Abstract
A novel multi-channel poly(methyl methacrylate) (PMMA) microfluidic biosensor with interdigitated ultramicroelectrode arrays (IDUAs) for electrochemical detection was developed. The focus of the development was a simple fabrication procedure and the realization of a reliable large IDUA that can provide detection simultaneously to several microchannels. As proof of concept, five microchannels are positioned over a large single IDUA where the channels are parallel with the length of electrode finger. The IDUAs were fabricated on the PMMA cover piece and bonded to a PMMA substrate containing the microfluidic channels using UV/ozone-assisted thermal bonding. Conditions of device fabrication were optimized realizing a rugged large IDUA within a bonded PMMA device. Gold adhesion to the PMMA, protective coatings and pressure during bonding were optimized. Its electrochemical performance was studied using amperometric detection of potassium ferri and ferro hexacyanide. Cumulative signals within the same chip showed very good linearity over a range of 0 - 38 μM (R2 = 0.98) and a limit of detection of 3.48 μM. The bonding of the device was optimized so that no cross-talk between the channels was observed which otherwise would have resulted in unreliable electrochemical responses. The highly reproducible signals achieved were comparable to those obtained with separate single-channel devices. Subsequently, the multi-channel microfluidic chip was applied to a model bioanalytical detection strategy, i.e. the quantification of specific nucleic acid sequences using a sandwich approach. Here probe-coated paramagnetic beads and probe-tagged liposomes entrapping ferri/ferro hexacyanide as the redox marker were used to bind to a single stranded DNA sequence. Flow rates of the non-ionic detergent n-octyl-β-D-glucopyranoside (OG) for liposome lysis were optimized and the detection of the target sequences was carried out coulometrically within 250 s and with a limit of detection of 12.5 μM. The robustness of the design and the reliability of the results obtained in comparison to previously published single-channel designs suggest that the multi-channel device offers an excellent opportunity for bioanalytical applications that require multi-analyte detection and high throughput assays.
Keywords: microfluidic chip, multi-channel, electrochemical biosensor, ultramicro electrode, PMMA
Introduction
Micro-Total Analysis System (μTAS) together with microfluidic technology has become a highly promising tool for protein and nucleic acid analysis [1-3]. This is due to a number of advantages arising from the detection of biomolecules in a miniaturized system. Those include reducing sample/reagent and power consumption, providing high assay sensitivity with shorter analysis time, increasing repeatability and reliability by system automation, and rendering the integration capability for multiple processes within a single device [4].
Electrochemical detection is an ideal technique to incorporate into microfluidic devices as it possesses the inherent ability to be miniaturized by existing microfabrication technology, and as it offers excellent analytical performance. In addition, the electrochemical response is independent from the optical path length or sample turbidity which is encountered in optical based techniques. Furthermore, low power consumption and cost effectiveness are additional advantages that makes miniaturized electrochemical detection systems highly desirable and sustainable [5-7].
Among the many electrochemical techniques, amperometry coupled with microelectrodes is ideally suited, and widely employed in microfluidic chips [8-14]. Microelectrodes are of interest due to their small iR drop, fast establishment of steady-state mass transfer and small capacitive charging currents [15]. In fact, the reduction of the electrode dimensions to micrometer size can greatly enhance their sensitivity, because a highly efficient collection of analyte species can be realized via non-planar diffusion characteristics at microelectrodes [5]. This provides in turn an increased signal-to-noise ratio which thus enables lower detection limits and improved sensitivity.
Interdigitated ultramicroelectrode arrays (IDUAs), a pair of microband array electrodes that mesh with each other, is a microelectrode geometry that is particularly well suited for reversible and quasi-reversible redox species determinations [15,16]. A very small gap size between adjacent electrode fingers is considerably favorable for achieving high signal amplification owing to the increase of the diffusion flux of redox species, which consequently leads to an enhanced redox cycling, a decreased equilibrium time, and an increased collection efficiency [17]. Previously, IDUAs have been characterized and studied as a transducer for electrochemical biosensors and have then been applied to a variety of bioanalytical systems in our research group [18-22].
The development of multi-channel microfluidic chips has received much attention in recent years [23-33] because it can increase the test throughput and can reduce production costs. Additionally, sample handling and analysis of multiple analytes within the same chip can substantially improve the analytical performance in terms of repeatability and reliability. However, there have been only few investigations towards the fabrication of multi-channel electrochemical microfluidic chips. For examples, Chen and co-worker developed a multi-channel microchip for simultaneously detecting multi-parameter of diabetes mellitus [24]. Moreira and co-worker have developed a poly(dimethyl) siloxane (PDMS)/glass hybrid multi-channel microfluidic device with integrated electrodes for amperometric detection of N-acetyl-p-aminophenol [34]. However, their design encountered problems of incomplete sealing over the gold surface which lead to leaking as soon as positive pressure was applied in the channels. This then considerably influenced the reproducibility of the electrochemical signals. While the authors have proposed a solution, their method is complicated and involves in hydrofluoric etching, which is an undesirable fabrication method due to its immense toxicity.
We consequently here describe the very robust design and fabrication of multi-channel PMMA microfluidic chips with integrated IDUAs. In our design, a single IDUA is shared by five micro-channels positioned in parallel with the length of the electrode finger. Five channels were chosen as model number only. Ten or more channels can easily fit across the same IDUA's surface. Hot embossing and UV/ozone-assisted thermal bonding were employed for microchannel fabrication and chip assembly, respectively. The fabrication process was optimized with respect to gold adhesion, protective coatings and bonding conditions. The electrochemical performance evaluation of the microfluidic chip was carried out with respect to the cumulative signal response as well as the detection sensitivity. Furthermore, the reproducibility of the multi-channel device was compared to those of individual single channel devices which were previously designed in our group [22]. Finally, we also demonstrated the capability of the multi-channel microfluidic chip for nucleic acid detection employing liposomes entrapping potassium ferri/ferro hexacyanide as a redox marker. Here, a well-developed bioanalytical detection strategy for the detection of RNA from Cryptosporidium parvum was chosen as model analyte [35-37].
