Abstract
Hepcidin, a peptide hormone that decreases intestinal iron absorption and macrophage iron release, is a potential drug target for patients with iron overload syndromes because its levels are inappropriately low in these individuals. Endogenous stimulants of Hepcidin transcription include bone morphogenic protein 6 (BMP6) and interleukin-6 (IL-6) by effects on mothers against decapentaplegic homolog (Smad)4 or signal transducer and activator of transcription (Stat)3, respectively. We conducted a small-scale chemical screen in zebrafish embryos to identify small molecules that modulate hepcidin expression. We found that treatment with the isoflavone, genistein, from 28-52 hours postfertilization in zebrafish embryos enhanced Hepcidin transcript levels, as assessed by whole-mount in situ hybridization and quantitative real-time reverse-transcriptase polymerase chain reaction. Genistein's stimulatory effect was conserved in human hepatocytes: Genistein treatment of HepG2 cells increased both Hepcidin transcript levels and promoter activity. We found that genistein's effect on Hepcidin expression did not depend on estrogen receptor signaling or increased cellular iron uptake, but was impaired by mutation of either BMP response elements or the Stat3-binding site in the Hepcidin promoter. RNA sequencing of transcripts from genistein-treated hepatocytes indicated that genistein up-regulated 68% of the transcripts that were up-regulated by BMP6; however, genistein raised levels of several transcripts involved in Stat3 signaling that were not up-regulated by BMP6. Chromatin immunoprecipitation and ELISA experiments revealed that genistein enhanced Stat3 binding to the Hepcidin promoter and increased phosphorylation of Stat3 in HepG2 cells. Conclusion: Genistein is the first small-molecule experimental drug that stimulates Hepcidin expression in vivo and in vitro. These experiments demonstrate the feasibility of identifying and characterizing small molecules that increase Hepcidin expression. Genistein and other candidate molecules may subsequently be developed into new therapies for iron overload syndromes. (Hepatology 2013;58:1315–1325)
Hepcidin is a transcriptionally regulated peptide hormone1 that is expressed primarily in the liver and excreted in urine. It is up-regulated in response to inflammation2,3 or iron overload4 and down-regulated in response to increased erythropoiesis, iron deficiency, or hypoxia.2 Hepcidin decreases intestinal iron absorption and macrophage iron release by causing internalization of the iron exporter, ferroportin1.5–7 Patients with hereditary hemochromatosis8 or thalassemia9–11 exhibit inappropriately low levels of hepcidin and increased intestinal iron absorption, despite the presence of systemic iron overload.12 Although treatment for iron overload is currently based on removal of blood or administration of iron chelators, it may be possible to prevent iron overload in patients with genetic predisposition if nontoxic small molecules can be administered that increase transcription of Hepcidin.
Iron overload4 and inflammation2,3 stimulate hepcidin expression by triggering the mothers against decapentaplegic homolog (Smad)-signaling or signal transducer and activator of transcription (Stat)-signaling pathways, respectively. It has been demonstrated that exposing human hepatocytes to bone morphogenic proteins (BMPs) up-regulates Hepcidin transcription by increasing Smad4 binding at Smad4-binding motifs, termed BMP response elements (BREs), in the Hepcidin promoter.13–16 BMPs are members of the transforming growth factor beta (TGF-β) family that signal by binding to transmembrane receptor complexes with serine-threonine kinase activity.17 Recent studies in mouse models18–21 indicate that BMP6 is the most likely physiologic regulator of hepcidin transcription in response to iron loading. Inflammatory stimuli, on the other hand, trigger increased serum interleukin-6 (IL-6) levels.22 IL-6 stimulates Hepcidin expression23 through increased Stat3 binding to a Stat3-responsive element in the Hepcidin promoter.24–27
We have developed the zebrafish embryo (Danio rerio) as an in vivo model to study hepcidin expression. Hepcidin expression begins at 36 hours postfertilization (hpf) in the zebrafish embryo and is responsive to iron levels and BMPs during embryonic development.28 To demonstrate that zebrafish embryos can be used to identify small-molecule modulators of Hepcidin expression, we screened a small number of naturally occurring isoflavones and related molecules for their effect on Hepcidin expression. We chose to evaluate isoflavones because they are nontoxic and are known to have kinase inhibitory actions.29 In this way, we identified genistein as the first small-molecule experimental drug to increase Hepcidin expression in vivo. We found that genistein also increased Hepcidin expression in cultured human hepatocytes (HepG2 cells). Using luciferase reporter assays, RNA sequencing (RNA-seq), and chromatin immunoprecipitation (ChIP), we demonstrated that genistein increases Hepcidin expression in a Smad4-dependent and Stat3-dependent manner.
Materials and Methods
Zebrafish Embryo Chemical Treatment, In Situ Hybridization, and Complementary DNA Preparation
Ethical approval was obtained from the institutional animal care and use committee of Beth Israel Deaconess Medical Center (Animal Welfare Assurance #A3153-01; Boston, MA) in accord with national and international guidelines. Zebrafish were maintained as previously described.30 Pools of 20 embryos were treated either with 7 μM of genistein, genistin, apigenin, daizdein, or estradiol and/or 40 μM of dorsomorphin (all obtained from Sigma-Aldrich, St. Louis, MO) or vehicle alone (1% dimethyl sulfoxide; DMSO) from 28 to 52 hpf. Embryos were then fixed in 4% paraformaldehyde in phosphate-buffered saline for whole-mount in situ hybridization using anti-sense zebrafish hepcidin or FOXA3 probes, as previously described.31 O-dianisidine staining for hemoglobin and flow cytometry were performed as described in the Supporting Materials. Representative embryos were photographed at 100× magnification with a BX51 compound microscope (Olympus, Center Valley, PA) and a Q-capture 5 digital camera (QImaging, Surrey, British Columbia, Canada). Expression analysis was conducted at the specified time points, by anesthetizing pools of 20 embryos with tricaine, followed by storage in RNAlater (Ambion, Life Technologies, Grand Island, NY). RNA extraction and generation of complementary DNA (cDNA) were performed as previously described.6,32
Cell Culture and Chemical Treatment
The human hepatocarcinoma cell line, HepG2 (American Type Culture Collection, Manassas, VA), was maintained in alpha-minimum essential medium (α-MEM)/10% certified endotoxin-free fetal bovine serum (FBS)/1% penicillin-streptomycin (Life Technologies) at 37°C in 5% CO2. HepG2 cells were seeded onto 12-well tissue-culture-treated plates at a density of 4 × 105 cells per well. Twenty-four hours later, the culture medium was changed to low-serum medium (α-MEM/1% FBS). After 8 hours of equilibration, cells were then treated for the specified time periods with genistein, genistin, apigenin, daizdein, dorsomorphin (each at 10 μM), estradiol (0.001-100 μM), ICI 182,780 (100 μM) (all from Sigma-Aldrich), IL-6 20 or 100 ng/mL, BMP6 50 ng/mL (both from R&D Systems, Minneapolis, MN), or vehicle only (1% DMSO). All the samples included 1% DMSO to control for any potential effects of the vehicle.
