Abstract
Many phenomena of fundamental importance to biology and biomedicine arise as a dynamic curve, such as organ growth and HIV dynamics. The genetic mapping of these traits is challenged by longitudinal variables measured at irregular and possibly subject-specific time points, in which case nonnegative definiteness of the estimated covariance matrix needs to be guaranteed. We present a semiparametric approach for genetic mapping within the mixture-model setting by jointly modeling mean and covariance structures for irregular longitudinal data. Penalized spline is used to model the mean functions of individual QTL genotypes as latent variables while an extended generalized linear model is used to approximate the covariance matrix. The parameters for modeling the mean-covariances are estimated by MCMC, using Gibbs sampler and Metropolis Hastings algorithm. We derive the full conditional distributions for the mean and covariance parameters and compute Bayes factors to test the hypothesis about the existence of significant QTLs. The model was used to screen the existence of specific QTLs for age-specific change of body mass index with a sparse longitudinal dataset. The new model provides powerful means for broadening the application of genetic mapping to reveal the genetic control of dynamic traits.
Keywords: Cholesky decomposition, genetic mapping, MCMC, penalized spline, quantitative trait loci
1 INTRODUCTION
Most traits of interest in biology and important to biomedicine are multifactorial, controlled by multiple genes displaying complex interactions with each other and with environmental factors [1]. For this reason, genetic analysis of these traits has been difficult, despite tremendous efforts for the development of genetic theory and methods over the past century. Recent advances of powerful molecular technologies have revolutionized the tools of analyzing multifactorial traits by dissecting them into the underlying quantitative trait loci (QTLs) using DNA-based markers. Lander and Botstein [2] proposed a statistical model for interval mapping of individual QTLs that contribute to a complex trait. A considerable body of literature on the methodological development of QTL mapping, which aims at improving the precision of QTL detection [3,4] broadens the scope of utility of this approach [5–7]. Specific statistical issues for QTL mapping have also been considered in several areas including the determination of critical thresholds [8,9], model selection [10], nonparametric mapping of QTLs [11], and asymptotic properties of QTL parameter estimates [12]. Because of its many favorable properties, the Bayesian approach has been introduced to map QTLs first by Satagopan et al. [13] and subsequently by a number of other researchers [14].
There is increasing recognition of the limitations of traditional QTL mapping approaches that only capitalize on single measurements of a complex trait at one time point, given that all biological traits or diseases undergo a dynamic process across time and spatial scales. To better describe the dynamic pattern of trait progression, it is crucial to measure phenotypic values of a trait longitudinally at multiple time points, which allows a comprehensive analysis of how genes govern time-specific changes of the trait. These so-called dynamic traits can now be mapped by a dynamic model that is equipped with a capacity to study the temporal pattern of QTL effects on trait progression [15]. This model approximates time-dependent mean vectors for individual QTL genotypes and tests their differences by using biologically relevant parametric curves, such as logistic equations for growth data [15], or nonparametrically when no explicit parametric equations exist for functional data [16–18]. It provides a general quantitative and testable platform for assessing the interplay between genetic actions and developmental pattern [19] and has now been used as a mapping tool in a number of areas such as allometric scaling, bird flight, thermal reaction norm, HIV-1 dynamics, tumor progression, biological clock, and drug response [20–24].
In many longitudinal trials, data are often collected at irregularly spaced time points and with measurement schedules specific to different subjects. The efficient estimation of covariance structure in this situation presents a major challenge for genetic mapping to study the genetic control of dynamic traits. The motivation of this study is to develop a robust approach for joint-modeling of the mean-covariance structures of subject-specific irregular longitudinal data. Within a generalized linear model framework, covariates were used to model the mean function by McCullagh and Nelder [25] and the covariance matrix by Pourahmadi [26]. Pan and Mackenzie [27] generalized Pourahmadi’s setting to sparse irregular longitudinal data and implemented iteratively re-weighted least squares algorithms (IRLS) to estimate the model parameters maximizing the likelihood. An alternative for modeling the covariance structure is to shrink the covariance matrix towards a specified structure using Bayesian hierarchical models. A popular and standard prior for the covariance matrix is the inverse Wishart distribution which is conjugate to a covariance matrix from a normal distribution. Daniels and Kass [28] showed that such a prior could enhance computing efficiency. Better priors for shrinking the covariance toward a known structure were proposed by Daniels and Pourahmadi [29]. All these methods guarantee the estimated covariance matrix to be positive definite.
In this article, we present a Bayesian approach for semiparametric modeling of mean and covariance structures for irregular longitudinal data within the framework of genetic mapping constructed by a finite mixture model. A Bayesian algorithm for genetic mapping of dynamic traits was already proposed by Liu and Wu [30] and Heuven and Janss [31]. Unlike their parametric modeling, however, we model the mean structure by a penalized spline [32] and the covariance structure by a generalized linear model [26]. This semiparametric modeling is particularly robust for dynamic trait mapping with irregular longitudinal data. Proper priors have been tested and chosen to get a smooth mean curve and meaningful covariance parameters. We derive the full conditional posterior distributions for the model parameters and then use Gibbs sampler and the Metropolis-Hastings algorithm to estimate the parameters. The new model was used to map QTLs responsible for age-specific changes of body mass index (BMI) in a random sample of 977 subjects from Framingham Heart Study [33]. As a heuristic measure of body fatness based on a person’s weight and height, BMI is the most widely used diagnostic tool to identify whether individuals are underweighted, overweighted, or obese and further examine their risk of developing obesity-related diseases, such as hypertension, type 2 diabetes, and cardiovascular diseases [34]. By mapping genes associated with BMI trajectories, we hope to diagnose and predict the timing of pathogenesis for these diseases based on individual patients’ genetic makeup. To validate the usefulness of the new model for QTL mapping, we performed simulation studies to investigate its statistical properties.
2 BAYESIAN GENETIC MAPPING
2.1 Genetic Design
Suppose there is a natural diploid population at Hardy-Weinberg equilibrium (HWE), from which a sample of n unrelated subjects are drawn randomly. In this study, a dynamic trait of interest is measured longitudinally at irregularly spaced time points, with measurement schedules uncommon to all subjects. Thus, it would be difficult to map these irregular data by a static mapping approach because only some of the subjects may be measured at the same time point. Also, traditional genetic mapping may not work because all the subjects are measured at a few number of time points, which does not allow parametric modeling of longitudinal data for individual subjects. Despite a high sparsity, a number of distinct time points can be yielded when all the subjects are combined.