Experimental
Materials and reagents
PMMA plates were purchased from Lucite International, Southampton, UK. Copper plates were purchased from McMaster Carr, Princeton, NJ. 2-Propanol, 2-(4-morpholino)-ethane sulfonic acid (MES), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), sodium carbonate, potassium ferrocyanide (K4Fe(CN)6·3H2O), and potassium ferricyanide (K3Fe(CN)6) were purchased from Sigma Aldrich, St. Louis, MO. N-hydroxysulfosuccinimide (Sulfo-NHS) was purchased from G-Biosciences, St. Louis, MO. Cystamine was purchased from MP Biomedicals, Solon, OH. Silver conductive epoxy was purchased from MG Chemicals, Burlington, Ont. Cyanoacrylate based adhesive was purchased from Henkel Consumer Adhesives, Avon, OH. Lipids and cholesterol were purchased from Avanti Polar Lipids, Alabaster, AL. n-Octyl-β-D-glucopyranoside (OG) was purchased from Alexis Corporation, Lausen, Switzerland. All oligonucleotides including cholesterol-tagged reporter probes were purchased from Operon, Huntsville, AL. Superparamagnetic beads (Dynabeads MyOne Streptavidin) were purchased from Dynal Biotech Inc., Lake Success, NY. Tygon® tubing and stainless steel blunt needle with luer polypropylene hub were purchased from Small Parts Inc., Miami Lakes, FL. Syringes were purchased from Hamilton Company, Reno, ND. Magnets were purchased from National Imports LLC, Vienna, VA.
Design and fabrication of IDUAs
The pattern of the interdigitated ultramicroelectrode arrays (IDUAs) was designed using L-Edit CAD Software and the 5″ quartz photomask containing IDUA pattern was fabricated using the 3600F PG Mask Writer. The total length and width of the IDUA pattern are 15 mm and 7 mm, respectively. The length and width of IDUA fingers are 3.9 mm and 10 μm, respectively, in the 10 mm × 4 mm central area of pattern. The gap between IDUA fingers is 5 μm. The fabrication of IDUAs on PMMA was performed following the procedure described in [22] with slight modifications. Specifically, the PMMA plate was cut into 50 mm × 30 mm piece and sonicated in 50% 2-propanol for 10 min to completely remove dust on the PMMA surface. The PMMA pieces were then washed in DI water and dried with nitrogen prior to the UV exposure. A UV/ozone stripper (SAMCO International, Inc., Sunnyvale, CA) was used for the oxidation of PMMA to generate carboxylic groups on the surface. The PMMA pieces were immersed in the stripper and treated with UV (10 mW/cm2 at 254 nm) for 5 min and then placed in agitated DI water for 30 min for a post exposure rinse before being dried with nitrogen. For the surface modification, 150 μL of a 0.05 M MES, pH 6.0 solution containing 300 mM EDC and 300 mM Sulfo-NHS was placed in the middle of two PMMA pieces with the UV-exposed side in contact with the liquid. Capillary action ensured coverage of the entire plates with the solutions. After 25 min, the PMMA pieces were rinsed and 150 μL of a 0.05 M sodium carbonate buffer, pH 9.0 solution containing 300 mM cystamine was placed in the middle of two PMMA pieces for cystamine conjugation. Following a 3-hour conjugation period, the surface thiolation was accomplished when the PMMA pieces were rinsed with DI water and dried with nitrogen again. These PMMA substrates were used within one or two days for gold evaporation.
The thiol-functionalized PMMA pieces were coated with gold using a CHA Mark 50 e-Beam Evaporator (CHA Industries, Freemont, CA). Briefly, the gold was deposited at 1 Å/s for a total thickness of 200 nm. After evaporation, the positive photoresist S1813 (Shipley Co., Marlborough, MA) was spun onto the gold-coated PMMA at 3000 rpm for 30 s in a manual resist spinner, and the pieces were baked in a 90 °C oven for 20 min to evaporate the remaining solvent on the surface. When the pieces were cooled down at room temperature, the photoresist was exposed under UV light for 12 s through the previously fabricated photomask containing the IDUA pattern using an ABM High Resolution Mask Contact Aligner. The pieces were then placed back in the 90 °C oven for an additional 5 min. Following the post-exposure bake, the exposed resist was removed and immersed in the developer MF-321 (Shipley Co., Marlborough, MA) for 2 min and then rinsed with DI water before being dried with nitrogen. The IDUAs were formed by submerging the pieces in gold etchant (Transene Co., Danvers, MA) for approximately 3 min. The PMMA and remaining photoresist on IDUAs were exposed to UV for 2 min and immersed in MF-321 for 2 min for complete removal of the photoresist on IDUAs. The PMMA with IDUA pieces were washed with DI water and dried with nitrogen for the bonding process.
Design and fabrication of multi-channel on PMMA
The multi-channel pattern illustrated in Fig. 1 and 2a (upper piece) was designed using L-Edit CAD Software and the photomask containing multi-channels was fabricated using the 3600F PG Mask Writer, followed by developing in 300MIF and chrome etching. The length and width of the multi-channel pattern are 50 mm and 25 mm, respectively. The width and height of channels are 400 μm and 50 μm, respectively, and five separated channels are in the pattern. Two inlet ports and one outlet ports allow for sample and OG solution additions and the mixture removal, respectively. The micromixers with sawtooth microstructures incorporated into channels are used to maximize the binding efficiency during the hybridization process.
Fig. 1.
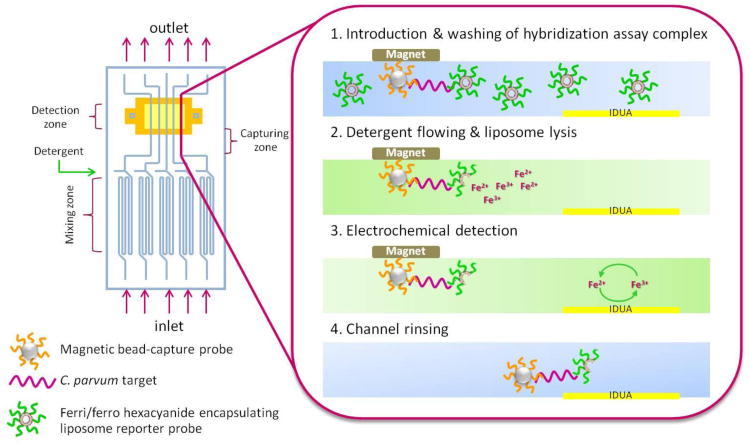
Multi-channel microfluidic biosensor principle: (1) A 4 μL sample solution is introduced through the inlet hole separating non-hybridized liposomes from the successful sandwiches at a capture zone. (2) After 8 min washing with running buffer, detergent is injected in order to lyse the liposomes and release ferri/ferro hexacyanide encapsulant. (3) The released redox marker is then detected over the IDUA generating a current signal corresponding to the C. parvum DNA target concentration. (4) Finally, the magnet is removed and channels rinsed with running buffer to prepare for the next assays.
Fig. 2.
