Abstract
Numerous insects have independently evolved the ability to feed on plants that produce toxic secondary compounds called cardenolides and can sequester these compounds for use in their defense. We surveyed the protein target for cardenolides, the alpha subunit of the sodium pump, Na+,K+-ATPase (ATPα), in 14 species that feed on cardenolide-producing plants and 15 outgroups spanning three insect orders. Despite the large number of potential targets for modulating cardenolide sensitivity, amino acid substitutions associated with host-plant specialization are highly clustered, with many parallel substitutions. Additionally, we document four independent duplications of ATPα with convergent tissue-specific expression patterns. We find that unique substitutions are disproportionately associated with recent duplications relative to parallel substitutions. Together, these findings support the hypothesis that adaptation tends to take evolutionary paths that minimize negative pleiotropy.
Evolutionary genetics has long aimed to understand both the limits of the rate of evolution and the extent to which evolutionary paths are predictable (1). Although it is often presumed that adaptive evolution is mutation-limited (2), others have emphasized the role of pleiotropic and epistatic effects in constraining evolution. In particular, it has been proposed that (i) adaptation may tend to involve regulatory, rather than protein, changes because the former may be less pleiotropic (3); (ii) for similar reasons, specific genes may be hotspots for adaptation (4); (iii) because of epistasis, major evolutionary transitions may be facilitated by small population sizes (4–6); and (iv) that gene duplication may play an important role in the evolution of novelties by releasing constraints imposed by pleiotropy (6–8). The relative importance of each of these factors in organismal evolution is still debated (1, 3, 4, 8, 9). Although there are specific examples for each of these effects, they are often difficult to compare directly because they occur in very different contexts (10–13). Evolutionary parallelisms in settings where different species repeatedly face a common selective pressure provide a natural experiment for addressing these questions, which allows one to assess the extent to which phenotypic changes are caused by the recruitment of a limited set of genes and to test whether the specific molecular changes observed involve identical (or highly similar) substitutions (9).
One example of convergence involves the ability of the larvae and adults of a variety of insect species to feed on cardenolide-producing plants of the family Apocynaceae, notably in the genera Asclepias (“milkweed”) and Apocynum (“dogbane”) (14). Among other antiherbivore adaptations (15), these plants produce a class of secondary metabolites called cardenolides that are toxic to most herbivores. Cardenolides bind to and inhibit the alpha subunit of the sodium pump, Na+-and K+-exchanging ATPase (Na+,K+-ATPase) (ATPα), part of a multisubunit active transporter of Na+ and K+ that is crucial for muscle contraction, neural function, and other animal cellular processes requiring cation transport. In humans and other mammals, naturally occurring cardenolides regulate the activity of Na+,K+-ATPase in the context of many physiological processes, and plant-derived cardenolides have been used medicinally for hundreds of years (16). Within insects, herbivores feeding on Apocynaceae include several taxonomic orders spanning 300 million years of insect evolution. Beyond obtaining nutrition from these plants, some of these insects sequester cardenolides for their own defense against predation and have evolved associated aposematic coloration to advertise their unpalatability (17). Perhaps the best-known example is monarch butterflies (Danaus plexippus), whose aposematic larvae feed on milkweed plants and sequester large amounts of cardenolide, which in turn provides protection to the larvae and adults from a variety of predators (18).
Insect-host specialization on Apocynaceae plants includes both behavioral and physiological adaptations (15, 19), including amino acid changes in ATPα. Monarch butterflies carry at least one amino acid substitution (N122H) in ATPα that confers resistance to the inhibitory effects of cardenolides (20, 21). Additionally, the same substitution (22), as well as a second substitution (L111V) (23), is shared between Danaid butterflies and Chrysomelid beetles, which suggests parallel molecular evolution in these taxa. However, resistant forms of the enzyme in other taxa (e.g., Rattus norvegicus, Hydra vulgaris, and Bufo spp.) lack these specific substitutions; this implies that parallel evolution of cardenolide insensitivity is not always due to the same amino acid substitutions (24–26).