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction
After 18 or 24 hours of treatment, cells were harvested for extraction of total RNA using the RNeasy Mini Kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. DNA was eliminated by on-column RNase-free DNase treatment. Reverse transcription was performed using RNA (300 ng), oligo-dT, and Superscript II reverse transcriptase (RT; Life Technologies) in a 20-μL reaction, which was later diluted to 100 μL.
Ten percent of each RT reaction was used for a single 50-μL quantitative real-time RT-PCR (polymerase chain reaction) assay using Taqman Universal PCR Master Mix or SYBR Green PCR Master Mix (both from Life Technologies). Primer and probe sequences are given in Supporting Table1. Transcript levels were calculated as fold increases over the control sample. The described reactions failed to amplify the nontemplate controls or RNA samples that were not treated with RT.
Table 1.
KEGG Pathway Analysis of Genes That Were Up-Regulated by Genistein
| Term | Count | P Value | Genes | Benjamini-Hochberg Statistic |
|---|---|---|---|---|
| hsa03010/ribosome | 37 | 0.00E-01 | RPL18, RPL13, RPS3AP47, RPL35, RPLP2, RPL36, RPL37, RPS19P3, RPL38, RPLP0P2, RPS27, RPL32, RPS11P5, RPLP1, RPL3, RPL29P26, RPL12, RPL7A, RPL10A, RPS20, RPS21, RPS13P2, RPS23, RPS15P5, RPSA, RPS16P10, RPS9, RPL27, RPL23A, RPSAP8, RPS3P3, RPS6, RPS5, RPL28, RPS8, RPL18A, RPL37A, UBA52 | 0.00E-01 |
| hsa04110/cell cycle | 36 | 0.00E-01 | MAD1L1, FZR1, E2F4, PKMYT1, ANAPC11, SFN, PTTG1, ZBTB17, TGFB1, RBX1, MCM7, CDKN2B, TFDP2, MYC, CCNA2, BUB3, CUL1, ANAPC2, CREBBP, CDC20, CDK7, CDC25C, MCM5, CDC25B, CCNB1, CDKN1A, CCND1, CDKN1B, YWHAH, CCNB2, PLK1, ANAPC7, ABL1, MAD2L2, GADD45B, GADD45A | 0.018 |
| hsa03040/spliceosome | 36 | 0.00E-01 | HNRNPA1L2, CHERP, CCDC12, NHP2L1, U2AF2, LSM7, SF3B5, BUD31, XAB2, SART1, CTNNBL1, SFRS4, DHX38, PRPF8, PCBP1, SFRS9, PQBP1, SNRNP70, ACIN1, LSM2, PRPF40B, DDX42, DHX8, SNRPA1, EFTUD2, SF3A2, SF3A1, EIF4A3, PPIE, SNRNP200, SNRPB, SNRPA, THOC4, PHF5A, PUF60, SNRPG | 0.014 |
| hsa05221/acute myeloid leukemia | 21 | 0.00E-01 | HRAS, PPARD, MAP2K1, MAP2K2, RELA, STAT5B, PIK3CD, PIM1, RPS6KB2, BAD, TCF7L2, STAT3, JUP, EIF4EBP1, CCND1, ARAF, IKBKG, RARA, RUNX1, MYC, AKT2 | 0.011 |
| hsa04142/lysosome | 33 | 0.01E-01 | SGSH, AP1M1, CLTB, AP1B1, LGMN, HEXA, HEXB, ATP6V0B, CTSL1, ATP6V0C, CD68, MAN2B1, ATP6V0D1, PSAP, ATP6V1H, AP4M1, CD63, AP4S1, CD164, LAMP1, NPC1, GLA, IGF2R, GAA, ATP6V0A1, CTSD, NEU1, CTSC, CTSB, GGA1, GGA2, CTSH, GGA3 | 0.021 |
| hsa05220/chronic myeloid leukemia | 23 | 0.00153 | HRAS, CTBP1, BCR, MAP2K1, MAP2K2, RELA, STAT5B, PIK3CD, BCL2L1, BAD, TGFB1, CCND1, CDKN1A, CDKN1B, GAB2, ARAF, IKBKG, SHC1, ABL1, RUNX1, SHC2, MYC, AKT2 | 0.046 |
Transferrin and Nontransferrin-Bound 55Fe Uptake Assays and Ferritin Enzyme-Linked Immunosorbent Assay
Methods for transferrin and nontransferrin-bound 55Fe uptake assays and ferritin enzyme-linked immunosorbent assay (ELISA) are provided in the Supporting Materials.
Dual Luciferase Assay
HepG2 cells were transfected with pGL4.17 Hepc, 3 kilobases (kb) of the human Hepcidin promoter upstream of the firefly luciferase reporter gene, or mutated versions of the construct with point mutations in the Stat3-binding site, the BREs, or both the BRE-binding and the Stat3-binding site.33 Additional details are provided in the Supporting Materials. The Hepcidin reporter constructs were donated by Drs. Ganz and Nemeth (Division of Pulmonary and Critical Care, University of California, Los Angeles). As a control, HepG2 cells were cotransfected with Renilla luciferase under control of the cytomegalovirus promoter (pRL-CMV; Promega, Madison, WI). Transiently transfected cells were treated for 6 hours with genistein (10 μM), BMP6 (50 ng/mL), or vehicle (1% DMSO) only. The Dual-Glo Luciferase Assay (Promega) was then performed according to the manufacturer's instructions.
Statistical Analysis for Quantitative PCR, Iron Uptake, Luciferase, ChIP, and ELISA
All data shown are means ± standard error from the mean. Data analysis was performed using Prism 5.0 (GraphPad Software, Inc., La Jolla, CA). For comparisons of two groups, two-tailed Student t tests were performed comparing experimental treatment to DMSO treatment. For comparisons of three or more groups, Kruskal-Wallis' test was performed on raw data or natural logs of raw data. If P < 0.05, pairwise comparisons were made with two-tailed Student t tests. Comparisons where P < 0.05 were deemed significant.