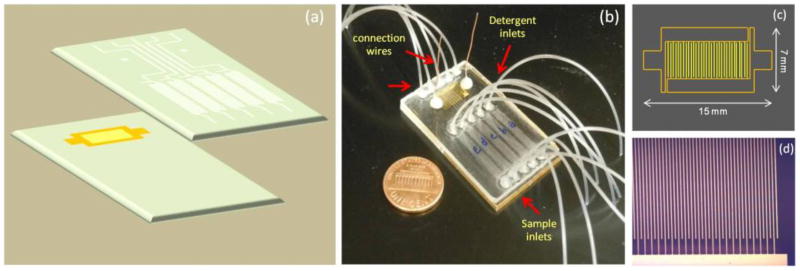
Chip fabrication and assembly. (a) Device configuration consists of IDUAs fabricated on PMMA substrate and micro-channel PMMA (400 μm channel wide and 50 μm channel high embedded in 1.5 mm PMMA thick) made by hot embossing, (b) Assembled multi-channel device (30 mm × 50 mm dimensions), (c) Layout of the IDUA, and (d) Microscopic image of IDUA fingers with 100× magnification (10 μm finger wide and 5 μm gap)
Copper electrodeposition was utilized to construct the master mold of multi-channel pattern. A copper plate with a polished mirror (1 mm thickness) was cut into 50 mm × 50 mm squares and cleaned with acetone and DI water. A negative resist, KMPR 1050 (MicroChem Corp., Newton, MA) was poured onto the center of the copper square and then was spun for 30 s at 3000 and 500 rpm/s to obtain a thickness of 50 μm using Karl Suss RC-8 wafer spinner. The plate was then baked on a 95 °C hot plate for 15 min followed by gradual cooling to room temperature. The resist was patterned by exposure to UV light for 60 s in ABM Contact Aligner through a 365 nm long pass filter to obtain vertical side walls. A post-exposure bake for 3 min at 95 °C was followed by developing for 8 min in SU-8 developer with mild agitation. Prior to electroplating, the KMPR patterned copper plate was de-scummed using 60 s of O2 plasma in Glen 1000 Resist Strip for stripping of photoresist and other organics. Electroplating was performed at room temperature using Microfab SC Copper Sulfamate solution (MicroFAB, Bremen, Germany), which at a current density of 0.21 mA/mm2 has a deposition rate of 0.105 mm3/mA h. Current was applied to the electroplating bath to produce 50 μm tall copper structures where KMPR did not cover the copper surface, resulting in the raised microchannel design. After plating, the KMPR was stripped off the copper plate by soaking in Microposit Remover 1165 (Shipley Co., Marlborough, MA) for 1 h at 70 °C. The master was then visually inspected for KMPR stripping and multi-channel pattern on the surface.
Hot embossing PMMA was used to replicate the multi-channel PMMA. The blank PMMA pieces were placed in an 80 °C vacuum oven (1 kPa) overnight to remove any volatiles that could result in out-gassing during the hot embossing process. The PMMA was then sandwiched between the copper master and a blank copper plate after they were preheated. A force of 5000 lbf was applied with an embossing temperature of 130 °C for 5 min in a Carver Laminating Hot Press (Fred S. Carver Inc., Summit, NJ). The sandwiched set-up was allowed to be hot embossed for another 3 min if there were air bubbles between PMMA plate and copper master. After that, the copper master and PMMA were removed from the hot press and cooled down to room temperature. The PMMA with multi-channel pattern was manually removed from the copper master and the inlet and outlet holes were drilled using steel bits. The PMMA was rinsed with DI water and dried with nitrogen, then put into an 80 °C vacuum oven for several hours to completely dry and evaporate any existing volatiles.
Multi-channel device assembly
The multi-channel structured PMMA and IDUA-attached PMMA pieces were treated under UV light in a UV/ozone stripper for 8 min for the photochemically induced main chain scission of PMMA to lower the glass transition temperature of the PMMA surface. The two PMMA pieces were aligned with the IDUA side in contact with the multi-channel side and then sandwiched between two blank copper plates after the copper plates were preheated. A force of 5000 lbf was applied with a bonding temperature of 80 °C for 5 min in a Carver Laminating Hot Press. The sandwiched set-up was allowed to be thermal bonded for another 3 min if air bubbles were present between two PMMA pieces. Subsequently, the bonded PMMA device and copper plates were removed from the hot press and cooled down to room temperature.
The inlet and outlet Tygon® tubing were glued in the drilled holes using cyanoacrylate based adhesive and the adhesive was solidified overnight. Stainless steel blunt needles with Luer polypropylene hub were inserted into tubing for connection with syringes containing assay solutions. Metal leads connecting the IDUA with the potentiostat were adhered to the IDUA contact pads with silver conductive epoxy; the epoxy was allowed to solidify for 2 hours.
Detection assay
Superparamagnetic beads coated with streptavidin were coupled with biotinylated capture probe 5′-biotin-AGA TTC GAA GAA CTC TGC GC-3′ according to the manufacturer's protocol. Briefly, 20 μL of bead stock (10 mg/mL) were washed twice in 2× concentrated binding and washing (B&W) buffer (10 mM Tris-HCl, 1 mM EDTA, 2 M sodium chloride, 0.01% sodium azide, pH 7.5) and resuspended in 19 μL of 1× concentrated B&W buffer. 1 μL of DNA capture probe (600 μM) were added to the beads and the resultant mixture was placed on a rotator for 15 min at room temperature to allow conjugation. After conjugation, the beads were washed 3 times with 1× B&W buffer and resuspended in the same buffer to a final volume of 20 μL. The beads with immobilized oligonucleotides could be stored at −20 °C for one week. At the same time, the liposomes were prepared following the protocol as described in [19] to encapsulate potassium ferri/ferrocyanide and coupled to cholesterol labeled reporter probe 5′-GTC CAA CTT TAG CTC CAG TT-cholesterol-3′. Briefly, 29.6 mg of dipalmitoyl phosphatidylcholine (DPPC), 15 mg dipalmitoyl phosphatidylglycerol (DPPG) and 20 mg cholesterol were dissolved in a mixture of 5 mL chloroform, 3 mL isopropyl ether and 0.3 mL methanol at 45 °C. Cholesterol labeled oligonucleotide reporter probe was added to the lipid mixture (0.013 mol% of total lipids). Four mL of 100 mM potassium ferricyanide and 100 mM potassium ferrocyanide in 0.01 M potassium phosphate buffer (PB), pH 7.0 was added to the lipid solution and the mixture was sonicated for 4 min at 45 °C. The organic solvents were evaporated in a rotary evaporator so that liposomes formed, spontaneously entrapping encapsulant solution. To obtain a uniform particle size, the liposomes were subsequently extruded through 2 μm, then 0.4 μm, filters (each 11 times) using the Avanti miniextruder and polycarbonate filters (Avanti Polar Lipids, Alabaster, AL). Liposomes were purified by gel filtration using a Sephadex G50 column and a 0.05 M PB, 0.15 M NaCl, pH 7.0, containing sucrose to increase the osmolarity to 500 mmol/kg. The liposomes were tagged with a reporter probe capable of hybridizing with the target sequence, a synthetic Cryptosporidium parvum target 5′-AAG GAC CAG CAT CCT TGA GTA CTT TCT CAA CTG GAG CTA AAG TTG CAC GGA AGT AAT CAG CGC AGA GTT CTT CGA ATC TAG CTC TAC TGA TGG CAA CTG A-3′. The sequences were chosen from previous biosensor development for the detection of C. parvum[36].
