Abstract
Rev-erbα is a ubiquitously expressed orphan nuclear receptor which functions as a constitutive transcriptional repressor and is expressed in vertebrates according to a robust circadian rhythm. We report here that two Rev-erbα mRNA isoforms, namely Rev-erbα1 and Rev-erbα2, are generated through alternative promoter usage and that both show a circadian expression pattern in an in vitro system using serum-shocked fibroblasts. Both promoter regions P1 (Rev-erbα1) and P2 (Rev-erbα2) contain several E-box DNA sequences, which function as response elements for the core circadian-clock components: CLOCK and BMAL1. The CLOCK–BMAL1 heterodimer stimulates the activity of both P1 and P2 promoters in transient transfection assay by 3–6-fold. This activation was inhibited by the overexpression of CRY1, a component of the negative limb of the circadian transcriptional loop. Critical E-box elements were mapped within both promoters. This regulation is conserved in vertebrates since we found that the CLOCK–BMAL1 heterodimer also regulates the zebrafish Rev-erbα gene. In line with these data Rev-erbα circadian expression was strongly impaired in the livers of Clock mutant mice and in the pineal glands of zebrafish embryos treated with Clock and Bmal1 antisense oligonucleotides. Together these data demonstrate that CLOCK is a critical regulator of Rev-erbα circadian gene expression in evolutionarily distant vertebrates and suggest a role for Rev-erbα in the circadian clock output.
Introduction
Circadian rhythms in physiology and behavior are observed throughout the animal and plant kingdoms as well as in fungi and bacteria. They are believed to enable organisms to anticipate and adapt to rhythmic changes in their environment (Lowrey & Takahashi 2000, Young & Kay 2001, Roenneberg & Merrow 2003). These daily oscillations persist under constant conditions; that is, in the absence of external time cues. In mammals, circadian rhythms influence many if not most aspects of physiology and behavior, including sleep/wake cycles, energy metabolism, heartbeat, blood pressure and body temperature.
Recently, genetic and biochemical approaches have identified genes that contribute to the generation of circadian rhythms in mammals as well as in Drosophila, zebrafish, Arabidopsis and Neurospora (Young & Kay 2001). The current model states that, in vertebrates, a transcriptional-translational feedback loop is established by the isochronal action of transcriptional regulators such as Period 1–3 (PER 1–3) and Cryptochrome 1–2 (CRY 1–2). These proteins enter the nucleus at a specific time of the day to block the interaction between the CLOCK–BMAL1 and the MOP4– BMAL1 heterodimers and their cognate E-box element, thereby interfering with the transcription of clock-regulated genes possibly by modifying histone acetylation (Chong et al. 2000, Lowrey & Takahashi 2000, Etchegaray et al. 2003). Several recent reports revealed the importance of the regulation of the nuclear entry of PER and CRY proteins. Indeed, it has been shown that phosphorylation by casein kinase I8 and I´ plays an important role in regulating the subcellular location and stability of these proteins (Lowrey & Takahashi 2000, Shearman et al. 2000, Lee et al. 2001, Vielhaber et al. 2001, Akashi et al. 2002, Yagita et al. 2002).
In mammals, the ‘master clock’ controlling circadian rhythms resides in the suprachiasmatic nuclei (SCN) of the hypothalamus (reviewed in Ripperger and Schibler 2001). To keep pace with the solar day/night cycle, the master clock can be entrained by light received through photoreceptors in the retina (Foster 1998, Abe et al. 1999). Recent evidence, however, suggests that other endogenous oscillators independent of the central pacemaker in the SCN exist and are widespread in peripheral tissues in mammals, Drosophila and zebrafish (Tosini and Menaker 1996, Plautz et al. 1997, Whitmore et al. 1998, Yamakazi et al. 2000; reviewed by Brown and Schibler 1999). The ultimate proof of the existence of peripheral oscillators has been the demonstration that circadian rhythms of clock gene expression can be induced by a serum shock administered to serum-starved immortalized rat fibroblasts in culture (Balsalobre et al. 1998, Yagita et al. 2001). The existence of these peripheral clocks can be extended to embryos since early zebrafish embryos contain a circadian clock that can be reset by light (Delaunay et al. 2000). All these data suggest that most cells, if not all, may have a circadian clock. It is believed that the SCN clock entrains the phase of peripheral clocks via chemical cues, such as rhythmically secreted hormones. Furthermore, peripheral clocks can be regulated independently from the master clock since in rodents restricted feeding during the day can completely reverse the phase of circadian oscillators in the liver but has apparently no effect on the central oscillator in the SCN (Damiola et al. 2000, Stokkan et al. 2001).
Although the core molecular pacemaker generating circadian rhythm has been defined both in the SCN and peripheral organs, the molecular outputs that ultimately regulate circadian control of cellular physiology, organ function and behavior are poorly understood (Jin et al. 1999). Specifically, the link between circadian transcriptional output and physiology under circadian control is missing. To decipher how the circadian clock is able to control output pathways it is important to identify clock-controlled genes, i.e. genes that are under the direct transcriptional control of the CLOCK– BMAL1 heterodimer.
Rev-erbα (NR1D1; Nuclear Receptors Nomenclature Committee 1999), a gene that encodes an orphan member of the nuclear receptor superfamily (Laudet & Gronnemeyer 2002) is a potential candidate as a clock-controlled gene. Indeed, we and others have shown that its expression exhibits a robust circadian rhythm in shocked fibroblasts or human hepatic cells, in rodent liver and also in zebrafish (Balsalobre et al. 1998, Delaunay et al. 2000, Torra et al. 2000, Grundschober et al. 2001). Rev-erbα appears particularly interesting as a putative clock-controlled gene because it was already known that its expression is under tight transcriptional control by a number of factors. The characterization of its promoter in humans lead to the demonstration that the expression of this gene is down-regulated by its own product, which behaves as a potent transcriptional repressor (Harding & Lazar 1995, Adelmant et al. 1996). In addition, human Rev-erbα promoter activity is inhibited by glucocorticoids (Torra et al. 2000) that are known to play an important role in the resetting of circadian time in peripheral tissues (Balsalobre et al. 2000). Finally, Rev-erbα expression is regulated by fibrates, hypolipidemic drugs that are ligands of the nuclear hormone receptor peroxisome proliferator-activated receptor a (PPARa; Fruchart et al. 1999). Interestingly, PPARa itself is expressed according to circadian rhythm (Lemberger et al. 1996), suggesting that the Rev-erbα gene can integrate several levels of regulation, both at the circadian and physiological levels. The link between Rev-erbα and circadian rhythm has been reinforced recently by the observation that Rev-erbα-deficient mice exhibit a circadian phenotype and that Rev-erbα controls the cyclic expression of Bmal1 (Preitner et al. 2002).
In this paper we report that the two promoters governing the expression of the Rev-erbα gene in mammals contain E-box DNA elements and generate circadian transcripts. We show that both promoters are activated by CLOCK– BMAL1 heterodimers in transient transfections and that CLOCK–BMAL1 heterodimers bind to E-box sequences and activate transcription through these E-boxes. Rev-erbα expression is strongly reduced in the livers of Clock mutant mice suggesting that the activation observed in transient transfections also occurs in vivo. In addition, we show that this regulation is evolutionarily conserved since CLOCK–BMAL1 also regulates the expression of the zebrafish Rev-erbα gene. Taken together, these data demonstrate clearly that the Rev-erbα gene is a clock-controlled gene.
Materials and methods
Plasmid constructs and vectors
All genomic Rev-erbα regions were subcloned in pGL2 luciferase reporter plasmids after PCR with oligonucleotides containing enzymatic restriction sites. The oligonucleotides were as follows. P1, including the E1A exon as a 1350 bp fragment: 1–24 MluI, 5’-ATTAACGCGTCTGCAGGGAG CAGACCCCCCTCTA-3’, top strand; 1331–1350 XhoI, 5’-ATTACTCGAGCTGTGTTGTTGTT GGAGTCTAG-3’, bottom strand. P2, including exon E1B as a 1852 bp fragment: 1353–1375 KpnI, 5’-ATTAGGTACCGTACTGAGATTCT TATCTTTGCT-3’, top strand; 3920–3942 XhoI, 5’-ATTACTCGAGCTAGGAAGAGAGCACGA GGGGAG-3’, bottom strand. P1+P2 as a 3142 bp fragment: 1–24 MluI and 3920–3942 XhoI.
Deletion constructs in P1 were generated by PCR using the following oligonucleotides. D400P1-Luc: 458–478 KpnI, 5’-ATTAAGGTACCCC AGGAATTCACATGCCCTTG-3’, top strand, and the 1331–1350 XhoI oligonucleotide for the bottom strand. Mutations of a1 and a2 sites in the P1 promoter were produced by PCR using the following oligonucleotides. a1*D400-P1-Luc: a1*458–478 KpnI, 5’-ATTAAGGTACCCCAGG AATTCACGGGCCCTTG-3’, top strand, and the 1331–1350 XhoI oligonucleotide for the bottom strand. a2*-P1-Luc: 1–24 MluI as top strand with a2*709–732 KpnI, 5’-ATTAGGTACCATTGAA TTCCAGGGAGCG-3’, bottom strand. Subsequently, PCR products were ligated into P1 at the KpnI site. Each construct was sequenced on both strands.
Three deletion constructs of the P2 promoter were generated: DistalP2-Luc, D800-P2-Luc and ProximalP2-Luc. The DistalP2-Luc region was obtained by PCR with the 1353–1375 KpnI oligonucleotide, top strand, and the 2215–2190 HindIII oligonucleotide, bottom strand, 5’-ATT AAAGCTTTGCCCCTCGCACGTGGCACC-3’. The D800-P2-Luc region was obtained by PCR amplification using a 2196–3218 KpnI, 5’-ATTA AGGTACCGATCCTCGTTGGGGTGCCACG T-3’ oligonucleotide, top strand, and a 3920–3942 XhoI oligonucleotide as bottom strand. The resulting PCR fragments were cloned in pGL2 vector and sequenced. The proximal P2 region was generated by enzymatic digestion of the P2-Luc vector with SmaI and PvuII before ligation.
The positive control vector (Per1E)3-Luc was generated by ligation in triplicate of the following oligonucleotide: 5’-AGATCCAAGTCCACGTG CAGGGC-3’ harboring the mPer1 E-box sequence, upstream of a synthetic TATA-box oligonucleotide (5’-AGATCTGCATCGGGTATA TAATAA-3’). The whole fragment was cloned as a XhoI–HindIII fragment in the pGL2 vector. An E-box-mutated version of this oligonucleotide (5’-AGATCCAAGTCCAATTGCAGGGC-3’) was used to generate the negative control vector (Per1Em)3-Luc.
Expression vectors were designed as follows: the coding region of mouse Cry1 was obtained by reverse transcriptase (RT)-PCR using the following oligonucleotides: top strand, 5’-GATCAAGCTT ACCATGGACTACAAGGACGAC-3’; bottom strand, 5’-CCAGCCTCCTTGGCCATCTTC AT-3’. The PCR product was cloned in the pCDNA3 vector and sequenced.
Expression vectors for the mouse Clock was provided by JST and the human BMAL1 generously provided by M Ikeda (Waseda University, Tokyo, Japan). For all vectors, PCR amplifications of cDNA were performed to replace the natural ATG by a consensus Kozak sequence to ensure translation with high yield. For this, oligonucleotides containing an efficient Kozak sequence (ACC) upstream of the ATG were used as 5’ primers and a 20 bp antisense oligonucleotide overlapping the stop codon as the 3’ primer. Oligonucleotides are available from G T upon request. The β-gal expression vector was constructed from the pCMV-SPORT-βgal vector (Life Technologies, Cergy Pontoise, France) by cloning an ApaI–KpnI fragment containing the entire open reading frame into the pCDNA3 vector.
Zebrafish Rev-erbα promoter fragments were obtained by PCR amplification on a genomic DNA clone (isolated from a Danio rerio DNA library) and subcloned into a pGL3 vector. The oligonucleotides were as follows. For the 3·2 kb promoter long fragment A: top strand, 5’-GAGAGCTCGC GGCCGCGAGCTC-3’; bottom strand, 5’-TCG AAGCTTCGCACCAAATACGTGCGC-3’. The resulting PCR product was cut by SalI and HindIII restriction enzymes and subcloned into XhoI– HindIII-digested pGL3 vector. For the 1·4 kb promoter long fragment B and the 0·4 kb promoter short fragment C: top strand (B fragment), 5’-GATGAGCTCCGAGGTAAATATCACCAC3’; top strand (C fragment), 5’-GATGAGCTCC TCTTTGACTTCGACTAC-3’; bottom strand (B and C fragments), 5’-TCGGGATCCCGCACC AAATACGTGCGC-3’. The resulting PCR products were digested by SacI and BamHI restriction enzymes and subcloned into SacI–BglII-digested pGL3 vector.