RNA-seq
As described above, HepG2 cells were cultured in 12-well plates, shifted to low-serum conditions, and treated with DMSO (1%), BMP6 (50 ng/mL), or genistein (10 μM) for 18 hours. The numbers of biological replicates were 3, 2, and 3, respectively. Details are provided in the Supporting Materials. Differential expression for BMP6-treated or genistein-treated biological replicates, compared to DMSO-treated controls, was determined using the Cufflinks tool, Cuffdiff. The criteria for differential expression were as follows: a log2 fold-change of >0.58 or <−0.58 and a Q value <0.05, when compared to the control condition. We also required that the differentially expressed genes used for downstream analysis had a fragments per kilobase of exon per million fragments mapped (FPKM) >0.1 in at least one of the conditions. Enrichments in particular biological processes, pathways, and functions were calculated statistically using the DAVID annotation database.34 Prism 5.0 was used to perform linear regression analysis of the correlation between the natural log of gene expression changes measured by RNA-seq and quantitative real-time RT-PCR. All RNA-seq data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo).
ChIP Followed by Quantitative PCR
HepG2 cells were seeded on 150-mm2 plates and cultured to reach a cell density of 5 × 107 cells per plate. Eight hours before treatment, cells were washed and shifted to low-serum media (α-MEM/1% FBS). Chemicals were added to achieve either 1% DMSO, 10 μM of genistein, 50 ng/mL of BMP6, or 20 ng/mL of IL-6. After 24 hours of treatment, cells were cross-linked by adding formaldehyde to reach a final concentration of 1%. After reversal of cross-linking, cells were lysed, sonicated, and chromatin was immunoprecipitated with antibody (Ab)-coated Dynabeads Protein G (Life Technologies). Additional details are provided in Supporting Materials. The Abs used were anti-Stat3 (sc-7154) and anti-rabbit immunoglobulin (Ig)G (sc-2027) (both from Santa Cruz Biotechnology, Santa Cruz, CA). DNA was eluted from the beads and used as a template in a quantitative PCR reaction using primers (Supporting Table1) to amplify the proximal BMP/Stat response element, located 84 nucleotides before the Hepcidin start site.25 The data were reported as fold increase in binding of the specific Ab over binding to nonspecific Ig.
Stat3 ELISA
Adherent HepG2 cells (2 × 104 cells per well of a 96-well plate) were incubated in serum-free α-MEM for 16 hours, followed by the addition of the following treatments: DMSO (1%); genistein (10 μM); or IL-6 (100 ng/mL). Cells were fixed in 4% formaldehyde after 0-60 minutes of treatment. Detection of Stat3 and phospho-Stat3 (pStat3) was performed using the Stat3 Colorimetric In-Cell ELISA Kit (Thermo Fisher Scientific Pierce, Rockford, IL), according to the manufacturer's instructions.
Results
Genistein Increases hepcidin Expression in Zebrafish Embryos
Because we had previously demonstrated evolutionarily conserved aspects of hepcidin regulation in zebrafish embryos,28 we used zebrafish embryos to evaluate naturally occurring isoflavones (genistein and daizdein), genistin (the inactive glucoside form of genistein), or a flavone (apigenin) as potential modulators of mammalian Hepcidin expression. Because genistein, daizdein, and apigenin have estrogenic effects, we also evaluated estradiol. Embryos were treated from 28 to 52 hpf with the chemicals, each at 7 μM, followed by fixation for whole-mount in situ hybridization (Fig. 1A) or quantitative real-time RT-PCR (Fig. 1B) to assess hepcidin RNA levels. We found that genistein treatment was associated with increased intensity of hepcidin expression (Fig. 1A,B) and an expanded domain of expression extending from the liver into the proximal intestine in 100% of 36 embryos, whereas treatment with apigenin, daizdein, estradiol, or genistin did not increase either intensity or extent of expression. However, treatment with the BMP receptor antagonist, dorsomorphin, abrogated genistein's effect on hepcidin expression (Fig. 1A).
Fig 1.
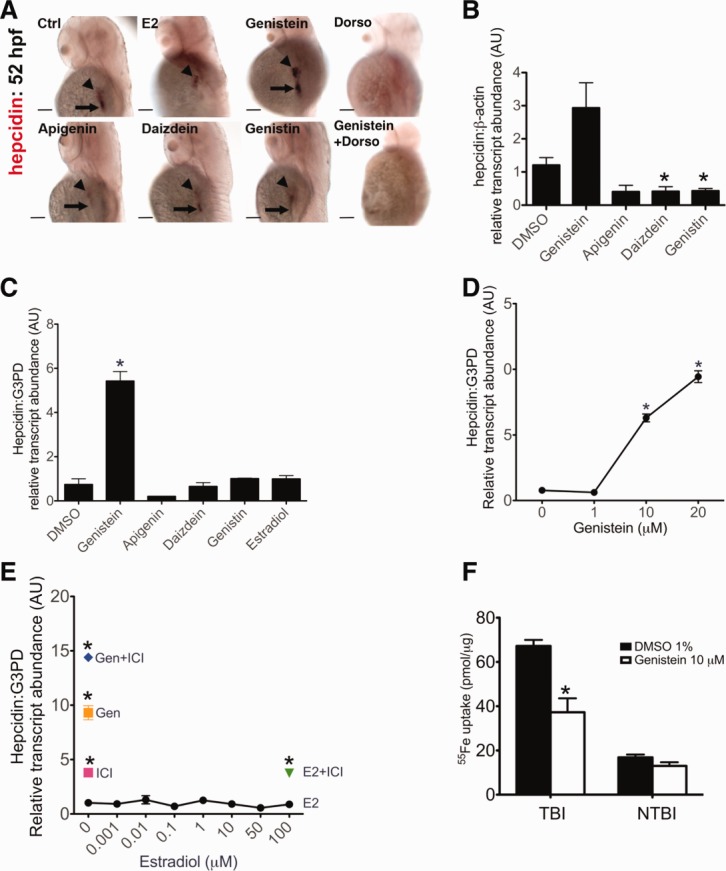
Effect of genistein treatment on Hepcidin expression. (A and B) Zebrafish embryos were treated from 28 to 52 hpf with either estradiol (E2), genistein, apigenin, daizdein, or genistin, each at 7 μM, and/or dorsomorphin (40 μM), followed by fixation for whole-mount in situ hybridization (A) or quantitative real-time RT-PCR (B) to assess hepcidin RNA levels. Hepcidin expression was consistently increased in intensity and extent in the genistein-treated embryos, compared to DMSO-treated controls, but this effect was abrogated by cotreatment with dorsomorphin (N = 36 per group). Arrowheads mark the liver and arrows mark the proximal intestine. Scale bar = 100 microns. For quantitative real-time RT-PCR, data shown are means ± standard error. N = 3 samples per group. *P < 0.05, compared to DMSO alone. (C-E) Hepcidin expression measured by quantitative real-time RT-PCR in human hepatoma (HepG2) cells after treatment with a variety of chemicals at 10 μM (C), with genistein from 0 to 20 μM (D), or (E) estradiol (E2) from 0 to 100 μM in the presence or absence of the estrogen antagonist, ICI 182,780 (ICI; 100 μM). N = 2-4 samples per condition. *P < 0.05, compared to DMSO treated. (F) Treatment with genistein for 24 hours failed to increase nontransferrin-bound iron (NTBI) uptake or transferrin-bound iron (TBI) uptake, as measured by 55Fe content per microgram of protein extracted from HepG2 cells. N = 2 per group. *P < 0.05, compared to DMSO treated.