To characterize the IDUAs and the multi-channel structure, various concentrations of potassium ferri/ferrocyanide were injected into the channels at a flow rate of 5 μL/min. Fluid flow through the multi-channel network was established by applying a positive pressure at the inlet using a syringe pump (KD Scientific Inc., Holliston, MA) connecting to a Luer polypropylene hub of stainless steel blunt needles and opening the outlet to atmospheric pressure. A 400 mV potential was applied across the IDUA and the resulting current was measured on an Epsilon potentiostat (Bioanalytical Systems, Inc., West Lafayette, IN).
For the detection assay, 1 μL of synthetic target sequence, 1 μL of superparamagnetic beads coupled with capture probe, 1 μL of 11.65 mM liposomes, and 1 μL of hybridization buffer (20% formamide, 4 × SSC, 0.2% Ficoll, 0.2 M sucrose, and 0.5% dextran sulfate) were mixed and incubated for 15 min at room temperature in a shaker. Following the incubation, the assay protocol was carried out following the scheme illustrated in Fig. 1. First, the sample solution was pumped into the multi-channel device in a reciprocating motion at a flow rate of 1 μL/min in order to aid in mixing through sample inlets. A rare earth magnet was placed over the channels in order to hold the beads in place while the sample solution was pumped out and replaced with a washing solution (20% formamide, 4× SSC, 0.2% Ficoll, 0.2 M sucrose, and 0.8% dextran sulfate) at 1.5 μL/min. The magnet was about 1 mm removed from the middle of the channels. The first magnet was then removed and a second rare earth magnet was placed directly over the IDUA. Second, the beads were then washed toward the IDUAs again at 1.5 μL/min for 8 min. The placement of the second magnet captured the majority of beads directly upstream of the IDUA. The washing step was needed to remove any unhybridized liposomes from the sample area. Third, a detergent, 60 mM OG (in 0.01 M PB, 0.15 M sodium chloride, 0.01 % sodium azide, pH 7.0) was injected into the channels at 2 μL/min (unless stated otherwise) in order to lyse the liposomes. Subsequently, released potassim ferri/ferrohexacyanide was detected on the IDUA positioned downstream of the capture zone. The resulting current from the redox reaction was detected amperomatrically. The resulting current peak height or peak area under the time was determined to be the assay signal. Finally, by removing the magnet the channels were rinsed with washing solution to retrieve the background current.
Results and discussion
Multi-channel microfluidic device fabrication and characterization
A multi-channel microfluidic device for the electrochemical detection of analytes was developed. Fig. 2 illustrates design and necessary components of the multi-channel microfluidic device system. The device composes of two major parts which are IDUAs fabricating on a PMMA substrate using standard photolithography processes and hot embossed micro-channel PMMA. Five micro-channels are aligned in parallel with the IDUA fingers as depicted in Fig. 2a. Strong sealing between the IDUA-bearing PMMA substrate and the channel piece was achieved using the UV/ozone-assisted thermal bonding approach. Special care has to be taken during this procedure to avoid sliding of the IDUA-chip over the channel-substrate in order to prevent short-circuiting of the delicate IDUA structure.
Also, appropriate bonding conditions need to be verified examining potential leaking of solution from the channels, as this would create varying surface coverage on the IDUA and result in varying signals between the channels. For this instance, 1 mM sulforhodamine B dye solution was injected through all channels and visualized using a microscope, verifying the appropriate channel dimensions prior to use with an electrochemical set up. In order to achieve well-bonded devices with functional IDUAs, the bonding conditions were optimized with special emphasis of keeping a simple protocol.
First, the adhesion of the gold layer on the PMMA was optimized. It was observed that intense bubble formation during the PMMA thiolation protocol led to non-uniform surface modifications which in turn led to unreliable gold adhesion upon gold evaporation and different gold thicknesses. During the etching process, not all fingers and gaps could be realized. Various PMMA modification strategies have been studied (see Electronic Supplementary Material Table S1) demonstrating that the addition of 0.1% Tween 20 (v/v) greatly reduces bubble formation, especially though in combination with Sulfo-NHS. As expected, these PMMA substrates enabled a highly uniform IDUA fabrication fingers and, more importantly, un-short IDUA circuits after etching.
Secondly, the UV/ozone assisted thermal bonding procedure needed to be optimized to avoid short-circuiting of the IDUAs. We optimized the applied bonding pressure, altered the UV/ozone treatment time and most importantly protected the IDUAs with a layer of photoresist, (see Electronic Supplementary Material Table S2). A final remaining factor was to avoid sliding of the electrode-bearing PMMA piece over a surface. This resulted in functioning and reliable large IDUAs bonded securely in microfluidic chips.
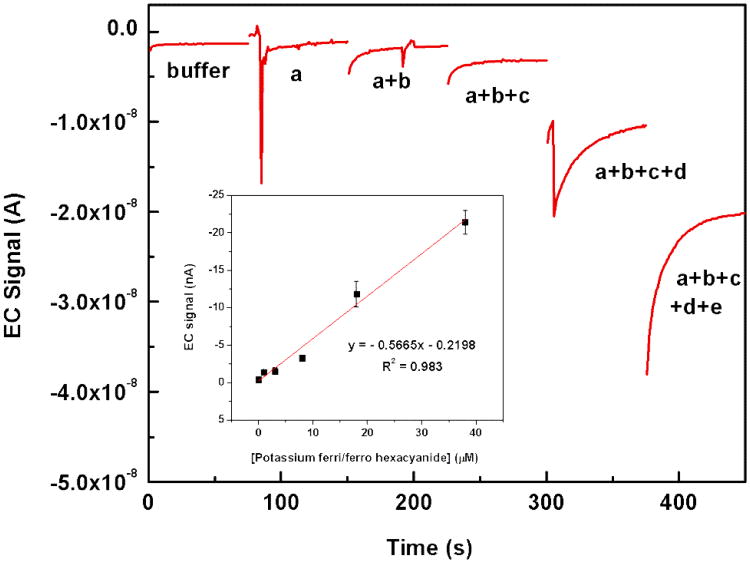
To evaluate the electrochemical performance of integrated IDUA in a multi-channel microfluidic chip, we determined the cumulative signal response upon injecting ferri/ferro hexacyanide consecutively into each channel; without removing the analytes from filled channels. Data were taken after steady-state current was reached. Since we utilized an IDUA shared by five microchannels, the total amperometric current is the sum of signals generated at the active area in each channel. In order to separate the signals obtained from the different channels, redox marker was first applied only to channel a, then channel a+b, then channels a+b+c etc. The remaining channels were filled with buffer. The concentrations injected in channel a, b, c, d, and e was 1, 2, 5, 10, and 20 μM respectively. As shown in Fig. 3, the current signals of ferri/ferro hexacynaide increase proportionally to the cumulative concentrations that were injected into each channel. An excellent linear relationship was found in the range of 0 – 38 μM with the linear equation of y (nA) = -0.5665× (μM) – 0.2198 (nA), R2 = 0.983. The calculated detection limit is thus 3.48 μM with a signal to noise ratio of 3 (background signal = -0.39 (nA) ± 0.08 (nA)).