Mutated sites of E-boxes were generated using two successive PCRs with the following oligonucleotides. Top strand E-box (ZFg1), 5’-TCAGGTG GACAATTGCGCGGGGGT-3’; bottom strand E-box(ZFg1), 5’-ACCCCCGCGCAATTGTCCA CCTGA-3’; top strand E-box(Zfa), 5’-AGTCGGG TCCATAGGACACATTT-3’; bottom strand E-box(Zfa), 5’-AAATGTGTCCTATGGACCCG ACT-3’.
Accession numbers for Rev-erbα genomic sequences are: AY336123, AY336124, AY336125 and AY336126 for human, mouse, rat and zebrafish species respectively.
Transient transfections and reporter assays in mammalian cells
Cos1, Ros17·2/8, 3Y1 and Rat-1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 5% fetal calf serum and 100 U/ml penicillin/streptomycin. Typically, 40 ng of the reporter vectors were co-transfected with 100 ng of each expression vector in 12-well plates. When necessary, the final DNA concentration was adjusted to 240 ng with the pCDNA3-βgal vector. Transfection was achieved using 2·5×104 cells in 0·5% fetal calf serum with 2 µl Exgen 500 transfection reagent (Euromedex, Souffelweyersheim, France) mixed with plasmids in 100 µl DMEM, according to the manufacturer’s protocol. Then, 48 h after transfection, cells were lysed in 200 µl 1xharvest buffer (50 mM Tris, pH 7·8, 1 mM dithiothreitol, 0·1% Triton X-100) on ice. Cell lysates were vortexed briefly, and cellular debris was pelleted by centrifugation. Then 50 µl lysate were mixed with 100 µl luciferase-assay cocktail containing 1 mM luciferin. Luciferase activity was measured in a Monolight 2010 luminometer (Analytical Luminescence Laboratory, Sparks, MD, USA).
To study CRY1 inhibition, 100 ng Clock and 100 ng Bmal1 expression vectors were co-transfected with pCDNA3-Cry1 vector and, when necessary, the pCDNA3-βgal vector, to adjust the DNA total amount.
Northern blots
Total RNA was extracted from the livers of Clock mutant or wild-type mice housed in constant darkness and killed at various circadian times. Ten µg were loaded and migrated on a 1% agarose gel. Hybridization was performed overnight in 50% formamide, 5xSSPE, 1xDenhardt’s solution, 0·1% SDS and 0·1 mg/ml denatured salmon-sperm DNA at 63 °C. The Rev-erbα probe used was the 230 bp fragment used in the RNase protection assay. Washing was carried out in 0·5xSSPE/0·1% SDS at 65 °C for 30 min.
Semi-quantitative RT-PCR analysis
Semi-quantitative PCR analyses were performed on RNA extracted from serum-shocked fibroblasts. RNAs were extracted using the Sigma GenElute Mammalian Total RNA Kit with 250 µl lysis buffer per well for cells cultivated in 12-well plates according to the manufacturer’s protocol. One-tenth of the extracted RNA was reverse-transcribed with avian myeloblastosis virus RT (Promega) at 42 °C for 1 h. 1–2 µl of the reverse-transcribed RNA was used for the PCR reaction with 200 ng primers, 2·5 mM Mg2+ and 1·5 U Taq Gold polymerase with appropriate buffer (Perkin-Elmer) in a final volume of 30 µl. For each gene analyzed, three different numbers of cycles were tested to reach linear phase in PCR reactions (see number of cycles on Fig. 2). PCR cycles were as follows: 94 °C for 5 min, 94 °C for 30 s, 57 °C for 40 s, 72 °C for 30 s and a final extension of 72 °C for 5 min. The oligonucleotides used for the PCR were as follows. Rev-erbα: E1A 5’-GGCTTCACTCGTCTCTCT CAGCC-3’, top strand; E1B 5’-TGAGTCTTAT CTCCATATCACA-3’, top strand; E2 5’-GCAC AGTGCCAAATGAGCGGGC-3’, bottom strand. Per1: 5’-ATGACTGGGGCAGAGGTTGAGCC TG-3’, top strand; 5’-TCATGCTTAGATCGTG AAATAGGG-3’, bottom strand. Per3: 5’-ATGA CATACCAGGTGCCGGAGAGG-3’, top strand; 5’-CTTCTGCAGTGTCACCAACTGAAC-3’, bottom strand. Cry1: 5’-CAGCAAAATGGAGC CCCTGG-3’, top strand; 5’-CACACCGCAGAG GACAAGCC-3’, bottom strand. Clock: 5’-GCG AGAACTTGGCATTGAAGAG-3’, top strand; 5’-TTTGCAGCTTGAGACATCGCTGGC-3’, bottom strand. 28S: 5’-GTGAAAGCGGGGCCT CACGATCC-3’, top strand; 5’-GTACTGAGCA GGATTACCATGGC-3’, bottom strand.
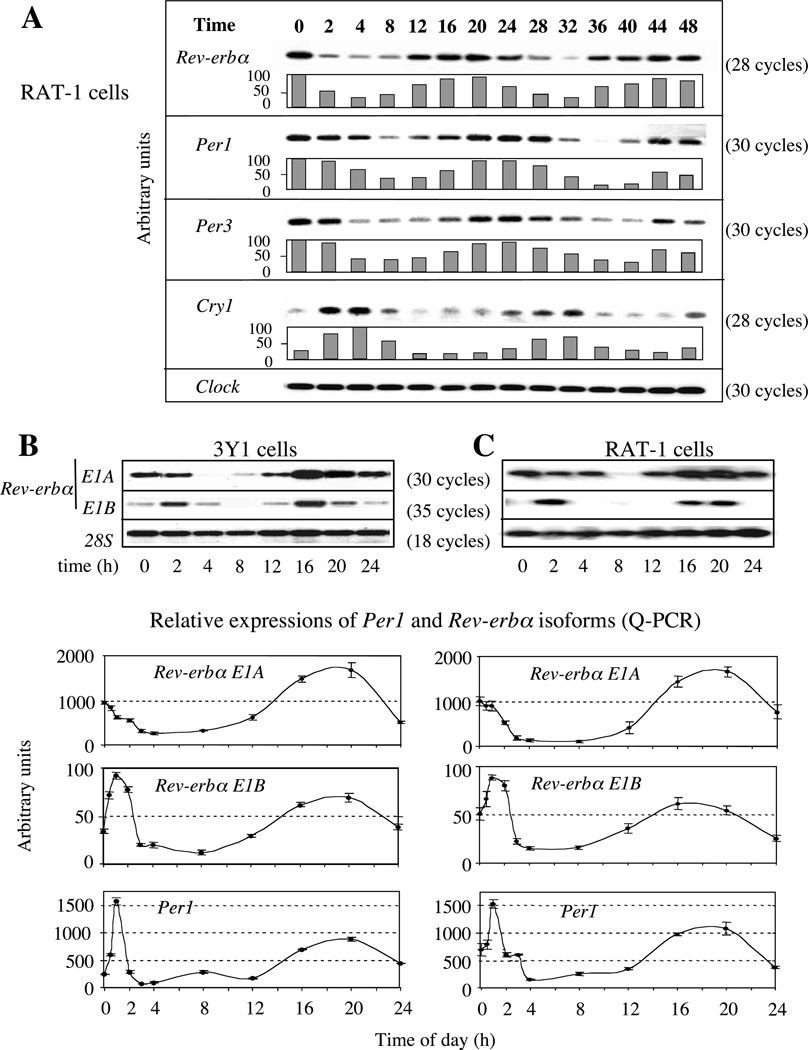
Figure 2.
Circadian expression of Rev-erbα transcripts in fibroblasts after 2 h horse serum-shock treatment. T0 corresponds to the serum-shock start. Expression was observed on RNA samples collected every 4 h with an additional point at 2 h during 48 h (A) or 24 h (B and C). (A) Expressions of Rev-erbα, Per1, Per3, Cry1 and Clock as a non-cyclic gene control in Rat-1 serum-shocked fibroblasts studied by semi-quantitative PCR. Each histogram is the average result of two experiments normalized on 28S analysis (not shown). For Cry1 two independent Q-PCRs were done on triplicates to refine the quantitation. (B, C) RT-PCR analysis of Rev-erbα1 (E1A) and Rev-erbα2 (E1B) transcripts in serum-shocked 3Y1 (B) and Rat-1 (C) fibroblasts. In each case the upper panel shows the result of a representative semi-quantitative RT-PCR assay whereas the lower curves are the averages from three Q-PCR experiments done on a 5700 GeneAmp Applied Biosystems apparatus. In Q-PCR experiments, a sharp analysis was done over the first 3 h. Note the difference of scales in the histograms (lower panels) and the number of PCR cycles (upper panels). All PCR products were obtained using an equal amount of reverse-transcribed mRNA amplified with primers located on two different exons hybridized with an internal specific probe. The signal obtained with 28S is a non-cycling control (upper B, C) and Q-PCR experiments were normalized on 28S values (lower B, C).
For the semi-quantification, half of the PCR products were loaded on 1% agarose gel. The gel was then incubated for 20 min in denaturation buffer (0·5 M NaOH/1·5 M NaCl) and transferred on to a nylon membrane (Hybond N; Amersham) in the same buffer for 3 h. Denatured DNA was fixed by ultraviolet treatment (254 nm; 0·36 J/cm2) with fluolink (Vilbert-Lourmat, Marne-La-Valle´e, France). Hybridizations with labeled oligonucleotides specific to each PCR fragment were performed overnight at 37 °C in 4xSSPE, 1xDenhardt’s solution, 0·1% SDS and 0·1 mg/ml denatured salmon-sperm DNA buffer. Each hybridization was performed with 500 ng of specific oligonucleotide labeled at the 5’ end using polynucleotide kinase in the presence of [γ-32P]dATP. Washing was carried out in 0·5xSSPE/0·1% SDS at 30 or 37 °C for 20 min depending on the length of the oligonucleotide used. The oligonucleotides used as probes were all designed within the PCR fragment and were as follows. Rev-erbα in E2, 5’-CACCTACATTGGCTCCAGCGGATCCT CCC-3’; Per1, 5’-TGCTGAAGTAGAGCCTGAA GTTC-3’; Per3, 5’-ATGACATACCAGGTGCC GGAGAGG-3’; Cry1, 5’-GACGTGATAGGGAA GTGCAC-3’; Clock, 5’-CAGTTTTCAGCTCA GTTAGGAGCC-3’; 28S, 5’-GGGATAACTGG CTTGTGGCGGCCAAGCG-3’.
Serum-shock treatment
Circadian induction of 3Y1 or Rat-1 fibroblasts was performed on 7-day-confluent cells, maintained in starvation conditions in 12-well plates before being shocked. At T0, 50% horse serum was added to the cells for 2 h. The cells were then grown in DMEM without serum for 2 days and harvested at various circadian times.
Electrophoretic mobility shift assays (EMSAs)
CLOCK and BMAL1 proteins were synthesized in vitro using the TNT T7 Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions. The two proteins were synthesized separately and mixed during the EMSA.
In other cases, either crude nuclear extracts of mouse fibroblast STO cells or nuclear extracts of STO cells transfected with Clock and Bmal1 expression vectors were used. All nuclear extracts were prepared using kit from Active Motif Europe (Rixensart, Belgium) according to the supplier’s protocol.
EMSAs were performed according to Vanacker et al. (1999) with the following modifications: 2 µl aliquots of each specific TNT reaction mixture were mixed with 20 fmol of acrylamide-purified, double-stranded DNA labeled probe in the presence or absence of the competitor and incubated for 30 min at 30 °C in incubation buffer (20 mM Tris-HCl, 50 mM NaCl, 5% glycerol, 5 mM magnesium sulfate, 1 µg poly(dIdC).poly(dIdC) and 1 mM dithiothreitol). The mixture was then loaded on to a 5% polyacrylamide gel.