The genistein-induced increase in hepcidin transcript levels was modest (Fig. 1B), related to decreased liver size and only a slight increase in hepatocyte-specific hepcidin expression (Supporting Figs. 3). This may explain why, despite the increase in hepcidin expression, staining for hemoglobin (Supporting Fig. 4) did not reveal anemia in any of the genistein-treated embryos (n = 50).
Fig 3.
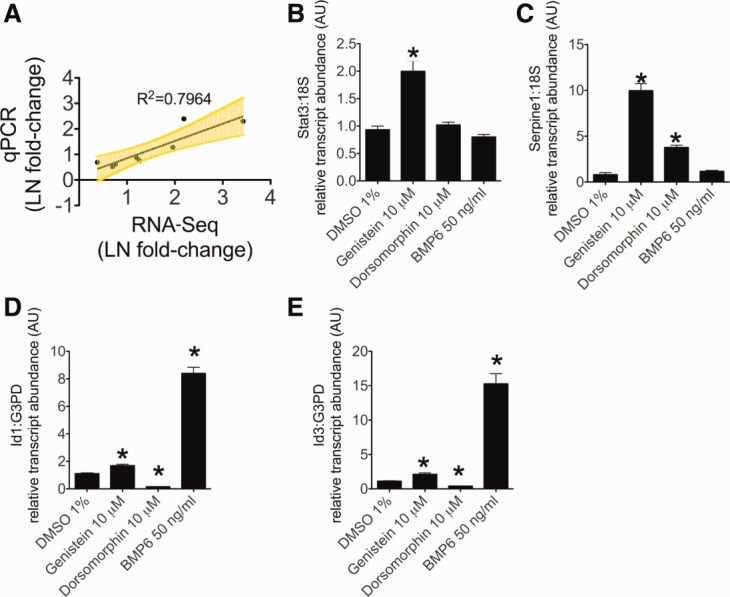
Validation of RNA-seq data by quantitative real-time RT-PCR. (A) Graph of correlation between the natural log of the fold-change in gene expression after genistein treatment for RNA-seq versus quantitative real-time RT-PCR analysis. Linear regression to determine the best-fit line and Pearson's coefficient R was performed using GraphPad 5.0 (GraphPad Software, Inc., La Jolla, CA). Yellow error bars include the 95% confidence interval from the best-fit line. (B-E) Quantitative real-time RT-PCR to measure the change in expression of genes that were up-regulated on RNA-seq analysis after treatment with genistein, including (B) Stat3, (C) Serpine1, (D) Id1, and (E) Id3. N = 3 per group. *P < 0.05, compared to DMSO treated.
Fig 4.
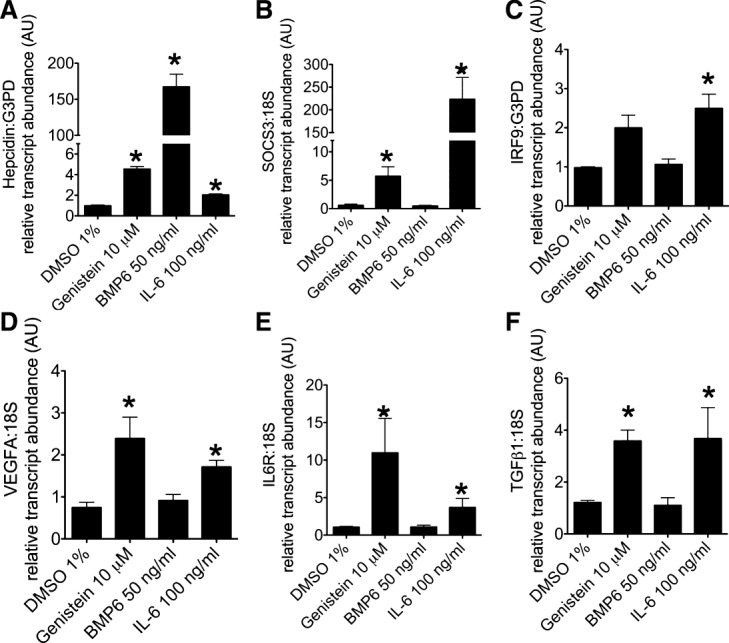
Effect of genistein on Stat3-related genes and TGF-β1 in HepG2 cells. (A-F) Quantitative real-time RT-PCR comparing changes in mRNA expression in HepG2 cells after treatment for 24 hours with genistein (10 μM), BMP6 (50 ng/mL), or IL-6 (100 ng/mL) in (A) Hepcidin, (B) SOCS3, (C) IRF9, (D) VEGFa, (E) IL6 receptor, and (F) TGFβ1. N = 2-6 per group. *P < 0.05, compared to DMSO treated.
Genistein Increases Hepcidin Expression in HepG2 Cells
To evaluate whether genistein exerted a conserved effect on human hepatocytes, we tested the compounds in human hepatoma (HepG2) cells. We found that genistein produced a five-fold increase in Hepcidin expression, as measured by quantitative real-time RT-PCR (Fig. 1C) that was not reproduced by treatment with the other small molecules. The effect was dose dependent (Fig. 1D), with higher increases in Hepcidin expression noted at 20 than 10 μM. In contrast, no significant increase in Hepcidin expression was noted after treatment with estradiol at a broad range of concentrations, ranging from 0.001 to 100 μM (Fig. 1E). The addition of the potent estrogen receptor antagonist, ICI 182,780 (Fig. 1E), failed to inhibit genistein's positive effects on Hepcidin transcript levels. In fact, the addition of the estrogen-receptor inhibitor, ICI 182,780, actually increased Hepcidin expression (Fig. 1E), suggesting that inhibition of estrogen receptor signaling may enhance Hepcidin transcript levels.
