Abstract
Eukaryotic cells compartmentalize biochemical reactions into membrane-enclosed organelles that must be faithfully propagated from one cell generation to the next. Transport and retention processes balance the partitioning of organelles between mother and daughter cells. Here we report the identification of an ER-peroxisome tether that links peroxisomes to the ER and ensures peroxisome population control in the yeast Saccharomyces cerevisiae. The tether consists of the peroxisome biogenic protein, Pex3p, and the peroxisome inheritance factor, Inp1p. Inp1p bridges the two compartments by acting as a molecular hinge between ER-bound Pex3p and peroxisomal Pex3p. Asymmetric peroxisome division leads to the formation of Inp1p-containing anchored peroxisomes and Inp1p-deficient mobile peroxisomes that segregate to the bud. While peroxisomes in mother cells are not released from tethering, de novo formation of tethers in the bud assists in the directionality of peroxisome transfer. Peroxisomes are thus stably maintained over generations of cells through their continued interaction with tethers.
Keywords: endoplasmic reticulum, organelle inheritance, organelle tether, protein complex, peroxisome
Introduction
Eukaryotic cells segregate biochemical functions into discrete membrane-enclosed organelles. To maintain the benefits of compartmentalization, cells have evolved elaborate mechanisms to faithfully transmit their organelles to future generations. The budding yeast Saccharomyces cerevisiae is well suited for the study of organelle inheritance, as yeast cells divide asymmetrically and therefore actively deliver their organelles to the growing bud. Transport of organelles occurs along the cytoskeletal tracks and is powered by molecular motors that connect to their cargo through organelle-specific adaptors. Movement of part of an organelle’s population to the bud is balanced by active retention of the remaining organelles in the mother cell. While common principles govern the partitioning of all organelles, inheritance factors that are specific for each type of organelle allow the cell to differentially regulate individual organelle populations (Fagarasanu et al, 2010).
Peroxisomes are organelles involved in the β-oxidation of fatty acids and the detoxification of reactive oxygen species. Peroxisomes are a specialized branch of the secretory pathway and have the ability to regenerate from the ER in cells that have lost their complement of peroxisomes (Hoepfner et al, 2005; Schekman, 2005). While the total number of peroxisomes in a cell is a result of both de novo formation and division of existing peroxisomes, the relative contribution of each of these processes is not equal in all cell types. In mammalian cells, an increase in peroxisome numbers is due primarily to the formation of new peroxisomes by ER-dependent pathways (Kim et al, 2006). In yeast cells, peroxisomes are routinely not made de novo but instead are inherited by the next cell generation. With each round of cell division, peroxisomes are duplicated and separated equitably between mother cell and bud (Motley and Hettema, 2007).
Unlike a single-copy organelle whose division is coupled to cell cycle progression, peroxisomes could divide either synchronously or asynchronously with the cell cycle. In vps1Δ and vps1Δ/dnm1Δ mutants lacking dynamin-like GTPases required for the final fission step of peroxisome division, the peroxisomal compartment collapses into a single organelle. This peroxisome is broken apart and apportioned between mother and daughter cells at cytokinesis (Hoepfner et al, 2001; Kuravi et al, 2006). But do peroxisomes divide in wild-type cells when they are partitioned? Yeast cells contain two types of peroxisomes, those that are mobile and those that are anchored to the cell cortex. During cell division, some peroxisomes are mobilized and travel to the bud, whereas others do not shift from their fixed cortical positions and are thus retained in the mother cell. The identity of the structure to which peroxisomes tether is unknown, but it is believed to be extensive, as peroxisomes scatter over the cell periphery. Understanding how peroxisomes anchor at the cell cortex is crucial to unravel how they transition from a fixed to a mobile state and whether peroxisome division contributes to this process.
The inheritance factors Inp1p and Inp2p, as well as the peroxin Pex3p (Fagarasanu et al, 2005, 2006; Chang et al, 2009; Munck et al, 2009), regulate peroxisome partitioning. Inp1p immobilizes peroxisomes at the cell cortex (Fagarasanu et al, 2005). In cells lacking Inp1p, all peroxisomes are mobile and eventually transported to the bud. Consistent with its role in peroxisome retention, overproduction of Inp1p causes peroxisomes to remain anchored at fixed cortical positions in the mother cell, thus preventing their inheritance. Notably, overproduced Inp1p decorates both peroxisomes and the cell cortex, suggesting that Inp1p has an affinity for a structure lining the cell periphery. Inp1p has also been implicated in the control of peroxisome abundance; however, it remains unknown how the two functions of Inp1p are linked (Fagarasanu et al, 2005).
Pex3p is a key regulator of peroxisome dynamics that controls peroxisome formation via interaction with Pex19p (Hettema et al, 2000; Fang et al, 2004), peroxisome retention via interaction with Inp1p (Munck et al, 2009), and peroxisome segregation via interaction with Myo2p (Chang et al, 2009).
Bud-directed motility of peroxisomes is accomplished by the actin-based class V myosin motor, Myo2p, which attaches to peroxisomes by recognizing its peroxisome-specific adaptor, Inp2p, on the surface of the organelle (Fagarasanu et al, 2006). In the yeast Yarrowia lipolytica, bud-directed motility of peroxisomes is carried out by Pex3Bp, one of two members of the Pex3 family of peroxisome biogenic factors (Chang et al, 2009).
Here we report the identification of a protein complex containing the peroxin Pex3p and the inheritance factor Inp1p, which together form an ER-peroxisome junction that tethers the two organelles at discrete foci at the cell cortex. The integral membrane protein Pex3p is present in both compartments and provides a membrane anchor for the tether, whereas Inp1p acts as a molecular hinge by connecting ER-bound Pex3p with peroxisomal Pex3p. We delineate mechanisms by which this tether assists in the maintenance of stable peroxisome numbers in a growing cell population. Our work demonstrates an intimate linkage between peroxisome biogenesis and retention through a division of labour by Pex3p.
Results
A Pex3p-point mutant defective in peroxisome retention
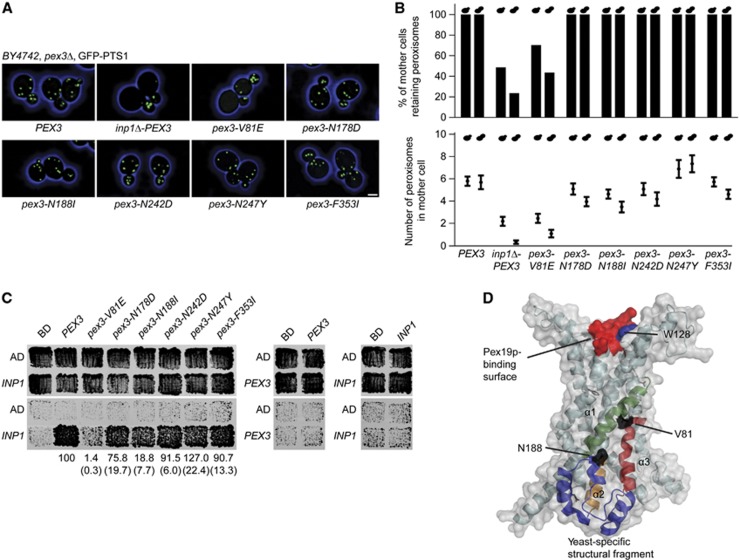
S. cerevisiae cells expressing the mutant allele pex3-1 exhibit a peroxisome segregation defect similar to that of inp1Δ cells, in which peroxisomes are depleted from mother cells. This defect of pex3-1 cells has been attributed to a failure of Pex3p to recruit Inp1p to the peroxisomal membrane (Munck et al, 2009). We functionally dissected the pex3-1 allele to examine the nature of the Inp1p–Pex3p interaction. Six pex3-mutant alleles, each encoding a single amino-acid change present in Pex3p-1, were introduced on centromeric plasmids into a pex3Δ strain expressing GFP-PTS1 as a peroxisomal reporter. All mutant alleles complemented the peroxisome biogenesis defect of the pex3Δ strain (Figure 1A). One mutant, pex3-V81E, exhibited a peroxisome retention defect comparable to that of an inp1Δ strain (Figure 1B). In contrast, cells expressing wild-type PEX3 or the other pex3-mutant alleles always retained peroxisomes in the mother cell, that is, they never lost their entire peroxisome population to the bud (Figure 1A and B).
Figure 1.
