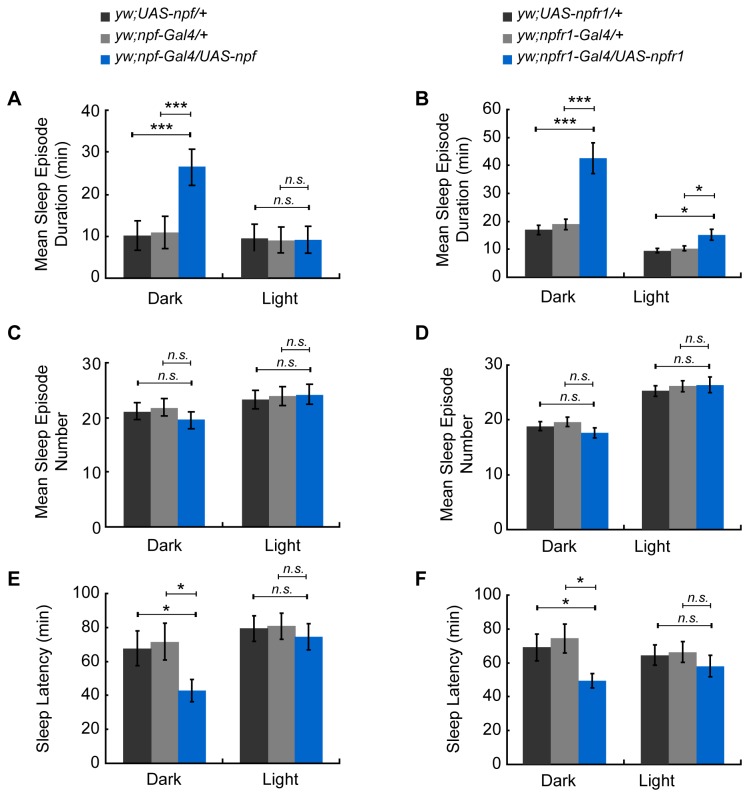
To investigate the role of NPF and NPFR1 in sleep regulation of D. melanogaster, we first used the Gal4-UAS system to increase or decrease npf and npfr1 expression in NPF- and NPFR1-expressing cells in the brain, and detected the sleep amount both in females and males. Under 12 hr L: 12 hr D conditions (LD), male flies with npf over-expression slept more, with a significant increase in whole nighttime sleep (mean±SEM: 392±22 min for npf over-expression line and 221±18 and 292±23 min for UAS and Gal4 controls) (p < 0.001) but not daytime sleep (Figure 1), and males with over-expressed NPFR1 also increased sleep both during daytime (mean±SEM: 350±24 min for npfr1 over-expression line and 241±11 and 243±13 min for UAS and Gal4 control lines) (p < 0.05) and nighttime (mean±SEM: 506±19 min for npfr1 over-expression line and 311±15 and 333±16 min for UAS and Gal4 control lines) (p < 0.001) (Figure 2). Furthermore, the sleep increase at nighttime was due to a significant increase in sleep episode duration in the males of overexpressing npf (mean±SEM: 26.56±4.16 min for npf over-expression line and 10.03±3.21 and 10.89±3.34 min for UAS and Gal4 control lines) (p < 0.001) but not in sleep bout number (Figure 3A, C) and the males of overexpressing npfr1 (mean±SEM: 42.68±5.49 min for npfr1 over-expression line and 17.33±1.79 and 19.86±2.01 min for UAS and Gal4 control lines) (p < 0.001) (Figure 3B & D). Further analysis showed that the sleep latency after lights off was significantly decreased in both the npf- and npfr1- over-expression lines, indicating a higher pressure to fall asleep (Figure 3E-F) (mean±SEM: 42.75±6.51 and 49.45±4.38 min for npf and npfr1 over-expression lines, 67.69±10.13 and 70.38±10.75 min for npf over-expression UAS and Gal4 control lines, and 68.92±7.75 and 74.83±7.26 min for npfr1 over-expression UAS and Gal4 control lines) (p < 0.05). However, sleep in males with npf and npfr1 down-regulated expression (Figure S2) and in females with both up- and down- regulated expression (Figure S3) was not affected in comparison with sleep in the controls. These data indicate that the NPF neuronal system regulates sleep in a sex-dependent manner.
Figure 1. Sleep in male flies with up-regulated npf.