We assume that these subjects differ in the dynamic trait due to specific genes or QTLs involved. These causal QTLs cannot be observed directly, but they can be inferred from molecular markers based on the random association between the markers and causal loci. Suppose there is such a marker that is associated with a putative QTL in the population sampled. We consider mostly commonly used single nucleotide polymorphism (SNP) markers which are mostly biallelic. Let W and w denote the two alternative alleles at the QTL, which have a frequency of q and 1 – q, respectively. Similarly, we use M and m to denote the two alternative alleles at the marker, with respective frequencies p and 1 – p. The original population is composed of diploid individuals, each with two haploids (i.e., a half set of chromosomes), one from the paternal parent and the other from the maternal parent. In the haploid population, the alleles at the marker and QTL combine randomly to form four haplotypes, MW, Mw, mW and mw, whose frequencies are expressed as p11 = pq + D, p10 = p(1 – q) – D, p01 = (1 – p)q – D, and p00 = (1 – p)(1 – q) + D, respectively, where D, called the linkage disequilibrium, describes the extent of random association between the QTL and marker. The four haplotypes are mating randomly to generate nine distinguishable genotypes with frequencies tabulated in Table 1. From Table 1, we can derive the conditional probabilities of a QTL genotype j (j = 0 for ww, 1 for Ww and 2 for WW), given a marker genotype of subject i, symbolized as ωj|i. Conditional probability ωj|i is a function of haplotype frequencies or (p, q, D).
Table 1.
Joint genotype frequencies at the marker and QTL in terms of gametic haplotype frequencies, from which the conditional probabilities of QTL genotypes given marker genotypes can be calculated according to Bayes’ theorem.
| WW | Ww | ww | |||||
|---|---|---|---|---|---|---|---|
| Genotype | Diplotype | W|w | W|w + w|W | w|w | Observations | ||
| MM | M|M | 2p11p10 | n1 | ||||
| Mm | M|m | 2p11p01 | 2p11p00 + 2p10p01 | 2p10p00 | n2 | ||
| mm | m|m | 2p01p00 | n3 | ||||
2.2 Mixture Model
Let yi = [y(ti1),…,y(tiTi)] be the vector of Ti measurements on subject i and let ti = (ti1,…,tiTi) be the corresponding vector of measurement times. These notations allow subject-specific differences in the number and interval of time points. The phenotypic value of subject i at time tiτ (τ = 1, …,Ti), affected by the QTL, is expressed as
| (1) |
where xij is an indicator variable for a possible QTL genotype of subject i and defined as 1 if a particular QTL genotype j is indicated and 0 otherwise, µij(tiτ) is the mean value of QTL genotype j for subject i at time tiτ, and ei(tiτ) is the residual error for subject i, assumed to be distributed as MVN(0, Σi). Note that equation (1) does not include a covariate because our focus here is on the explanation of the new model.
The mixture model-based likelihood of subjects with longitudinal measurements y and marker information M is formulated as
| (2) |
where Ω is the vector for unknown parameters, ωj|i is the proportion of mixture component j, expressed as the conditional probability of QTL genotype j for subject i given its marker genotype, which is calculated from Table 1, fj(yi) is a multivariate normal distribution with QTL genotype-specific mean vectors μij (from equation (1)) and a subject-specific covariance matrix Σi. The unknown vector Ω contains marker-QTL haplotype frequencies and the parameters that model the mean-covariance structures.
2.3 Joint Mean-Covariance Regression Model
The central theme of genetic mapping lies in modeling the mean vector and covariance structure. Here, we will incorporate a regression approach to jointly model the mean-covariance structures. Consider a particular subject measured at T different time points. For a simple description, we ignore the subscript for the moment. Without loss of generality, assume the response vector y = (y1, …,yT) has mean vector 0 and covariance matrix Σ. The response at time t, yt, can be predicted by its predecessors as follows:
| (3) |
where ϕt,t′ is the corresponding regression coefficient. Let εt be the prediction error with 0 mean and be its variance. Assuming that εt’s are uncorrelated [26], we get cov(ε) = E, a diagonal matrix with being the t-th diagonal element, where ε = (ε1, …, εT)′, the vector of prediction errors. Hence, the matrix representation of the above autoregression becomes
| (4) |
where L is a lower triangular matrix with 1’s in diagonal elements and −ϕt,t′ in the (t,t′)th position. The above equation simply gives,
| (5) |
which is related to the modified Cholesky decomposition of Σ.
Equation (5) will be considered as the basis for modeling the covariance structure, since this guarantees the estimated covariance matrix to be positive definite. In an irregular setting where different subjects are measured at different time points, we model subject-specific error covariance matrix as .
Following Pourahmadi [26], we model the unconstrained dependence parameters log and ϕt,t′ with a suitable ordered polynomial function of time t, expressed as
| (6) |
| (7) |
The order of each of the above polynomials (g and h) is determined by comparing the BIC values for different orders. We will estimate variance parameters and the dependence parameters in a Bayesian framework using the MCMC method as discussed later. In order to have maximal smoothness in the mean curve and study the overall features of the unknown mean curve properly, we use a penalized spline. Recently, Chen and Wang [35] have used penalized spline for functional mixed effects model analysis.
2.4 Penalized Spline
A polynomial spline of degree r, with knots (𝒯1,…,𝒯K), has continuous derivatives upto (r – 1) times and a discontinuous r-th order derivative. QTL genotype-specific means at different time points in equation (1) are fitted by a nonparametric regression model,
| (8) |
where r is the degree of the spline, and (𝒯1 < 𝒯2 < … < 𝒯K) is a fixed set of knots. Usually, r is kept relatively small (between 1 and 3). Here, bj = (bj0, bj1,…,bjr) and cj = (bj(r+1), bj(r+2), …, bj(r+K)) represent the coefficients of the parametric and spline portion of the model, respectively. The P-spline regression model for the mean of a QTL genotype j described above can be described for irregular longitudinal data in matrix notation:
| (9) |
where
In order to smooth-out the fit of the resulting model, a roughness penalty is placed on the above parameters. This is done by minimizing the following expression:
| (10) |
where λ* is the smoothing parameter which plays a crucial role in the smoothing process since it controls the goodness of fit and roughness of the fitted model. Here we will consider a Bayesian version of ridge regression (d = 2) by choosing normal priors on bj(r+s)’s followed by a gamma prior on λ*. Note that d=1 gives LASSO, which has a literature for QTLs in a different context.