Cells expressing pex3-V81E have a defect in peroxisome retention. (A) Strain BY4742 lacking the PEX3 gene and expressing the peroxisomal reporter GFP-PTS1 was transformed with plasmids encoding either wild-type PEX3 or mutant pex3 sequences. The strain inp1Δ-PEX3 also carried a deletion of the INP1 gene. Images were acquired by confocal fluorescence microscopy and flattened into maximum intensity projections. Bar, 1 μm. (B) Mother cells were scored for the presence or absence of peroxisomes (upper panel) or total peroxisome numbers (mean±s.e.m., lower panel). Small and large bud size categories are presented in the left and right bars. Quantification was done on at least 100 budded cells of each strain. (C) Yeast two-hybrid analysis to score for interaction between Inp1p and Pex3p. Upper panels show total growth of strains, while bottom panels show growth arising from protein interaction. Strength of interaction between mutant Pex3 proteins and Inp1p in β-galactosidase assays is presented as mean±s.e.m. (brackets) of three independent experiments. (D) Model of Pex3p. Secondary elements likely involved in interacting with Inp1p are highlighted (helices α1, green; α2, orange; and α3, red; yeast-specific structure, blue). The surface of the model is displayed in grey, while the mutated residues V81 and N188 are in black. The approximate binding site for Pex19p is shown in red. Mutagenesis of W128 is described below (Figure 5). See Supplementary Movie 1.
We performed yeast two-hybrid analysis to determine if the Pex3p mutants were compromised in their ability to interact with Inp1p. Neither Pex3p nor Inp1p self-interacted (Figure 1C). Strong interaction between wild-type Pex3p and Inp1p contrasted with near lack of interaction between Pex3p-V81E and Inp1p. Pex3p-N188I was also compromised in its interaction with Inp1p, while the other mutants were not adversely affected (Figure 1C).
The crystal structure of the cytosolic tail of human Pex3p in complex with a short α-helical fragment of Pex19p has been solved (Sato et al, 2010; Schmidt et al, 2010). Using this information, we modelled yeast Pex3p to position the amino acids whose mutation causes a defect in its interaction with Inp1p. V81 is found at a surface-exposed location in α-helix 1 (Figure 1D). N188 is present in a yeast-specific structure (Figure 1D, blue helices) in proximity to α-helix 1, which may, together with α-helices 1, 2, and 3, form a surface patch required for interaction with Inp1p. N242, N247, and F353 are on the opposite side of Pex3p (Supplementary Movie 1).
For further analysis, we introduced the pex3-V81E point mutation into the genomic locus of PEX3 by in vivo site-directed mutagenesis (Storici et al, 2001).
Pex3p and Inp1p interact at the ER
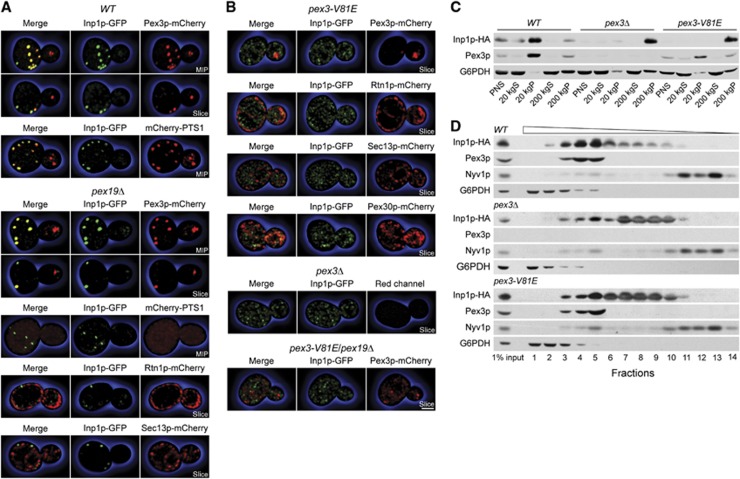
We examined the localization of Inp1p and Pex3p to unravel how these proteins orchestrate the tethering of peroxisomes to the cell cortex. Confocal fluorescence microscopy showed that Inp1p and Pex3p targeted to discrete foci in wild-type cells (Figure 2A). In maximum intensity projections, Inp1p colocalized with Pex3p in the mother cell but was mostly excluded from the bud. Optical sections showed Inp1p and Pex3p in close proximity at the cell periphery of wild-type mother cells. Inp1p also colocalized with the peroxisomal matrix protein mCherry-PTS1, thus demonstrating that Inp1p foci overlap with peroxisomes in wild-type cells (Figure 2A).
Figure 2.
Pex3p and Inp1p segregate to distinct membranous compartments. (A, B) Wild-type BY4742 cells (WT), as well as pex19Δ, pex3-V81E, pex3Δ, and pex19Δ/pex3-V81E mutant cells, were imaged by confocal fluorescence microscopy. Cells expressed Inp1p-GFP and one of Pex3p-mCherry, mCherry-PTS1, Sec13p-mCherry, Pex30p-mCherry, and Rtn1p-mCherry. Maximum intensity projections (MIP) and optical sections (slice) are shown. Bar, 1 μm. (C) PNS from WT, pex3Δ, and pex3-V81E cells was separated by differential centrifugation into 20 and 200 kg supernatant and pellet fractions. Equal amounts of protein were resolved by SDS–PAGE, and immunoblots were probed with antibodies against HA, Pex3p, and G6PDH. (D) Vesicles in PNS prepared from the same strains as in (C) were floated in a step gradient of sucrose solutions of decreasing density. Equal portions of fractions were analysed by immunoblotting for the indicated proteins. Wedge depicts fraction density.
Source data for this figure is available on the online supplementary information page.
Pex19p is indispensable for peroxisome formation (Götte et al, 1998), as Pex3p cannot exit the ER in a pex19Δ mutant (Hoepfner et al, 2005). In pex19Δ cells, Inp1p and Pex3p formed foci at the ER that resemble closely the foci seen in wild-type cells (Figure 2A). The foci do not correspond to functional peroxisomes in pex19Δ cells, as is evident by the cytosolic mislocalization of mCherry-PTS1 (Figure 2A). These foci were found in close proximity to the cortical ER (cER) marker, Rtn1p, but were not overlapping with ER exit sites (as decorated by Sec13p, Figure 2A, bottom panel).
Interaction between Pex3p and Inp1p is impaired in pex3-V81E cells. The distribution of Inp1p in this mutant and in pex3Δ cells, which lack peroxisomes, was considerably different from that observed in cells containing wild-type Pex3p (Figure 2B). In pex3-V81E cells, Inp1p did not target to foci but was present in diffuse reticular elements extending over the entire cell. Pex3p was found in puncta that were predominantly localized to the bud, consistent with the peroxisome retention defect observed in this strain. Inp1p was not recruited to these puncta. Inp1p in pex3-V81E cells displayed partial overlap with the polytopic membrane protein Pex30p, which has previously been shown to target both to the ER and peroxisomes (Yan et al, 2008), but did not colocalize with other ER markers. In a pex3-V81E/pex19Δ double mutant, neither Inp1p nor Pex3p sorted to foci but instead were present in non-overlapping reticular elements (Figure 2B, bottom panel).
Pex3p and Inp1p segregate to distinct membrane compartments in pex3-V81E cells
The diffuse distribution of Inp1p in pex3-V81E cells could indicate that Inp1p is either cytosolic or that it associates with a non-peroxisomal membrane. We performed biochemical analyses to differentiate between these possibilities. Postnuclear supernatant (PNS) from wild-type, pex3Δ, and pex3-V81E cells was separated into low-speed (20 000 g) and high-speed (200 000 g) supernatant and pellet fractions. Pex3p enriched in the 20 kgP fraction and to a lesser extent in the 200 kgP fraction in samples from wild-type and pex3-V81E cells (Figure 2C). Inp1p cosedimented with Pex3p and was found predominantly in the 20 kgP fraction of wild-type cells. However, in pex3Δ and pex3-V81E mutants, Inp1p enriched in the 200 kgP fraction and thus did not exhibit the sedimentation profile of either a peroxisomal protein or of a cytosolic protein (glucose-6-phosphate dehydrogenase, (G6PDH)) (Figure 2C).
Sedimentation of Inp1p at 200 000 g could be due to either aggregation or its presence in a small vesicle. If Inp1p is membrane associated, it should exhibit low buoyant density, enabling it to float in sucrose gradients. In flotation-gradient centrifugation of samples from wild-type cells, Inp1p peaked with Pex3p in fractions of intermediate density, but a significant portion of Inp1p was also present in fractions of lower density (Figure 2D). Pex3p and Inp1p were separated from the vacuolar protein Nyv1p at the top and the cytosolic G6PDH at the bottom of the gradients. In samples from pex3-V81E and pex3Δ mutants, Inp1p shifted to fractions of lower density (Figure 2D). Inp1p and Pex3p thus segregate into separate membrane compartments in the pex3-V81E mutant.