Genistein Does Not Increase Hepatocyte Iron Uptake
We considered the possibility that genistein may increase Hepcidin expression by promoting cellular iron loading. To evaluate this hypothesis, we tested the effect of genistein on 55Fe uptake in HepG2 cells. After 24 hours of treatment with genistein, transferrin-bound iron uptake was significantly reduced, compared to vehicle alone (37.27 ± 6.29 versus 67.21 ± 2.755; P < 0.05), whereas nontransferrin-bound iron uptake was not significantly changed from vehicle-treated controls (Fig. 1F). To evaluate effects on hepatocyte iron stores (Supporting Fig. 5), we measured cellular ferritin levels after 24 hours of treatment with genistein or DMSO. We found no significant difference in ferritin levels (0.171 ± 0.058 versus 0.165 ± 0.026; P = 0.92), suggesting that the genistein-induced decrease in transferrin-bound iron uptake is offset by a hepcidin-induced decrease in ferroportin-dependent iron export.
Fig 5.
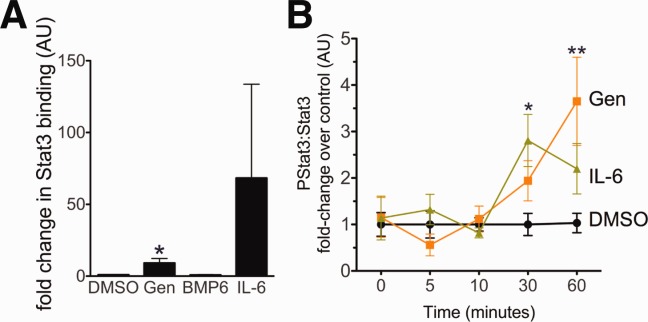
Effect of genistein on Stat3 binding to the Hepcidin promoter and Stat3 phosphorylation. (A) HepG2 cells were treated for 24 hours with DMSO (1%), BMP6 (50 ng/mL), IL-6 (20 ng/mL), or genistein (10 μM) in media containing 1% FBS, followed by ChIP with an Ab specific for Stat3 (Santa Cruz Biotechnology, Santa Cruz, CA) or with nonspecific IgG. After reversal of cross-linking, quantitative PCR with primers specific for the Stat3-binding site was performed to determine the fold change in Stat3 binding to the Hepcidin promoter, relative to nonspecific IgG binding. Data shown are means ± standard error (SE). N = 3-4 samples per group. *P < 0.05, compared to DMSO treated. (B) ELISA was used to quantify the ratio of pStat3 to Stat3 protein in HepG2 cells in serum-free media after exposure to IL-6 (100 ng/mL, light green triangles) or genistein (10 μM, orange squares), normalized to vehicle (1% DMSO, black circles) from 0 to 60 minutes. Data shown are means ± SEs. *P < 0.05 for IL-6 versus DMSO; **P < 0.05 for genistein versus DMSO. N = 7 per time point per group.
Genistein Increases Hepcidin Promoter Activity
Because genistein did not appear to cause increased Hepcidin expression by enhancing cellular iron uptake, we theorized that it could enhance the effect of either of the major transcription factor pathways that have been implicated in Hepcidin's regulation: Smad4 or Stat3 signaling. To interrogate these pathways, we transfected HepG2 cells with reporter constructs encoding 3 kb of the human Hepcidin promoter upstream of the firefly luciferase reporter gene. The promoter sequence was either wild type (WT), mutated in the Stat3-binding site, mutated in the Smad4-binding BREs, or mutated in both the Stat3-binding motif and the BREs.33 We found that treatment with either genistein or BMP6 for 6 hours enhanced Hepcidin-luciferase activity by two-fold and three-fold, respectively (Fig. 2A). Mutation of the Smad4-binding BREs, with or without mutation of the Stat3 motif, decreased Hepcidin promoter activity below basal levels in all conditions. However, mutation of the Stat3 motif significantly impaired induction of Hepcidin promoter activity in the genistein-treated, but not the BMP6-treated or vehicle-treated, cells. Thus, the genistein-induced increase in Hepcidin promoter activity required Stat3 as well as Smad4 binding.
Fig 2.
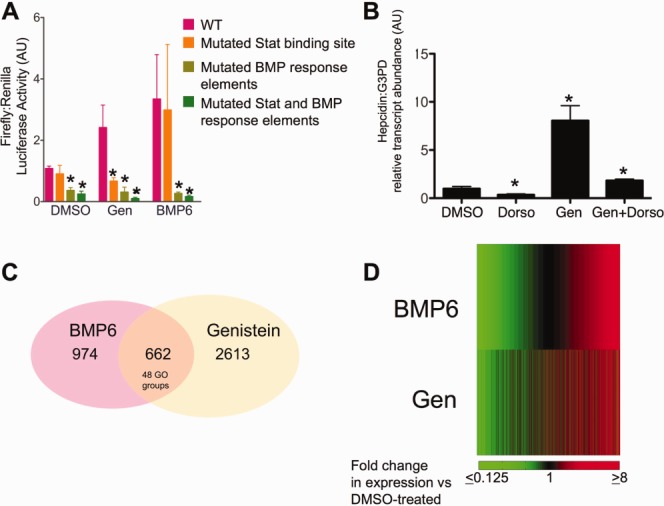
Effect of genistein and BMP6 on Hepcidin promoter activity and transcriptional profile. (A) Hepcidin promoter luciferase assay. HepG2 cells were cotransfected with a Renilla control vector (pRL-CMV) and either a construct containing a 3-kb WT human Hepcidin promoter upstream of a firefly luciferase reporter (pink) or the same construct with inactivating mutations in the Stat3-binding site (orange), in the BMP response elements (light green), or in both the Stat3-binding site and the BMP response elements (dark green). One day after transfection, cells were treated for 6 hours with vehicle (1% DMSO), genistein (10 μM), or BMP6 (50 ng/mL), followed by lysis and measurement of luciferase activity. Hepcidin promoter activity was reported as the ratio of firefly to Renilla activity and normalized to the WT Hepcidin promoter activity in the presence of vehicle only. Data shown are means ± standard error. N = 4 samples per group. (B) The BMP receptor antagonist, dorsomorphin, significantly reduces genistein's effect on Hepcidin expression, as measured by quantitative real-time RT-PCR in HepG2 cells treated for 18 hours with vehicle (1% DMSO), dorsomorphin (10 μM), genistein (10 μM), or genistein and dorsomorphin combined (10 μM of each). N = 3 samples per group. *P < 0.05, compared to DMSO treated. (C) Venn diagram illustrating results of RNA-seq. Nine hundred and seventy-four transcripts were up-regulated in HepG2 cells after treatment with BMP6 (50 ng/mL), and 2,613 transcripts were up-regulated by genistein (10 μM), compared to treatment with 1% DMSO. Of the genes up-regulated by BMP6, 662 were also up-regulated after treatment with genistein (10 μM), corresponding to 48 GO groups. All treatments were performed for 18 hours before harvesting cells for RNA extraction. The numbers of biological replicates were 3 (DMSO), 3 (genistein), and 2 (BMP6). (D) Heat maps displaying average fold-change in gene expression, relative to DMSO-treated cells, in the RNA-seq experiment after treatment with BMP6 (top) or genistein (bottom). In each heat map, genes were ordered from lowest to highest intensity of expression in BMP6-treated cells. Green corresponds to ≤0.125 fold-change, whereas red corresponds to ≥8-fold change in expression over DMSO-treated controls. Gene expression was quantified as FKPM using the Cufflinks algorithm.