A limitation in examining cardenolide-resistant forms of ATPα, with the exception of Danaid butterflies and Chrysomelid beetles, is that resistance did not arise in the same, or even a similar, ecological context. Additionally, previous work has focused on the 12–amino acid H1-H2 extracellular domain of ATPα (20–23, 26, 27), yet mutagenesis studies have revealed at least 23 residues scattered throughout the ∼1000–amino acid ATPα protein at which amino acid substitutions confer some degree of reduced sensitivity to cardenolides (fig. S1). Structural studies have identified 17 residues that likely directly interact with the cardenolide ouabain or stabilize its binding (28–31), including 12 not detected in mutagenesis studies. Moreover, the states for most residues outside of the H1-H2 extracellular domain have not previously been investigated in cardenolide-sequestering insects (with the exception of the monarch butterfly) (32).
Recurrent gene duplication in Apocynaceae-associated taxa
We investigated the evolution of the ATPα protein among diverse taxa feeding on Apocynaceae plants, which are subject to similar selective pressures driving the evolution of insensitivity to cardenolides. The genomes of the fruit fly (Drosophila melanogaster), pea aphid (Acyrthosiphon pisum), monarch butterfly, and flour beetle (Tribolium castaneum) contain two, two, three, and four copies of ATPα, respectively, representing two ancient, distinct lineages (which we refer to as ATPα1 and ATPα2). This duplication likely dates back to the diversification of arthropods (fig. S2). However, the current assembly of the silk moth (Bombyx mori) genome contains just one detectable copy of ATPα that falls in the ATPα1 clade, which suggests that either the silk moth genome is incomplete or that these ancient duplicates have not persisted in all taxa. In monarch and fruit fly, ATPα2 genes have relatively low levels of expression (32) (http://flybase.org/reports/FBgn0040037.html).
We focused on the evolution of ATPα1, the copy common to all of the insects surveyed here, which is evolutionarily constrained at sites implicated in ouabain-binding in outgroup taxa. We performed a survey based on mRNA-Seq transcriptome shotgun sequencing of 26 taxa [including 14 that feed on Apocynaceae (table S1) (33)], as well as an analysis of the complete genomes of pea aphid, silk moth, monarch butterfly, and red flour beetle (33), and we detected single copies of ATPα1 in all but four taxa. Two copies of ATPα1 (designated ATPα1A and ATPα1B) [see (Fig. 1)] were identified in dogbane beetles (Chrysochus auratus) and milkweed stem weevils (Rhyssomatus lineaticollis), and we detected three copies (designated ATPα1A to C) (Fig. 1) in large and small milkweed bugs (Oncopeltus fasciatus and Lygaeus kalmii, respectively). Our results imply that at least four recent duplications of ATPα1 have occurred in lineages that feed on Apocynaceae hosts, and none have occurred in lineages that do not [the duplication in the dogbane beetle was overlooked in a previous study (22)]. We estimate that the most ancient of these duplication events, found in large and small milkweed bugs, occurred ∼60 million years ago (Mya) for copies A and B and ∼125 Mya for copies C and the ancestor of A and B (33). The estimated divergence time for the gene copies A to C exceeds the divergence between the large and small milkweed bugs (∼30 Mya), which implies that all three duplications are likely shared by many species within the Hemipteran subfamily Lygaeidae. The duplications of ATPα1 observed in the dogbane beetle and the milkweed stem weevil are considerably younger (∼24 and ∼9 Mya, respectively) but may also include multiple species (33).