Selection of a reasonable number of knots is quite subjective in a P-spline regression model. In practice, researchers predetermine the optimum number of knots by looking at the independent variable. Locations of knots are usually determined by the equally spaced sample quantiles of the independent variable.
2.5 Estimation Procedure
By specifying their prior distributions, we obtain the posterior distributions of those parameters that model the mean-covariance structures given the data and the priors. Let λ and δ denote the vector of coefficients for the variance structure and the dependence structure, respectively, in models (6) and (7). For bj of QTL genotype j, we place a multivariate normal prior with zero mean and covariance matrix Σb. A multivariate normal prior with mean 0 and covariance matrix Σc is placed for cj, where Σc is a diagonal matrix with diagonal element . A gamma prior with parameters(α*,β*) is taken for λ* in order to shrink the parameters of the spline part of the mean function. Priors for λ and δ are taken as MVN(0,Σλ) and MVN(0,Σδ), respectively.
With the above priors and likelihood function, we get the posterior density of b, c, λ, δ, and λ* as
Assuming that priors for different genotypes are independent, we can express the above posterior distribution as
| (11) |
where π(bj) and π(cj) are priors described in equation (10), π(λ) and π(δ) are priors described in equations (6) and (7), and π(λ*) is a prior described in equation (9).
A Markov chain Monte Carlo (MCMC) technique was used to estimate the parameters that specify QTL genotype-specific normal distributions by drawing samples from the joint posterior distributions of these parameters with explicit expressions (A1) – (A5). A simple Gibbs sampler was used to update b, c, and λ*, whereas a Metropolis-Hastings algorithm used to update λ and δ. A multivariate normal with mean as the current λ and covariance matrix Σλ was taken as the proposal for simulating λ. A similar proposal was considered for the simulation of δ. Parameter estimates are obtained from the mean of the posterior distributions.
Once the distribution parameters are updated, we update the parameters (p, q, D) that specify the (co)segregation of the marker and QTL at each state by implementing the EM algorithm. Wang and Wu [36] give detailed derivation for the EM estimation of these population genetic parameters.
2.6 Hypothesis Testing
After we obtain the estimates for the population parameters and distribution parameters, we need to test the following hypotheses. First, we need to test the existence of a QTL for the longitudinal trait. This hypothesis is formulated as
| (12) |
In a frequentist framework, one can compute the likelihood ratio test statistic LR = −2 log Λ, where Λ is the ratio of the value of the likelihood function under null to that under the alternative. However, since linkage disequilibrium (D) is not identifiable under the null, it is difficult to get an approximate distribution of the test statistic. Permutation tests are suggested to get the critical threshold.
A similar approach in a Bayesian framework, known as posterior predictive p-values, described by Gelman et al. [37], can also be used. Since the hypothesis testing above can be viewed as a model selection problem, we perform posterior predictive checks to decide which of the two models (suggested by the null and the alternative, respectively) is better advocated by the data. Let Θ be the set of all parameters and yrep be the replicated data. Then, given the data y, the posterior predictive distribution of yrep is given by p(yrep|y) = ∫ p(yrep|Θ)p(Θ|y)dΘ. From their respective posterior densities, we first simulate the model parameters and, then, using those parameters, draw yrep, the replicated data. Having a large number of such simulations, the posterior predictive p-value is estimated as the proportion of the simulations for which LR(yrep, Θ) ≥ LR(y, Θ).
Second, we will test whether this QTL can be detected by the marker that is associated with it by testing the significance of linkage disequilibrium (D). We have
An informal way to test this hypothesis is to construct credible intervals having equal tails. A 100(1 – α) percent credible interval is constructed using and percentiles of the posterior distribution of D. The evidence for or against the null hypothesis can be justified by checking whether the credible interval contains 0 or not. Since there is no identifiability problem in this case, we compute bayes factor: . We follow the scale given by Jeffreys for the interpretation of Bayes factor.
3 ANALYSIS OF REAL DATA
3.1 Background
We used the newly developed method to analyze a longitudinal trait with an irregular and sparse measurement structure. The data was collected from a Framingham Heart Study (FHS), aimed to study the genetic control of cardiovascular diseases. Beginning in 1948 with 5,209 healthy men and women aged 30 to 60 from Framingham, MA [33], the FHS project is now in its third generation of participants and has played a central role in advancing our understanding of the epidemiological cause of hypertensive or arteriosclerotic cardiovascular disease. All subjects underwent a medical history and physical examination (including body height and weight), laboratory tests, and electrocardiography. Examinations have been repeated every two or more years, with different subjects having different numbers and time intervals of measurements. Thus, almost all these phenotypic measurements were collected on an irregular schedule. The data we used is composed of 977 subjects (all Caucasians) in a range of ages from 29 to 104 years. All these subjects were measured for body mass index (BMI) and other variables at multiple time points. The number of serial measurements for a subject can be as low as 5, but the times and intervals of measurement are highly variable among the subjects. Thus, despite high sparsity, this data set contain a long range of time points when all the subjects are projected on a time scale.
Marker data consist of approximately 3.5 million single nucleotide polymorphisms (SNPs) genotyped from the human genome. As usual, the SNPs with minor allele frequencies < 10% were excluded from the analysis. Traditional genome-wide association studies (GWAS) based on a direct relationship between genotype and phenotype were used to identify significant SNPs associated with BMI phenotypes. This approach is simple, but is not able to detect QTLs that affect the phenotypes. Since these significant SNPs present some signals, we will use them as markers to map QTL for BMI trajectories. The GWAS approach identifies four significant SNPs, rs4451518 on chromosome 1, rs2171168 on chromosome 3, rs13124340 on chromosome 4, and rs3903759 on chromosome 6, for females and eight significant SNPs, rs3903759 on chromosome 6, rs17782554 and rs11783045 on chromosome 8, rs7903156 on chromosome 10, rs7309679 on chromosome 12, rs9915696 on chromosome 17, rs948716 on chromosome 18, and rs747911 on chromosome 20, for males. Of these SNPs, rs3903759 on chromosome 6 was detected for both sexes, whereas the others are sex-specific.