Fig. 3.
Cumulative amperometric response example of ferri/ferro hexacynaide in 0.1 M PB, pH 7.0 for five channels. The analyte solution was injected consecutively into the different channels. First, only channel a, then channel a+b, then channels a+b+c etc. contained the redox marker. The concentrations injected in channel a, b, c, d, and e was 1, 2, 5, 10, and 20 μM respectively. All measurements were carried out at 400 mV (stagnant condition). Inset: dose response curve from triplicate measurements.
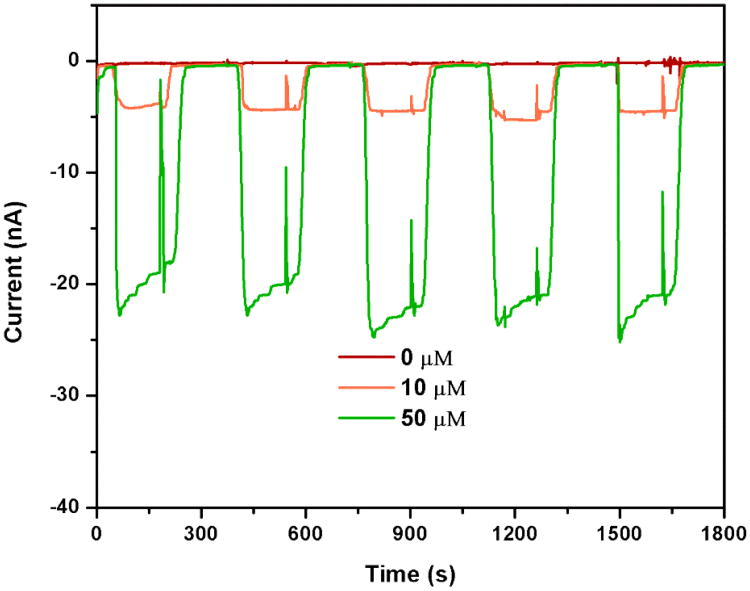
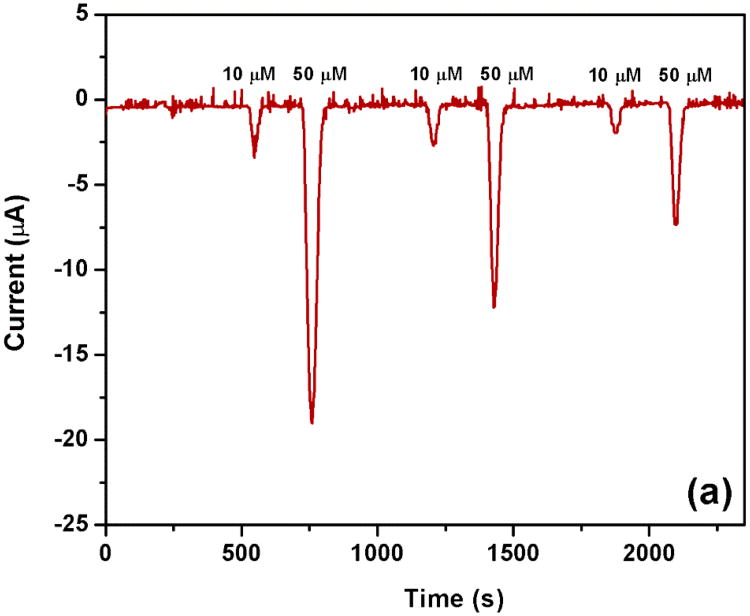
The reproducibility of the multi-channel device was tested for each channel and between channels. In the case of in-channel reproducibility, triplicate measurements of 5 μM ferri/ferro hexacyanide were performed in each channel. The result showed that all of five channels provided highly reproducible signals with relative standard deviations below 5%. Additionally, the reproducibility of the multi-channel design between different channels was compared to separate single-channel devices. Solutions of 10 μM and 50 μM ferri/ferro hexacyanide were tested. High reproducibility was obtained in both cases, which was a powerful proof of the feasibility of different electrochemical detection schemes considering the different size and thus complexity of the two different systems. Specifically, signals obtained for the multi-channel device are shown in Fig. 4 and Table 1, signals obtained for the single IDUAs are summarized in Table 2. In contrast to the multi-channel IDUA, background normalization was required for the single IDUA chips in order to obtain reproducible results.
Fig. 4.
Replication testing of multi-channel microfluidic device. Ferri/ferro hexacyanide in 0.1 M PB pH 7.0 was detected at 5 μl/min flow rate in each channel for 3 min. Current background was achieved by rinsing the channel with 0.1 M PB pH 7.0 for 3 min at the same flow rate of analyte measurement.
Table 1. Replication testing data of multi-channel device.
| Ferri/ferro hexacyanide concentration (μM) | Current signal (nA) | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Channel | Average | %RSD | |||||
|
|
|||||||
| a | b | c | d | e | |||
| 10 | 4.49 | 4.97 | 4.46 | 4.38 | 4.13 | 4.49 | 6.88 |
| 50 | 21.39 | 21.28 | 22.92 | 20.60 | 19.60 | 21.16 | 5.74 |
Table 2. Replication testing data of different single channel devices.