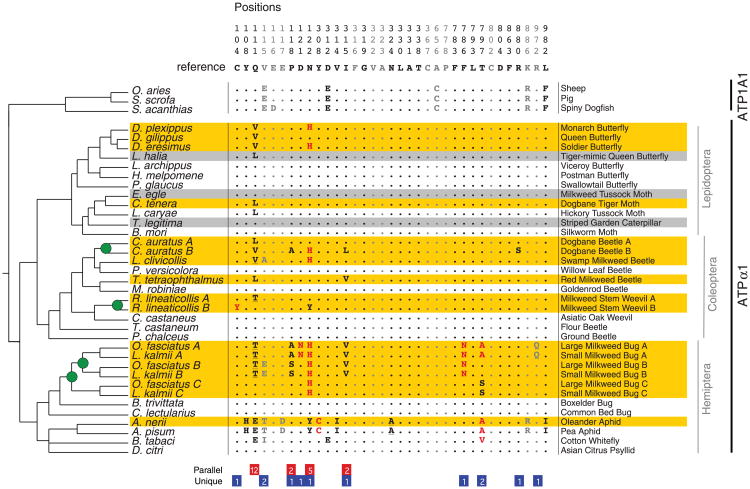
Fig. 1.
The phylogenetic pattern of amino acid substitution at sites implicated in cardenolide-binding for ATPα1 in taxa that feed on Apocynaceae and outgroups. Numbered columns correspond to sites for which site-directed mutagenesis or protein structure analysis has suggested a role in cardenolide-binding (fig. S1). Gray sites correspond to those with only structural evidence for a role in cardenolide-binding. Dots represent identity with the consensus sequence. Letters in red are specific substitutions characterized in functional assays (fig. S1). Underlined are substitutions that require at least two nonsynoymous substitutions relative to the ancestral sequence. Orange-shaded rows correspond to specialists known to sequester cardenolides. Gray-shaded rows represent species that either are not known to sequester cardenolides and/or are nonspecialists only occasionally found on Apocynaceae. The cladogram represents the accepted species branching order (table S6), and branch lengths are not meaningful. Green circles represent inferred duplications of ATPα1. Numbers below correspond to the number of inferred substitutions associated with use of Apocynaceae. Red boxes correspond to parallel substitutions (observed in more than one independent lineage). Blue boxes correspond to unique substitutions (observed in only one lineage).
Accelerated protein evolution in Apocynaceae-associated taxa
A signature of adaptive protein evolution is apparent in patterns of substitution at sites of ATPα1 implicated in ouabain binding (Fig. 1). Specifically, 33 of the 49 inferred amino acid substitutions occur in lineages that use Apocynaceae (including feeding ability and sequestration of cardenolides) (P = 1.7 × 10−7, accounting for branch lengths sampled) (33). Note that aphids show a distinct pattern (Fisher's exact test, P = 1.5 × 10−7), with all 11 ATPα1 substitutions having accumulated before the diversification of the oleander aphid and the pea aphid (∼65 Mya) (33). If we exclude the aphid-whitefly lineage, the association between amino acid substitutions and use of Apocynaceae therefore becomes stronger, with 33 of 34 substitutions occurring on lineages that use Apocynaceae (Lepidoptera: 5 of 6 substitutions; Coleoptera: 16 of 16 substitutions; Hemiptera: 12 of 12 substitutions; P = 7.9 × 10−10, accounting for branch lengths sampled) (33).
We also see considerable parallelism at the level of amino acid substitution in species that use Apocynaceae. In particular, glutamine at position 111 has experienced 12 substitutions, all of which are observed in at least one other lineage (Fig. 1). Also, position 122 has a replacement of asparagine by histidine in five independent lineages. These observations suggest that sites 111 and 122 are hotspots for substitution in Apocynaceae-feeding lineages (binomial test P = 6.0 × 10−11 and P = 3.1 × 10−4, respectively) (33). Overall, substitutions are significantly clustered in the H1-H2 extracellular domain (i.e., 24 of 33 substitutions occur at positions 111 through 122, binomial test, P = 9.5 × 10−11).