Inp1p anchors peroxisomes in trans
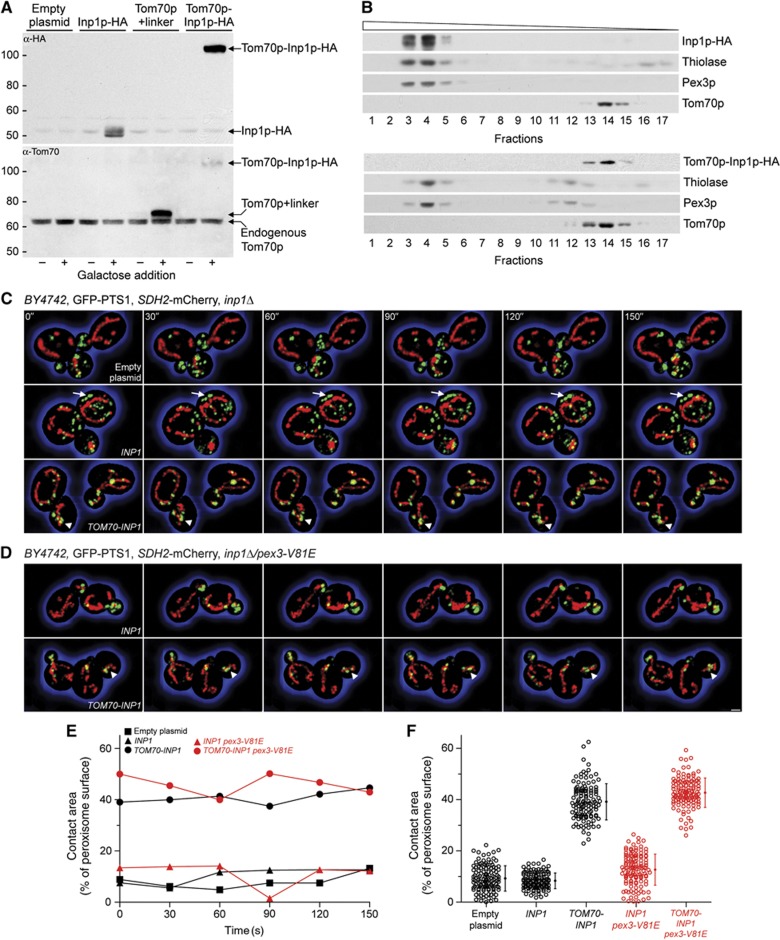
Pex3p and Inp1p have been reported to interact in cis at the peroxisomal membrane (Munck et al, 2009). As Pex3p is integral to both the ER and the peroxisome, we also tested for possible interaction between Inp1p and Pex3p in trans. We ectopically expressed Inp1p on the surface of the mitochondria as a fusion to the outer mitochondrial membrane protein Tom70p, and queried whether under these conditions peroxisomes tether to the mitochondria. Using a galactose-inducible expression system, we produced Inp1p-HA, Tom70p+linker (the N-terminal portion of the fusion construct), and Tom70p-Inp1p-HA in cells (Figure 3A).
Figure 3.
Ectopic expression of Inp1p on the surface of the mitochondria tethers peroxisomes to mitochondria. (A) Production of Inp1p-HA, Tom70p+linker, and Tom70p-Inp1p-HA from the GAL1 promoter. Samples were collected before and 2 h after galactose addition. Immunoblots of whole-cell lysates were probed with antibodies against HA and Tom70p. Endogenous Tom70p is an internal loading control. Numbers at left represent molecular mass markers. (B) Cells transformed with plasmids coding for Inp1p-HA (top panel) or Tom70p-Inp1p-HA (bottom panel) were grown for 15 h in oleate-containing medium. Recombinant proteins were induced for 90 min prior to fractionation. The peroxisomal and mitochondrial fractions were identified by immunodetection of thiolase, Pex3p, and Tom70p, respectively. Wedge depicts fraction density. (C, D) inp1Δ (C) and inp1Δ/pex3-V81E (D) cells expressing GFP-PTS1 and Sdh2p-mCherry were transformed with empty plasmid or plasmid expressing INP1 or TOM70-INP1 (inserts), and transgenes were induced for 30 min. Six consecutive frames from a time-lapse series are shown. Arrows depict static peroxisomes. Arrowheads show clumped peroxisomes and mitochondria. Bar, 1 μm. (E) The peroxisomal and mitochondrial surfaces were computed using Imaris software. Contact area is expressed as a percentage of the total peroxisome surface. Quantification was done on the images presented in (C) and (D). (F) Association of peroxisomes with the mitochondria. Recordings of all individual time points and their means±s.e.m. are presented. See Supplementary Movies 2–4.
Source data for this figure is available on the online supplementary information page.
Targeting of the fusion proteins was examined by fractionation. Isopycnic density gradient centrifugation of 20 kgP fractions revealed cofractionation of Inp1p-HA with Pex3p and the peroxisomal enzyme thiolase but not with Tom70p. Tom70p-Inp1p-HA exhibited the opposite pattern, being present in the mitochondrial, but excluded from the peroxisomal fractions (Figure 3B).
Peroxisome dynamics were recorded by 3D time-lapse confocal video microscopy in inp1Δ cells expressing GFP-PTS1 and succinate dehydrogenase 2-mCherry (Sdh2p-mCherry) as peroxisomal and mitochondrial reporters. To avoid overexpression of recombinant proteins due to high-level transcription from the GAL1 promoter, we exposed cells to a 30-min pulse of galactose followed by a chase in glucose medium. When cells were transformed with empty plasmid, we observed the characteristic inp1Δ phenotype of mobile peroxisomes that were transported to the bud (Figure 3C; Supplementary Movie 2, top panel). Expression of INP1 immobilized peroxisomes at the cell cortex (Figure 3C, arrows) and led to an increase in peroxisome numbers (Figure 3C; Supplementary Movie 2, middle panel). In cells expressing Tom70p-Inp1p, peroxisomes no longer tethered to the cell cortex but instead attached to the mitochondria (Figure 3C; Supplementary Movie 2, bottom panel). Sometimes peroxisomes and mitochondria clumped (Figure 3C and D, arrowheads) or peroxisomes appeared ‘to roll’ along the surface of the mitochondria (Supplementary Movies 2–4). Only during bud growth did peroxisomes occasionally detach from their mitochondrial tethers and enter the bud independently of mitochondria, but they were recaptured as soon as a mitochondrion entered the bud (Supplementary Movies 2 and 4, lower panels). Expression of Inp1p with a C-terminal mitochondrial membrane anchor in the Inp1p-Tom22p chimera also led to peroxisome tethering to the mitochondria (Supplementary Movie 3).
When Inp1p was introduced into the inp1Δ/pex3-V81E strain, peroxisomes remained mobile and were transferred to buds (Figure 3D; Supplementary Movie 4, upper panel). Expression of Inp1p alone therefore could not rescue the inp1Δ phenotype of the pex3-V81E mutant. In contrast, when Tom70p-Inp1p was expressed in inp1Δ/pex3-V81E cells, peroxisomes tethered to the mitochondria (Figure 3D; Supplementary Movie 4, lower panel). Quantification revealed an approximately four-fold increase in the association between the peroxisomes and mitochondria in the presence of Tom70p-Inp1p regardless of whether the cells expressed Pex3p or Pex3p-V81E (Figure 3E and F).