The oligonucleotides used for EMSA were as follows. mPer1 E-box: 5’-GATCCAGCACCCAA GTCCACGTGCAGGGATGTGTGA-3’, top strand; 5’-GATCTCACACATCCCTGCACGTG GACTTGGGTGCTG-3’, bottom strand. g1: 5’-GATCCCAGTTCTGCAATCACGTGAAGCTC TCACGTA-3’, top strand; 5’-GATCTACGTG AGAGCTTCACGTGATTGCAGAACTGG-3’, bottom strand. g2: 5’-GATCCCAGAGCCGGGC CCACGTGCTGCATTTGTTTA-3’, top strand; 5’-GATCTAAACAAATGCAGCACGTGGGCC CGGCTCTGG-3’, bottom strand. g3: 5’-GATC CTCGTTGGGGTGCCACGTGCGAGGGGCA CACA-3’, top strand; 5’-GATCTGTGTGCCC CTCGCACGTGGCACCCCAACGAG-3’, bottom strand. g4: 5’-GATCCGTGCGAGGGGCAC ACGTGGAGCGGGGACGTA-3’, top strand; 5’-GATCTACGTCCCCGCTCCACGTGTGCCC CTCGCACG-3’, bottom strand. g5: 5’-GATC CTGTCAGCTCCCACACGTGTCTGGGGATC CTA-3’, top strand; 5’-GATCTAGGATCCCC AGACACGTGTGGGAGCTGACAG-3’, bottom strand. g3+4: 5’-GATCCTCGTTGGGGTGCCA CGTGCGAGGGGCACACGTGGAGCGGGGA CGTGA-3’, top strand; 5’-GTACTACGTCCCCGCTCCACGTGTGCCCCTCGCACGTGGC ACCCCAACGAG-3’, bottom strand. a1: 5’-GA TCCTCCCCAGGAATTCACATGCCCTTGCC ATACA-3’, top strand; 5’-GATCTGTATGGCAA GGGCATGTGAATTCCTGGGGAG-3’, bottom strand. a2: 5’-GATCCGCTCCCTGGAATCAC ATGGTACCTGCTCCAA-3’, top strand; 5’-GA TCTTGGAGCAGGTACCATGTGATTCCAGG GAGCG-3’, bottom strand. a3: 5’-GATCCCCG GGAAGGGCTCACATGGCTGCAGAGCCGA −3’, top strand; 5’-GATCTCGGCTCTGCAGCC ATGTGAGCCCTTCCCGGG-3’, bottom strand.
Mutated versions of the E-box elements were obtained by substitution of two bases inside the core sequence (CACGTG to CAATTG, or CAC ATG to CCATAG) in all the relevant oligonucleotides. a2 g and Per1a substitutions were obtained with oligonucleotides bearing a CACGTG and a CACATG respectively. ZFg1, 5’-TCAGGTGGA CACGTGCGCGGGGGT-3’ E-box (G), top strand; ZFg1, 5’-ACCCCCGCGCACGTGTCCA CCTGA-3’ E-box (G), bottom strand. ZFg1*, 5’-TCAGGTGGACAATTGCGCGGGGGT-3’ E-box (mutant), top strand; ZFg1*, 5’-ACCCCC GCGCAATTGTCCACCTGA-3’ E-box (mutant), bottom strand. ZFa, 5’-AGTCGGGTCACATG GACACATTT-3’ E-box (A), top strand; ZFa, 5’-AAATGTGTCCATGTGACCCGACT-3’ E-box (A), bottom strand. ZFa*, 5’-AGTCGGG TCCATAGGACACATTT-3’ E-box (A), top strand; ZFa*, 5’-AAATGTGTCCTATGGACCC GACT-3’ E-box (A), bottom strand.
Quantitative (Q)-PCR experiments
To verify the semi-quantitative PCR and to complete the analysis of Rev-erbα isoforms, Q-PCR was performed with the ABI Prism SYBR Green Reagents (Applied Biosystems, Courtaboeuf, France). cDNAs were synthesized by reverse transcription as below. Samples contained 1xSYBR Green Master Mix, 0·5 µM primers and 1/40 synthesized cDNA in a 25 µl final volume. PCR conditions were as follows: 10 min at 95 °C, then 50 cycles of 15 s at 95 °C, 60s at 60 °C and a final elongation cycle of 1 min at 60 °C. Absolute abundance of cDNA was calculated using the standard curve obtained with the two ranges done by serial dilutions from pure to 1/64. For the 28S RT products were diluted 1/300 before use.
Oligonucleotides used for rat samples were as follows. E1A top strand, 5’-CACGGGGCGAGA GAGGGCACC-3’; E1B top strand, 5’-TGAG TCTTATCTCCATATCACA-3’; E2 bottom strand, 5’-GAAGGGGAGCTATCATCACTG-3’; 28S top strand, 5’-GTGAAAGCGGGGCCTCA CGATCC-3’; 28S bottom strand, 5’-GTACT GAGCAGGATTACCATGGC-3’; Per1 top strand, 5’-ATGACTGGGGCAGAGGTTGAGC CTG-3’; Per1 bottom strand, 5’-TCATGCTT AGATCGTGAAATAGGG-3’. Cry1, 5’-CAGCA AAATGGAGCCCCTGG-3’, top strand; 5’-CAC ACCGCAGAGGACAAGCC-3’, bottom strand. Oligonucleotides used for mouse samples were as follows. E1A top strand, 5’-CAGGGGGCGAGA GAGGCCATCAC-3’; E1B top strand, 5’-AGG AAGGGGAATTCCTAAATCCC-3’; E2 bottom strand, 5’-GGCGCACTGCCAAAAGAGCGG GC-3’; 28S oligonucleotides were the same as for rat experiments.
RNA in situ hybridization
Zebrafish (Danio rerio) were kept at 28·5 °C in a 14-h light/10-h dark cycle. Embryos were collected after spawning to perform the morpholino injection. To prevent pigmentation, 0·2 mM 1-phenyl-2-thiourea (Sigma) was added to the water at 12 h post-fertilization. At 48 h post-fertilization, embryos were fixed in 4% paraformaldehyde in PBS overnight at 4 °C. Whole-mount in situ hybridization was performed using antisense digoxigeninlabeled Rev-erbα probe and embryos were incubated at 70 °C in a 50% formamide hybridization solution. Probes were detected using alkaline phosphatase-conjugated antibodies and visualized by 4-Nitro Blue Tetrazolium and 5-bromo-4chloro-3-indolyl-phosphate staining (for a protocol reference, see Thisse et al. 1993).
Morpholinos
We obtained morpholinos from GeneTools (Philomath, OR, USA). The morpholinos sequences were as follows: clock1, 5’-CATCCCGG TCTATGCTGGAGGTCAT-3’; clock2, 5’-GAT AACTCGGTCTCATGGATCAGTC-3’; bmal1, 5’-TATCCATTCTTTGGTCTGCCATTAG-3’; bmal2, 5’-CAGATTTCATTTCCAGGTTGTC CAT-3’. For the control, we used standard control morpholino provided by GeneTools: 5’-CCUCUU ACCUCAGUUACAAUUUAUA-3’. We injected wild-type embryos at the one–two cell stage with 1–2 µl morpholino in 1xDanio buffer at 0·25 or 0·5 mg/ml.
Results
Circadian expression of the two Rev-erbα isoforms
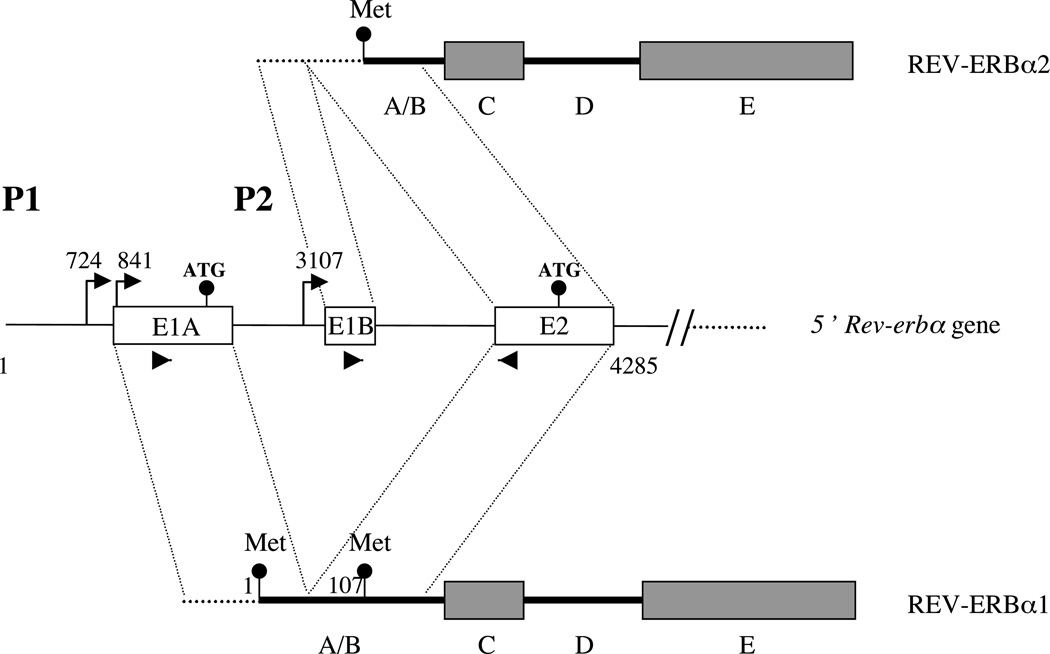
The human and rat Rev-erbα genes generate two mRNA isoforms, Rev-erbα1 and Rev-erbα2, with different 5’ regions (Lazar et al et al89, Miyajima et al. 1989, Laudet et al. 1991, Dumas et al. 1994, G T Triqueneaux, B Staels&VL Laudet, unpublished observations; see Fig. 1). The resulting proteins differ only in their N-terminal A/B domain. The long isoform, Rev-erbα1, is generated from the P1 promoter located upstream of exon E1A which contains a start codon (Adelmant et al. 1996). The short isoform Rev-erbα2 is expressed at lower levels and can be generated from a newly identified promoter called P2 located upstream of the non-coding exon E1B (G T Triqueneaux, B Staels &VL Laudet, unpublished observations; see supplementary Fig. 1 at http://jme.endocrinologyjournals.org/content/vol33/issue3/). This exon is spliced to exon 2, which contains an in-frame start codon. Of note, the transcripts generated at P1 can encode the two protein isoforms by alternative start-codon usage. The relative importance of the two mechanisms (alternative promoter usage and splicing or alternative start-codon selection) used to generate Rev-erbα1 and Rev-erbα2 is, at present, unknown.
Figure 1.
The Rev-erbα gene generates two N-terminal distinct isoforms, Rev-erbα1 and Rev-erbα2, from two different promoters, P1 and P2. Transcription start sites are indicated by arrows. Untranslated sequences are shown as dotted lines whereas translated ones are represented as thick lines. The start codons are indicated by circles showing that the Rev-erbα1 mRNA can generate both the Rev-erbα1 and Rev-erbα2 protein isoforms by alternative initiator codon usage. The A/B, C and E domains are indicated. The 58 and 38 primers used to detect specifically Rev-erbα1 and Rev-erbα2 transcripts are indicated by arrowheads on E1A, E1B and E2 exons respectively.
Rev-erbα gene expression has been shown to be under circadian regulation in vivo and in serum-shocked tissue-culture cells (Balsalobre et al. 1998, Delaunay et al. 2000, Torra et al. 2000, Grundschober et al. 2001). We observed this rhythmic expression in serum-shocked Rat-1 or 3Y1 rat fibroblasts using a strategy that did not discriminate between the two Rev-erbα isoforms (Fig. 2A and results not shown). To determine whether expression of both Rev-erbα1 and Rev-erbα2 was circadian, we designed a Q-PCRbased assay using isoform-specific primers for exon 1 (Figs 1, 2B and 2C). Analysis of serum-shocked fibroblastic 3Y1 and Rat-1 cells showed that both isoforms exhibited a circadian expression. Interestingly, this experiment showed that whereas circadian expression of both transcripts was induced by serum, the initial serum response was different. Transcripts initiated at P1 were repressed whereas those transcribed from P2 were induced. This suggests that transcription of these two isoforms is differentially regulated by serum. It is important to note that, in both cell lines, there are P1-generated transcripts much more than those formed at P2. Indeed we detected in those cells only the Rev-erbα1 isoform using an antibody directed against the ligand binding domain (LBD) (from N Preitner, Department of Molecular Biology, NCCR Frontiers of Genetics, Sciences II, University of Geneva, Switzerland; see supplementary Fig. 2A at http://jme.endocrinologyjournals.org/content/vol33/issue3/). In every Q-PCR experiment, five additional cycles were needed for P2 transcript detection compared with transcripts initiated at P1. Taken together, these data suggest that (1) the activity of both promoters is circadian, (2) the two promoters display different responses to serum and (3) the respective phase of Per transcripts and Rev-erbα transcripts is compatible with a down-regulation of Rev-erbα by the negative components of the circadian clock, PER and CRY.