Unique substitutions are associated with duplications
Under a model in which pleiotropic effects constrain evolution, gene duplications may facilitate substitutions that would be deleterious in organisms with a single copy (7, 8). Consistent with this hypothesis, of the substitutions that have occurred on lineages associated with the use of Apocynaceae, unique substitutions are almost exclusively observed in lineages that carry a recent duplication of ATPα1 (11:1, P = 0.017, accounting for branch lengths sampled) (33), whereas parallel substitutions show no such pattern (11:10, P = 0.67, accounting for branch lengths sampled) (33). Comparing the ratios above confirms that parallel and unique substitutions show significantly different patterns of substitution with respect to duplication status of ATPα1 (Fisher's exact test, P = 0.027).
The molecular basis for insensitivity to cardenolides
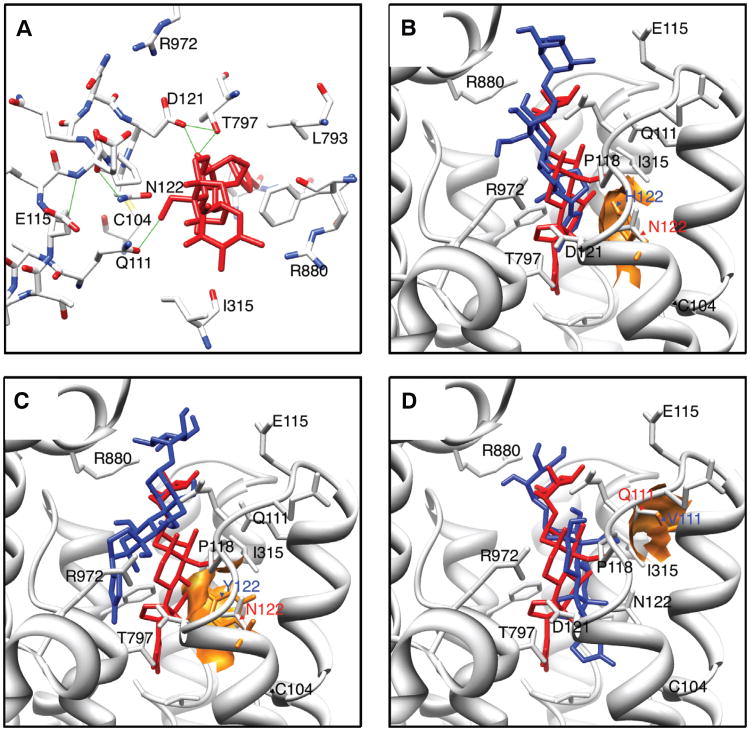
In a handful of cases (e.g., C104Y, D121N, N122H, Y308C, F786N, and T797A in Fig. 1), we know the effects of particular substitutions on cardenolide-binding affinity from site-directed mutagenesis studies (fig. S1). However, for most substitutions associated with the use of Apocynaceae, we lack this type of detailed information. To examine the likely effects of substitutions in ATPα1 across insects, we used molecular docking simulations with the crystal structure of the pig ortholog of ATPα bound to the cardenolide ouabain (Fig. 2A) (33). We compared ouabain docking onto the wild-type pig protein with docking onto a protein modified to incorporate individual amino acid substitutions that occurred in lineages associated with use of Apocynaceae (Fig. 1). We show that N122H, which has arisen separately in five lineages and is known from mutagenesis studies to confer resistance to ouabain in cell lines (21), likely acts by sterically blocking ouabain from entering the steroid-binding pocket of ATPα (Fig. 2B). Our docking simulations suggest that four other substitutions (C104Y, N122Y, R880S, and R972Q) have similar effects on ouabain binding (see Figs. 1 and 2C, fig. S3, and table S2). Of these, C104Y is known from functional studies to reduce the affinity of ATPα for ouabain to one-sixth of normal (fig. S1). Thus, although these amino acid substitutions differ, they appear to be functionally parallel. Most other observed substitutions are predicted to have more subtle effects (table S2). For example, the substitution Q111V, which appears in three independent lineages, appears to allow ouabain to fully penetrate the binding pocket (Fig. 2D), although it likely disrupts a key hydrogen bond between the protein and the steroid core of ouabain (Fig. 2A).