3.2 Results
In an exploratory data analysis, we found that there was no significant improvement of the mean curve in terms of smoothness if more than six knots were used. Hence, K = 6 was chosen as a reasonable number of knots for our modeling and analysis. To get the optimum (r, g, h), we compute BIC values for different orders for (r, g, h) using Pourahmadi’s [26] approach,
where n is the number of subjects, K is the number of knots, (r, g, h) are the orders of modeling the mean function (equation 8) and covariance function (equations 6 and 7). Table 2 provides the corresponding BIC values for different orders from which (2,2,2) was found to give the smallest BIC value and therefore, taken as the optimum order.
Table 2.
BIC values for selecting the optimum triple (r, g, h) to model the mean-covariance structure in the analysis of BMI data.
| Triple | BIC(male) | BIC(female) |
|---|---|---|
| (3,3,3) | 4.17 | 5.19 |
| (3,2,3) | 3.89 | 2.67 |
| (3,3,2) | 2.46 | 3.12 |
| (3,2,2) | −1.89 | 1.75 |
| (2,2,2) | −4.41 | 1.31 |
| (2,3,2) | 2.51 | 2.07 |
| (2,2,3) | 3.14 | 2.38 |
| (2,3,3) | −2.65 | 3.33 |
Next, we consider one SNP at a time as a marker and test for the existence of a QTL having a significant effect on BMI trajectories. The following priors were placed for the unknown parameters. We assume that Σb is a diagonal matrix with all the diagonal elements equal to 1000, λ* has a Gamma distribution with parameters (α* = 0.001, β* = 1000) so that the distribution has mean 1 and high variance 1000 (a diffuse prior, so to speak), and Σλ and Σδ are diagonal matrices with all diagonal elements equal to 10 and 5, respectively. Gibbs sampler works for the parameters b, c, and λ*, whereas for λ and δ, a Metropolis Hastings algorithm was implemented with the proposal density being multivariate normal with mean as the parameter value in the current state and Σλ and Σδ, respectively, as the covariance matrix. This choice of proposals results in a reasonably good acceptance rates (above 0.20). We run 60,000 iterations and initial 10,000 burn-in iterations were discarded. The convergence of the chains is assessed by computing Multivariate Potential Scale Reduction Factor (MPSRF) proposed by Brooks and Gelman [38] by considering 5 different chains with different staring values. We consider the posterior mean as the parameter estimate by taking into account a squared error loss function.
To test the existence of a QTL, we computed posterior predictive p-values for genetic effects of a QTL by considering 5,000 replications yrep of the data and counting the number for which the test statistic LR for the replicated data is higher than or equal to that for the observed data. The proportion gives an estimate of the posterior predictive p-value for the corresponding test. Table 3 provides these p-values for each significant SNP identified from traditional GWAS that can reflect the effect of a QTL on age-specific changes in BMI. For females, all four SNPs may each be associated with a QTL, whereas, for males, six of eight SNPs each associated with a QTL. In Supplement 1, we provide MCMC estimates of the parameters and their standard errors for significant QTLs in females and males, from which it can be seen that all estimates display reasonably good precision.
Table 3.
Posterior predictive p-values for testing the existence of a QTL for 11 significant SNPs identified by a traditional GWAS approach
| Chr | SNP | Position | Alleles | MAF | p-value |
|---|---|---|---|---|---|
| Male-specific | |||||
| 8 | rs17782554 | 11059516 | G/C | 0.3984(C) | 0.0104* |
| 8 | rs11783045 | 11093585 | A/G | 0.3843(G) | 0.0197* |
| 10 | rs7903156 | 133498454 | A/G | 0.1197(G) | 0.7627ns |
| 12 | rs7309679 | 96235821 | A/G | 0.1402(G) | 0.0233* |
| 17 | rs9915696 | 2688526 | G/C | 0.2596(C) | 0.0082** |
| 18 | rs948716 | 73293243 | T/C | 0.1435(C) | 0.0115* |
| 20 | rs747911 | 22580931 | C/T | 0.2505(T) | 0.6819ns |
| Female-specific | |||||
| 1 | rs4451518 | 180384945 | T/C | 0.1907(C) | 0.0024** |
| 3 | rs2171168 | 122054294 | C/T | 0.2728(T) | 0.0053** |
| 4 | rs13124340 | 125989067 | G/T | 0.1306(T) | 0.0072** |
| Males/Females | |||||
| 6 | rs3903759 | 131562687 | C/A | 0.1645(A) | 0.0096** (Male) 0.0135* (Female) |
MAF = minor allele frequencies;
Significant at the 5% and 1% significance level;
Nonsignificant.
For those SNPs that reflect the effects of a QTL, we now examine whether each of them can be used to detect the QTL by testing the significance of linkage disequilibrium, D. First, we constructed 95 percent credible intervals for D and checked whether the interval contains zero. However, this should be taken as an informal way just like the classical frequentist approach. To compute Bayes factors, we run the Markov chain again under the null hypothesis and compute the value of the likelihood function. We made the interpretation of the computed Bayes factor according to the scale suggested by Jefferys. B01 < 1 gives evidence against the null hypothesis, 3 < B01 < 10 gives substantial evidence for the null hypothesis, and B01 > 10 provides strong evidence for the null hypothesis. Table 4 shows that for females SNP rs2171168 on chromosome 3 cannot be used to detect the significant QTL and for the males the same is true for SNP rs7309679 on chromosome 12 and SNP rs948716 on chromosome 18. Similar results can be seen from the credible intervals.
Table 4.