Sleep during the nighttime was significantly increased when npf expression was enhanced (n=60 for controls, and 62 for yw; npf-Gal4/UAS-npf). (A) Daily sleep profile that showed increased sleep during the nighttime in npf over-expressing line. (B) Average daily sleep profile over 4 days that revealed increased sleep during the night. (C) Statistical analysis of sleep. Black and gray histograms indicate the controls, and blue histograms indicate the up-regulated npf line. Black and gray lines indicate controls, and the blue line represents the up-regulated npf line. Error bars indicate the SEM. White bars indicate lights on; black bars indicate lights off. ***, p<0.001; *, p<0.05; n.s., no significant differences.
Figure 2. Sleep in male flies with up-regulated npfr1.
Sleep during the nighttime was significantly increased when npfr1 expression was enhanced (n=61 for controls, and 51 for yw; npfr1-Gal4/UAS-npfr1). (A) Daily sleep profile that showed increased sleep during the nighttime and the daytime in an npfr1 over-expressing line. (B) Average daily sleep profile over 4 days that revealed increased sleep during the night and the day. (C) Statistical analysis of sleep. Labeling is the same as that described in Figure 1.
Figure 3. Analysis of sleep quality and quantity in male flies with up-regulated npf and npfr1.
(A–B) Analysis of sleep episode duration for npf and npfr1. (C–D) Analysis of sleep episode number for npf or npfr1. (E–F) Analysis of sleep latency for npf and npfr1. Labeling is the same as that described in Figure 1.
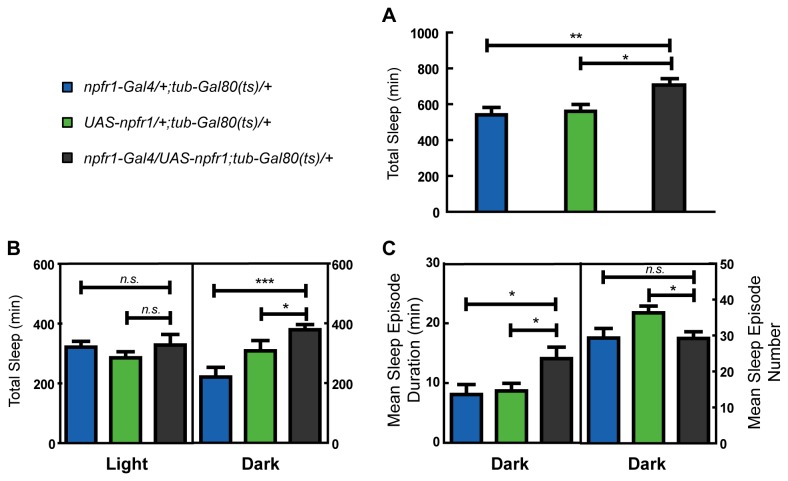
To exclude any impact of increased NPF signaling on embryonic and larval development, npfr1 was over-expressed only in adults. Larvae and newly emerged adults were reared at 18°C to express an active Gal80 protein that represses the activity of Gal4. Newly emerged flies were entrained for three LD cycles at 18oC and then transferred to 30°C to block Gal80, in order to activate Gal4-driven gene expression. Results showed that over-expression of npfr1 only in adults can increase total sleep time, caused by a significant increase in sleep during the night (Figure 4A & B). Further analysis showed that the increase in nighttime sleep was mainly derived from an increased sleep episode duration (Figure 4C), consistent with the results above.
Figure 4. Analysis of sleep in male flies with up-regulated npfr1 during adulthood.
Larvae and newly emerged adults were reared at 18°C, and newly emerged flies were entrained for three LD cycles at 18oC and then transferred to 30°C to block Gal80, in order to activate Gal4-driven gene expression.
(A) Total sleep was increased in npfr1 over-expression flies (n=15 for both the controls and n=13 for npfr1-Gal4/UAS-npfr1;tub-Gal80(ts)). (B) Daily sleep profile showed increased sleep during nighttime in an npfr1 over-expressing line. (C) Sleep increase during nighttime was mainly derived from increased sleep episode duration. Error bars and significant difference are as described above.