Fig. 2.
Cardenolide-binding pocket structure and molecular docking–simulation results. (A) Structure of the cardenolide-binding pocket of ATP1A1 for Sus scrofa bound to the cardenolide ouabain (in red) from (31). Green lines represent possible hydrogen bonds between the protein and ouabain. (B to D) Docking simulation results for the substitutions (B) N122H, (C) N122Y, and (D) Q111V. In each case, we show the best docking position for ouabain docked to the native protein (in red) versus the protein with one of three simulated substitutions (in blue). The best docking position is defined as that closest to the coordinates of ouabain in the published cocrystal structure (31). The atomic surfaces of the derived amino acid side chains at position 111 and 122 are detailed in gold. N122H and N122Y sterically block ouabain from entering the binding pocket [root mean square deviations (RMSDs) from cocrystal position are 5.5 and 6.9 Å, respectively; RMSDs from the best wild-type docking position are 5.6 and 6.9 Å, respectively]. Three other substitutions (C104Y, R880S, and R972Q) are predicted to have similar effects (table S2). In contrast, Q111V has more subtle effects on the position of ouabain (RMSD is 2.98 Å from the cocrystal structure and 0.14 Å from the best wild-type docking position) but likely disrupts a stabilizing H-bond (A).
Because the ATPα of vertebrates shares only ∼77% amino acid identity with insects, it is possible that, in insect lineages, substitutions that ameliorate the ouabain-inhibitory effects of some of the substitutions have arisen elsewhere in the protein. To address this possibility, we performed ouabain-docking simulations on predicted structures for the 19 native proteins of the taxa associated with Apocyanacae (fig. S3) (33). We found a significant positive correlation between the number of substitutions at sites implicated in ouabain-binding (Fig. 1) and the extent of displacement of the best ouabain docking (Spearman rank correlation test, P = 0.007) (table S3). Further, the 11 native proteins with one or more substitutions predicted to have a large effect (table S2) exhibit significantly greater displacement of ouabain than the eight native proteins lacking such substitutions (Mann-Whitney U test, P = 8 × 10–4) (table S3). Finally, we show that the best dockings for ouabain to predicted native protein structures with one or more large effect substitutions are further displaced relative to revertant forms of the native proteins in all 11 cases (Sign test, P = 0.001) (table S4).
Differential expression of ATPα1 duplicates
The docking simulations that we performed allow us to predict the extent of ouabain sensitivity of duplicate copies of ATPα1. Of these duplicates, we expect higher expression of the putatively less sensitive copy in the gut, as this is the primary location of cardenolide processing in these species. In contrast, the copy with the highest level of sensitivity to cardenolides should occur in the brain, because a glial sheath surrounding nervous tissue likely functions as a barrier that prevents cardenolides from reaching Na+,K+-ATPase in the ventral nerve cord (34). In support of this hypothesis, in the Asian grasshopper Poekilocerus bufonius, which has tissue-specific variation in cardenolide sensitivity, Na+,K+-ATPase in the brain was found to be the most sensitive to ouabain inhibition compared with Na+,K+-ATPase of the gut and excretory system (35).