Rev-erbα P1 and P2 promoters contain circadian clock-response elements
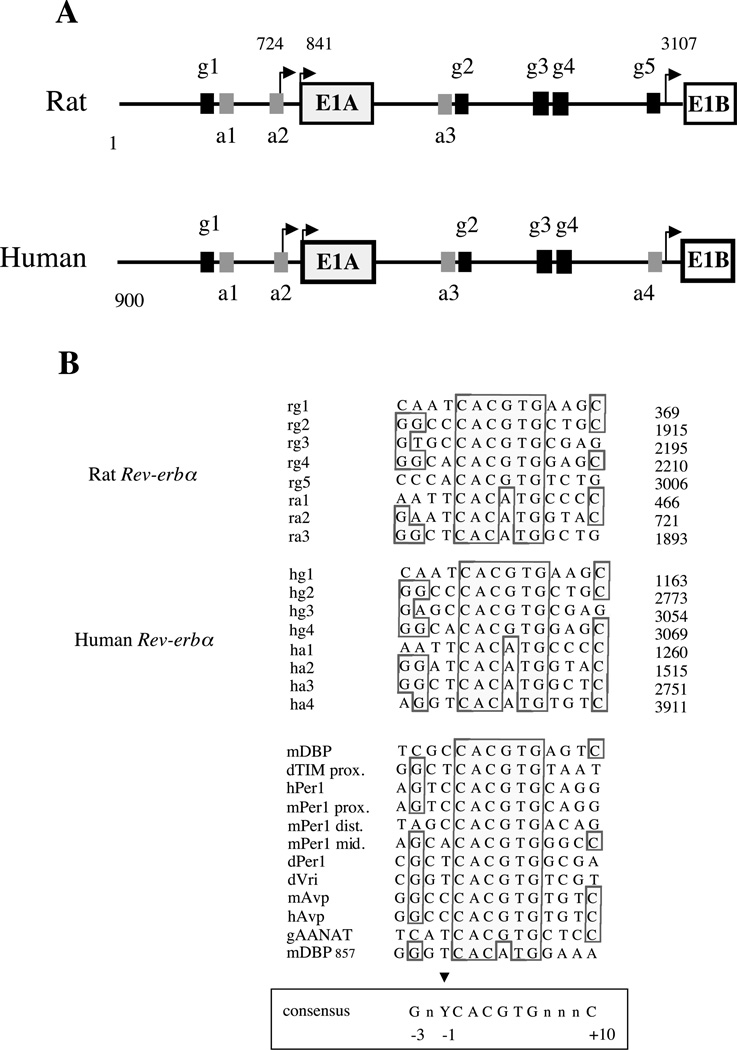
The circadian regulation of P1 and P2 prompted us to search within these promoter regions for E-box sequences, which are known response elements for the circadian transcriptional activating heterodimer CLOCK–BMAL1 (Gekakis et al. 1998). Comparative analysis of the human, rat (Rattus norvegicus) and mouse (Mus musculus) P1 promoters revealed one canonical E-box element (CACGTG; g1 in Fig. 3A) and two divergent E-boxes (CACATG; a1 and a2). We also found three conserved canonical E-boxes (g2, g3 and g4 in Fig. 3A) and one divergent E-box (a3) in the human and rat P2 region. These E-boxes were also conserved in the mouse P2 promoter (results not shown). An additional canonical E-box (g5) was found only in the rodent promoters instead of a divergent one in human (a4).
Figure 3.
(A) Genomic structure of Rev-erbα promoters in rat (Rattus norvegicus) and human. P1 and P2 are the two promoters. E1A and E1B are the first alternative exons spliced to exon 2. The numbered E-boxes, g1–g5 (black boxes), correspond to classical E-boxes (CACGTG) and a1–a4 (grey boxes) to the divergent ones (CACATG). Transcription start sites are indicated by arrows. (B) Alignment of the E-boxes found in P1 and P2 rat and human promoters with E-boxes found in various circadian promoters. Numbers on the right indicates the E-boxes position in each promoter. The alignment was obtained by Seqvu software analysis (The Garvan Institute of Medical Research, Sydney, Australia). Boxed bases show conserved positions and the arrow indicates the base similarity at position −1. At −3 and +10 positions were found with a higher frequency of G and C respectively. A consensus sequence derived from this comparison is boxed on the bottom of the Figure.
Alignment of these E-box sequences with those shown to bind CLOCK–BMAL1 heterodimers and to confer circadian regulation of other promoters (see Fig. 3B) showed that Rev-erbα E-boxes contain flanking sequences that are reminiscent of those ‘circadian’ E-boxes with a pyrimidine base at position −1 and, a G at −3 and often a C at position +10 (Fig. 3B). Thus, Rev-erbα promoters contain all the necessary DNA elements required for a regulation by CLOCK–BMAL1 and these elements are conserved in mammals.
The CLOCK–BMAL1 heterodimer regulates Rev-erbα promoters
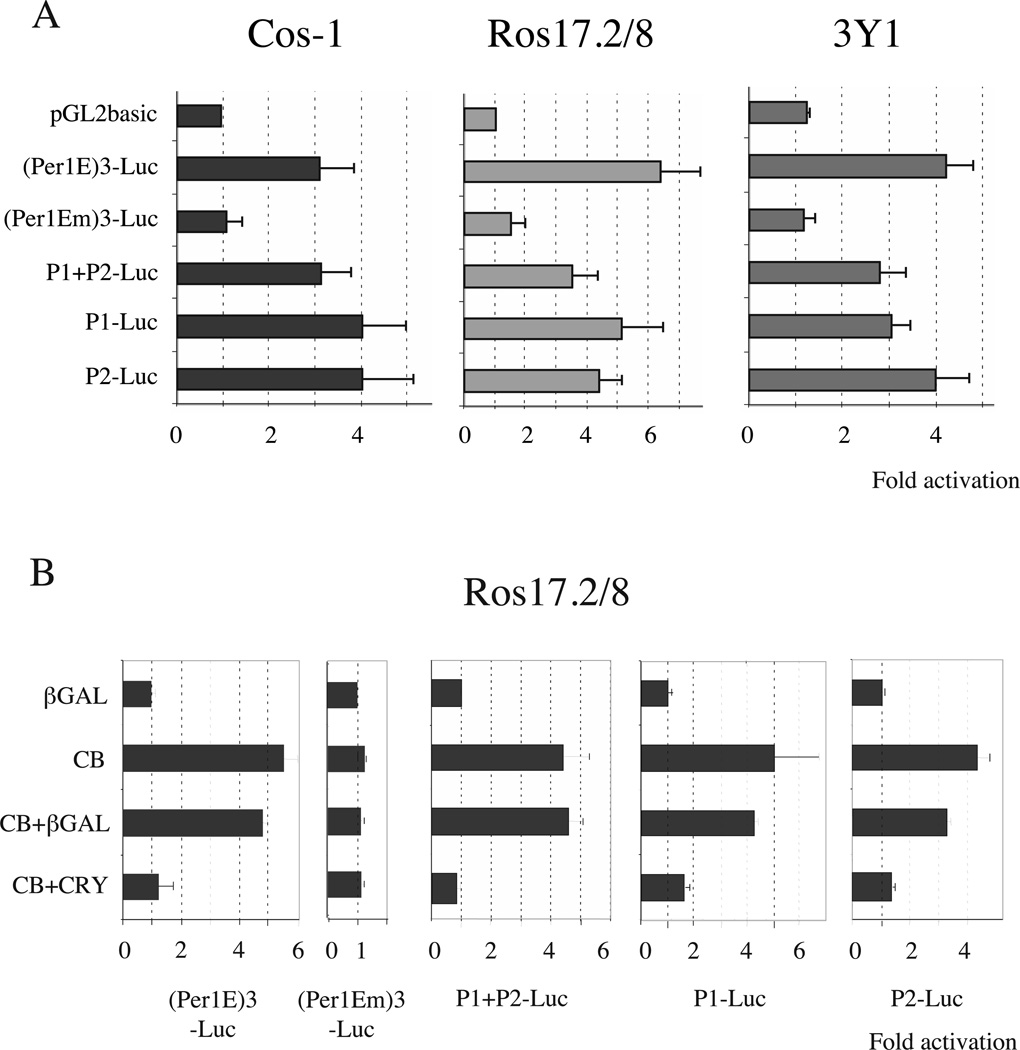
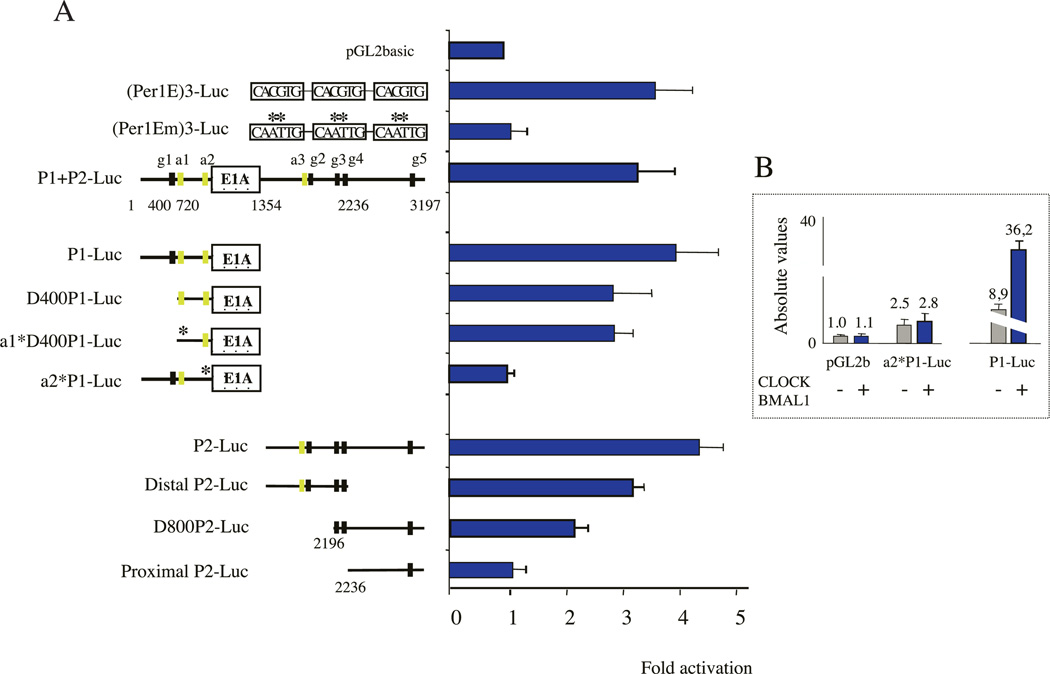
We next explored whether the CLOCK– BMAL1 heterodimer was able to activate P1, P2 or a construct containing P1+P2 in transient co-transfection assays. Since CLOCK–BMAL1 activity was shown to be tissue-specific in certain cases (Chen & Baler 2000), we performed these experiments in three different cell lines: Cos-1 cells, human osteosarcoma Ros17·2/8 cells and rat fibroblastic 3Y1 cells. As a positive control we used a construct in which we cloned three canonical E-boxes derived from the mouse Per1 promoter upstream of a TATA box and the luciferase gene to generate the (Per1E)3-Luc construct. The mutated version of these E-boxes from this construct give rise to (Per1Em)3-Luc, which was used as a negative control. In all three cell lines the CLOCK–BMAL1 heterodimer activated the (Per1E)3-Luc construct between 4-and 6-fold and was inactive on the mutated version (Fig. 4A). CLOCK alone or BMAL1 alone didn’t affect the promoter activity (supplementary Fig. 3). Interestingly, CLOCK–BMAL1 was able to activate 3–5-fold the activity of P1, P2 or P1+P2 in the three different cell lines (Fig. 4A). The association of low amounts of Cry1 expression vector in of P1 and P2 in the same vector did not confer an co-transfection experiments virtually abolished increased sensitivity to CLOCK–BMAL1. CLOCK–BMAL1-induced activation of P1+P2-,
Figure 4.
CLOCK–BMAL1 heterodimers activate both the P1 and P2 promoters. (A) Transactivation assays were performed on Rev-erbα promoters in Cos-1, Ros17·2/8 and 3Y1 growing cells. Results are shown in fold activation and are the means±SEM from at least three independent experiments. 40 ng pGL2-luciferase reporter plasmid were used and mixed with 100 ng pCDNA3-Clock and 100 ng pCDNA3-Bmal1 or 200 ng pCDNA3-gal as a neutral vector. 48 h later cells were harvested and luciferase activity was analyzed. (Per1E)3-Luc and (Per1Em)3-Luc are positive and negative controls, containing wild-type or mutated E-boxes respectively in front of a minimal promoter. (B) Cry1 inhibits CLOCK–BMAL1-induced activation of the Rev-erbα promoters. The left panel shows the effect of Cry1 on the positive (Per1E)3-Luc and the negative (Per1Em)3-Luc reporter vectors. The right-hand panel shows the effect of Cry1 on P1+P2, P1 and P2 promoters. This experiment was performed on Ros17·2/8 cells but similar results were obtained on Cos-1 cells. 20 ng of pCDNA3-Cry1 were used and the total amount of DNA was adjusted to 300 ng; CB, CLOCK–BMAL1.