Inp1p binds Pex3p at both its N- and C-terminal domains
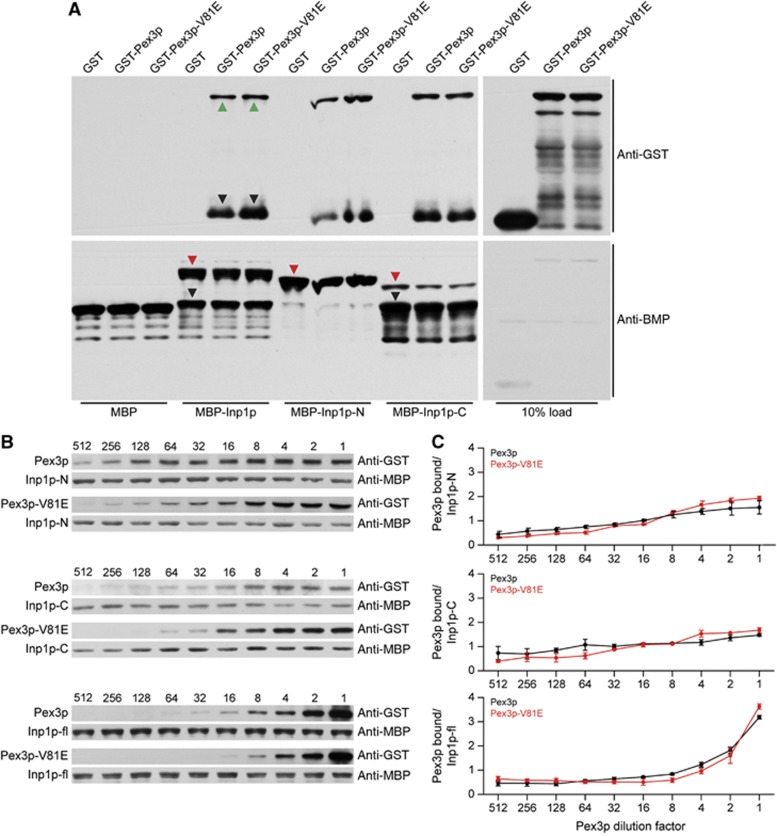
The ability of Pex3p-V81E to function as efficiently as Pex3p in tethering peroxisomes to the mitochondria was unexpected, as Inp1p and Pex3p-V81E do not interact by yeast two-hybrid assay (Figure 1C). Wild-type Pex3p and Inp1p were previously shown to bind each other directly (Munck et al, 2009). In vitro binding assays between glutathione S-transferase-Pex3p-V81E (GST-Pex3p-V81E) or GST-Pex3p and maltose binding protein–Inp1p (MBP–Inp1p) fusion proteins made in E. coli showed that full-length Inp1p bound Pex3p and Pex3p-V81E equally well (Figure 4A).
Figure 4.
The V81 residue of Pex3p is not required for direct binding of Inp1p. (A) MBP alone or MBP–Inp1p fusions immobilized to amylose beads were incubated with extracts of E. coli synthesizing GST, GST-Pex3p, or GST-Pex3p-V81E. Bound proteins were detected by immunoblotting with anti-GST antibody (upper panel). Total MBP fusion proteins were visualized by immunoblotting with anti-MBP antibody (lower panel). Inp1p-N and Inp1p-C are N-terminal (a.a. 1–280) and C-terminal (a.a. 281–420) fragments of Inp1p. Red triangles indicate full-length MBP fusions, green triangles indicate full-length GST fusions, and black triangles indicate degradation products. (B) Equimolar amounts of purified recombinant MBP–Inp1p, MBP–Inp1p-N, and MBP–Inp1p-C were coupled individually to amylose beads and incubated with serial dilutions of E. coli lysates containing GST-Pex3p or GST-Pex3p-V81E. Dilution factors are denoted above immunoblots. GST- and MBP-fusion proteins were detected as in (A). (C) The ratio of bound Pex3p or bound Pex3p-V81E to Inp1p obtained by densitometric analysis of the bands in (B) was plotted against the Pex3p-dilution factor. The means±s.e.m. of three independent experiments are presented.
Source data for this figure is available on the online supplementary information page.
Secondary structure analysis ( http://www.predictprotein.org/) predicts Inp1p to be a modular protein comprised of globular N- and C-terminal domains separated by a flexible loop rich in aspartate residues. The termini of Inp1p (Inp1p-N and Inp1p-C) also individually bound Pex3p and Pex3p-V81E (Figure 4A).
We incubated serial dilutions of E. coli lysates containing either Pex3p or Pex3p-V81E with defined amounts of immobilized Inp1p, Inp1p-N, or Inp1p-C (Figure 4B). Binding of Pex3p to Inp1p-N and Inp1p-C began at low concentrations of Pex3p and increased linearly with increasing concentration of Pex3p. Conversely, no binding to full-length Inp1p was detected at low Pex3p concentrations, but strongly increased at higher concentrations. Quantification showed that full-length Inp1p can bind at least twice as much Pex3p than either its N- or C-terminal domain individually (Figure 4C). Our data suggest that the N- and C-termini of Inp1p cooperate in binding Pex3p and, moreover, that the region of Pex3p around V81 does not directly bind Inp1p, as otherwise Pex3p-V81E would consistently fail to interact with either full-length Inp1p or one of its domains.
Inp1p foci are docking sites for peroxisomes at the ER membrane
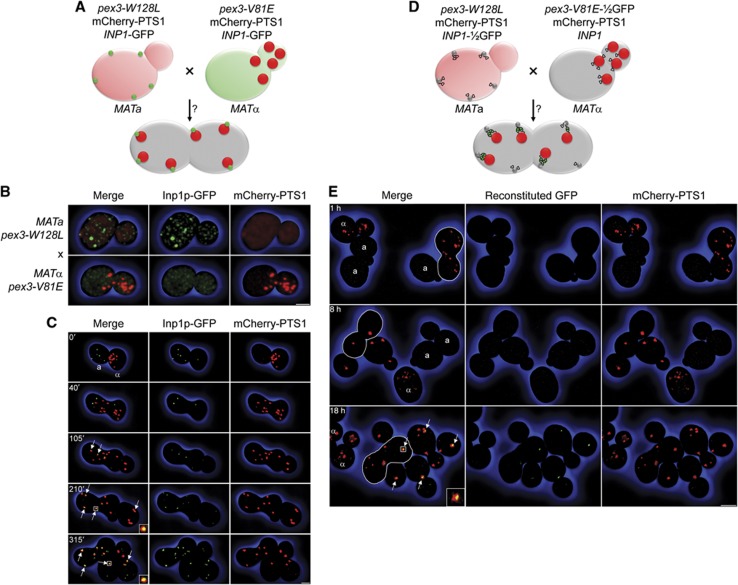
Reasoning that Inp1p-containing cortical foci, which form independently of peroxisome biogenesis (Figure 2A), could represent docking sites for peroxisomes at the ER membrane, we dissected genetically the peroxisome assembly and tethering functions of Pex3p. We mated the peroxisome biogenic but retention deficient pex3-V81E strain with a second strain expressing a Pex3p-point mutant capable of recruiting Inp1p to foci but not of exiting the ER due to its inability to bind Pex19p, and tested whether peroxisome tethering was reconstituted in the diploid cell (Figure 5A).
Figure 5.
Inp1p acts as a molecular hinge between ER and peroxisomes. (A) Mating assay. Cells expressing mCherry-PTS1 and Inp1p-GFP and either Pex3p-W128L (MATa) or Pex3p-V81E (MATα) were mated to evaluate reconstitution of peroxisome tethering in the diploid cell. (B) Haploid cells used for mating in (A). Bar, 1 μm. (C) Time-lapse series of images of cells mated as depicted in (A). Time 0′ denotes cell fusion. MATa and MATα cells are labelled. Arrows highlight tethered peroxisomes in the zygote (105′) and its progeny (210′, 315′). Inserts show tethering complexes at high magnification. Bar, 3 μm. (D) Combined mating and split-GFP assay. Cells expressing mCherry-PTS1 and either Pex3p-W128L and Inp1p-½GFP (MATa) or Pex3p-V81E-½GFP and Inp1p (MATα) were mated to evaluate reconstitution of GFP fluorescence via interaction between Inp1p-½GFP in foci and Pex3p-V81E-½GFP on the surface of peroxisomes. (E) Images of cells at different times following the mating depicted in (D). Haploid cells are designated a and α. Zygotes are outlined. Diploid cells are unlabelled. Arrows highlight reconstitution of GFP fluorescence in tethering complexes. The insert shows a tethering complex at high magnification. Bar, 3 μm. See Supplementary Movie 5.
Tryptophan 104 of human Pex3p is found in an α-helix in its Pex19p-binding region (Supplementary Figure S1). Its mutagenesis to alanine abrogates Pex19p binding and peroxisome formation (Sato et al, 2008). Mutagenesis of the homologous residue W128 of S. cerevisiae Pex3p (Figure 1D) to alanine likewise resulted in a peroxisome biogenesis defect but also eliminated formation of Inp1p foci. However, a mutant carrying a pex3-W128L allele could still recruit Inp1p. Consequently, pex3-W128L cells mislocalized the peroxisomal reporter mCherry-PTS1 to the cytosol but formed Inp1p foci, whereas pex3-V81E cells sequestered mCherry-PTS1 in peroxisomes but mislocalized Inp1p (Figure 5B).