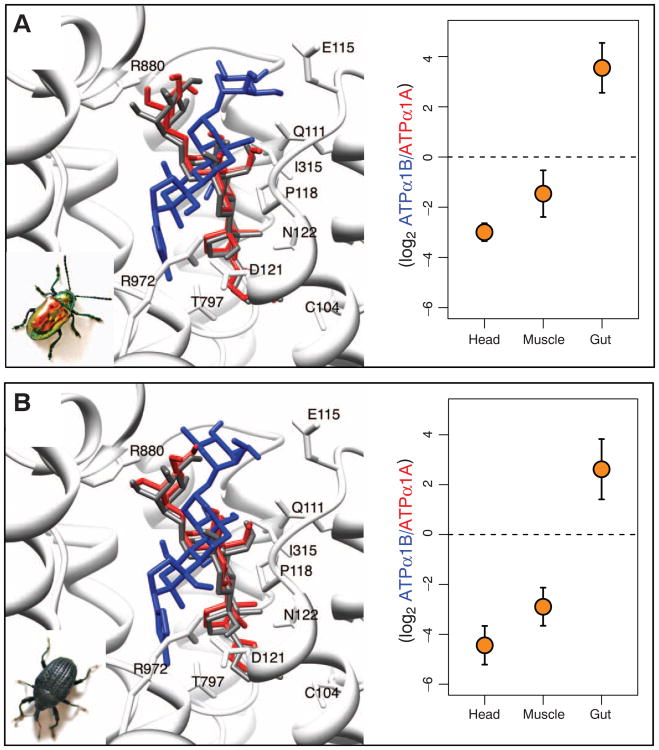
In the dogbane beetle, ATPα1B has accumulated five substitutions at sites in the protein that are predicted to determine ouabain-binding affinity (Fig. 1), including N122H and R880S. Docking simulations of ouabain on the predicted structure of native ATPα1B suggest significant displacement of ouabain relative to ATPα1A (Fig. 3A and table S3) and imply that ATPα1B is likely to be less sensitive to ouabain inhibition than ATPα1A. These two copies are differentially expressed among tissues (Fig. 3A). The relatively cardenolide-insensitive ATPα1B of the dogbane beetle is expressed seven times as much as ATPα1A in the digestive tract (Fig. 3A). Conversely, the sensitive ATPα1A has three to six times the expression of the less-sensitive ATPα1B in the head and thoracic muscle.
Fig. 3.
Native protein–docking simulations and tissue-specific gene expression for ATPα1 duplicates in (A) dogbane beetles and (B) milkweed stem weevils. (Left) Best docking results for ouabain onto pig ATP1A1 (gray ligand) and estimated structures for ATPα1A (red ligand) and ATPα1B (blue ligand) for each species (33). For both species, the best dockings of ouabain to the predicted structure of native ATPα1A are within 1 Å RMSD of the best docking to pig ATP1A1 (table S3). In contrast, the best dockings of ouabain to putatively resistant ATPα1B copies for dogbane beetles (A) and milkweed stem weevils (B) are displaced by 5.9 and 6.2 Å RMSD, respectively. (Right) Tissue-specific expression means (with two standard errors) of estimates for three unrelated individuals. Positive values indicate higher expression of the putatively resistant copy, ATPα1B. Analysis of variance (ANOVA) reveals significant variation in expression pattern among tissues (dogbane beetles: F = 74.3, P = 6 × 10−5; milkweed stem weevils: F = 21.1, P = 4 × 10−4). A Tukey-Kramer test reveals significant differences in all pairwise comparisons except head versus muscle (dogbane beetles, gut-head: P = 6 × 10−5; gut-muscle: P = 3 × 10−4; head-muscle: P = 0.08; weevils, gut-head: P = 4 × 10−4; gut-muscle: P = 3 × 10−3; head-muscle: P = 0.36).
Similarly, ATPα1B of the milkweed stem weevil harbors two substitutions (C104Y and N122Y) that are predicted to diminish its sensitivity to cardenolides. Our docking simulations imply that this copy of ATPα1 is also likely to be less sensitive to ouabain inhibition than ATPα1A, and the two copies have a pattern of differential expression that is almost identical to that of dogbane beetles (Fig. 3B). We also found the same rank-order of tissue-specific expression of putatively less sensitive to more-sensitive copies of ATPα1 in large and small milkweed bug, where the sensitivity of the three duplicates of ATPα1 can be ranked by the total number of substitutions at sites implicated in ouabain-binding and the number of substitutions with large effects identified in docking simulations (table S5 and fig. S4).