We then tested whether CRY1, a repressor of the master molecular oscillator (Kume et al. 1999), was able to inhibit this activation. The addition of low amounts of Cry1 expression vector in co-transfection experiments virtually abolished CLOCK-BMAL1- induced activation of P1+P2-, P1-or P2-driven reporter gene (Fig. 4B). This effect was specific as the control expression vector (β-gal) induced only a marginal effect on CLOCK– BMAL1-mediated activation of both promoters. This specific inhibitory effect was comparable to that observed using the positive-control reporter vector (Per1E)3-Luc. These data suggest that CLOCK and BMAL1 are directly involved in the circadian regulation of both Rev-erbα isoforms.
The CLOCK–BMAL1 heterodimer binds to and transactivates Rev-erbα E-box elements
To map the regions involved in CLOCK–BMAL1 binding and trans-activation, we designed a series of P1 and P2 deletion constructs that were all tested for their ability to be activated by CLOCK– BMAL1 in transient assay in Cos-1 cells.
Deletion of the canonical E-box (g1) from P1 sequence (D400-P1-Luc construct) decreased but did not abolish its ability to be activated by CLOCK–BMAL1 (Fig. 5A), most likely due to the presence of two divergent CACATG E-boxes (a1 and a2). Trans-activation was not affected by introducing the mutation into a1 E-boxes (a1*D400-P1-Luc construct), strongly suggesting a critical role for the a2 site in P1 circadian regulation. This site also corresponds to the major transcriptional start site of the human P1 promoter (Adelmant et al. 1996) but to a minor one in rat (results not shown). Mutation of this site (a2*-P1Luc construct) in the context of the complete P1 promoter totally abolished CLOCK–BMAL1 activation but not the basal activity of the promoter (Fig. 5B). These data indicate that CLOCK– BMAL1 activates the P1 promoter through the a2 site.
Figure 5.
(A) Mapping of the E-box mediating CLOCK–BMAL1 activation on the Rev-erbα promoters. Trans-activation assays were performed on mutated Rev-erbα promoter fragments in Cos-1 cells. Results are shown in fold activation and are the means±SEM from at least three independent experiments. 40 ng pGL2-luciferase reporter plasmid were used and mixed with 100 ng pCDNA3-Clock and 100 ng pCDNA3-Bmal1 or 200 ng pCDNA3-gal as a neutral vector. (Per1E)3-Luc and (Per1Em)3-Luc are positive and negative controls containing wild-type or mutated E-boxes respectively in front of a minimal promoter. Cells were harvested 48 h after transfection and luciferase activity was analyzed with a luminometer. The E-boxes discussed in the text are shown as black boxes (classical E-box CACGTG) or green boxes (divergent E-box CACATG). An asterisk indicates a mutated E-box. (B) Absolute activities of P1-Luc and a2*P1-Luc (mutated a2) promoters (grey bars) and their CLOCK–BMAL1 activation responses (blue bars) indicated the specificity of the a2 E-box element.
For P2, we identified a cluster of closely spaced E-boxes (three canonical, one divergent) in the distal part of the promoter and one downstream E-box in the proximal region. Deletion of the distal promoter region led to a construct (ProximalP2-Luc) containing only the latter isolated site (g5 in the rat promoter). This construct still exhibited a strong constitutive activity of the promoter (results not shown) but was not activated by CLOCK–BMAL1 (Fig. 5A). This suggests that the distal region of the P2 promoter is the target of CLOCK–BMAL1. This was confirmed using an internal deletion mutant lacking the P2 proximal region (DistalP2-Luc) which was fully activated by CLOCK–BMAL1. Using a new construct (D800-P2-Luc) adding g3 and g4 to the P2 proximal region we restored a partial activation (about 2-fold), indicating that they were active.
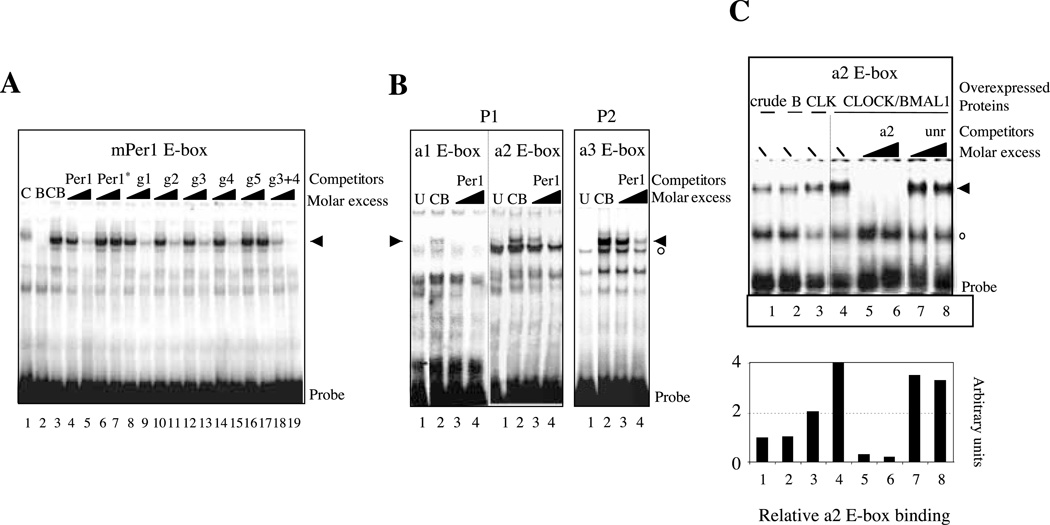
To determine whether the CLOCK and BMAL1 proteins were able to bind the identified Rev-erbα E-boxes, we set up a gel-shift assay using in vitro-translated proteins and the mouse Per1 proximal promoter E-box sequence as a probe together with competitor oligonucleotides corresponding to each of the Rev-erbα E-boxes. As expected, the CLOCK–BMAL1 heterodimer bound strongly to the mPer1 E-box probe (Fig. 6A). This binding is reduced by adding a molar excess of unlabeled mPer1 E-box but not by the mutated version of the Per1 E-box (Per1*). Surprisingly, g1 site, which was not sufficient for CLOCK–BMAL1 activation of the P1 promoter in our transient transfection experiments, competed strongly with CLOCK–BMAL1 binding to the mPer1 E-box. When using the a1 site-specific oligonucleotide as a probe, we observed very weak, if any, binding of CLOCK–BMAL1 consistent with the fact that mutation of this site does not modify CLOCK– BMAL1 activation of the Rev-erbα gene (Fig. 6B, left-hand panel). In contrast, we detected a specific, albeit weak, binding of CLOCK–BMAL1 using the a2 probe (Fig. 6B, middle panel). We then performed similar gel-shift assay (Fig. 6C) using nuclear extracts of mouse STO fibroblasts in which the CLOCK and BMAL1 proteins were transiently expressed. If BMAL1 alone only marginally affected the amount of the shifted complex, the overexpression of CLOCK increased the binding strongly, suggesting that endogenous CLOCK is a limiting factor in the crude nuclear extract. As shown in Fig. 6C, CLOCK–BMAL1 binding to an a2-specific probe was strongly reduced by addition of an a2 unlabeled competitor but not by an unrelated oligonucleotide or by the mutated a2 site (a2*; results not shown).
Figure 6.
CLOCK–BMAL1 heterodimers bind to the E-boxes of the Rev-erbα promoters. EMSAs were performed on E-box from the mouse Per1 promoter (A) or E-boxes from the Rev-erbα promoter (B and C) as probes. For A and B, the CLOCK and BMAL1 proteins were produced independently in reticulocytes lysates whereas for C the EMSA was performed using nuclear extracts from mouse STO fibroblasts in which Clock and Bmal1 expression vectors were transiently transfected. In each case, specific complexes are indicated by black arrows whereas non-specific ones are shown by empty circles. (A) The classical E-boxes of the Rev-erbα promoters compete with the binding of CLOCK–BMAL1 to the mPer1 sequence. Lane 1, C, CLOCK protein alone; lane 2, B, BMAL1 protein alone; lanes 3–19, CB, CLOCK–BMAL1 heterodimer. Competition experiments were done with a molar excess of 10-and 100-fold of the relevant E-box as indicated by the triangles. g3+4 indicates the competition by an oligonucleotide containing both the g3 and g4 E-boxes that are adjacent in the P2 promoter. (B) Direct binding of CLOCK–BMAL1 to the divergent E-boxes. The a1, a2 and a3 E-boxes were used as probes in these EMSA. Lane 1, U, untranslated reticulocyte lysates; lanes 3 and 4, CB, CLOCK–BMAL1 heterodimer; the triangle indicates increasing amount of competitor (10-and 100-fold) which in each case is the mPer1 E-box. (C) EMSA performed with nuclear-extracts of STO fibroblasts producing BMAL1 (B, lane 2), CLOCK (CLK, lane 3) or CLOCK–BMAL1 (lanes 4–8). Nuclear extracts of cells producing the -galactosidase protein were used as controls (crude, lane 1). The probe used was the a2 element; the competitors added with a molar excess (10-or 100-fold, indicated by triangles) are indicated above each lane. Bottom panel, quantitation of the specific binding of the a2 probe.
Using a similar competition gel-shift assay we observed that all E-boxes of the P2 promoter, except the g5 site located in its distal region, were able to bind the CLOCK–BMAL1 heterodimer (Fig. 6A). This is consistent with our transient transfection data, which showed the importance of the distal region for CLOCK–BMAL1 activation. Since the g3 and g4 sites were adjacent, we tested these two sites together with a single oligonucleotide in a competition experiment and detected a strong competition ability of this DNA fragment in good agreement with their activity in transient transfection. As for the P1 promoter, the divergent a3 site competed with the binding of CLOCK– BMAL1 to the mPer1 E-box probe and displayed direct specific binding activity when labeled as a probe (Fig. 6B, right-hand panel). All these data suggest that the four E-boxes of the P2 promoter distal region are able to bind CLOCK– BMAL1 heterodimer to mediate its transcriptional activation.
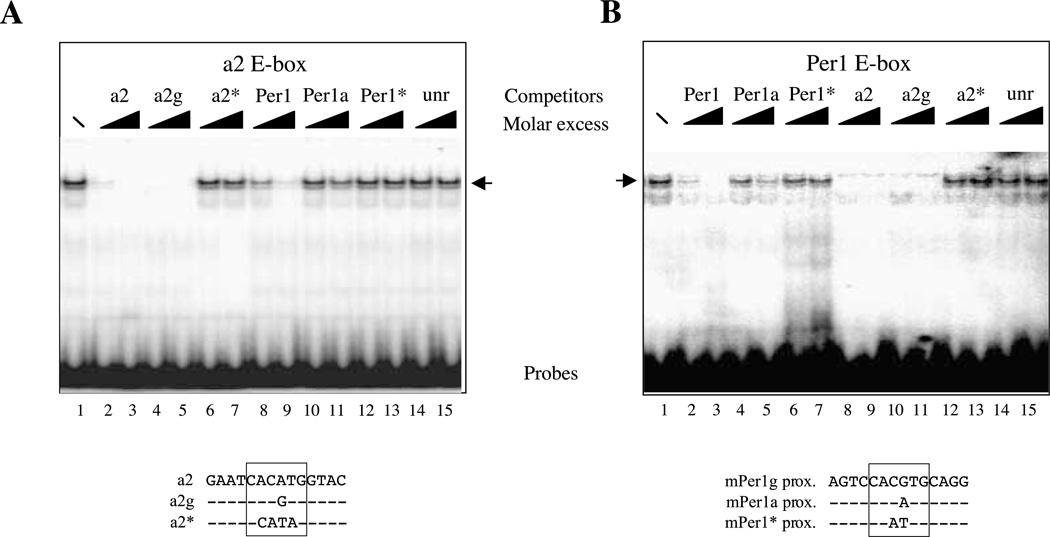
Since in P1 the main E-box responsible for CLOCK–BMAL1 activation is the divergent CACATG E-box (a2), we decided to study in more detail the importance of the core sequence of the E-box compared to the adjacent sequences. As shown in Fig. 7, using crude nuclear extracts of STO fibroblasts we obtained a specific retarded complex at the same level as the bona fide CLOCK–BMAL1 complex with the divergent a2 E-box probe (Fig. 6C), as well as the canonical CACGTG E-box from Per1 promoter. The complex was competed out by an excess of specific competitors but not by unrelated sequences (see Fig. 7, lanes 14 and 15) or by the mutated version of the probes (a2* and Per1*; see Fig. 7, lanes 6, 7, 12 and 13). In all cases, it is clear that the a2 unlabeled competitor was more efficient at decreasing the binding than the Per1 unlabeled competitor (compare lanes 2 and 3 with lanes 8 and 9 on Fig. 7A and B). Interestingly, when we replaced the core sequence CACATG of the a2 site with a canonical CACGTG (a2 g element) we observed that the resulting sequence competed equally well for the binding to either the a2 or the Per1 probe (see Fig. 7A, lanes 4 and 5 and Fig. 7B, lanes 10 and 11). This was not the case when the Per1 canonical sequence was mutated to a CACATG-type element (Per1a) since this sequence competed poorly for the binding to the a2 probe (Fig. 7A, lanes 10 and 11) and only moderately for the binding to the Per1 probe (Fig. 7B, lanes 4 and 5). All these data clearly suggest that the context of the E-box is important for CLOCK– BMAL1 binding: the adjacent positions allow the divergent a2 element to bind CLOCK–BMAL1 whereas the context is less important for canonical E-boxes.