Influence of NPF and NPFR1 on sleep homeostasis
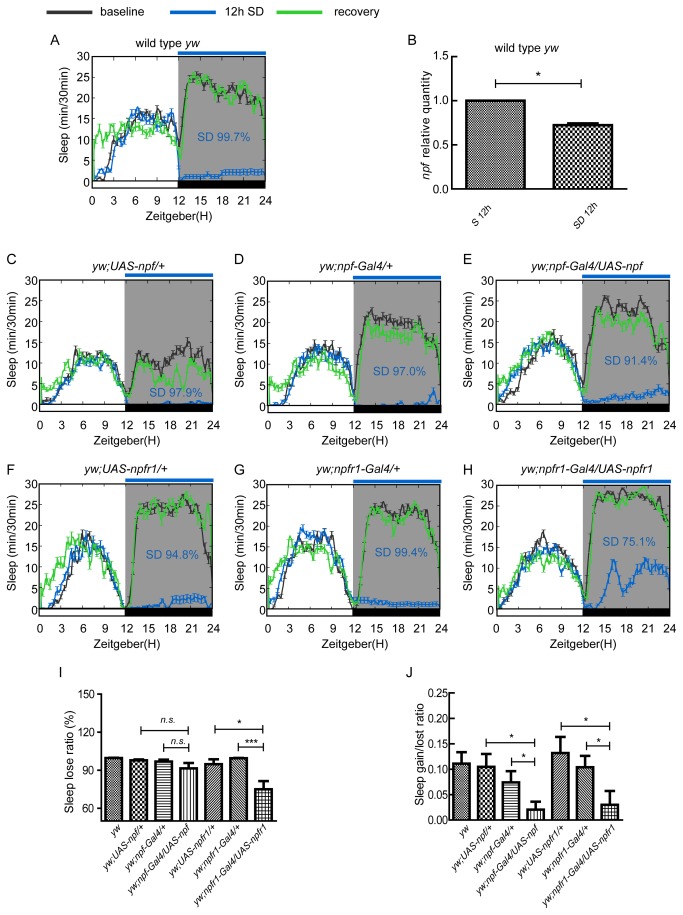
Homeostatic regulation is a key feature of sleep [28]. To investigate whether NPF and NPFR1 have a function in sleep homeostasis, we employed sleep deprivation experiments using a mechanical sleep deprivation method as previously mentioned [24]. One of the key features for sleep homeostatic regulation is that an obvious sleep rebound occurs after sleep deprivation (Figure 5A). We measured the NPF transcript in flies both with and without sleep deprivation, and found that npf expression in the heads of adult flies was down-regulated after sleep deprivation for 12h (Figure 5B), suggesting that NPF signaling plays an important role in sleep homeostasis. Consistent with such a role, when npf expression was up-regulated, flies exhibited less sleep loss (Figure 5I: 91.4% in an npf up-regulated line compared to 97.9% and 97.0% in the UAS and Gal4 control lines), and sleep loss was significantly reduced in npfr1up-regulated lines (Figure 5I: 75.1% in npfr1 up-regulated line compared to 94.8% in UAS control line (p = 0.03) and 99.4% in Gal4 control line (p = 0.0003)). Correspondingly flies with up-regulated npf and npfr1 expression showed smaller sleep rebound after sleep deprivation than did the controls (Figure 5 C–H). The sleep rebound for npf up-regulated flies was 13.24±4.93, which was smaller than UAS (25.18±5.81, p = 0.13) and Gal4 controls (29.00±9.31, p = 0.12) although the differences were not statistically significant. The sleep rebound for npfr1 up-regulated flies was 15.63±6.93, which was significantly smaller than UAS (57.00±10.831, p = 0.024) and Gal4 controls (32.90±7.31, p = 0.038). The sleep gain/loss ratio in flies with up-regulated npf and npfr1 expression was also lower (0.02±0.01 for the npf up-regulation line, and 0.03±0.02 for the npfr up-regulation line) than the controls (0.10±0.03, 0.07±0.02 for npf up-regulation UAS and Gal4 controls, and 0.13±0.03, 0.10±0.02 for npfr1 up-regulation UAS and Gal4 controls) (Figure 5J). These results indicate that NPF and NPFR1 play an important role in the homeostatic regulation of sleep.
Figure 5. The homeostatic sleep response regulated by NPF signal in males.