95 percent credible intervals (CI) and Bayes Factors (BF) for linkage disequilibrium (D) between the SNP and QTL.
| Male |
Female |
||||
|---|---|---|---|---|---|
| Chr | SNP | CI | BF | CI | BF |
| 8 | rs17782554 | (0.0078,0.1603) | 0.7085 | ||
| 8 | rs11783045 | (0.0201,0.1484) | 0.8960 | ||
| 12 | rs7309679 | (−0.0216,0.1023) | 9.32 | ||
| 17 | rs9915696 | (0.0511,0.1868) | 0.2397 | ||
| 18 | rs948716 | (−0.0364,0.0911) | 11.94 | ||
| 1 | rs4451518 | (0.0507,0.1406) | 0.5985 | ||
| 3 | rs2171168 | (−0.0356,0.0672) | 14.97 | ||
| 4 | rs13124340 | (0.0116,0.1847) | 0.8609 | ||
| 6 | rs3903759 | (0.0226,0.1143) | 0.3584 | (0.0174,0.1597) | 0.8343 |
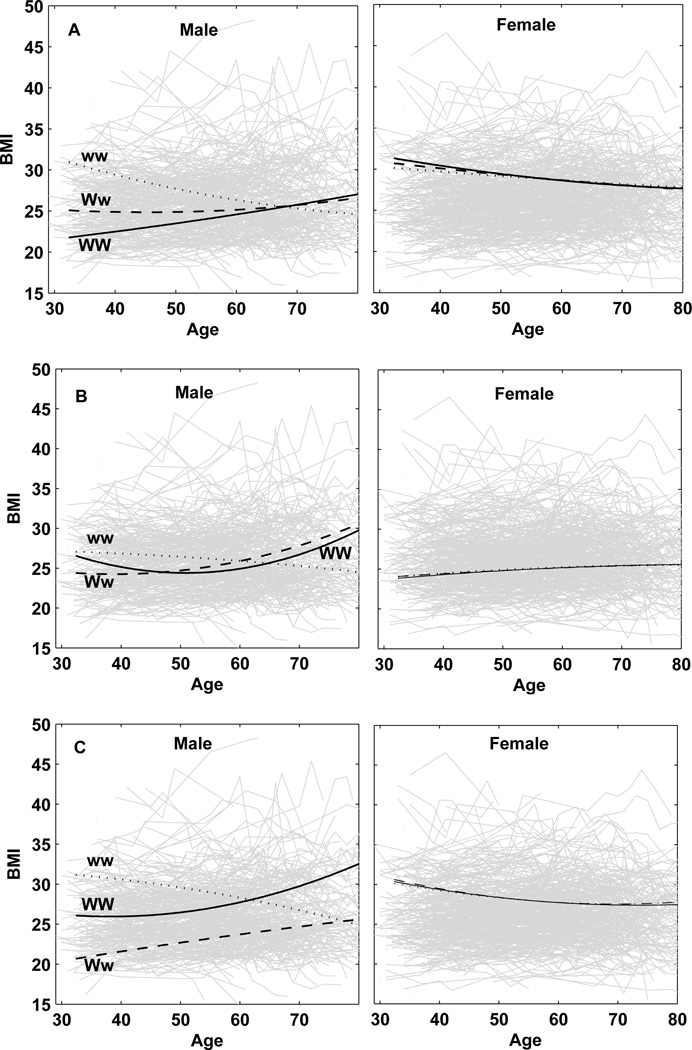
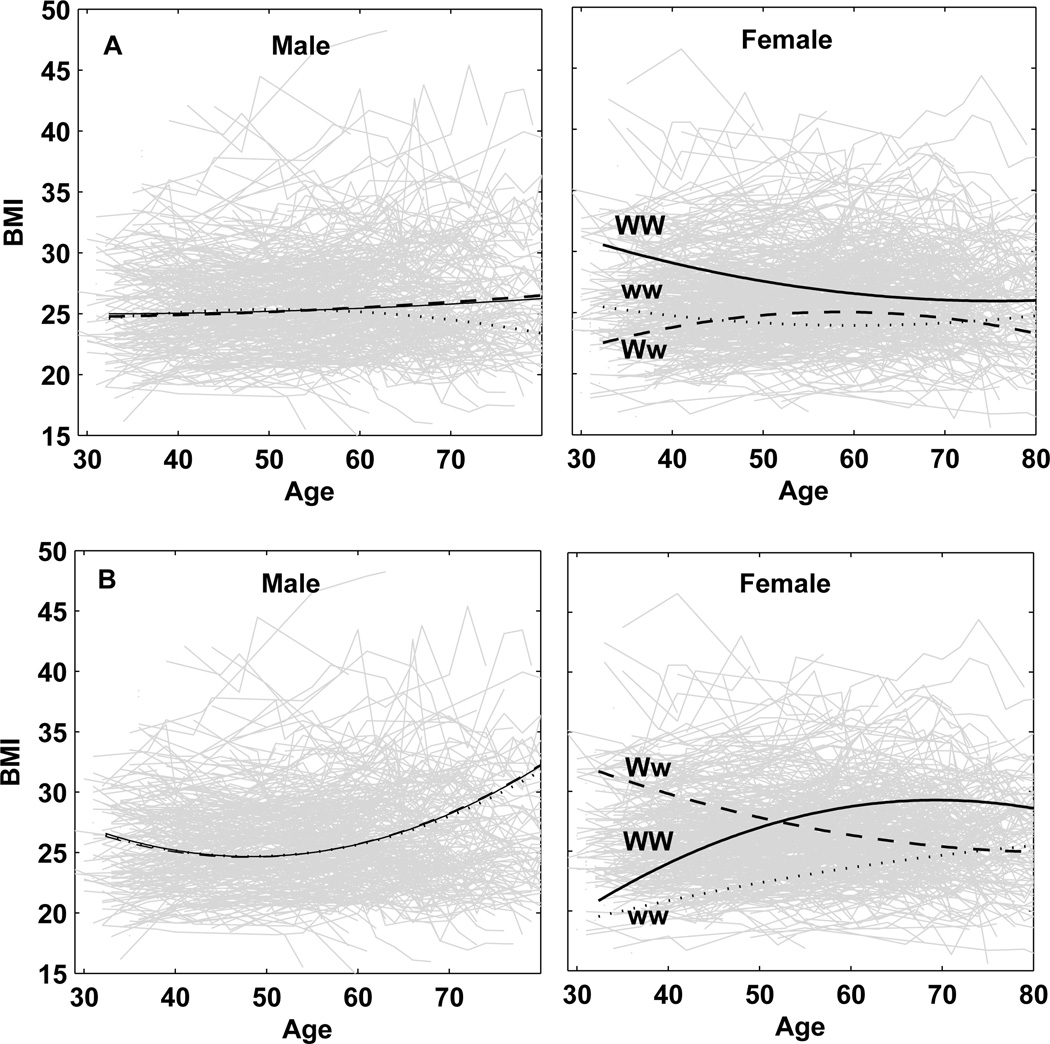
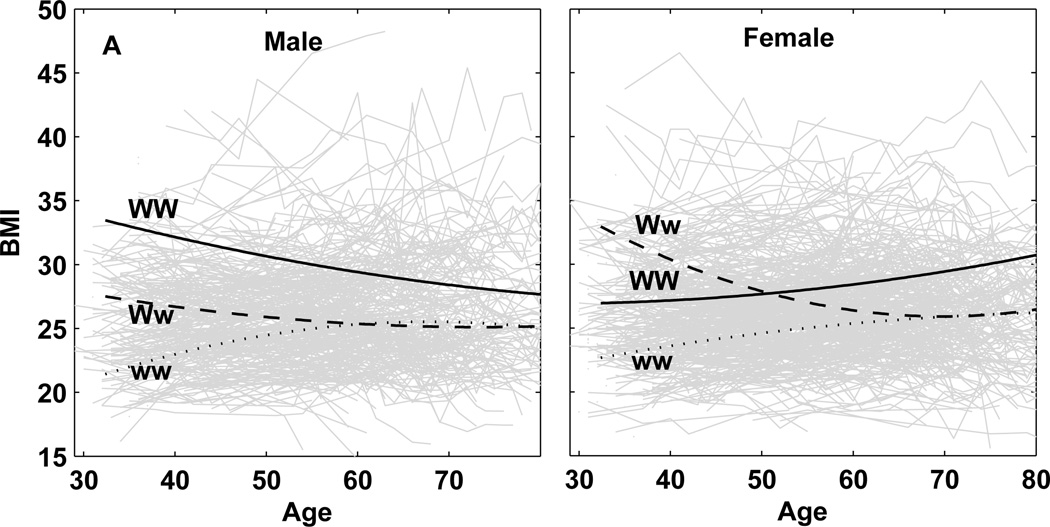
Our model can have now detected QTLs based on their associations with a SNP. Using the estimates of curve parameters in Supplement 1, we draw age-dependent curves of BMI for three different genotypes at a QTL that is detected by a SNP. SNPs rs4451518 and rs13124340 are associated with a QTL that controls the temporal change of BMI only in females (Fig. 1), whereas SNPs rs17782554, rs11783045, and rs9915696 are associated with a QTL that is only expressed in males (Fig. 2). The QTL associated with SNP rs3903759 is expressed in both females and males (Fig. 3), but with different extents and patterns. All these QTLs detected display genotype × sex interactions for age-specific changes of BMI, although these are more pronounced for sex-specific QTLs (Figs. 1 and 2) than the sex-biased QTL (Fig. 3).
Figure 1.
Age-specific change of BMI for three different genotypes at the QTLs expressed in males. Upper: a QTL associated with SNP rs17782554 on chromosome 8 with QTL allele W in a coupling phase with SNP allele G and QTL allele w in a coupling phase with SNP allele C. Middle: a QTL associated with SNP rs11783045 on chromosome 8 with QTL allele W in a coupling phase with SNP allele A and QTL allele w in a coupling phase with SNP allele G. Bottom: a QTL associated with SNP rs9915696 on chromosome 17 with QTL allele W in a coupling phase with SNP allele G and QTL allele w in a coupling phase with SNP allele C.
Figure 2.
Age-specific change of BMI for three different genotypes at the QTLs expressed in females. Upper: a QTL associated with SNP rs4451518 on chromosome 1 with QTL allele W in a coupling phase with SNP allele T and QTL allele w in a coupling phase with SNP allele C. Bottom: a QTL associated with SNP rs13124340 on chromosome 4 with QTL allele W in a coupling phase with SNP allele G and QTL allele w in a coupling phase with SNP allele T.
Figure 3.