Figure 7.
Influence of the E-box core sequence for CLOCK–BMAL1 binding. EMSAs were performed on the divergent a2 E-box from the rat P1 promoter (A) or the canonical E-box from the Per1 promoter (B) as probes with crude nuclear extracts from STO fibroblasts. In each case, competition experiments were performed with two different molar excess (10-or 100-fold, indicated by triangles) of the Per1 E-box sequence (Per1), the Per1 E-box in which the core sequence has been changed to CACATG (Per1a), the mutated Per1 E-box (CAATTG; Per1*), the rat a2 E-box (CACATG; a2), the a2 element that has been changed toward CACGTG (a2 g), the mutated a2 E-box (CCATAG; a2*) or an unrelated oligonucleotide (unr).
Rev-erbα is a target gene of Clock in vivo
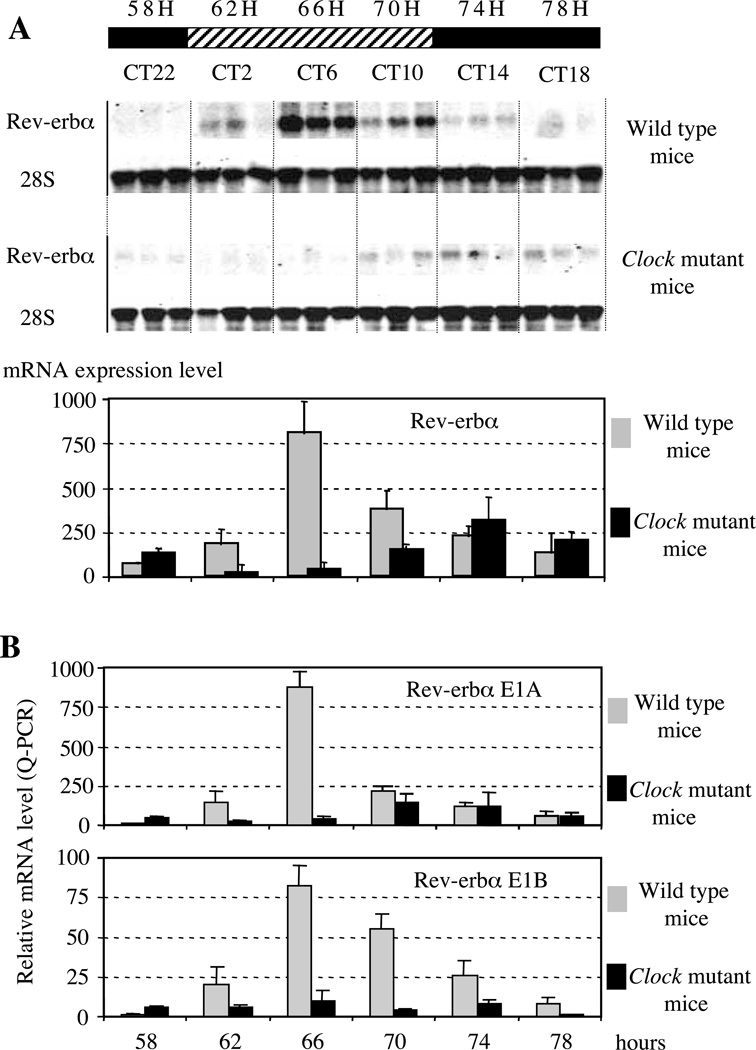
If Rev-erbα is a target of the circadian pacemaker as suggested by our transfection and gel-shift data, then an alteration of the circadian clock function should impair Rev-erbα gene expression. To address this question, we first compared Rev-erbα circadian gene expression profiles in the livers of wild-type and Clock mutant mice. Clock mutant mice express a dominant-negative version of CLOCK, which is defective in transactivation (King et al. 1997, Gekakis et al. 1998). At the behavioral level, they exhibit lengthening of an endogenous period followed by complete arhythmicity after long exposure to constant darkness conditions (Vitaterna et al. 1994). Northern blot analysis of Rev-erbα expression levels in wild-type and Clock mutant mice kept under conditions of constant darkness was performed using total RNA samples extracted from livers collected at 4-h intervals during the third cycle of constant darkness. In the livers of wild-type mice Rev-erbα transcript demonstrates robust oscillation with a peak of expression occurring at 66 h of constant darkness, which corresponds to circadian time (CT) 6 (Fig. 8A). In contrast, mutant mice maintained under the same conditions showed a marked decrease of Rev-erbα expression level with significantly reduced amplitude and the phase of expression delayed by approximately 8 h. Therefore, this experiment indicates that Rev-erbα is a target gene of CLOCK in vivo.
Figure 8.
(A) Northern blot analysis of Rev-erbα expression in Clock mutant mice. (A, B) The scale above the Figure indicates subjective day (hatched) or night (black), respectively. Time is indicated either as the number of hours after constant darkness (above the scale), or circadian time (CT below the scale): CT0 is the time when light would normally be switched on. Each lane was loaded with 10 µg total liver RNA. Animals were maintained in dark/dark conditions and three animals were killed at each indicated time. (A) Membranes were first probed with a Rev-erbα probe (detecting the two isoforms), exposed, washed and rehybridized with 28S probe to normalize the signal. Normalized Rev-erbα expression levels are shown as histograms at the bottom of the Figure. These histograms are the results of two independent Northern blot experiments. (B) Differential analysis of transcripts emanating from transcripts starting from P1 (E1A, upper panel) or P2 (E1B, lower panel) in wild-type or Clock mutant mice by Q-PCR experiments. Note the different scales of the histograms.
Since our transient transfection and gel-shift assays suggest that the two Rev-erbα promoters are direct targets of CLOCK–BMAL1, we checked whether the circadian expression of the transcripts emanating from each promoter was altered in Clock mice. Using Q-PCR analysis we observed that the transcripts starting at P1 as well as those from P2 are altered in the Clock mice suggesting that the two promoters are regulated independently by CLOCK–BMAL1 (Fig. 8B). These data clearly indicate that, in vivo, Rev-erbα expression is under the control of the Clock gene and that the Rev-erbα gene is a Clock-controlled gene.
The zebrafish Rev-erbα gene is also a Clock-controlled gene
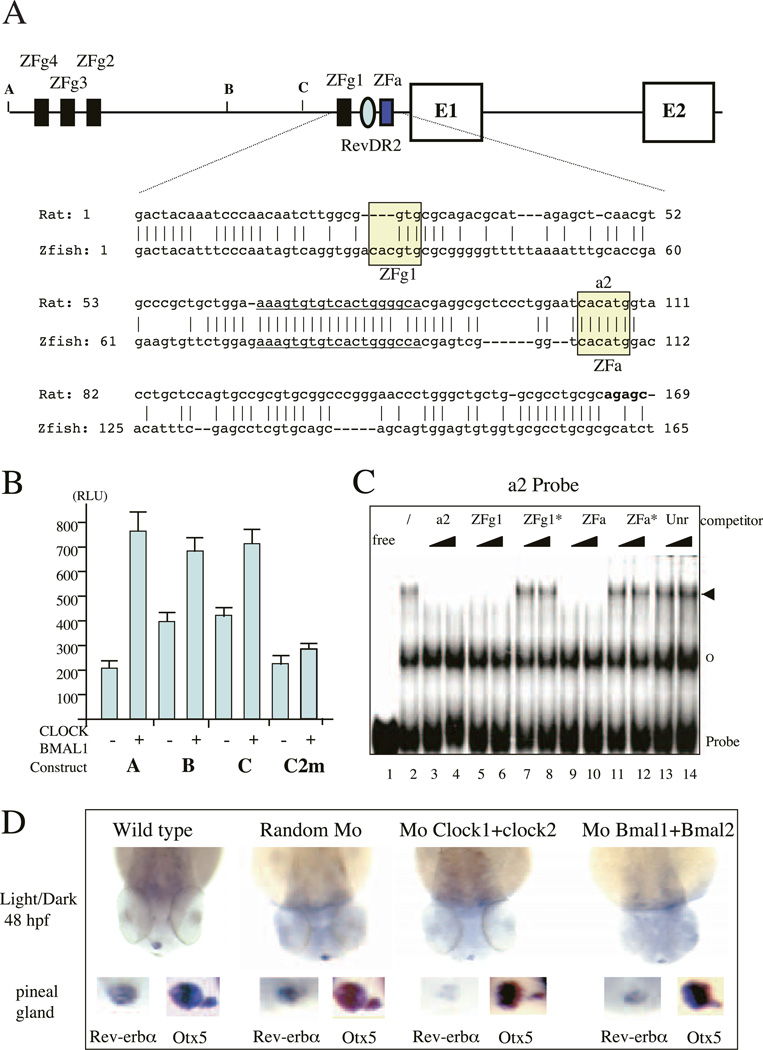
We observed previously that Rev-erbα expression is circadian in zebrafish, as it is in mammals (Delaunay et al. 2000). To determine if Rev-erbα is regulated by the CLOCK–BMAL1 heterodimer in zebrafish, we isolated 3·2 kb of the 5’ flanking region of the zebrafish Rev-erbα gene. Interestingly, just upstream of exon 1, we noticed a region harboring 61% sequence identity with rat P1 promoter over 165 bp (Fig. 9A). Of note, this region contains two E-boxes, one of which appears to be homologous to the a2 E-box. The other one is a canonical E-box that is not present in rat or human. In addition, we observed that three canonical E-boxes are present in the 5’ region of the 3·2 kb P1 region (Fig. 9A). Between exon 1 and 2 we did not find any exon reminiscent of exon 1B that may be under the control of a specific promoter.
Figure 9.
(A) Genomic structure of Rev-erbα promoter in zebrafish. The two first coding exons E1 and E2 are indicated. The numbered E-boxes ZFg1–ZFg4 (black boxes) correspond to classical E-boxes (CACGTG) and ZFa (grey boxes) to the unique divergent one (CACATG) which is homologous to the rat and human a2 element. The Rev-DR2 element is indicated by an oval. The sequence of the zebrafish proximal promoter region compared with the rat proximal P1 sequence is shown under the genomic structure. E-boxes are boxed whereas the Rev-DR2 element is underlined. The start of the rat exon 1A is emboldened. The various constructs A, B and C used in transient transfections (see panel B) are indicated. (B) CLOCK–BMAL1 heterodimers activates the zebrafish Rev-erbα promoter in COS-1 cells. The construct used, A, B, C or C mutated in the two E-boxes ZFg1 and ZFa (C2 m) are indicated below the histogram. Results are the means±SEM from at least three independent experiments. 40 ng pGL2-luciferase reporter plasmid were used and mixed with 100 ng pCDNA3-Clock and 100 ng pCDNA3-Bmal1 or 200 ng pCDNA3 as a neutral vector. Cells were harvested after 48 h and luciferase activity was analyzed. (C) CLOCK–BMAL1 binds to the zebrafish E-boxes. EMSAs were performed on the a2 E-box from the rat P1 promoter as probe with crude nuclear extract from STO fibroblasts. In each case, competition experiments were performed with two different molar excess (10-or 100-fold, indicated by triangles) of either the a2 E-box (lanes 3 and 4), the ZFg1 E-box (lanes 5 and 6), the mutated version of the ZFg1 E-box (CAATTG; ZFg1*; lanes 7 and 8), the divergent ZFa E-box (lanes 9 and 10), the mutated version of the ZFa E-box Per1 E-box sequence (CCATAG; ZFa*; lanes 11 and 12) or an unrelated oligonucleotide (unr; lanes 13 and 14). Specific complexes are indicated by an arrowhead whereas non-specific ones are shown by an empty circle. (D) Whole-mount in situ hybridization analysis of Rev-erbα expression in the zebrafish epiphysis at 48 h post-fertilization (hpf). All analyses were performed on at least 25 embryos. Wild-type and injected embryos were shown as follows: random morpholino (Mo; 0·5 mg/ml), CLOCK1+CLOCK2 morpholinos (0·25 mg/ml+0·25 mg/ml) and BMAL1+BMAL2 morpholinos (0·25 mg/ml+ 0·25 mg/ml). Rev-erbα probe was labeled with UTP-digoxigenin for overnight at 70 °C. Higher magnification at the pineal gland level is depicted below each stage in the left-hand inset. In each case the right-hand inset shows the expression of Otx5, a non-circadian gene expressed in the pineal gland, the parapineal and the retina (results not shown). For both probes, all the staining was performed under precisely identical conditions.