| Ferri/ferro hexacyanide concentration (μM) | Normalized peak area with corresponding background signal | ||||
|---|---|---|---|---|---|
|
| |||||
| Microfluidic chip | Average | %RSD | |||
|
|
|||||
| A | B | C | |||
| 10 | 3.81±0.38 | 3.44±0.13 | 3.67±0.45 | 3.64±0.15 | 4.09 |
| 50 | 20.53±2.10 | 16.57±1.47 | 17.71±1.19 | 18.27±1.66 | 9.09 |
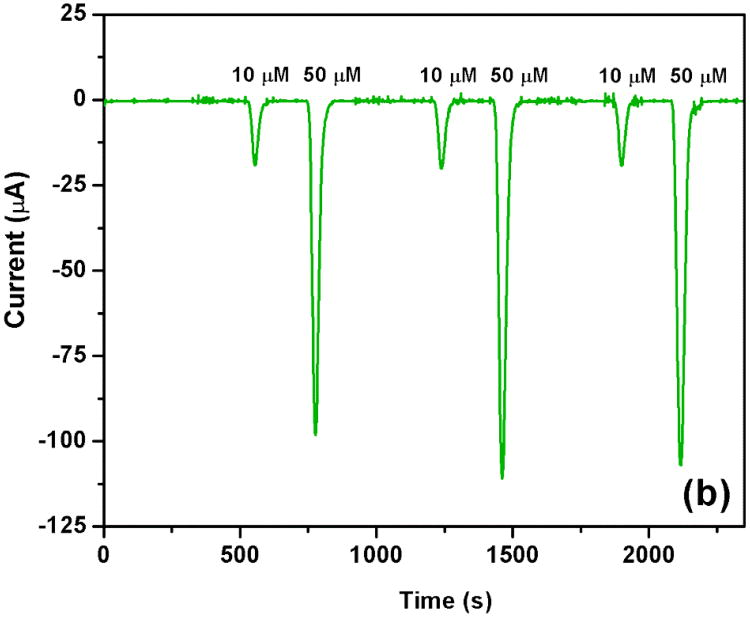
Finally, the cleaning of the IDUAs packaged inside the microfluidic device in between reactions was investigated. As can be seen in Fig. 5a, a chip without IDUA cleaning procedures other than rinsing the channels with DI water or buffer exhibits declining signals for both 10 and 50 μM ferri/ferro hexacyanide in a consecutive measurement. A drop in signal was observed in a series of just three measurements which is due to the complexity of buffer solutions used leading to electrode fouling. The signals dropped 21 and 27% for the 10 μM solution and 36 and 40% for the 50 μM solution. A variety of cleaning procedures amenable for in-device cleaning were investigated including electrochemical stripping of the electrode surface, isopropanol, acetone and acids and bases. Finally, it was found that a treatment with 0.1 M NaOH at 5 μL/min flow rate for 10 min cleaned the IDUA surfaces sufficiently and enabled highly reproducible signals as shown in Fig. 5b. In addition, using this cleaning procedure prior to a first use of the IDUA ensures stripping of any remaining photoresist and leads to significantly higher currents and signal to noise ratios.
Fig. 5.
Effect of electrode cleaning in channel on current responses of 10 and 50 μM potassium ferri and ferro hexacyanide in 0.1 M PB, pH 7.0. (a) without cleaning IDUA in channel and, (b) clean IDUA in channel before electrochemical measurement with 0.1 M NaOH at 5 μL/min flow rate for 10 min.
Application to nucleic acid detection
Detergent flow rate optimization
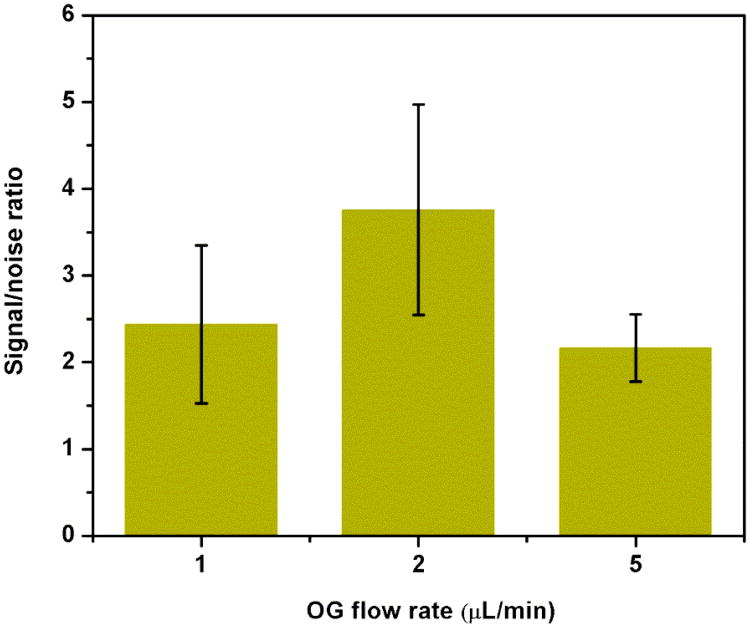
Single-channel electrochemical microfluidic sensors were used [22] to determine the dependency of the nucleic acid detection assay on the detergent flow rate. The ultimate goal is the identification of flow rates that will enable time-dependent detection within the multi-channel system where signals are resolved due to different lengths of the detergent supply channels. Thus, a time-resolved multianalyte detection will be possible. Here, C. parvum DNA was detected using a hybridization assay between capture probe-coupled paramagnetic beads and reporter probe-tagged liposomes [19,20,22]. The hybridization complexes were captured via an external magnet located upstream of the IDUA. Upon lysis of the liposome using the non-ionic detergent n-octyl-β-D-glucopyranoside (OG), the released potassium ferri/ferrohexacyanide was detected on the IDUA. Detergent flow rates were varied at 1, 2 and 5 μL/min to investigate the dependency of the signal-to-noise ratio (SNR) from a 4 μL sample containing 300 pmol synthetic DNA target (75 μM).
As can be seen from Fig. 6, the flow rate of 2 μL/min shows the highest SNR whereas 1 and 5 μL/min are comparable to each other. We assume that this is due to a combination of detergent contact time resulting in liposome lysis, diffusion of the redox couple away from the electrode and length of time spent over the electrode area. Overall, full signal development for quantification ranges from 400- 120 seconds depending on the flow rates.
Fig. 6.
Optimization of detergent flow rate for liposome lysis. Signal to noise ratio was obtained from normalized peak area of 300 pmol target by negative control signal in which nuclease free water was used instead of target.
Cryptosporidium parvum assay in multi-channel devices
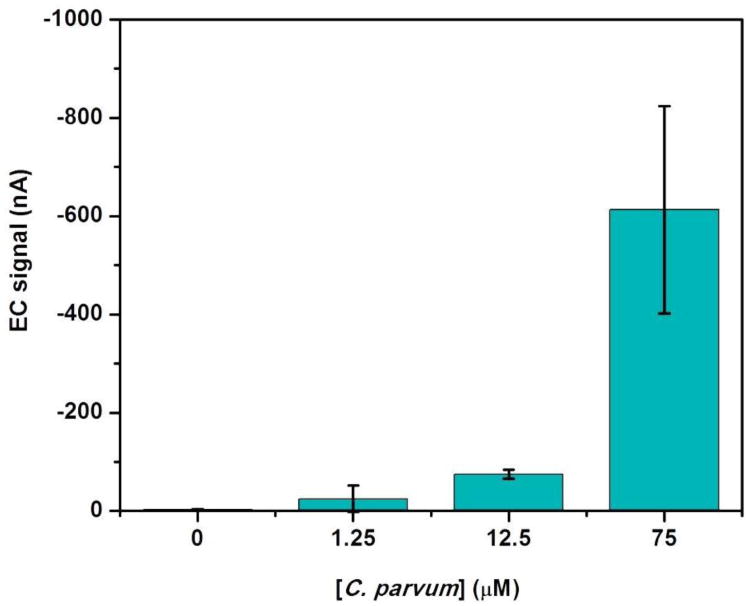
Finally, a dose response curve for the detection of the synthetic C. parvum DNA [36] was obtained using the multi-channel microfluidic chip. Fig. 7 displays the electrochemical responses upon the addition of 0, 1.25, 12.5 and 75 μM target injected into four different channels of the device in which time-dependent signal generations were accomplished after a consecutive injection of OG solution through the channels. The background signal obtained from 0 μM DNA target was found to be -2.51 (nA) ± 1.33 (nA). Even though the average signal of 1.25 μM target is -24.99 nA, the variation of current signal at this concentration was too high to reliable distinguish from the background signal. As a result, the detection limit of 12.5 μM target is reported as the lowest concentration demonstrated for our multi-channel microfluidic device.