Age-specific change of BMI for three different genotypes at the QTL expressed in both males and females. This QTL is associated with SNP rs3903759 on chromosome 6 with QTL allele W in a coupling phase with SNP allele C and QTL allele w in a coupling phase with SNP allele A.
Considering the marker gene rs3903759 (on chromosome 6) for which we have detected QTL for both male and female population, we compare our joint modeling approach with the other traditional methods. Since the trait trajectories for different genotypes seem to be quadratic functions of time, we consider polynomial functions of order 2 to estimate the mean functions. For modeling the longitudinal covariance function, we consider the traditional autoregressive order 1 (AR(1)) and compound symmetry (CS) structures. Model parameters are estimated in a Bayesian framework using MCMC algorithm as discussed earlier. Table 5 and 6 show the Residual Sum of Squares (RSS) and Deviance Information Criterion (DIC), two standard measures of model selection for each combination for male and female population respectively. In terms of DIC the proposed method performs significantly better than other two, although in terms of RSS they are quite comparable for the male population. However, as table 6 shows, for the female population the proposed method performs much better than other two methods both in terms of RSS and DIC. Since DIC is preferred in Bayesian literature for some of its desirable properties, we have numerical evidence to believe that the proposed method would perform better than the traditional methods in reality.
Table 5.
RSS and DIC values for different combinations for male population.
| Methods | RSS | DIC |
|---|---|---|
| Quadratic mean with AR(1) cov. | 213.56 | 145.12 |
| Quadratic mean with CS cov. | 218.71 | 152.67 |
| Proposed joint modeling | 216.19 | 113.05 |
Table 6.
RSS and DIC values for different combinations for female population.
| Methods | RSS | DIC |
|---|---|---|
| Quadratic mean with AR(1) cov. | 336.15 | 130.55 |
| Quadratic mean with CS cov. | 327.91 | 142.72 |
| Proposed joint modeling | 289.56 | 108.98 |
3.3 Computer Simulation
In order to evaluate the statistical properties of our model and estimation procedure, we undertook simulation studies by mimicking the data structure of FHS. We used seven SNPs detected from the FHS data as a marker to simulate their associated QTLs, which are rs17782554, rs11783045, rs9915696 and rs3903759 for males and rs4451518, rs13124340 and rs3903759 for females. By using allele frequencies and genotype distribution of these SNPs, we simulate longitudinal phenotypes of each subject separately for different sexes based on the model parameter estimates as given in Supplement 1. We kept the total number of subjects 977, the same as the original data. Also, the number of longitudinal measurements from each subject was kept the same as the original data.