When tested in COS-1 cells, the zebrafish Rev-erbα upstream sequence (construct A) clearly exhibited a promoter activity and was activated 3–4-fold by co-transfection of the Clock and Bmal1 expression vectors (Fig. 9B). When we tested deletion constructs (B and C) we observed that if the three canonical E-boxes clearly play a role in CLOCK–BMAL1 activation the two downstream E-boxes were also important. Indeed, mutation of these two E-boxes (C2 m construct) abolished promoter activation (Fig. 9B) whereas mutation of each of them is not sufficient (results not shown). In accordance with these findings, these two sequences compete efficiently for the binding of nuclear extracts containing CLOCK–BMAL1 to the rat a2 probe (Fig. 9C). The three upstream E-boxes are also able to bind the CLOCK– BMAL1 heterodimer (results not shown).
To provide an independent confirmation of the relevance of these data in vivo, we used morpholinos (Nasevicius & Ekker 2000) to knock-down the expression of either CLOCK or BMAL1 proteins during zebrafish embryogenesis. Due to extensive gene duplication, the zebrafish genome contains at least two Clock genes and two Bmal1 genes (F Delaunay & V Laudet, unpublished observations), we decided to co-inject two morpholinos able to block the protein synthesis of both genes (results not shown). We checked that these morpholinos effectively block the translation of the relevant protein in an in vitro expression system. In zebrafish, Rev-erbα is expressed specifically in the pineal gland at 48 h of development (Fig. 9D; Delaunay et al. 2000). Interestingly, the injection of a control morpholino at the one-cell stage did not modify this expression (Fig. 9D). In contrast, the injection of morpholinos that blocked either Clock or Bmal1 mRNA translation strongly decreased the expression of the endogenous Rev-erbα in the pineal gland at 48 h post-fertilization. The effect of these morpholinos was specific since they did not affect the expression of Otx5, a gene that is not circadian in the pineal gland (Fig. 9D; Gamse et al. 2002). Taken together these data are in accordance with those obtained using the Clock mutant mice and suggest that in vivo the Clock and Bmal1 genes regulate Rev-erbα expression effectively.
Discussion
Two Rev-erbα isoforms are under circadian regulation
Rev-erbα is an orphan nuclear hormone receptor that was cloned more than 10 years ago in several mammalian species and for which no well-defined physiological role has been found (reviewed in Laudet & Gronemeyer 2000). Importantly, this receptor lacks the C-terminal AF2 domain that is required for nuclear receptor ligand-dependent transcriptional activation and consequently behaves as a constitutive repressor. This suggests that Rev-erbα may be a true orphan receptor, the activity of which might be regulated through mechanisms other than ligand (Renaud et al. 2000). In this study, we show that the Rev-erbα gene encodes two transcripts, namely Rev-erbα1 and Rev-erbα2, that differ only in their 5’ region and which are both regulated in a circadian manner. Our functional data suggest that the short Rev-erbα2 isoform is generated from an internal promoter located in the first intron. This finding suggests that these two Rev-erbα isoforms could repress the transcription of target genes in a manner dependent on the time of day and independent of ligand. N-terminus isoforms are commonly found within the nuclear receptor superfamilly and they have been shown, in some instances, to result in proteins with different target gene specificity (for examples see Mulac-Jericevic et al. 2000 and Laudet & Gronemeyer 2002).
The circadian regulation of Rev-erbα could be controlled at either the transcriptional or posttranscriptional levels. However, since this rhythmic expression was observed in serum-shocked fibroblasts, an in vitro system, which mimicks in vivo free-running conditions (Balsalobre et al. 1998, Grundschober et al. 2001), we hypothesize that it is under the transcriptional control of the circadian clock pacemaker. In addition, analysis of the rat and human Rev-erbα 5’ flanking regions revealed several E-box DNA motifs, which are circadian system-response elements. Our paper presents two main lines of evidence strongly suggesting that the Rev-erbα gene is effectively under direct control of CLOCK and BMAL1: (1) functional data showing that the Rev-erbα promoters are activated by the CLOCK– BMAL1 heterodimer through binding to specific E-boxes in mammals as well as in zebrafish and (2) genetic evidence showing that Rev-erbα expression is decreased and the amplitude of its rhythmic expression severely affected in Clock mutant mice, as well as zebrafish embryos injected with CLOCK or BMAL1 morpholinos.
Rev-erbα is a target of the circadian pacemaker
The phase of Rev-erbα circadian expression both in vivo and in serum-shocked fibroblasts is consistent with control of this gene by the circadian oscillator. CLOCK and BMAL1 protein expression cycle with peak values at CT21–CT3 and a minimal value at CT6–CT12 in mouse liver (Lee et al. 2001). This expression pattern is exactly antiphasic to PER1, PER2, CRY1 and CRY2 expression as well as that of Rev-erbα, which peaks in the liver at CT8 (Balsalobre et al. 1998, Oishi et al. 1998, Torra et al. 2000). Interestingly, the pattern of Per2, Per3, Rev-erbα and Clock rhythmic expression is inverted in zebrafish when compared with nocturnal rodents but the relative phases are identical (Whitmore et al. 1998, Delaunay et al. 2000, 2003). Together, these expression data are strongly suggestive of an activation of Rev-erbα expression by the positive limb of the clock (CLOCK– BMAL1 heterodimer) and a repression by the negative limb including the PER and CRY proteins. Indeed, it has recently been shown that the phase of Rev-erbα mRNA accumulation is considerably advanced in Per2Brdm1 mutant mice (Preitner et al. 2002). This observation is further supported by the near-complete loss of Rev-erbα circadian expression in Clock mice livers and in ‘anti-CLOCK’ morpholino-treated and serum-shocked fibroblasts. These experiments provide the genetic evidence that Rev-erbα is an output of the circadian clock system. Because Rev-erbα is a transcriptional regulator, it is likely to act as a molecular link between the circadian oscillator and downstream target genes. For instance, Rev-erbα has been shown to bind to the promoter of the cellular retinol-binding protein I (CRBP1) for which mRNA circadian oscillation in the mouse liver has been described (Harding et al. 1995, Zheng et al. 2001). In addition, a direct role of Rev-erbα in the circadian pacemaker, at least in part through repression of Bmal1 gene expression, has been proposed recently (Preitner et al. 2002).
Our functional data strongly suggest that the CLOCK–BMAL1 heterodimer controls Rev-erbα expression directly. Both promoters were activated by CLOCK–BMAL1 and this activation was abolished by CRY protein. Interestingly, deletion and mutation analysis pointed to a divergent CACATG E-box in the P1 promoter, which is located in one of the transcription initiation sites of this promoter (Adelmant et al. 1996). In the P2 promoter, a complex of four E-boxes have been found (three canonical and one divergent) to respond to CLOCK– BMAL1. Gel-shift assays clearly showed that all these elements, including the divergent E-boxes, were bound by CLOCK–BMAL1. Interestingly, such divergent E-boxes have already been identified in the regulatory region of the liver transcription factor albumin-D site-binding protein (DBP) and they were shown to be recognized by CLOCK– BMAL1 (Ripperger et al. 2000). Our gel-shift experiments led us to propose that the context of these divergent E-boxes, and not only the core sequence itself, is important in determining their efficacy for CLOCK–BMAL1 binding. Indeed we show that E-boxes are less tolerant to variations in the core sequence than divergent E-boxes. These data suggest that CLOCK–BMAL1 can regulates genes that are devoid of canonical E-boxes and suggest that the promoter sequences of putative target genes of CLOCK–BMAL1 should be studied for divergent CACATG E-boxes with adjacent bases favorable for CLOCK–BMAL1 binding. In that respect, we note that all the divergent E-box of the Rev-erbα contains a T at position −1. This TCACATG motif would be interesting to use in a bioinformatics search for putative CLOCK–BMAL1 targets.
All the identified elements in the rat Rev-erbα promoter are conserved in the human and mouse sequences, stressing their functional relevance. In addition, we provide evidence suggesting that the structure of the P1 promoter and its regulation by CLOCK–BMAL1 is also conserved in the zebrafish Rev-erbα gene, suggesting that Rev-erbα may be under the control of the master oscillator in vertebrate species that have been diverged for more than 300 million years. This conservation suggests that the role of Rev-erbα in circadian regulation is under strong positive selection and thus really important for the organism’s fitness. This is in accordance with the circadian phenotype of the Rev-erbα-deficient mice. Experiments are under way in our laboratory to better delineate the role played by Rev-erbα and its paralogue Rev-erb in circadian clocks in both mammals and zebrafish.
Although a similar response to CLOCK– BMAL1 was observed for both Rev-erbα promoters, in agreement with the rhythmicity of both Rev-erbα transcripts, the serum-shock experiments suggest that these two promoters are differentially serum-regulated. It is likely that the transient and rapid differential regulation of P1 and P2 activities found after a serum shock is not the result of a circadian regulation but rather the action of serum-induced factors such as AP1. The basis for these different early effects of serum is unknown and may result from a promoter-specific response to growth factors. Of note, both P1 and P2 contain AP1 sites conserved between human and rodent promoters and it is possible that these sites do not respond identically to the serum treatment.
Multiple interlocked loops in circadian rhythm
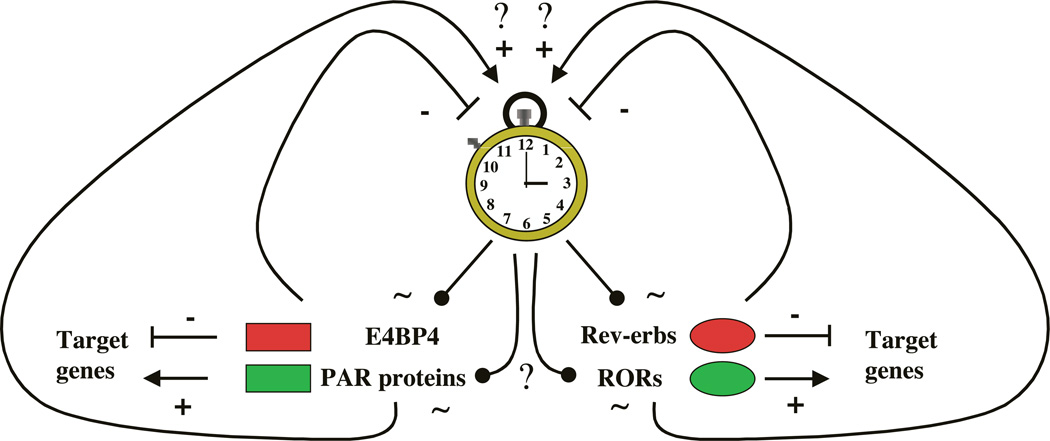
Interestingly, a recent report shows that Rev-erbα mRNA accumulation is considerably advanced in Per2Brdm1 mutant mice, an observation that also suggests that Rev-erbα expression is under the control of the circadian pacemaker (Preitner et al. 2002). This report also demonstrates that Rev-erbα controls the cyclic expression of BMAL1, since in Rev-erbα-knockout mice BMAL1 expression remains constant. These data, together with our demonstration that Rev-erbα is a target of the molecular oscillator, suggest that Rev-erbα is part of a regulatory loop which plays an important role in generating circadian rhythm. This situation is reminiscent of the case of DBP, which is also controlled by the clock and able to regulate Clock gene expression (Ripperger et al. 2000, Yamaguchi et al. 2000; reviewed in Roenneberg & Merrow 2003). Interestingly, in the case of Rev-erbα, as for DBP, other closely related genes also appear to play a role in circadian rhythm (Fig. 10). The three paralogues DBP, hepatic leukemia factor (HLF) and thyrotroph embryonic factor (TEF), which are all transcriptional activators, appear to cycle with identical phase, whereas the related gene, adenovirus E4 promoter-binding protein (E4 BP4), encoding a transcriptional repressor which does not contain the proline and acidic amino acid rich (PAR) activation domain, cycles in an opposite phase (Mitsui et al. 2001). E4 BP4 has been shown to compete with DBP, HLF and TEF for binding on the same DNA target sequences. In the case of Rev-erbα, it is interesting to observe that its paralog, Rev-erb , also displays circadian expression, whereas two of the three closely related Retinoid-related Orphan Receptor (ROR) genes, ROR and RORy, have been shown to cycle in brain and/or liver with a phase opposed to that of Rev-erbα (Andre et al. 1998, Panda et al. 2002, Preitner et al. 2002, Storch et al. 2002). RORs are transcriptional activators that bind to the same target sequence as Rev-erbs, namely the RevRE elements, and activate genes that are negatively regulated by Rev-erbα. Thus, DBP/E4 BP4 as well as RORs/Rev-erbs are paired components of a reciprocal mechanism that may complement each other in regulating the circadian oscillatory loop. It is also possible that other factors such as the basic helix-loop-helix transcription factors Differentially Expressed in Chondrocytes (DEC1) and DEC2, which both interact negatively with CLOCK– BMAL1-mediated transcriptional activation, are also able to form another independent loop (Honma et al. 2002; reviewed in Roenneberg & Merrow 2003). From all these data it is tempting to speculate that these loops, regulating the central oscillator, are interconnected but this remains to be addressed experimentally.