(A) Sleep rebound after 12 hour sleep deprivation in wild type flies (yw) is depicted. (B) npf expression was reduced after 12 hour sleep deprivation. (C, D and E) NPF regulated sleep homeostasis, in which C and D are controls for E. Sleep rebound was reduced when npf expression was enhanced (n=19-22 for controls, and n=20 for yw; npf-Gal4/UAS-npf). (F, G and H) NPFR1 regulated sleep homeostasis, in which F and G are controls for H. Sleep rebound was reduced when npfr1 expression was enhanced (n=21-24 for controls, and n=23 for yw; npfr1-Gal4/UAS-npfr1). Error bars and significant differences are as described above. (I) Sleep loss is reduced after sleep deprivation in npf and npfr1 up-regulated lines. (J) Sleep gain/lost ratio is reduced after sleep deprivation in npf and npfr1 up-regulated lines. Error bars and significant difference are as described above.
Npf levels in male and female flies during nighttime
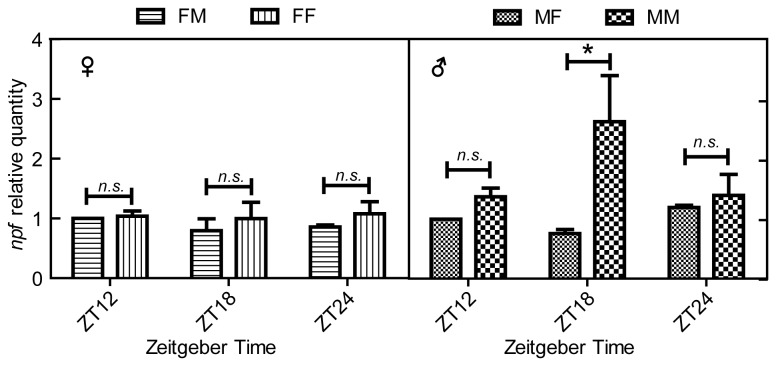
Previous studies showed that the nocturnal phase of Drosophila courtship behavior is dependent on the male and not the female circadian clock [29], indicating there exists a sex-specific regulation pathway to balance sleep and mating behavior in male flies. Might NPF signaling in males participate in this pathway? Is NPF signaling decreased to suppress sleep for mating behavior in males when females are present or increased to promote sleep in males when females are not present? To explore these questions, we analyzed the npf expression levels in wild type males and females flies during nighttime with the presence of opposite sex or same sex couples. Results showed that the npf expression levels in female flies were not changed whether male flies were present or not (Figure 6). In contrast, when females were present, male flies expressed a lower npf level at midnight (ZT18) that could promote increased activity for courtship behavior, and when females were not present males expressed a higher level of npf that could promote sleep (Figure 6).
Figure 6. Brain npf levels in male and female flies.
(A) npf levels in female flies with the presence of males (FM) and female flies without the presence of males (FF): (B) npf levels in male flies with the presence of females (MF) and male flies without the presence of females (MM). Error bars and significant differences are as described above.
The neuronal localization of NPF
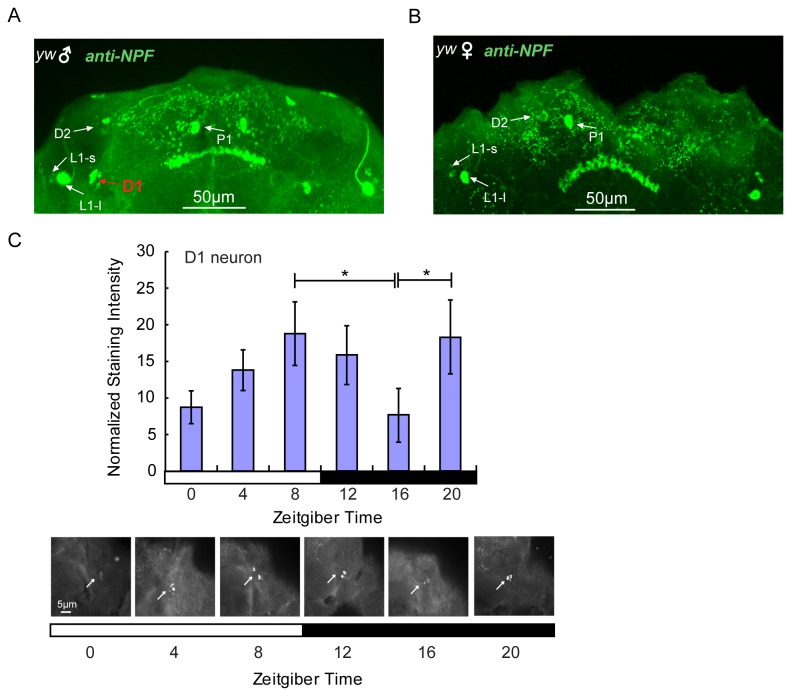
We found in this study that sleep regulation by NPF in flies is sex-dependent. What are the structures that mediate sex-dependent regulation of sleep? Previous reports showed that NPF was expressed in two groups of dorsal neurons (D1 and D2), two groups of dorsomedial neurons (P1 and P2), two groups of dorsolateral neurons (L1-l and L1-s), one lateroventral group (L2), one subesophageal group (L2 and S) and the fan-shaped body (FB) [20,27,30]. Interestingly, NPF expression was male-specific in 2-4 D1 noncircadian neurons (Figure 7A & B). Furthermore, daily expression of NPF in D1 neurons exhibited two daily peaks - one at ZT8 and another at ZT20 (Figure 7C). The peak at ZT20 is consistent with the npf mRNA peak at ZT18 (Figure 6) in males like these in Figure 7C, which are isolated from females at this time.