We ran our MCMC method to estimate the model parameters for males for each of the 4 markers mentioned above and did the same thing for females for each of the 3 markers. The estimates and the Monte Carlo standard errors of the model parameters for these simulation studies are shown in Supplement 2. The accuracy and precision of the estimation show that the results for the original FHS project with 977 subjects are reasonably convincing. The simulation based on seven SNPs show that the estimation is quite accurate, which suggests that the proposed model detects the plausible QTLs controlling the trait of interest in practice.
We performed an additional simulation to examine the power and false positive rate (FPR) of our model. Power analysis was based on the same simulated data as described above by assuming that there exists a significant QTL. The analysis of FPR was conducted by simulating phenotypic data that involve no significant QTL. On the basis of 5,000 simulations, across all 7 SNPs, the power of gene detection of our model is found about 0.8, whereas the FPR of the model is less than 0.10 for a sample size of 1,000. Because our simulations were conducted by mimicking the FHS, it is likely that our power and FPR analyses reflect an actual case.
4 DISCUSSION
Because of their paramount importance in biology and biomedicine, a dynamic model has been derived to map QTLs for complex dynamic traits [15–24]. This model was founded on rigorous biological principles of trait formation and development and allows the formulation of numerous quantitative hypothesis tests on the interplay between genes and development, showing an increasing implication for quantitative, developmental, and ecological genetic studies. In many practical trials, longitudinal data are often collected at irregularly spaced time points and with measurement schedules specific to different subjects. Despite high sparsity, the subjects combine together to display a long window of points on the time scale. The efficient estimation of covariance structure in this situation will be a significant concern for genetic mapping to map the QTLs that control dynamic traits. In this article, we have extended the previous dynamic model to handle a challenge in modeling the mean-covariance structures of subject-specific irregular longitudinal data by integrating different statistical approaches.
First, the new model is based on a nonparametric modeling of the mean vector by a penalized spline and a parametric modeling of the covariance structure. This semiparametric treatment shows an advantage of combining the flexibility of a spline approach for modeling a long window of time points and efficiency of a parametric approach for modeling subject-specific variances and correlations with a limited number of time points. Second, we implemented a Bayesian algorithm to take its advantage of obtaining the estimation of a high-dimensional space of parameters. Meanwhile, the EM algorithm which has been shown to be efficient for estimating haplotype frequencies [36] was embedded into the Bayesian framework. Third, different from a traditional association study based on a direct phenotype-marker association, our model allows the identification of QTLs that determine phenotypic expression. Because QTLs are directly involved in pathways of trait formation and development, results from our model should be more biologically relevant.
It should be pointed out that our model was based on a simplified one-marker/one-QTL assumption, but its principle can be considered to model multiple QTLs and their epistatic interactions by involving multiple markers. If two QTLs are modeled, the likelihood (2) should be changed as
| (13) |
where j1 and j2 denote a genotype at the first and second QTL, respectively, ωj1j2 is the conditional probability of two-QTL genotypes given marker information, modeled by linkage disequilibria of different orders, and fj1j2(yi) contains genotype-specific mean vectors modeled by the additive, dominant, and epistatic effects of the QTLs. The complexity of multiple QTLs is being considered in the development of computer software for the proposed model.
Lastly, the model was used to analyze a real data set, validating its utilization and usefulness. Through simulation studies, the statistical properties of the model have been studied, including the precision of parameter estimates, model power, and false positive rates. All these have evidenced the application of the new model to practical problems. In particular, the new model is meritorious in tackling longitudinal data with high sparsity through semiparametric modeling of the mean-covariance structures.
More recently, Fan et al. [39] proposed a semiparametric model for the covariance structure of irregular longitudinal data, in which they approached the time-dependent correlation by a parametric function and the time-dependent variance by a nonparametric kernel function. The advantage of Fan et al.’s [39] model lies in the combination between the flexibility of nonparametric modeling and parsimony of parametric modeling. The establishment of a robust estimation procedure and asymptotic properties of the estimators will make this semiparametric model useful in the practical estimation of covariance function.
Supplementary Material
Supplement 1: Posterior estimates of the model parameters for significant QTLs for BMI trajectories from the FHS project. (DasFunMap Sup1.pdf)
Supplement 2: Simulation results by mimicking the data structure of the FHS project. (DasFunMap Sup2.pdf)
Acknowledgements
This work is partially supported by grant NSF/IOS-0923975 and NIH/UL1RR0330184 to RW, and NIDA, NIH grants R21 DA024260 and R21 DA024266 to RL. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIDA or the NIH.
APPENDIX: FULL CONDITIONAL DISTRIBUTIONS
In what follows, we give the full conditional distributions for the unknown parameters, bj, cj (j = 2, 1, 0), λ, δ, and λ*. Denote b−j = (bj′: j′ = 0, 1, 2; j′ ≠ j) and c−j = (cj′: j′ = 0, 1, 2; j′ ≠ j). Let mj be the number of subjects with QTL genotype j. The full conditional distribution of bj is expressed as
Hence, we have
| (A1) |
Similarly, the full conditional distributions for the other parameters can be derived as follows:
| (A2) |
| (A3) |
| (A4) |
| (A5) |
REFERENCES
- 1.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 2.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao CH, Zeng Z-B, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152:1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu S, Atchley WR. Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics. 1996;143:1417–1424. doi: 10.1093/genetics/143.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jannink JL, Wu XL. Estimating allelic number and identity in state of QTLs in interconnected families. Genetical Research. 2003;81:133–144. doi: 10.1017/s0016672303006153. [DOI] [PubMed] [Google Scholar]