Inhibition of Type I BMP Receptor Activity Decreases Genistein's Effect on Hepcidin Expression
Dorsomorphin is a specific inhibitor of type I BMP receptors that has previously been shown to impair Hepcidin expression.35 Although treatment with dorsomorphin alone greatly reduces Hepcidin expression, as measured by quantitative real-time RT-PCR in HepG2 cells, cotreatment with genistein and dorsomorphin allows the Hepcidin transcript level to remain significantly above the basal level of expression (1.8 ± 0.15 versus 0.99 ± 0.39; P < 0.05; Fig. 2B). These observations imply that genistein activates both BMP-dependent and BMP-independent pathways.
Genistein Treatment Is Associated With Increased Transcript Levels of Many Genes That Are Up-Regulated by BMP6
To compare the effects of genistein and BMP6 on transcription in HepG2 cells, we performed high-thoughput RNA-seq and identified the transcripts up-regulated in HepG2 cells after treatment with genistein (10 μM) or BMP6 (50 ng/mL) for 24 hours, in comparison to treatment with vehicle alone. We found that genistein treatment significantly up-regulated 2613 messenger RNA (mRNA) transcripts, whereas BMP6 treatment significantly increased expression of 974 RNA transcripts. Using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, we identified genistein-induced increases in transcripts of genes affecting the ribosome, cell cycle, spliceosome, acute and chronic myeloid leukemia, and the lysosome (Table1). Among the genes in the leukemic pathway were the transcription factors, Stat3 and TGFβ1, which are known to affect Hepcidin expression.24–27,36 KEGG pathways that were down-regulated after genistein treatment (Supporting Table 2) included those affecting valine, leucine, and isoleucine catabolism, fatty acid metabolism, steroid biosynthesis, and the peroxisome proliferator-activating receptor-signaling pathway.
We found that approximately two thirds of the transcripts that were up-regulated by BMP6 were also up-regulated by genistein, as illustrated in a Venn diagram (Fig. 2C) and heat map (Fig. 2D). Gene Ontology (GO) classification using the DAVID algorithm indicated that genistein and BMP6 both affect 48 fundamental cellular processes (Supporting Table 3), including translation and stress response, whereas KEGG pathway analysis demonstrated that both genistein and BMP6 increase transcript levels of ribosomal proteins (Supporting Table 4).
To validate the results of the RNA-seq analysis, we performed quantitative real-time RT-PCR to detect changes in transcript levels in genes that exhibited increased expression on RNA-seq after genistein treatment (Fig. 3A-E). We found a significant correlation between the natural log of the fold change in gene expression, as measured by quantitative PCR and RNA-seq (R2 = 0.7964; Fig. 3A). Genistein significantly increased transcript levels of Stat3 and the TGFβ1-responsive37,38 gene, Serpine1, also known as plasminogen activator inhibitor type 1. In contrast, BMP6 treatment did not induce expression of either Stat3 or Serpine1 (Fig. 3B,C). Genistein treatment produced significant increases in the BMP-regulated17,39,40 transcripts, Id1 and Id3 (1.68 ± 0.11 [P = 0.009] and 2.11 ± 0.21 [P = 0.01], respectively), but these increases (Fig. 3D,E) were far lower than the increases produced by BMP6 (8.4 ± 0.44 [P < 0.001] and 15.3 ± 1.49 [P < 0.001], respectively). As expected, the BMP receptor antagonist, dorsomorphin, repressed transcript levels of Id1 and Id3.
To evaluate genistein's effect on the Stat3-signaling pathway, in comparison to IL-6 and BMP6, we used quantitative real-time RT-PCR to assess changes in gene expression of Stat3-related genes that were up-regulated by genistein on RNA-seq (Fig. 4A-E). Genistein, BMP6, or IL-6 each produced significantly increased transcript levels of Hepcidin; however, only genistein and IL-6 increased transcript levels of the Stat3 target genes, suppressor of cytokine signaling 3 (SOCS3) and vascular endothelial growth factor A (VEGFa), the Stat2-interacting protein, interferon regulatory factor 9 gene (IRF9), and IL6 receptor alpha. In addition, genistein and IL-6 each up-regulated TGFβ1 (Fig. 4F).
Genistein Increases Stat3 Binding to the Hepcidin Promoter
Because genistein up-regulated several Stat-related genes and mutation of the Stat3-binding site in the Hepcidin promoter impaired genistein's effect on Hepcidin promoter activity, we hypothesized that genistein would increase Stat3 binding to the Hepcidin promoter. We performed immunoprecipitation with a Stat3-specific Ab or with nonspecific IgG, followed by quantitative RT-PCR, to amplify the Stat3-binding site. We found that genistein significantly increased Stat3 binding to the Hepcidin promoter (9.2 ± 3.16 versus 0.94 ± 0.11; P = 0.004), whereas BMP6 did not (Fig. 5A).