Mating between pex3-W128L (MATa) and pex3-V81E (MATα) cells was recorded by live-cell imaging (Figure 5C). At cell fusion (0 min), each haploid cell contained either Inp1p foci (pex3-W128L) or peroxisomes (pex3-V81E), which initially remained separate in the zygote (40 min). Upon bud emergence, several peroxisomes were recruited to foci (105 min, arrows). Later, peroxisomes tethered to foci in not only the zygote but also its progeny (210 min, 315 min, arrows). Equal distribution of peroxisomes between mother and daughter cells confirmed that peroxisome retention had been restored (Figure 5C; Supplementary Movie 5).
Inp1p connects ER-bound Pex3p with peroxisomal Pex3p into an ER-peroxisome tether
Our observation that the N- and C-termini of Inp1p can bind Pex3p independently (Figure 4) raised the possibility that Inp1p might link Pex3p molecules across two membranes. We used bimolecular fluorescence complementation (Wilson et al, 2004) to determine if Inp1p forms a molecular bridge between ER-bound Pex3p and peroxisomal Pex3p. Tagging the endogenous INP1 and PEX3 genes at their 3′ ends with sequence coding for the N- and C-terminal portions of GFP, respectively, we first confirmed the previously reported reconstitution of GFP fluorescence at sites overlapping peroxisomes (Munck et al, 2009). However, we could not ascertain whether Inp1p interacted with ER-bound or peroxisomal Pex3p or both in wild-type cells. We tested for binding in trans between Inp1p at foci and Pex3p-V81E at the peroxisomal membrane by mating cells expressing mCherry-PTS1 and either Pex3p-V81E-½-GFP (MATα), or Pex3p-W128L and Inp1p-½-GFP (MATa) (Figure 5D). ½-GFP molecules do not fluoresce until brought together by interaction of the proteins to which they are fused. No GFP signal was therefore detected in haploid cells or early after cell mating (Figure 5E). Later, GFP fluorescence appeared specifically at peroxisomes in both the zygote and its progeny (Figure 5E, arrows). Peroxisome tethering in the diploid cell was thus restored through interaction between Pex3p-W128L and Inp1p at the ER membrane and interaction between Inp1p and Pex3p-V81E at the peroxisomal membrane. We conclude that Inp1p bridges ER-bound Pex3p and peroxisomal Pex3p into an ER–peroxisome tethering complex.
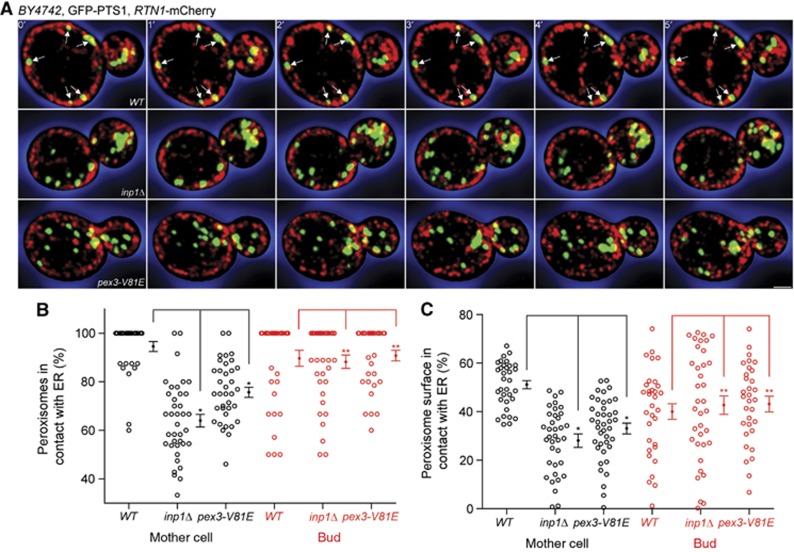
Contacts between peroxisomes and the ER are transient in inp1Δ and pex3-V81E cells
Having identified the molecular components of the ER-peroxisome tether, we assessed the extent of attachment of peroxisomes to the cER. Wild-type, inp1Δ, and pex3-V81E cells expressing GFP-PTS1 and Rtn1p-mCherry as peroxisomal and cER markers, respectively, were imaged over time. Optical sections showed peroxisomes immobilized at the cER in wild-type mother cells (Figure 6A, arrows). The cell interior, which is essentially devoid of cER, was also essentially devoid of peroxisomes in wild-type mother cells (Figure 6A). Three-dimensional reconstruction of images revealed that peroxisomes remained attached to the cER of wild-type mother cells and formed large areas of contact, appearing as flattened disks (Supplementary Movie 6). Conversely, peroxisomes were not immobilized at the cER in inp1Δ and pex3-V81E mother cells; instead they moved in and out of the focal plane, appearing both in the cell interior and periphery (Figure 6A). Three-dimensional reconstruction underscored that ER-peroxisome contacts were transient in inp1Δ and pex3-V81E mother cells (Supplementary Movie 6). Quantification over all time points showed that 95% of peroxisomes in wild-type mother cells versus 64 and 76% of peroxisomes in inp1Δ and pex3-V81E mother cells made contact with the cER, respectively (Figure 6B). Moreover, peroxisomes in wild-type mother cells maximize their area of attachment with the ER, as the contact surface between peroxisomes and the ER was almost twice than that observed for mutant mother cells (Figure 6C).
Figure 6.
Peroxisomes interact transiently with the ER in inp1Δ and pex3-V81E cells. (A) Wild-type, inp1Δ, and pex3-V81E cells expressing GFP-PTS1 and Rtn1p-mCherry were visualized by 3-D confocal video microscopy. 1 μm optical midsections of six consecutive frames of a time-lapse series are presented. Arrows show static peroxisomes. Bar, 1 μm. (B) The percentage of all peroxisomes in contact with the ER is shown for every time point (dot) of six independent recordings, of which one is depicted in (A). Bars represent means±s.e.m. Quantifications were done independently for mother cell and bud. (C) Peroxisomal and ER surfaces were computed using Imaris software. Peroxisome surface in contact with the ER is expressed as a percentage of total peroxisome surface. Quantification of six independent recordings, of which one is shown in (A), was done separately for mother cell and bud. Significant (*)/not significant (**) difference at the 99% confidence interval using a pairwise t-test. See Supplementary Movie 6.
It is noteworthy that although peroxisomes came into contact with the cER in the buds of wild-type, inp1Δ, and pex3-V81E cells alike, these peroxisomes did not appear to be tethered to the ER and moved freely in relation to the ER (Figure 6A). Neither the percentage of peroxisomes in contact with the ER nor the extent of their contact area with the ER was significantly different in the buds of any of the strains (Figure 6B and C). Overall, our observations suggest that a delay normally occurs between the insertion of peroxisomes into the bud and their immobilization at the cER.