Figure 7. Male specific expression of NPF in male brain.
(A) anti-NPF labeled NPF localization in male brains in which D1 is male-specific. (B) NPF localization in female brains in which D1 is not detected. (C) NPF oscillation in male-specific D1 neurons. The arrows in the images represent the staining intensity of NPF at ZT0, ZT 4, ZT8, ZT12, ZT16 and ZT20. The ZT8 and ZT20 are compared with ZT16 for statistical difference analysis (*, p < 0.05). Scale bar: 5 µm for neuron figures.
Discussion
In this study, we analyzed the function of NPF and NPFR1 on the sleep phenotype in Drosophila and found that NPF and NPFR1 both promote sleep during the night in male flies, in a similar fashion to NPY’s effect in humans. Either NPF or NPFR1 over-expression causes an increase in nighttime sleep by increasing average sleep episode duration and reducing sleep latency during the night. NPF and NPFR1 also play an important role in the homeostatic regulation of sleep. NPF and NPFR1 may regulate sleep in Drosophila because NPFR1 activated by NPF inhibits cAMP formation [15]. The activity of cAMP-dependent PKA has been reported to be involved in sleep regulation in Drosophila [3,31].
Sexually dimorphic behaviors are widely observed in invertebrate and vertebrate species. Locomotor activity in Drosophila was found to show sexual dimorphism [32,33]. However, sexually dimorphic regulation of sleep has rarely been reported. Sleep in the cyc 01 mutant fly is sexually dimorphic with reduced or absent locomotion in males and exaggerated locomotion in compensatory rebound after sleep deprivation in females [34]. Fujii et al. have found a male-specific function in courtship, indicating a male-specific signal circuit might function in this sexually dimorphic behavior [30]. The NPF signal system is shown to participate in the circuit in this paper. In Drosophila , sexual differentiation is ultimately dependent on a chromosomal signal that is different between males (XY) and females (XX), resulting in repression or activation of the key gene Sexlethal (Sxl), responsible for dosage compensation, somatic sexual development, oogenesis and sexually dimorphic neural development. The subordinate gene transformer (tra) is one of the downstream regulatory genes of Sxl that controls sex determination [35]. The expression of npf in Drosophila is regulated in both sex-nonspecific and male-specific (ms) manners, and the ms-npf expression is controlled by the tra-dependent sex-determination pathway [27], indicating NPF is regulated by the sexual differentiation pathway. The male-specific NPF brain neurons (D1) may provide the neuronal locus for male-specific regulation of sleep.
The circadian system is involved in fly sleep regulation, and the circadian pacemaker neurons participate in regulating sleep. Some NPF in fly brains is found in clock neurons including the LN vs, LN ds and DN1s, among which the LN vs have been shown to promote wakefulness [4,5,6], while the dorsal circadian neurons modulate sleep suppression during starvation [7]. We found that NPF and NPFR1 promote sleep quality especially during the night by prolonging average sleep episode duration and reducing night sleep latency in males. While these effects are associated with D1-specific expression in males, it is possible that the clock-neuron specific expression in males also contributes to these effects, which are absent in females because the neural circuit is incomplete (i.e., clock-neuron specific expression is present but D1 expression is absent.).
In humans, sleep is restorative and plays a crucial role in long-term memory storage and learning. Sleep disorders cause a series of health problems associated with cognitive deficiency, poor job performance and low productivity [36]. In Drosophila , aging and diet are associated with sleep change [37,38]. Starvation suppresses sleep and sleep deprivation is known to affect longevity [7,39]. Therefore, it is likely that the function of NPF involves regulation of interactions among feeding, sleep, copulation and even longevity.