Genistein Increases Stat3 Phosphorylation
To assess the effect of genistein on Stat3 activation in human hepatocytes, we treated HepG2 cells with genistein or IL-6 and assessed the ratio of pStat3 to Stat3 protein from 0 to 60 minutes after the start of treatment (Fig. 5B). We found that genistein significantly increased pStat3/Stat3 over the vehicle control after 60 minutes of treatment (3.65 ± 0.95 versus 1.03 ± 0.21; p = 0.02). As expected, IL-6 also significantly increased pStat3/Stat3 (2.81 ± 0.56 versus 1.00 ± 0.24; P = 0.01), but the effect peaked after 30 minutes of treatment.
Discussion
In our study, we identified genistein as the first small-molecule experimental drug to up-regulate Hepcidin transcript levels in vivo and in vitro. Genistein is known to cause both estrogen receptor-dependent and receptor-independent effects. Genistein exhibits estrogenic effects on gene transcription,41–43 scavenges free radicals,44 and inhibits numerous protein tyrosine kinases.29 We found that genistein's effect on Hepcidin expression was estrogen receptor independent. Estradiol failed to increase Hepcidin expression in either zebrafish embryos or HepG2 cells, and the potent estrogen receptor inhibitor, ICI 182,780, failed to block genistein's effect. In fact, we found that ICI 182,780 stimulated Hepcidin expression. These observations agree with recently published data indicating that estradiol does not increase Hepcidin expression, and that ICI 182,780 increases Hepcidin transcript levels in HepG2 cells in the presence or absence of estradiol.45
Hepcidin exerts control over iron homeostasis by decreasing intestinal iron absorption and macrophage iron release in response to excessive accumulation of iron in vital organs. Hepatic iron overload increases hepcidin transcript levels in normal mice46 and humans.46 Thus, we hypothesized that genistein might increase Hepcidin expression by enhancing hepatocyte iron uptake. We found that genistein failed to increase either transferrin-bound or nontransferrin-bound iron uptake. Thus, genistein does not appear to increase cellular iron overload, which is encouraging in a potential therapy for hemochromatosis.
Our in vivo zebrafish and human hepatocyte data (Figs. 1 and 2) indicate that BMP signaling is required for hepcidin expression, and that genistein's effect on hepcidin expression is diminished by the BMP antagonist, dorsomorphin. Although our RNA-seq data indicate that genistein up-regulated 68% of the genes that were up-regulated by BMP6, genistein's mode of action differed from BMP6's. Genistein increased Hepcidin promoter activity in a Stat3-dependent manner, whereas BMP6 did not. We also identified several genes in the Janus kinase/Stat-signaling pathway that were up-regulated by genistein, but not by BMP6. For example, Stat3 itself, IL6 receptor, SOCS3, Serpine1, and VEGFa, all of which are up-regulated in response to Stat3 activation,47 and IRF9, which interacts with Stat2 (reviewed elsewhere48). Increasing IL6 receptor expression would be expected to increase sensitivity to IL6, which, in turn, could promote Hepcidin expression. Genistein also increased the expression of TGF-β1, a Smad4-signaling protein that has been shown to increase Hepcidin expression.36
Previously, genistein has been reported either to promote or inhibit Stat3 phosphorylation. This apparently paradoxical effect appears to be dose dependent. At lower concentrations, such as those used in our study, genistein promotes Stat3 phosphorylation and cell proliferation,49 whereas at high concentrations (40-50 μM) genistein inhibits Stat3 phosphorylation and cell proliferation49,50 and promotes apoptosis.49 Consistent with these observations, we found that 10 μM of genistein enhanced Stat3 phosphorylation and Stat3 binding to the Hepcidin promoter in cultured human hepatocytes.
We have demonstrated that genistein increases hepcidin expression in human hepatocytes in a Stat3-dependent and Smad4-dependent manner. The screening technique that we have developed may be used to identify other Hepcidin regulatory molecules with different modes of action. Genistein and other candidate molecules may subsequently be tested in preclinical models of iron overload syndromes and used to develop new therapies for iron overload.
Acknowledgments
The authors thank Eric Samarut for his assistance with the zebrafish experiments, Anthony Soltis for his assistance with the heat map, and Dr. Nancy Berliner for her critical review of the manuscript.
Glossary
- Abs
antibodies
- BMP
bone morphogenic protein
- BRE
BMP response element
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- DMSO
dimethyl sulfoxide
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- FKPM
fragments per kilobase of exon per million fragments mapped
- GO
Gene Ontology
- hpf
hours postfertilization
- Ig
immunoglobulin
- IL-6
interleukin-6
- IRF9
interferon regulatory factor 9 gene
- kb
kilobase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MEM
minimal essential media
- mRNA
messenger RNA
- pRL-CMV
Renilla luciferase under control of the cytomegalovirus promoter
- pStat3
phospho-Stat3
- RNA-seq
RNA sequencing
- RT-PCR
reverse-transcriptase polymerase chain reaction
- Smad
mothers against decapentaplegic homolog
- SOCS3
suppressor of cytokine signaling 3 gene
- Stat3
signal transducer and activator of transcription
- TGF-β
transforming growth factor beta
- VEGFa
vascular endothelial growth factor A gene
- WT
wild type
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- 1.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 2.Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shike H, Shimizu C, Lauth X, Burns JC. Organization and expression analysis of the zebrafish hepcidin gene, an antimicrobial peptide gene conserved among vertebrates. Dev Comp Immunol. 2004;28:747–754. doi: 10.1016/j.dci.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 5.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 6.Fraenkel PG, Traver D, Donovan A, Zahrieh D, Zon LI. Ferroportin1 is required for normal iron cycling in zebrafish. J Clin Invest. 2005;115:1532–1541. doi: 10.1172/JCI23780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Anderson GJ, Frazer DM. Iron metabolism meets signal transduction. Nat Genet. 2006;38:503–504. doi: 10.1038/ng0506-503. [DOI] [PubMed] [Google Scholar]
- 9.Papanikolaou G, Tzilianos M, Christakis JI, Bogdanos D, Tsimirika K, MacFarlane J, et al. Hepcidin in iron overload disorders. Blood. 2005;105:4103–4105. doi: 10.1182/blood-2004-12-4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kattamis A, Papassotiriou I, Palaiologou D, Apostolakou F, Galani A, Ladis V, et al. The effects of erythropoetic activity and iron burden on hepcidin expression in patients with thalassemia major. Haematologica. 2006;91:809–812. [PubMed] [Google Scholar]