Peroxisomes that partition to the bud lack Inp1p
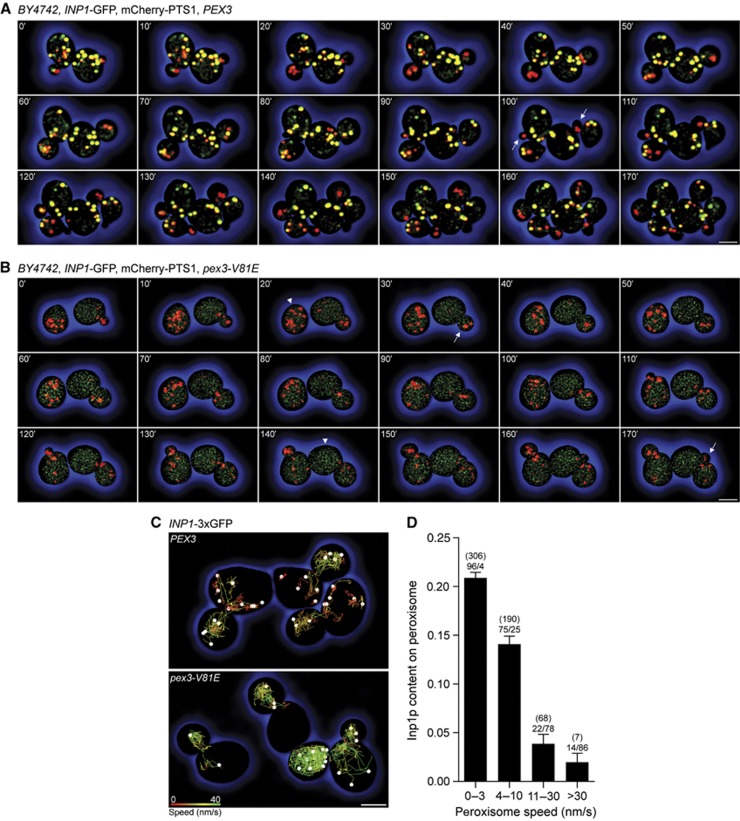
We next imaged Inp1p-GFP and peroxisomes labelled with mCherry-PTS1 over multiple cell generations to determine the dynamics of Inp1p’s association with peroxisomes during peroxisome inheritance. In budding wild-type cells, Inp1p exhibited marked asymmetry along the cell division axis (Figure 7A). Most mother cell peroxisomes contained Inp1p and were yellow in merged images of the red and green channels. Conversely, peroxisomes that were polarized towards the bud contained little or no Inp1p and therefore appeared red. Early in bud growth (10–40 min), the peroxisome populations segregated from one another. Peroxisomes lacking Inp1p transferred to buds, whereas Inp1p-containing peroxisomes remained in mother cells. Later, Inp1p was recruited to bud-localized peroxisomes, which as a result changed from red to yellow (60–90 min). When mother cells formed second generation buds (100 min, arrows), again only Inp1p-deficient, that is, red, peroxisomes moved to the buds. This cycle repeated when the first generation buds had grown sufficiently to become mother cells. Each new mother cell now had yellow and red peroxisomes, of which only the red ones segregated (130–160 min). By 170 min, eight cells had arisen from the initial two, each containing a balanced number of peroxisomes.
Figure 7.
Inp1p exhibits a polarized distribution along the cell division axis. (A, B) Peroxisome inheritance observed in budding cells expressing Inp1p-GFP, mCherry-PTS1, and either Pex3p (A) or Pex3p-V81E (B). Merged images of the red and green channels are shown. Bar, 3 μm. (C) Peroxisomes in cells expressing either Pex3p or Pex3p-V81E were tracked over the first 100 frames of Supplementary Movie 7. Peroxisomes are presented as white spheres and their trajectories as colour-coded lines (red to green, 0–40 nm/s). Bar, 3 μm. (D) Inp1p content and peroxisome speed were computed for each peroxisome and each time point for the top panel of Supplementary Movie 7. Peroxisomes are grouped into speed categories and plotted against their Inp1p content. Bars show mean±s.e.m. Figures in brackets denote the number of individual recordings per category, while the percentages of mother cell- to bud-localized peroxisomes are displayed below the bracketed numbers.
Cells expressing pex3-V81E exhibited a different pattern of peroxisome partitioning (Figure 7B). Inp1p was distributed throughout the cell and was neither polarized along the cell division axis nor recruited to peroxisomes, which therefore appeared red irrespective of their location. During cell division, the entire peroxisome population was inserted into the bud (30, 170 min, arrows), but sometimes peroxisomes later returned to the mother cell. This segregation defect resulted in an uneven peroxisome distribution. While some cells contained many peroxisomes (20 min, arrowhead), others contained only few (140 min, arrowhead).
Individual peroxisome movements were tracked by live-cell video microscopy in cells expressing Pex3p or Pex3p-V81E, mCherry-PTS1, and Inp1p-3 × GFP to enhance the intensity of Inp1p fluorescence (Supplementary Movie 7). A clear distinction could be made between mother cell and bud-localized peroxisomes in wild-type cells. Mother cell peroxisomes maintained essentially fixed positions, whereas peroxisomes in buds sampled the entire cell (Figure 7C, upper panel). The mobility of a peroxisome and its Inp1p content were inversely related (Figure 7D). Immobile peroxisomes (0–3 nm/s) contained the most Inp1p, whereas highly mobile peroxisomes (>30 nm/s) contained little or no Inp1p. Ninety-six percent of peroxisomes with the lowest speed were localized to mother cells, while highly mobile peroxisomes were nearly always found in the bud (Figure 7D). In pex3-V81E cells, all peroxisomes displayed rapid movements (Figure 7C, lower panel).
Equitable partitioning of peroxisomes between mother cell and bud requires peroxisome division and transport
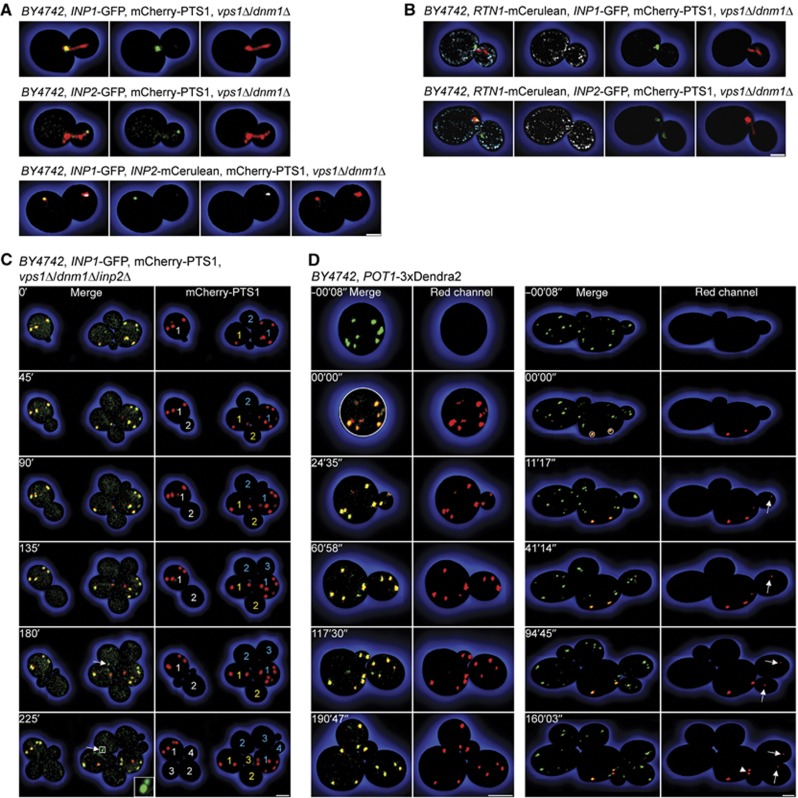
Cells lacking the dynamin-related GTPases Vps1p and Dnm1p contain a single enlarged peroxisome that projects a tubular extension into the bud and is eventually split between mother and daughter cells (Hoepfner et al, 2001; Kuravi et al, 2006). This altered peroxisome morphology enabled us to show an uneven distribution of inheritance factors on peroxisomes. Inp1p-GFP and Inp2p-GFP localized to opposite ends of the peroxisome in a vps1Δ/dnm1Δ mutant. Inp1p was confined to that part of the peroxisome that was anchored in the mother cell, while Inp2p enriched at the tip of the tubule that protruded into the bud (Figure 8A, top and middle panels). When Inp1p-GFP and Inp2p-mCerulean were coexpressed, they separated from each other late in the cell cycle, with Inp1p exclusively found on the mother cell peroxisome and Inp2p restricted to the bud-localized peroxisome (Figure 8A, bottom panel).
Figure 8.
Peroxisome inheritance requires peroxisome division. (A) Inp1p and Inp2p were imaged in vps1Δ/dnm1Δ cells expressing mCherry-PTS1 and Inp1p-GFP (top panels), Inp2p-GFP (middle panels), or Inp1p-GFP and Inp2p-mCerulean (bottom panels). Panels at extreme left present merged images of the panels at right. Inp2p-mCerulean fluorescence is shown in white. Bar, 1 μm. (B) Rtn1p-mCerulean and mCherry-PTS1 were coexpressed with Inp1p-GFP (top panels) or Inp2p-GFP (bottom panels) in vps1Δ/dnm1Δ cells. Panels at extreme left present the merged images of the panels at right. Rtn1p-mCerulean fluorescence is shown in white. Images are 0.6 μm optical midsections. Bar, 1 μm. (C) vps1Δ/dnm1Δ/inp2Δ cells expressing Inp1p-GFP and mCherry-PTS1 were tracked over time. Left panels present merged images of the red and green channels. Arrows depict Inp1p-GFP foci. Right panels show mCherry-PTS1 fluorescence and numbered cell generations. Bar, 3 μm. (D) Peroxisome dynamics in wild-type cells analysed by photoconversion of the peroxisomal reporter Pot1p-3 × Dendra2. Select peroxisomes were photoconverted from green to red, and their movements were followed by time-lapse video microscopy. One of three (left) and ten (right) recordings is shown. Left panels present merged images of the red and green channels, whereas right panels present the red fluorescence channel only. Photoconverted peroxisomal material transferred to the bud is depicted by arrows. Arrowhead shows macroscopic peroxisome division. Bars, 3 μm. See Supplementary Movies 8–10.