Thus, in addition to parallelisms in gene copy number and at the level of individual amino acid substitutions, we also observe parallel evolution of expression patterns for ATPα1 duplicates arising in distantly related taxa. Differential expression of ATPα1 duplicates in these taxa may have facilitated the functional specialization of what is otherwise a highly conserved protein.
Discussion
Our results suggest that adaptive evolutionary paths are, at least to some extent, predictable. ATPα1 is clearly a hotspot for molecular evolution associated with insect adaptation to using Apocynaceae, as seen by repeated duplication of ATPα1, parallel changes in gene expression, and parallel amino acid substitutions. The basis for this parallel evolution seems most readily explained by negative pleiotropic effects of most amino acid substitutions in this protein. In particular, we observed that unique substitutions are mostly confined to lineages with duplicate copies of ATPα1 and that 9 of 11 amino acid substitutions in lineages with one copy occur at only two sites (111 and 122). This level of parallelism is consistent with the hypothesis that negative pleitropic effects prevent changes at other amino acid sites that could theoretically contribute to reduced sensitivity of ATPα1 to cardenolides.
Despite the large number of parallelisms observed in the evolution of this system, the pattern in aphids (Fig. 1) indicates that not all substitutions conferring reduced sensitivity to cardenolides at ATPα1 are necessarily related to adaptation to cardenolide-containing host plants. Notably, the ATPα2 of aphids is ancestral at many sites implicated in cardenolide binding, contrary to all other insects surveyed here (fig. S5). In addition, ATPα2 of aphids lacks introns, whereas the fruit fly, flour beetles, and monarchs retain them, which suggests that the ATPα2 of aphids may represent a retrotransposed copy of the original duplicate. On this basis, we speculate that ATPα1 and ATPα2 may perform different roles in aphids than in other insects.
A decade-old debate centers around the relative importance of protein versus regulatory changes and the role of gene duplication in the evolution of novelties [e.g., (3, 8)]. Although our findings point to a certain level of predictability in evolution, they also reveal a plurality of evolutionary responses to a similar selective pressure, which demonstrates how adaptive evolution can leverage a combination of gene duplication, regulatory changes, and protein evolution. The genetic basis of repeated adaptations can be further elucidated by considering contexts where diverse groups of species are faced with a common selective pressure, such as insect-host specialization; cryptic melanism; or environmental gradients in oxygen, salinity, temperature, or acidity.
Supplementary Material
Acknowledgments
Thanks to M. and K. Kardestuncer for collecting the hickory tussock moths; A. Briscoe for providing queen, solider, and tiger-mimic queen butterfly tissue samples; D. Stern for the suggestion of trying de novo assembly of RNA-seq data; and M. Przeworski for helpful discussions and comments on the manuscript. Funded in part by an International Centre for Genetic Engineering and Biotechnology (ICGEB) Research Grant (No. CRP/COL09-01) to E.M.M. and NSF grant DEB-0946398 and NIH grant R01-GM083228 to P.A. All raw sequence data generated in this study can be downloaded at http://genomics-pubs.princeton.edu/insect_genomics/data.shtml, and assemblies of ATPα have been submitted to GenBank under accession nos. JQ771496 to 527.