Supporting Information
Detection of expression in the transgenic genotypes for npf and npfr1 mRNAs by qRT-PCR. The Gal4-driven expression of npf and npfr1 increased transcripts by 1.5 to 2 fold. Double strand RNA (dsRNA) for npf and npfr1 expression caused about a 50% decrease in expression.
(TIF)
Sleep in male flies with down-regulated npf and npfr1. (A) Average daily sleep profile over 4 days in male flies with down-regulated npf (n=31 for each control, and 32 for yw; npf-Gal4/UAS-npf dsRNA). (B) Statistical analysis of sleep. (C) Average daily sleep profile over 4 days in male flies with down-regulated npfr1 (n=32 for each control, and 36 for yw; npf-Gal4/UAS-npf dsRNA). (D) Statistical analysis of sleep. Labeling is the same as that described in Figure 1.
(TIF)
Sleep is unchanged when npf or npfr1 expression is up- or down-regulated in female flies. (A) Total sleep in npf over-expressing females is maintained at a similar level with sleep in the control flies except for a small increase before lights on (n=32 for controls, and 34 for yw; npf-Gal4/UAS-npf). (B) The total sleep in npf-down-regulated females is not changed either (n=32 for controls, and 36 for yw; npf-Gal4/UAS-npf dsRNA). (C) Total sleep in females with npfr1 over-expression is similar to that in control flies (n=54 for controls and 49 for yw; npfr1-Gal4/UAS-npfr1). (D) The total sleep in npfr1-down-regulated females is not changed (n=59 for controls, and 58 for yw; npfr1-Gal4/UAS-npfr1 dsRNA). Linear graphs, error bars, white and black bars, and white and gray background are as indicated in Figure 1.
(TIF)
Acknowledgments
UAS- fly lines were kindly provided by P. Shen (University of Georgia, Athens, GA USA) and npf and npfr1 Gal4- fly lines by Y. Rao (Peking University, Beijing, China).
Funding Statement
This work was supported by the National Basic Research Program from the Ministry of Science and Technology of the People’s Republic of China (“973” Program Grant number 2012CB114100), the National Natural Science Foundation of China (Grant number 31272371), the Doctoral Scientific Fund Project of the Ministry of Education of China (Grant number 20110008110018) to Z. Zhao, and the Innovation Fund for Graduate Students of China Agricultural University (Grant number KYCX2011004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cirelli C, Bushey D (2008) Sleep and wakefulness in Drosophila melanogaster. Molecular and Biophysical Mechanisms of Arousal, Alertness, and Attention 1129: 323-329. PubMed: 18591491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cirelli C (2006) Sleep disruption, oxidative stress, and aging: New insights from fruit flies. Proc Natl Acad Sci U S A 103: 13901-13902. doi:10.1073/pnas.0606652103. PubMed: 16966597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joiner WJ, Crocker A, White BH, Sehgal A (2006) Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441: 757-760. doi:10.1038/nature04811. PubMed: 16760980. [DOI] [PubMed] [Google Scholar]
- 4. Parisky KM, Agosto J, Pulver SR, Shang YH, Kuklin E et al. (2008) PDF Cells Are a GABA-Responsive Wake-Promoting Component of the Drosophila Sleep Circuit. Neuron 60: 672-682. doi:10.1016/j.neuron.2008.10.042. PubMed: 19038223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bushey D, Tononi G, Cirelli C (2011) Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332: 1576-1581. doi:10.1126/science.1202839. PubMed: 21700878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shang Y, Haynes P, Pírez N, Harrington KI, Guo F et al. (2011) Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14: 889-895. doi:10.1038/nn.2860. PubMed: 21685918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keene AC, Duboué ER, McDonald DM, Dus M, Suh GSB et al. (2010) Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol 20: 1209-1215. doi:10.1016/j.cub.2010.05.029. PubMed: 20541409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larhammar Dan, Blomqvist AG, Söderberg C (1993) Evolution of neuropeptide Y and its related peptides. Comp Biochem Physiol 106C: 743-752. PubMed: 7905810. [DOI] [PubMed] [Google Scholar]