We also analysed the positions of Inp1p and Inp2p on the enlarged peroxisomes of the vps1Δ/dnm1Δ mutant in relation to the cER protein, Rtn1p. The portion of the peroxisome containing Inp1p overlapped with Rtn1p at the mother–bud neck interface (Figure 8B, upper panels), whereas the tip of the peroxisomal tubule enriched for Inp2p that was inserted into the bud was devoid of Rtn1p (Figure 8B, lower panels).
In vps1Δ/dnm1Δ/inp2Δ cells, neither the peroxisome divisional machinery nor Myo2p can act on peroxisomes. We monitored the peroxisome population in this mutant over several cell generations, expecting a random distribution of peroxisomes between mother and daughter cells if peroxisome inheritance involves the release of peroxisomes from tethering. Three cells expressing mCherry-PTS1 and Inp1p-GFP multiplied to a total of 11 cells over a 4-h recording (Figure 8C). Although peroxisomes contained variable levels of Inp1p, no peroxisome transferred to daughter cells, that is, first generation mother cells retained 100% of the peroxisome population. As de novo formation of peroxisomes is a slow process (Motley and Hettema, 2007), most of the cells remained peroxisome deficient. Inp1p foci appeared in younger cells (Figure 8B, arrows), thus demonstrating that peroxisome inheritance and formation of ER-docking sites for peroxisomes are independent processes. Of note, peroxisomes in a vps1Δ/dnm1Δ/inp1Δ/inp2Δ quadruple mutant occasionally broke apart and were inserted into daughter cells (Supplementary Figure S2).
To elucidate whether peroxisome division is a component of peroxisome inheritance in wild-type cells, we tracked individual peroxisomes using photoconversion of the peroxisomal reporter Pot1p-3 × Dendra2 (Supplementary Movies 8–10). When all peroxisomes of an interphase cell were photoconverted and the cell was allowed to grow and divide, its progeny contained red fluorescent peroxisomes (Figure 8D, left panels; Supplementary Movie 8). Daughter cells thus receive peroxisomes by inheritance from mother cells. After ∼60 min, some peroxisomes containing only green Dendra2 formed; however, most of the peroxisomes contained red fluorescent reporter. As the total number of red peroxisomes also increased over time, these organelles had to have arisen from the division of photoconverted peroxisomes. Division of pre-existing peroxisomes therefore dominates over de novo peroxisome formation (Motley and Hettema, 2007). When individual peroxisomes anchored at the mother cell cortex were photoconverted, they shifted in their relative positions but were not released, even if the mother cell grew multiple buds (Figure 8D, right panels; Supplementary Movie 9). Buds, on the other hand, always received red peroxisomal material, which originated from the photoconverted peroxisomes in the mother cell (Figure 8D, arrows; Supplementary Movies 9 and 10). As macroscopic division of these mother cell peroxisomes was only occasionally observed (Figure 8D, arrowhead), vesicular transport may contribute to the transfer of peroxisomal material to the bud. This mechanism of sharing ensures the maintenance of peroxisome populations and a balanced distribution of peroxisomes between mother and daughter cells.
Discussion
Here we report the identification of an ER-peroxisome tether required for maintenance of uniform peroxisome numbers in yeast cells. The tether assembles when peroxisomes attach to a macromolecular structure in the cell cortex—the peroxisome anchor—via binding between proteins on the anchor and proteins on the surface of the peroxisome.
The ER is well suited to serve as a peroxisome anchor not only because it provides an extensive surface of interconnected tubules and cisternae (West et al, 2011) to which peroxisomes potentially could dock, but also because it acts as the site of de novo peroxisome biogenesis. Peroxisomes assemble from ER-derived precursor vesicles (Titorenko et al, 2000; van der Zand et al, 2012), and many peroxisomal membrane proteins are either dually localized to the ER and peroxisomes (Yan et al, 2008) or traffic through the ER en route to peroxisomes (Titorenko and Rachubinski, 1998; van der Zand et al, 2010). The ER thus provides the link between newly formed and established peroxisomes.
Pex3p, a protein required for peroxisome biogenesis and, as we show here, peroxisome tethering to the ER, is inserted into the ER membrane to initiate peroxisome formation (Hoepfner et al, 2005; Tam et al, 2005; Thoms et al, 2012). Interaction between Pex3p and the inheritance factor Inp1p was previously shown to be necessary for peroxisome retention (Munck et al, 2009). That study concluded that Pex3p acts as a receptor for Inp1p on the surface of peroxisomes and that recruitment of Inp1p to peroxisomes is required to dock them to a yet unidentified cortical anchor. Our findings present a more complex geometry for the Pex3p–Inp1p interaction. Pex3p and Inp1p assemble into cortical foci in peroxisome-deficient mutants in which all Pex3p is trapped in the ER. These foci represent local enrichments of two ER proteins that bind one another, as becomes evident when the interaction between Pex3p and Inp1p is impaired. In the pex3-V81E mutant, Inp1p and Pex3p segregate to distinct membrane compartments, and Pex3p presents the morphology of a conventional ER protein when its egress from the ER is blocked. By expressing Inp1p on the surface of the mitochondria and tethering peroxisomes artificially to this compartment, we demonstrate that Inp1p does not need to be in the peroxisomal membrane to anchor peroxisomes. In effect, it can bridge two membrane compartments in trans. Functional reconstitution of the ER-peroxisome tether by mating cells expressing either its ER or peroxisomal part shows that Inp1p foci are docking sites for peroxisomes at the ER membrane. Finally, using the split-GFP assay, we confirm that Inp1p in foci interacts not only with ER-bound Pex3p but also with Pex3p in the peroxisomal membrane.
Collectively, our data reveal that the core of the ER-peroxisome tether is made by the Inp1p-mediated linkage of ER-bound Pex3p with peroxisomal Pex3p. While the integral membrane protein Pex3p localizes to both compartments and builds the membrane component of the tether, Inp1p is not a classical peroxisomal protein but a hinge protein situated at the ER–peroxisome interface and specifically required for connecting both compartments. A similar distinction has been made for Mmm1p, a component of the ER-mitochondrion tether. Mmm1p, formerly considered to be a bona fide mitochondrial protein, is actually an ER-resident membrane protein that bridges the mitochondria and the ER through its binding of a protein complex in the outer mitochondrial membrane (Kornmann et al, 2009).
How does the ER-peroxisome tether assemble? Inp1p and Pex3p make cortical foci in peroxisome-null mutants, in which Pex3p is confined to the ER. In cells with peroxisomes, Inp1p is found only on static, that is, ER-bound, and not mobile peroxisomes. Its exclusion from mobile peroxisomes suggests that Inp1p first needs to enrich in foci before it can dock peroxisomes. If Inp1p discriminates between ER-bound and peroxisomal Pex3p, a change in Inp1p concentration could modulate the equilibrium between the two Pex3p forms. Moderate overexpression of Inp1p causes an increase in the number of ER-docked peroxisomes. These likely result from de novo formation, as more of Pex3p is kept in its ER-bound, that is, peroxisome biogenic, state. Extreme overexpression of Inp1p leads to a collapse of the peroxisomal compartment, probably because the entire Pex3p pool is captured by Inp1p in the ER (Supplementary Figure S3).
Full-length Inp1p, as well as its N- and C-termini individually, bind directly to Pex3p and Pex3p-V81E in vitro. Inp1p’s divalency for Pex3p could enable it to link many Pex3p molecules into a supramolecular lattice capable of stably docking peroxisomes at the ER membrane (Figure 9). Our data also suggest that the V81 surface on Pex3p does likely not act as a site for direct binding of Inp1p, as otherwise Pex3p-V81E should consistently fail to interact with either full-length Inp1p or one of its domains. Instead, we observed that only the initial recruitment of Inp1p to foci, but not the subsequent tethering of peroxisomes to these foci, is dependent on an intact V81-site on Pex3p in vivo. This region of Pex3p may therefore modulate Inp1p, perhaps inducing a conformational change to make it competent for binding Pex3p. When Inp1p is artificially immobilized on the mitochondria, peroxisomes attach to the mitochondria regardless of whether the peroxisomes contain Pex3p or Pex3p-V81E. Because Inp1p can bind Pex3p by both its N- and C-termini, Pex3p may contain more than one binding site for Inp1p. These interfaces are currently uncharacterized but are expected to be structurally distinct from the region containing V81.