Footnotes
Supplementary Materials: www.sciencemag.org/cgi/content/full/337/6102/1634/DC1
Materials and Methods
References (36–80)
References and Notes
- 1.Crow JF. Annu Rev Genet. 2008;42:1. doi: 10.1146/annurev.genet.42.110807.091612. [DOI] [PubMed] [Google Scholar]
- 2.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ. Press; Cambridge: 1983. [Google Scholar]
- 3.Carroll SB. Cell. 2008;134:25. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Stern DL. Evolution, Development, and the Predictable Genome. Roberts and Company Publishers; Greenwood Village, CO: 2011. [Google Scholar]
- 5.Wright S. Evolution and the Genetics of Populations. Univ. of Chicago Press; Chicago: 1968. pp. 1–4. [Google Scholar]
- 6.Lynch M. The Origins of Genome Architecture. Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- 7.Taylor JS, Raes J. Annu Rev Genet. 2004;38:615. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 8.Hoekstra HE, Coyne JA. Evolution. 2007;61:995. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 9.Christin PA, Weinreich DM, Besnard G. Trends Genet. 2010;26:400. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Sucena E, Delon I, Jones I, Payre F, Stern DL. Nature. 2003;424:935. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 11.Prud'homme B, et al. Nature. 2006;440:1050. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 12.Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Philos Trans R Soc London Ser B. 2010;365:2439. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riehle MM, Bennett AF, Long AD. Proc Natl Acad Sci USA. 2001;98:525. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobler S, Petschenka G, Pankoke H. Phytochemistry. 2011;72:1593. doi: 10.1016/j.phytochem.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal AA. Am Nat. 2004;164:113. doi: 10.1086/420980. [DOI] [PubMed] [Google Scholar]
- 16.Prassas I, Diamandis EP. Nat Rev Drug Discov. 2008;7:926. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 17.Duffey SS. Annu Rev Entomol. 1980;25:447. [Google Scholar]
- 18.Nishida R. Annu Rev Entomol. 2002;47:57. doi: 10.1146/annurev.ento.47.091201.145121. [DOI] [PubMed] [Google Scholar]
- 19.Vaughan GL, Jungreis AM. J Insect Physiol. 1977;23:585. [Google Scholar]
- 20.Holzinger F, Frick C, Wink M. FEBS Lett. 1992;314:477. doi: 10.1016/0014-5793(92)81530-y. [DOI] [PubMed] [Google Scholar]
- 21.Holzinger F, Wink M. J Chem Ecol. 1996;22:1921. doi: 10.1007/BF02028512. [DOI] [PubMed] [Google Scholar]
- 22.Labeyrie E, Dobler S. Mol Biol Evol. 2004;21:218. doi: 10.1093/molbev/msg240. [DOI] [PubMed] [Google Scholar]
- 23.Aardema ML, Zhen Y, Andolfatto P. Mol Ecol. 2012;21:340. doi: 10.1111/j.1365-294X.2011.05379.x. [DOI] [PubMed] [Google Scholar]
- 24.Shull GE, Greeb J, Lingrel JB. Biochemistry. 1986;25:8125. doi: 10.1021/bi00373a001. [DOI] [PubMed] [Google Scholar]
- 25.Canfield VA, Xu KY, D'Aquila T, Shyjan AW, Levenson R. New Biol. 1992;4:339. [PubMed] [Google Scholar]
- 26.Moore DJ, Halliday DCT, Rowell DM, Robinson AJ, Keogh JS. Biol Lett. 2009;5:513. doi: 10.1098/rsbl.2009.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mebs D, Zehner R, Schneider M. Chemoecology. 2000;10:201. [Google Scholar]
- 28.Qiu LY, et al. J Biol Chem. 2005;280:32349. doi: 10.1074/jbc.M505168200. [DOI] [PubMed] [Google Scholar]
- 29.De Pont JJHHM, et al. Pfluegers Archiv Eur J Physiol. 2009;457:623. [Google Scholar]
- 30.Ogawa H, Shinoda T, Cornelius F, Toyoshima C. Proc Natl Acad Sci USA. 2009;106:13742. doi: 10.1073/pnas.0907054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yatime L, et al. J Struct Biol. 2011;174:296. doi: 10.1016/j.jsb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Zhan S, Merlin C, Boore JL, Reppert SM. Cell. 2011;147:1171. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Material and methods and supplementary text are available as supplementary materials on Science Online
- 34.Freeman MR, Doherty J. Trends Neurosci. 2006;29:82. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 35.al-Robai AA. Comp Biochem Physiol B. 1993;106:805. doi: 10.1016/0305-0491(93)90034-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.