- 9. Vezzani A, Sperk G, Colmers WF (1999) Neuropeptide Y: emerging evidence for a functional role in seizure modulation. Trends Neurosci 22: 25-30. doi:10.1016/S0166-2236(98)01284-3. PubMed: 10088996. [DOI] [PubMed] [Google Scholar]
- 10. Thorsell A, Heilig M (2002) Diverse functions of neuropeptide Y revealed using genetically modified animals. Neuropeptides 36: 182-193. doi:10.1054/npep.2002.0897. PubMed: 12359508. [DOI] [PubMed] [Google Scholar]
- 11. Herzog H (2003) Neuropeptide Y and energy homeostasis: insights from Y receptor knockout models. Eur J Pharmacol 480: 21-29. doi:10.1016/j.ejphar.2003.08.089. PubMed: 14623347. [DOI] [PubMed] [Google Scholar]
- 12. Pedrazzini T, Pralong F, Grouzmann E (2003) Neuropeptide Y: the universal soldier. Cell Mol Life Sci 60: 350-377. doi:10.1007/s000180300029. PubMed: 12678499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Antonijevic IA, Murck H, Bohlhalter S, Frieboes RM, Holsboer F et al. (2000) Neuropeptide Y promotes sleep and inhibits ACTH and cortisol release in young men. Neuropharmacology 39: 1474-1481. doi:10.1016/S0028-3908(00)00057-5. PubMed: 10818263. [DOI] [PubMed] [Google Scholar]
- 14. Tóth A, Hajnik T, Záborszky L, Détári L (2007) Effect of basal forebrain neuropeptide Y administration on sleep and spontaneous behavior in freely moving rats. Brain Research Bulletin 72: 293-301. doi:10.1016/j.brainresbull.2007.01.006. PubMed: 17452289. [DOI] [PubMed] [Google Scholar]
- 15. Holliday ND, Michel MC, Cox HM (2004) NPY Receptor Subtypes and Their Signal Transduction. Neuropeptide Y and related peptides: 45-73. [Google Scholar]
- 16. Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW (2002) Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides 23: 773-780. doi:10.1016/S0196-9781(01)00647-7. PubMed: 11897397. [DOI] [PubMed] [Google Scholar]
- 17. Wu Q, Wen TQ, Lee G, Park JH, Cai HN et al. (2003) Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39: 147-161. doi:10.1016/S0896-6273(03)00396-9. PubMed: 12848939. [DOI] [PubMed] [Google Scholar]
- 18. Wen TQ, Parrish CA, Xu D, Wu Q, Shen P (2005) Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A 102: 2141-2146. doi:10.1073/pnas.0406814102. PubMed: 15677721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dierick HA, Greenspan RJ (2007) Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet 39: 678-682. doi:10.1038/ng2029. PubMed: 17450142. [DOI] [PubMed] [Google Scholar]
- 20. He C, Cong X, Zhang R, Wu D, An C et al. (2013) Regulation of circadian locomotor rhythm by neuropeptide Y-like system in Drosophila melanogaster. Insect Mol Biol, 22: 376–88. doi:10.1111/imb.12027. PubMed: 23614491. PubMed: 23614491 [DOI] [PubMed] [Google Scholar]
- 21. Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451-461. doi:10.1016/S0896-6273(00)80701-1. PubMed: 10197526. [DOI] [PubMed] [Google Scholar]
- 22. Lingo PR, Zhao Z, Shen P (2007) Co-regulation of cold-resistant food acquisition by Insulin- and neuropeptide Y-like systems in Drosophila melanogaster. Neuroscience 148: 371-374. doi:10.1016/j.neuroscience.2007.06.010. PubMed: 17658221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilestro GF, Cirelli C (2009) pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics 25: 1466-1467. doi:10.1093/bioinformatics/btp237. PubMed: 19369499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishimoto H, Kitamoto T (2010) The steroid molting hormone ecdysone regulates sleep in adult Drosophila melanogaster. Genetics 185: 269-281. doi:10.1534/genetics.110.114587. PubMed: 20215472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiu JC, Low KH, Pike DH, Yildirim E, Edery I (2010) Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp 43: 2157 PubMed: 20972399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M et al. (2008) Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 11: 354-359. doi:10.1038/nn2046. PubMed: 18223647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee GH, Bahn JH, Park JH (2006) Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc Natl Acad Sci U S A 103: 12580-12585. doi:10.1073/pnas.0601171103. PubMed: 16894172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G et al. (2004) Sleep homeostasis in Drosophila melanogaster. Sleep 27: 628-639. PubMed: 15282997. [DOI] [PubMed] [Google Scholar]