Figure 10.
Hypothetical model for the transcriptional loops regulating the central oscillator. Two transcription factor families, the PAR and the nuclear orphan receptors, contain activators and repressors that might form two loops regulating the circadian clock. Transcriptional activators are in green and repressors in red. Activation is shown by +, whereas repression is depicted by -. Cycling regulation is shown by | and curved arrows. CLOCK and BMAL1 have been shown to regulate some PAR proteins such as DBP (Ripperger et al. 2000, Yamaguchi et al. 2000) and Rev-erbα (this study). Whether other PAR proteins and Rev-erb are also controlled by the central clock remains to be shown. DBP regulates in turn the central clock by activating Per1 expression (Yamaguchi et al. 2000) whereas Rev-erbα represses BMAL1 expression (Preitner et al. 2002). The expression of E4 BP4 is cycling (Mitsui et al. 2001) as is the expression of at least two of the ROR genes; (Andre et al. 1998) and y (Panda et al. 2002, Preitner et al. 2002, Storch et al. 2002). Nevertheless, it is still unknown if these genes are directly controlled by the central oscillator (? in the scheme). It is likely that E4 BP4 and RORs also regulate the oscillator, even though this remains to be shown. Not depicted in the scheme is the attractive possibility that the PAR and nuclear receptor loops are interconnected in some way.
Rev-erbα as a model for integrating circadian and physiological regulation
Previous studies have shown that Rev-erbα gene expression is regulated by multiple signaling pathways. For instance, adipocyte differentiation and fibrates upregulate Rev-erbα expression while myocyte differentiation and glucocorticoids repress its expression (Chawla & Lazar 1993, Downes et al. 1995, Gervois et al. 1999, Torra et al. 2000). We already reported that Rev-erbα gene expression is inhibited by its own product as well as by the Rev-erb gene product through a Rev-DR2 element located in the P1 promoter (Adelmant et al. 1996). This site also allows an up-regulation of Rev-erbα by the related transcriptional activator orphan receptor, RORa (Raspe et al. 2002). ROR is circadian in the mouse SCN and pineal gland and the corresponding knockout mice exhibit a circadian phenotype (Andre et al. 1998). RORy expression is circadian in liver (Panda et al. 2002). We recently found that the second Rev-erbα promoter is also downregulated by Rev-erbs and activated by RORs through a monomeric RevRE (S Thenot and V Laudet, unpublished observations). The opposite phase of Rev-erbα and ROR gene expression suggests that the balance of their products is also regulated in a circadian manner and that this regulation might contribute to some of the circadian patterns of Rev-erbα expression in tissues such as the SCN, pineal gland and liver. The Rev-DR2 site, which is located in the P1 promoter and conserved in all mammals examined to date, is also the target for PPARa, which mediates the biological effects of fibrates (Gervois et al. 1999). Interestingly, the PPARa gene was also shown to be regulated according to a circadian rhythm with a peak at zeitgeber 10 (ZT10) (Lemberger et al. 1996). Finally, the P1 promoter is also downregulated by glucocorticoids through an unidentified non-Glucocorticoid Responsive Element (GRE) site (Torra et al. 2000). Plasma glucocorticoid levels oscillate with a peak around the onset of the activity phase and they participate in the glucocorticoid receptor-mediated resetting of peripheral clocks (Balsalobre et al. 2000, Le Minh et al. 2001).
Our finding that the CLOCK–BMAL1 heterodimer is a positive regulator of Rev-erba expression and that disruption of CLOCK activity in vitro and in vivo abolishes Rev-erbα circadian expression extends the repertoire of pathways that regulate Rev-erbα. Moreover, the data obtained with Clock mutant mice suggest that the circadian clock pacemaker is a master control point for Rev-erbα circadian expression. Furthermore, Rev-erbα circadian expression was observed in liver, heart and SCN suggesting that this gene is a critical output of the clock in multiple tissues (Panda et al. 2002, Storch et al. 2002; reviewed in Delaunay & Laudet 2002). How Rev-erbα integrates this circadian regulation with other regulatory pathways awaits further investigation. Interestingly, it was shown recently that the feeding schedule is a strong synchronizer of peripheral clocks (Damiola et al. 2000). The mechanisms by which the timing of food intake regulates peripheral clocks are still poorly understood, yet several hypotheses including roles for temperature and redox cofactors have been proposed (Rutter et al. 2001). The analysis of the mechanisms by which Rev-erbα integrates circadian and physiological inputs using mutant mice and nuclear receptor ligands will undoubtedly shed light on the function of circadian clocks.
Supplementary Material
Acknowledgement
We thank Michael Schubert for critical reading of the manuscript. S T holds a fellowship from the Association pour la Recherche contre le Cancer (ARC) and from the CNRS, T K from Région Rhône-Alpes and from the CNRS, R S from Ligue Nationale Contre le Cancer. We thank CNRS, MENRT (ACI program), ARC and Région Rhône-Alpes for financial support. JST is an Investigator of the Howard Hughes Medical Institute.
References
- Abe H, Honma S, Namihira M, Tanahashi Y, Ikeda M, Yu W, Honma K. Phase-dependent induction by light of rat Clock gene expression in the suprachiasmatic nucleus. Brain Research Molecular Brain Research. 1999;66:104–110. doi: 10.1016/s0169-328x(99)00031-5. [DOI] [PubMed] [Google Scholar]
- Adelmant G, Begue A, Stehelin D, Laudet V. & A functional Rev-erb alpha responsive element located in the human Rev-erb alpha promoter mediates a repressing activity. PNAS. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Tsuchiya Y, YoshinoT&Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Molecular and Cellular Biology. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M. Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO Journal. 1998;17:3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, DamiolaF&Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, SchutzG&Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Brown SA, Schibler U. The ins and outs of circadian timekeeping. Current Opinion in Genetics and Development. 1999;9:588–594. doi: 10.1016/s0959-437x(99)00009-x. [DOI] [PubMed] [Google Scholar]
- Chawla A, Lazar MA. Induction of Rev-erbα alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. Journal of Biological Chemistry. 1993;268:16265–16269. [PubMed] [Google Scholar]
- Chen W, Baler R. The rat arylalkylamine N-acetyltransferase E-box differential use in a master vs a slave oscillator. Brain Research Molecular Brain Research. 2000;81:43–50. doi: 10.1016/s0169-328x(00)00160-1. [DOI] [PubMed] [Google Scholar]
- Chong NW, Bernard M, Klein DC. Characterization of the chicken serotonin N-acetyltransferase gene. Activation via clock gene heterodimer/E box interaction. Journal of Biological Chemistry. 2000;275:32991–32998. doi: 10.1074/jbc.M005671200. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nature Neuroscience. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes and Development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay F, Laudet V. Circadian clock and microarrays mammalian genome gets rhythm. Trends in Genetics. 2002;18:595–597. doi: 10.1016/s0168-9525(02)02794-4. [DOI] [PubMed] [Google Scholar]
- Delaunay F, Thisse C, Marchand O, Laudet V, Thisse B. An inherited functional circadian clock in zebrafish embryos. Science. 2000;289:297–300. doi: 10.1126/science.289.5477.297. [DOI] [PubMed] [Google Scholar]
- Delaunay F, Thisse C, Thisse B, Laudet V. Differential regulation of Period 2 and Period 3 expression during development of the zebrafish circadian clock. Gene Expression Patterns. 2003;3:319–324. doi: 10.1016/s1567-133x(03)00050-4. [DOI] [PubMed] [Google Scholar]
- Downes M, Carozzi AJ, Muscat GE. Constitutive expression of the orphan receptor, Rev-erbα alpha, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Molecular Endocrinology. 1995;9:1666–1678. doi: 10.1210/mend.9.12.8614403. [DOI] [PubMed] [Google Scholar]
- Dumas B, Harding HP, Choi HS, Lehmann KA, Chung M, Lazar MA, Moore DD. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Molecular Endocrinology. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Foster RG. Shedding light on the biological clock. Neuron. 1998;20:829–832. doi: 10.1016/s0896-6273(00)80464-x. [DOI] [PubMed] [Google Scholar]
- Fruchart JC, Duriez P, Staels B. Peroxisome proliferator-activated receptor-alpha activators regulate genes governing lipoprotein metabolism, vascular inflammation and atherosclerosis. Current Opinion in Lipidology. 1999;10:245–257. doi: 10.1097/00041433-199906000-00007. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Shen Y-C, Thisse C, Thisse B, Raymond PA, Halpern ME, Liang JE. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nature Genetics. 2002;30:117–121. doi: 10.1038/ng793. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gervois P, Chopin-Delannoy S, Fadel A, Dubois G, Kosykh V, Fruchart JC, Najib J, Laudet V, Staels B. Fibrates increase human REV-erbαlpha expression in liver via a novel peroxisome proliferator-activated receptor response element. Molecular Endocrinology. 1999;13:400–409. doi: 10.1210/mend.13.3.0248. [DOI] [PubMed] [Google Scholar]
- Grundschober C, Delaunay F, Puhlhofer A, Triqueneaux G, Laudet V, Bartfai T, Nef P. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. Journal of Biological Chemistry. 2001;276:46751–46758. doi: 10.1074/jbc.M107499200. [DOI] [PubMed] [Google Scholar]
- Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Molecular and Cellular Biology. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Laudet V, Gronemeyer H. The nuclear receptors -Facts Book. San Diego, San Francisco, New York, Boston, London, Sydney: Tokyo Academic Press; 2002. [Google Scholar]
- Laudet V, Begue A, Henry-Duthoit C, Joubel A, Martin P, Stehelin D, Saule S. Genomic organization of the human thyroid hormone receptor alpha (c-erbα-1) gene. Nucleic Acids Research. 1991;19:1105–1112. doi: 10.1093/nar/19.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbα alpha transcriptional unit. Molecular and Cellular Biology. 1989;9:1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. Journal of Biological Chemistry. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO Journal. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system. Photic entrainment, circadian pacemaker mechanisms, and post-translational regulation. Annual Review of Genetics. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4 BP4 and PAR proteins in the circadian oscillatory mechanism. Genes and Development. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima N, Horiuchi R, Shibuya Y, Fukushige S, Matsubara K, Toyoshima K, Yamamoto T. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell. 1989;57:31–39. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- Mulac-Jericevic B, Mullinax RA, DeMayo FJ, Lydon JP, Conneely OM. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science. 2000;289:1751–1754. doi: 10.1126/science.289.5485.1751. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genetics. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochemical and Biophysical Research Communications. 1998;253:199–203. doi: 10.1006/bbrc.1998.9779. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-erbαlpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Raspe E, Mautino G, Duval C, Fontaine C, Duez H, Barbier O, Monte D, Fruchart J, Fruchart JC, Staels B. Transcriptional regulation of human Rev-erb alpha gene expression by the orphan nuclear receptor retinoic acid-related orphan receptor alpha. Journal of Biological Chemistry. 2002;277:49275–49281. doi: 10.1074/jbc.M206215200. [DOI] [PubMed] [Google Scholar]
- Renaud JP, Harris JM, Downes M, Burke LJ, Muscat GE. Structure-function analysis of the Rev-erbα and RVR ligand-binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Molecular Endocrinology. 2000;14:700–717. doi: 10.1210/mend.14.5.0444. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes and Development. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Circadian regulation of gene expression in animals. Current Opinion in Cell Biology. 2001;13:357–362. doi: 10.1016/s0955-0674(00)00220-9. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. The network of time understanding the molecular circadian system. Current Biology. 2003;13:R198–R207. doi: 10.1016/s0960-9822(03)00124-6. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Circadian and glucocorticoid regulation of Rev-erb alpha expression in liver. Endocrinology. 2000;141:3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- Vanacker JM, Pettersson K, Gustafsson JA, Laudet V. Transcriptional targets shared by estrogen receptor-related receptors (ERRs) and estrogen receptor (ER) alpha, but not by ERbeta. EMBO Journal. 1999;18:4270–4279. doi: 10.1093/emboj/18.15.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber EL, Duricka D, Ullman KS, Virshup DM. Nuclear export of mammalian PERIOD proteins. Journal of Biological Chemistry. 2001;276:45921–45927. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nature Neuroscience. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO Journal. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Mitsui S, Yan L, Yagita K, Miyake S, Okamura H. Role of DBP in the circadian oscillatory mechanism. Molecular and Cellular Biology. 2000;20:4773–4781. doi: 10.1128/mcb.20.13.4773-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones a comparative genetics of circadian clocks. Nature Review in Genetics. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.