- 7.Wu RL, Ma CX, Casella G. Joint linkage and linkage disequilibrium mapping of quantitative trait loci in natural populations. Genetics. 2002;160:779–792. doi: 10.1093/genetics/160.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou F, Fine JP, Hu J, Lin DY. An efficient resampling method for assessing genome-wide statistical significance in mapping quantitative trait loci. Genetics. 2004;168:2307–2316. doi: 10.1534/genetics.104.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broman KW, Speed TP. A model selection approach for the identification of quantitative trait loci in experimental crosses (with discussion) Journal of the Royal Statistical Society B. 2002;64:641–656. [Google Scholar]
- 11.Zou F, Nie L, Wright FA, Sen PK. A robust QTL mapping procedure. Journal of Statistical Planning and Inference. 2009;139:978–989. doi: 10.1016/j.jspi.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z. The full EM algorithm for the MLEs of QTL effects and positions and their estimated variances in multiple interval mapping. Biometrics. 2005;61:474–480. doi: 10.1111/j.1541-0420.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 13.Satagopan JM, Yandell BS, Newton MA, Osborn TC. A Bayesian approach to detect quantitative trait loci using Markov chain Monte Carlo. Genetics. 1996;144:805–816. doi: 10.1093/genetics/144.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi N, Xu S, Allison DB. Bayesian model choice and search strategies for mapping interacting quantitative trait loci. Genetics. 2003;165:867–883. doi: 10.1093/genetics/165.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma CX, Casella G, Wu RL. Functional mapping of quantitative trait loci underlying the character process a theoretical framework. Genetics. 2002;161:1751–1762. doi: 10.1093/genetics/161.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W, Wu RL. Wavelet-based nonparametric functional mapping of longitudinal curves. Journal of the American Statistical Association. 2008;103:714–725. [Google Scholar]
- 17.Yang RQ, Gao HJ, Wang X, Zhang J, Zeng Z-B, Wu RL. A semiparametric model for composite functional mapping of dynamic quantitative traits. Genetics. 2007;177:1859–1870. doi: 10.1534/genetics.107.077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang RQ, Xu SZ. Bayesian shrinkage analysis of quantitative trait loci for dynamic traits. Genetics. 2007;176:1169–1185. doi: 10.1534/genetics.106.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu RL, Lin M. Functional mapping - how to study the genetic architecture of dynamic complex traits. Nature Reviews Genetics. 2006;7:229–237. doi: 10.1038/nrg1804. [DOI] [PubMed] [Google Scholar]
- 20.Wu RL, Ma CX, Lin M, Wang ZH, Casella G. Functional mapping of growth quantitative trait loci using a transform-both-sides logistic model. Biometrics. 2004;60:729–738. doi: 10.1111/j.0006-341X.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin M, Zhao W, Wu RL. A statistical framework for genetic association studies of power curves in bird flight. Biological Proceedings Online. 2006;8:164–174. doi: 10.1251/bpo125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao W, Chen YQ, Casella G, Cheverud JM, Wu RL. A non-stationary model for functional mapping of longitudinal quantitative traits. Bioinformatics. 2005;21:2469–2477. doi: 10.1093/bioinformatics/bti382. [DOI] [PubMed] [Google Scholar]
- 23.Wu RL, Hao W, Cui YH, Li HY, Liu T, Wu S, Ma CX, Zeng YR. Mapping the genetic architecture of complex traits with molecular markers. Recent Patents on Nanotechnology. 2007;1:41–49. doi: 10.2174/187221007779814835. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Wu RL. Functional mapping of growth and development. Biological Reviews. 2010;85:207–216. doi: 10.1111/j.1469-185X.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 25.McCullagh P, Nelder JA. Generalized Linear Models. London: Chapman and Hall; 1989. [Google Scholar]
- 26.Pourahmadi M. Joint mean-covariance models with applications to longitudinal data: Unconstrained parameterisation. Biometrika. 1999;86:677–690. [Google Scholar]
- 27.Pan J, Mackenzie G. On modelling mean-covariance structures in longitudinal studies. Biometrika. 2003;90:239–244. [Google Scholar]
- 28.Daniels MJ, Kass RE. Shrinkage estimation for covariance matrices. Biometrics. 2001;57:1173–1184. doi: 10.1111/j.0006-341x.2001.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniels MJ, Pourahmadi M. Bayesian analysis of covariance matrices and dynamic models for longitudinal data. Biometrika. 2002;89:553–566. [Google Scholar]
- 30.Liu T, Wu RL. A Bayesian algorithm for functional mapping of dynamic complex traits. Algorithms. 2009;2:667–691. [Google Scholar]
- 31.Heuven HCM, Janss LLG. Bayesian multi-QTL mapping for growth curve parameters. BMC Proc. 2010;4(Suppl 1):S12. doi: 10.1186/1753-6561-4-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durban M, Harezlak J, Wand MP, Carroll RJ. Simple fitting of subject-specific curves for longitudinal data. Statistics in Medicine. 2005;24:1153–1167. doi: 10.1002/sim.1991. [DOI] [PubMed] [Google Scholar]
- 33.Dawber TR, Meadors GF, Moore FE., JR Epidemiological approaches to heart disease: The Framingham Study. American Journal of Public Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirier P, Giles T, Bray G, et al. Obesity and cardiovascular disease: Pathophysiology, evaluation, and effect of weight-loss. Circulation. 2006;113:898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Wang Y. A penalized spline approach to functional mixed effects model analysis. Biometrics. 2011 doi: 10.1111/j.1541-0420.2010.01524.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang ZH, Wu RL. A statistical model for high-resolution mapping of quantitative trait loci determining HIV dynamics. Statistics in Medicine. 2004;23:3033–3051. doi: 10.1002/sim.1870. [DOI] [PubMed] [Google Scholar]
- 37.Gelman A, Carlin J, Stern H, Rubin DB. Bayesian Data Analysis. Chapman and Hall; 1995. [Google Scholar]
- 38.Brooks SP, Gelman A. General Methods for Monitoring Convergence of Iterative Simulations. Journal of Computational and Graphical Statistics. 1998;7:434–455. [Google Scholar]
- 39.Fan J, Huang T, Li RZ. Analysis of longitudinal data with semiparametric estimation of covariance function. Journal of the American Statistical Association. 2007;35:632–641. doi: 10.1198/016214507000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang Y, Wang Y. Testing for familial aggregation of functional traits. Statistics in Medicine. 2009;28:3611–3625. doi: 10.1002/sim.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang C, Zeng Z-B. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics. 1995;140:1111–1127. doi: 10.1093/genetics/140.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solberg LC, Baum AE, Ahmadiyeh N, Shimomura K, Li R, Turek FW, Churchill GA, Takahashi JS, Redei EE. Sex and lineage-specific inheritance of depression-like behavior in the rat. Mamm Genome. 2004;15:648–662. doi: 10.1007/s00335-004-2326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1: Posterior estimates of the model parameters for significant QTLs for BMI trajectories from the FHS project. (DasFunMap Sup1.pdf)
Supplement 2: Simulation results by mimicking the data structure of the FHS project. (DasFunMap Sup2.pdf)