- 29. Fujii S, Krishnan P, Hardin P, Amrein H (2007) Nocturnal male sex drive in Drosophila. Curr Biol 17: 244-251. doi:10.1016/j.cub.2006.11.049. PubMed: 17276917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hermann C, Yoshii T, Dusik V, Helfrich-Förster C (2012) The neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol 520: 970-987. doi:10.1002/cne.22742. PubMed: 21826659. [DOI] [PubMed] [Google Scholar]
- 31. Wu MN, Ho K, Crocker A, Yue Z, Koh K et al. (2009) The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci 29: 11029-11037. doi:10.1523/JNEUROSCI.1653-09.2009. PubMed: 19726661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belgacem YH, Martin JR (2002) Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc Natl Acad Sci U S A 99: 15154-15158. doi:10.1073/pnas.232244199. PubMed: 12399547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gatti S, Ferveur JF, Martin JR (2000) Genetic identification of neurons controlling a sexually dimorphic behaviour. Curr Biol 10: 667-670. doi:10.1016/S0960-9822(00)00517-0. PubMed: 10837249. [DOI] [PubMed] [Google Scholar]
- 34. Hendricks JC, Lu SM, Kume K, Yin JCP, Yang ZH et al. (2003) Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms 18: 12-25. doi:10.1177/0748730402239673. PubMed: 12568241. [DOI] [PubMed] [Google Scholar]
- 35. Schütt C, Nöthiger R (2000) Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127: 667-677. PubMed: 10648226. [DOI] [PubMed] [Google Scholar]
- 36. Caylak E (2009) The genetics of sleep disorders in humans: narcolepsy, restless legs syndrome, and obstructive sleep apnea syndrome. Am J Med Genet A 149A: 2612-2626. doi:10.1002/ajmg.a.33087. PubMed: 19876894. [DOI] [PubMed] [Google Scholar]
- 37. Bushey D, Hughes KA, Tononi G, Cirelli C (2010) Sleep, aging, and lifespan in Drosophila. BMC Neurosci 11: 56. doi:10.1186/1471-2202-11-56. PubMed: 20429945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Catterson JH, Knowles-Barley S, James K, Heck MM, Harmar AJ et al. (2010) Dietary modulation of Drosophila sleep-wake behaviour. PLOS ONE 5: e12062. doi:10.1371/journal.pone.0012062. PubMed: 20706579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald DM, Keene AC (2010) The sleep-feeding conflict: Understanding behavioral integration through genetic analysis in Drosophila. Aging-Us 2: 519-522. PubMed: 20689154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of expression in the transgenic genotypes for npf and npfr1 mRNAs by qRT-PCR. The Gal4-driven expression of npf and npfr1 increased transcripts by 1.5 to 2 fold. Double strand RNA (dsRNA) for npf and npfr1 expression caused about a 50% decrease in expression.
(TIF)
Sleep in male flies with down-regulated npf and npfr1. (A) Average daily sleep profile over 4 days in male flies with down-regulated npf (n=31 for each control, and 32 for yw; npf-Gal4/UAS-npf dsRNA). (B) Statistical analysis of sleep. (C) Average daily sleep profile over 4 days in male flies with down-regulated npfr1 (n=32 for each control, and 36 for yw; npf-Gal4/UAS-npf dsRNA). (D) Statistical analysis of sleep. Labeling is the same as that described in Figure 1.
(TIF)
Sleep is unchanged when npf or npfr1 expression is up- or down-regulated in female flies. (A) Total sleep in npf over-expressing females is maintained at a similar level with sleep in the control flies except for a small increase before lights on (n=32 for controls, and 34 for yw; npf-Gal4/UAS-npf). (B) The total sleep in npf-down-regulated females is not changed either (n=32 for controls, and 36 for yw; npf-Gal4/UAS-npf dsRNA). (C) Total sleep in females with npfr1 over-expression is similar to that in control flies (n=54 for controls and 49 for yw; npfr1-Gal4/UAS-npfr1). (D) The total sleep in npfr1-down-regulated females is not changed (n=59 for controls, and 58 for yw; npfr1-Gal4/UAS-npfr1 dsRNA). Linear graphs, error bars, white and black bars, and white and gray background are as indicated in Figure 1.
(TIF)