Fig. 7.
Electrochemical responses from C. parvum assays with analyte concentrations from 0, 1.25, 12.5 and 75 μM. All measurements were performed at 2 μL/min flow rate. Error bars show the standard deviation of triplicate data.
Conclusions
In summary, we have presented the development of a multi-channel PMMA microfluidic chip with integrated IDUAs and its application to electrochemical nucleic acid detection. Our simple fabrication approach enables strong sealing for all channels of the multi-channel microfluidic chip. In addition, the chip shows desirable electrochemical performance with respect to analytical sensitivity, linearity, and reproducibility. The multi-channel design will be especially desirable when more complex fluidics and higher degree multi-analysis are to be used that can otherwise not be realized with single small IDUAs.
Supplementary Material
Acknowledgments
The authors acknowledge financial support which enabled the research presented. This publication was developed under the auspices of the Cornell University Center for Life Science Enterprise, a New York State Center for Advanced Technology supported by New York State and industrial partners. Also, this publication was supported by a subcontract with Rheonix, inc. and 1U01 A1082448-01 from the NIH. Any opinions, findings and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of Rheonix nor those of the National Institutes of Health. This work was performed in part at the Cornell NanoScale Science and Technology Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECS-0335765). NW and WS thank for financial support from the Thailand Research Fund through the Royal Golden Jubilee Ph.D. program (Grant No. PHD/0319/2548).
Abbreviations
- DPPC
dipalmitoyl phosphatidylcholine
- DPPG
dipalmitoyl phosphatidylglycerol
- EDC
1 -ethyl-3 -(3 -dimethylaminopropyl) carbodiimide
- EDTA
2-({2- [Bis(carboxymethyl)amino]ethyl}(carboxymethyl)amino)acetic acid
- MES
2-(4-morpholino)-ethane sulfonic acid
- OG
n-Octyl-β-D-glucopyranoside
- PB
potassium phosphate buffer
- SSC
saline sodium citrate
- Sulfo-NHS
N-hydroxysulfosuccinimide
References
- 1.Choi S, Goryll M, Sin LYM, Wong PK, Chae J. Microfluidic-based biosensors toward point-of-care detection of nucleic acids and proteins. Microfluid Nanofluid. 2011;10:231–247. doi: 10.1007/s10404-010-0638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lien KY, Lee GB. Miniaturization of molecular biological techniques for gene assay. Analyst. 2010;135:1499–1518. doi: 10.1039/c000037j. [DOI] [PubMed] [Google Scholar]
- 3.Selvaganapathy PR, Carlen ET, Mastrangelo CH. Recent progress in microfluidic devices for nucleic acid and antibody assays. Proc IEEE. 2003;91:954–975. [Google Scholar]
- 4.Noh J, Kim HC, Chung TD. Biosensors in microfluidic chips. Top Curr Chem. 2011;304:117–152. doi: 10.1007/128_2011_143. [DOI] [PubMed] [Google Scholar]
- 5.Hervás M, López MA, Escarpa A. Electrochemical immunosensing on board microfluidic chip platforms. TrAC - Trend Anal Chem. 2012;31:109–128. [Google Scholar]
- 6.Mir M, Homs A, Samitier J. Integrated electrochemical DNA biosensors for lab-on-a-chip devices. Electrophoresis. 2009;30:3386–3397. doi: 10.1002/elps.200900319. [DOI] [PubMed] [Google Scholar]
- 7.Sassa F, Morimoto K, Satoh W, Suzuki H. Electrochemical techniques for microfluidic applications. Electrophoresis. 2008;29:1787–1800. doi: 10.1002/elps.200700581. [DOI] [PubMed] [Google Scholar]
- 8.Amatore C, Da Mota N, Sella C, Thouin L. General concept of high-performance amperometric detector for microfluidic (bio)analytical chips. Anal Chem. 2008;80:4976–4985. doi: 10.1021/ac800227t. [DOI] [PubMed] [Google Scholar]
- 9.Chen SP, Wu J, Yu XD, Xu JJ, Chen HY. Preparation of metal nanoband microelectrodeon poly(dimethylsiloxane) for chip-based amperometric detection. Anal Chim Acta. 2010;665:152–159. doi: 10.1016/j.aca.2010.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Do J, Ahn CH. A polymer lab-on-a-chip for magnetic immunoassay with on-chip sampling and detection capabilities. Lab Chip. 2008;8:542–549. doi: 10.1039/b715569g. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Bhattacharya S, Chen X, Barizuddin S, Gangopadhyay S, Gillis KD. A microfluidic cell trap device for automated measurement of quantal catecholamine release from cells. Lab Chip. 2009;9:3442–3446. doi: 10.1039/b913216c. [DOI] [PubMed] [Google Scholar]
- 12.Ordeig O, Ortiz P, Muñoz-Berbel X, Demming S, Büttgenbach S, Fernández-Sánchez C, Llobera A. Dual photonic-electrochemical lab on a chip for online simultaneous absorbance and amperometric measurements. Anal Chem. 2012;84:3546–3553. doi: 10.1021/ac203106x. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Luo J, Chen H, He Q, Gan N, Li T. A microchip-based flow injection-amperometry system with mercaptopropionic acid modified electroless gold microelectrode for the selective determination of dopamine. Anal Chim Acta. 2008;625:180–187. doi: 10.1016/j.aca.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi A, Jin P, Tsuchiyama H, Masuda T, Sun K, Matsuo S, Misawa H. Rapid fabrication of electrochemical enzyme sensor chip using polydimethylsiloxane microfluidic channel. Anal Chim Acta. 2002;468:143–152. [Google Scholar]
- 15.Niwa O, Morita M, Tabei H. Electrochemical behavior of reversible redox species at interdigitated array electrodes with different geometries: consideration of redox cycling and collection efficiency. Anal Chem. 1990;62:447–452. [Google Scholar]