- 11.Camberlein E, Zanninelli G, Detivaud L, Lizzi AR, Sorrentino F, Vacquer S, et al. Anemia in beta-thalassemia patients targets hepatic hepcidin transcript levels independently of iron metabolism genes controlling hepcidin expression. Haematologica. 2008;93:111–115. doi: 10.3324/haematol.11656. [DOI] [PubMed] [Google Scholar]
- 12.Finch C. Regulators of iron balance in humans. Blood. 1994;84:1697–1702. [PubMed] [Google Scholar]
- 13.Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med (Berl) 2008;86:531–540. doi: 10.1007/s00109-008-0313-7. [DOI] [PubMed] [Google Scholar]
- 14.Island ML, Jouanolle AM, Mosser A, Deugnier Y, David V, Brissot P, Loreal O. A new mutation in the hepcidin promoter impairs its BMP response and contributes to a severe phenotype in HFE related hemochromatosis. Haematologica. 2009;94:720–724. doi: 10.3324/haematol.2008.001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med (Berl) 2009;87:471–480. doi: 10.1007/s00109-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 16.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115:2657–2665. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 17.Conidi A, Cazzola S, Beets K, Coddens K, Collart C, Cornelis F, et al. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Andriopoulos B, Jr, et al. Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41:478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 20.Arndt S, Maegdefrau U, Dorn C, Schardt K, Hellerbrand C, Bosserhoff AK. Iron-induced expression of bone morphogenic protein 6 in intestinal cells is the main regulator of hepatic hepcidin expression in vivo. Gastroenterology. 2010;138:372–382. doi: 10.1053/j.gastro.2009.09.048. [DOI] [PubMed] [Google Scholar]
- 21.Kautz L, Besson-Fournier C, Meynard D, Latour C, Roth MP, Coppin H. Iron overload induces BMP6 expression in the liver but not in the duodenum. Haematologica. 2011;96:199–203. doi: 10.3324/haematol.2010.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemna E, Pickkers P, Nemeth E, van der Hoeven H, Swinkels D. Time-course analysis of hepcidin, serum iron, and plasma cytokine levels in humans injected with LPS. Blood. 2005;106:1864–1866. doi: 10.1182/blood-2005-03-1159. [DOI] [PubMed] [Google Scholar]
- 23.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, et al. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem. 2002;277:41163–41170. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- 25.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–3209. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 27.Matak P, Chaston TB, Chung B, Srai SK, McKie AT, Sharp PA. Activated macrophages induce hepcidin expression in HuH7 hepatoma cells. Haematologica. 2009;94:773–780. doi: 10.3324/haematol.2008.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraenkel PG, Gibert Y, Holzheimer JL, Lattanzi VJ, Burnett SF, Dooley KA, et al. Transferrin-a modulates hepcidin expression in zebrafish embryos. Blood. 2009;113:2843–2850. doi: 10.1182/blood-2008-06-165340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan GR, Xiao CL, He GW, Yin XF, Chen NP, Cao Y, He QY. Global phosphoproteomic effects of natural tyrosine kinase inhibitor, genistein, on signaling pathways. Proteomics. 2010;10:976–986. doi: 10.1002/pmic.200900662. [DOI] [PubMed] [Google Scholar]
- 30.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio. Eugene, OR: University of Oregon Press; 1994. [Google Scholar]
- 31.Gibert Y, Lattanzi VJ, Zhen AW, Vedder L, Brunet F, Faasse SA, et al. BMP signaling modulates hepcidin expression in zebrafish embryos independent of hemojuvelin. PLoS One. 2011;6:e14553. doi: 10.1371/journal.pone.0014553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 33.Maes K, Nemeth E, Roodman GD, Huston A, Esteve F, Freytes C, et al. In anemia of multiple myeloma, hepcidin is induced by increased bone morphogenetic protein 2. Blood. 2010;116:3635–3644. doi: 10.1182/blood-2010-03-274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang daW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama N, Nakamura T, Okada H, Iwaki S, Sobel BE, Fujii S. Modulators of induction of plasminogen activator inhibitor type-1 in HepG2 cells by transforming growth factor-beta. Coron Artery Dis. 2011;22:468–478. doi: 10.1097/MCA.0b013e32834a3817. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem. 2002;277:3176–3185. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- 40.Kersten C, Dosen G, Myklebust JH, Sivertsen EA, Hystad ME, Smeland EB, Rian E. BMP-6 inhibits human bone marrow B lymphopoiesis—upregulation of Id1 and Id3. Exp Hematol. 2006;34:72–81. doi: 10.1016/j.exphem.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Ise R, Han D, Takahashi Y, Terasaka S, Inoue A, Tanji M, Kiyama R. Expression profiling of the estrogen responsive genes in response to phytoestrogens using a customized DNA microarray. FEBS Lett. 2005;579:1732–1740. doi: 10.1016/j.febslet.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 42.Satih S, Chalabi N, Rabiau N, Bosviel R, Fontana L, Bignon YJ, Bernard-Gallon DJ. Gene expression profiling of breast cancer cell lines in response to soy isoflavones using a pangenomic microarray approach. OMICS. 2010;14:231–238. doi: 10.1089/omi.2009.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jefferson WN, Couse JF, Padilla-Banks E, Korach KS, Newbold RR. Neonatal exposure to genistein induces estrogen receptor (ER)alpha expression and multioocyte follicles in the maturing mouse ovary: evidence for ERbeta-mediated and nonestrogenic actions. Biol Reprod. 2002;67:1285–1296. doi: 10.1095/biolreprod67.4.1285. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee S, Li Y, Wang Z, Sarkar FH. Multi-targeted therapy of cancer by genistein. Cancer Lett. 2008;269:226–242. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q, Jian J, Katz S, Abramson SB, Huang X. 17beta-estradiol inhibits iron hormone hepcidin through an estrogen responsive element half-site. Endocrinology. 2012;153:3170–3178. doi: 10.1210/en.2011-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolondi G, Garuti C, Corradini E, Zoller H, Vogel W, Finkenstedt A, et al. Altered hepatic BMP signaling pathway in human HFE hemochromatosis. Blood Cells Mol Dis. 2010;45:308–312. doi: 10.1016/j.bcmd.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 48.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 49.Chau MN, El Touny LH, Jagadeesh S, Banerjee PP. Physiologically achievable concentrations of genistein enhance telomerase activity in prostate cancer cells via the activation of STAT3. Carcinogenesis. 2007;28:2282–2290. doi: 10.1093/carcin/bgm148. [DOI] [PubMed] [Google Scholar]
- 50.Kim EK, Kwon KB, Song MY, Seo SW, Park SJ, Ka SO, et al. Genistein protects pancreatic beta cells against cytokine-mediated toxicity. Mol Cell Endocrinol. 2007;278:18–28. doi: 10.1016/j.mce.2007.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.