- 16.Hintsche R, Paeschke M, Woflenberger U, Schnakenberg U, Wagner B, Lisec T. Microelectrode arrays and application to biosensing devices. Biosens Bioelectron. 1994;9:697–705. [Google Scholar]
- 17.Heo JI, Shim DS, Teixidor GT, Oh S, Madou MJ, Shin H. Carbon Interdigitated Array Nanoelectrodes for Electrochemical Applications. J Electrochem Soc. 2011;158:J76–J80. [Google Scholar]
- 18.Min J, Baeumner AJ. Characterization and optimization of interdigitated ultramicroelectrode arrays as electrochemical biosensor transducers. Electroanalysis. 2004;16:724–729. [Google Scholar]
- 19.Goral VN, Zaytseva NV, Baeumner AJ. Electrochemical microfluidic biosensor for the detection of nucleic acid sequences. Lab Chip. 2006;6:414–421. doi: 10.1039/b513239h. [DOI] [PubMed] [Google Scholar]
- 20.Kwakye S, Goral VN, Baeumner AJ. Electrochemical microfluidic biosensor for nucleic acid detection with integrated minipotentiostat. Biosens Bioelectron. 2006;21:2217–2223. doi: 10.1016/j.bios.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Bunyakul N, Edwards KA, Promptmas C, Baeumner AJ. Cholera toxin subunit B detection in microfluidic devices. Anal Bioanal Chem. 2009;393:177–186. doi: 10.1007/s00216-008-2364-6. [DOI] [PubMed] [Google Scholar]
- 22.Nugen SR, Asiello PJ, Connelly JT, Baeumner AJ. PMMA biosensor for nucleic acids with integrated mixer and electrochemical detection. Biosens Bioelectron. 2009;24:2428–2433. doi: 10.1016/j.bios.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Benhabib M, Chiesl TN, Stockton AM, Seherer JR, Mathies RA. Multichannel capillary electrophoresis microdevice and instrumentation for in situ planetary analysis of organic molecules and biomarkers. Anal Chem. 2010;82:2372–2379. doi: 10.1021/ac9025994. [DOI] [PubMed] [Google Scholar]
- 24.Chen SP, Wu J, Yu XD, Xu JJ, Chen HY. Multi-parameter detection of diabetes mellitus on multichannel poly(dimethylsiloxane) analytical chips coupled with nanoband microelectrode arrays. Electrophoresis. 2010;31:3097–3106. doi: 10.1002/elps.201000181. [DOI] [PubMed] [Google Scholar]
- 25.Ding X, Lin SCS, Lapsley MI, Li S, Guo X, Chan CY, Chiang IK, Wang L, McCoy JP, Huang TJ. Standing surface acoustic wave (SSAW) based multichannel cell sorting. Lab Chip. 2012;12:4228–4231. doi: 10.1039/c2lc40751e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Shen Z, Wang H, Dai Z, Lin B. Chiral separations on multichannel microfluidic chips. Electrophoresis. 2005;26:4774–4779. doi: 10.1002/elps.200500283. [DOI] [PubMed] [Google Scholar]
- 27.Irawan R, Tjin SC, Fang X, Fu CY. Integration of optical fiber light guide, fluorescence detection system, and multichannel disposable microfluidic chip. Biomed Microdevices. 2007;9:413–419. doi: 10.1007/s10544-007-9052-8. [DOI] [PubMed] [Google Scholar]
- 28.Kim SJ, Gobi KV, Iwasaka H, Tanaka H, Miura N. Novel miniature SPR immunosensor equipped with all-in-one multi-microchannel sensor chip for detecting low-molecular-weight analytes. Biosens Bioelectron. 2007;23:701–707. doi: 10.1016/j.bios.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Pan X, Jiang L, Liu K, Lin Bingcheng B, Qin Jianhua J. A microfluidic device integrated with multichamber polymerase chain reaction and multichannel separation for genetic analysis. Anal Chim Acta. 2010;674:110–115. doi: 10.1016/j.aca.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Shadpour H, Hupert ML, Patterson D, Liu C, Galloway M, Stryjewski W, Goettert J, Soper SA. Multichannel microchip electrophoresis device fabricated in polycarbonate with an integrated contact conductivity sensor array. Anal Chem. 2007;79:870–878. doi: 10.1021/ac0612168. [DOI] [PubMed] [Google Scholar]
- 31.Siew Bang C, Skinner CD, Taylor J, Attiya S, Lee WE, Picelli G, Harrison DJ. Development of a multichannel microfluidic analysis system employing affinity capillary electrophoresis for immunoassay. Anal Chem. 2001;73:1472–1479. doi: 10.1021/ac0007938. [DOI] [PubMed] [Google Scholar]
- 32.Wu MS, Shi HW, He LJ, Xu JJ, Chen HY. Microchip device with 64-site electrode array for multiplexed immunoassay of cell surface antigens based on electrochemiluminescence resonance energy transfer. Anal Chem. 2010;84:4207–4213. doi: 10.1021/ac300551e. [DOI] [PubMed] [Google Scholar]
- 33.Ymeti A, Kanger JS, Greve J, Besselink GAJ, Lambeck PV, Wijn R, Heideman RG. Integration of microfluidics with a four-channel integrated optical Young interferometer immunosensor. Biosens Bioelectron. 2005;20:1417–1421. doi: 10.1016/j.bios.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Moreira NH, De Jesus De Almeida AL, De Oliveira Piazzeta MH, De Jesus DP, Deblire A, Gobbi AL, Fracassi Da Silva JA. Fabrication of a multichannel PDMS/glass analytical microsystem with integrated electrodes for amperometric detection. Lab Chip. 2009;9:115–121. doi: 10.1039/b807409g. [DOI] [PubMed] [Google Scholar]
- 35.Nugen SR, Asiello PJ, Baeumner AJ. Design and fabrication of a microfluidic device for near-single cell mRNA isolation using a copper hot embossing master. Microsyst Technol. 2009;15:477–483. [Google Scholar]
- 36.Connelly JT, Nugen SR, Borejsza-Wysocki W, Durst RA, Montagna RA, Baeumner AJ. Human pathogenic Cryptosporidium species bioanalytical detection method with single oocyst detection capability. Anal Bioanal Chem. 2008;391:487–495. doi: 10.1007/s00216-008-1967-2. [DOI] [PubMed] [Google Scholar]
- 37.Baeumner AJ, Humiston MC, Montagna RA, Durst RA. Detection of viable oocysts of Cryptosporidium parvum following nucleic acid sequence-based amplification. Anal Chem. 2001;73:1176–1180. doi: 10.1021/ac001293h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.