Figure 9.
A model for peroxisome population control. Multiple Inp1p molecules connect ER-bound Pex3p and peroxisomal Pex3p into an ER-peroxisome tethering complex that anchors a peroxisome to the cER of the mother cell. Recruitment of Inp1p, but not peroxisome tethering, to foci depends on the integrity of the patch containing V81 on the surface of Pex3p (A). Pulling forces exerted by Myo2p and constriction forces exerted by the peroxisome divisional machinery lead to elongation, constriction, and ultimate rupture of the peroxisome. The division process is asymmetric and may trigger the release of larger and smaller peroxisomal fragments, which contain Inp2p and are transported to the bud (B). After its release from Myo2p, the bud-localized peroxisome can attach to a tether that is newly formed by passage of Pex3p through the ER and recruitment of Inp1p by Pex3p (C).
We expected the proteins in the ER-peroxisome tether to interact transiently and in coordination with the cell cycle to enable repetitive rounds of association and dissociation of peroxisomes and their anchors during peroxisome inheritance. This is seemingly the case as Inp1p is absent from peroxisomes that partition to the bud. Inp1p’s concentration gradient along the cell division axis is opposite to that of Inp2p, which enriches on bud-localized peroxisomes (Fagarasanu et al, 2006). The segregation of inheritance factors could be achieved by differential regulation of Inp1p and Inp2p on individual peroxisomes. But how would these peroxisomes be selected, how would Inp1p and Inp2p rebalance and how could the cell monitor its peroxisome population as a whole? Considering that peroxisome inheritance depends on force, which is exerted by the peroxisome divisional machinery and Myo2p, a combination of division and transport processes could lead to the separation of Inp1p and Inp2p. Using photoconversion of a peroxisomal matrix protein to track the fate of individual peroxisomes, we showed that peroxisomes in the mother cell are not fully released from their tethers, while bud-localized peroxisomes contain reporter originally photoconverted in the mother cell and are therefore the product of a division event. As macroscopic peroxisome division occurs infrequently, our findings provide supporting evidence for the trafficking of peroxisomal proteins by vesicular carriers, which have been characterized biochemically (Titorenko et al, 2000; Agrawal et al, 2011; Lam et al, 2011).
Our findings lead us to revise the view of peroxisomes as entities that are transferred independently of each other from mother cell to bud. The steady state of this multicopy organelle is rather established by the number of peroxisomes attached to the cell cortex and reset once per cell cycle. The number of tethers is determined by the abundance of Inp1p, which traps Pex3p in its ER-bound state. While existing tethers in the mother cell are not deconstructed, directionality of peroxisome transfer is assisted by the de novo formation of docking sites in the bud via passage of Pex3p through the ER. By combining peroxisome biogenic and retention functions in a single protein, Pex3p, cells achieve an exacting level of control over their peroxisome populations. Peroxisome inheritance and formation of ER-docking sites are independent processes. In wild-type cells, new docking sites in the bud are the recipients of peroxisomes that have been transferred from the mother cell. However, if peroxisome inheritance is blocked, Pex3p could enrich at a docking site, escape its association with Inp1p and begin to form a new peroxisome, thus tilting its activity in favour of peroxisome biogenesis over peroxisome retention. We summarize our data in a model (Figure 9). All major requirements for such a system, that is the interaction of peroxisomes and their anchor in trans, the dynamic nature of this interaction, the directionality of peroxisome transfer and the control of peroxisome numbers, are satisfied.
In closing, we have identified the ER as the site of peroxisome anchoring in the yeast cell cortex, defined the protein complex required for ER-peroxisome tethering and elucidated the system by which uniform peroxisome numbers are maintained in a growing cell population. Our findings provide a mechanistic understanding of how the cell achieves an equitable sharing of a multicopy organelle with its progeny.
Materials and methods
Yeast strains and culture conditions
Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table S1. Unless otherwise noted, all strains were grown in YPD at 30°C. If fatty acid induction of peroxisomes was required, cells were first grown to mid-log phase in YPD and then incubated in SCIM for 15 h. Media components were: YPD, 1% yeast extract, 2% peptone, 2% glucose; SCIM, 0.67% yeast nitrogen base without amino acids (YNB), 0.5% yeast extract, 0.5% peptone, 3.3% Brij 35, 0.3% glucose, 0.3% oleic acid, 1 × complete supplement mixture (CSM); synthetic minimal medium (SM), 0.67% YNB, 2% glucose, 1 × CSM; nonfluorescent medium, 6.61 mM KH2PO4, 1.32 mM K2HPO4, 4.06 mM MgSO4·7H2O, 26.64 mM (NH4)SO4, 1 × CSM, 2% glucose, 1% agarose; 5-FOA, 0.67% YNB, 1 × CSM-uracil, 0.1% 5-fluoroorotic acid, 0.0012% uracil, 2% glucose; SM-arg+can, 0.67% YNB, 1 × CSM-arginine, 0.006% canavanine, 2% glucose.
Confocal fluorescence microscopy
Live-cell imaging was performed with a LSM710 confocal fluorescence microscope (Carl Zeiss) equipped with a 63 × 1.4 NA Plan-Apo chromate objective and a piezoelectric stage to allow for rapid acquisition of z-stacks. Images were collected with a z-resolution between 0.1 μm and 0.4 μm to a total stack height of 5 μm. GFP was excited with a 488 nm laser, and its emission was collected within a range of 493–610 nm. In strains expressing both GFP and mCherry, the GFP emission was collected within a range of 493–552 nm, while mCherry was excited with a 561 nm laser and its emission collected between 574–735 nm. For triple channel acquisition, fluorophores were excited sequentially with 440, 488, and 561 nm lasers for mCerulean, GFP, and mCherry, respectively. The emission bandwidths were 444–493 nm for mCerulean, 493–552 nm for GFP, and 567–638 for mCherry. The excitation lasers were adjusted to minimum levels to avoid cross-excitation and photobleaching. Photoconversion of the 3 × Dendra2 reporter was accomplished by 20 iterative scans with 405 nm beams of a 30 mW diode laser set at 1.5% intensity. The fate of photoconverted peroxisomes was tracked over a period of 2–4 h. All images were captured at 30°C with the confocal microscope pinhole adjusted to 1 Airy unit of the longest emission wavelength used.
Cells used for live-cell imaging were diluted 1:100 from an overnight culture into fresh YPD medium and grown for 4–5 h. Cells were washed with water, and 2 μl of the cell suspension were spotted onto a thin agarose pad prepared from hot nonfluorescent medium, covered with a cover slip and sealed with VALAP (Fagarasanu et al, 2009). Haploid cells used in mating assays were grown and washed as above. Cells from each strain were mixed and spotted onto an agarose pad, and cell matings were viewed immediately in the microscope. For galactose induction of recombinant genes, cells were grown for 4–5 h in YPD medium containing 0.5% glucose. Galactose was then added to a final concentration of 0.5% for an additional 30 min before cells were washed and mounted. For experiments using the galactose-inducible reporter POT1-3 × Dendra2, 2 ml from an overnight culture of cells were seeded into 50 ml of YPD medium containing 0.5% glucose and 0.5% galactose, and grown for 3–4 h. To turn off expression of the reporter and allow its uptake into peroxisomes, cells were pelleted, resuspended in the same volume of YPD medium and cultured for an additional 2 h before washing and mounting.
Supplementary Material
Acknowledgments
We thank Arvonn Tully (Bitplane) for image analysis; Frank Nargang, Randy Schekman and Karsten Weiss for reagents; Nicolas Touret for help with photoconversion experiments; and Vladimir Titorenko for advice on subcellular fractionation. The technical assistance of Hanna Kroliczak, Elena Savidov and Dwayne Weber is acknowledged. This work was supported by grant 9208 from the Canadian Institutes of Health Research to RAR and the Howard Hughes Medical Institute.
Author contributions: BK, AF and RAR provided a conceptual framework for the study, interpreted data and wrote the manuscript. BK also performed the experiments. NC provided the structural analysis of yeast Pex3p. XS provided confocal microscopy support and expertise. RLP constructed recombinant plasmids and yeast strains.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agrawal G, Joshi S, Subramani S (2011) Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci USA 108: 9113–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Mast FD, Fagarasanu A, Rachubinski DA, Eitzen GA, Dacks JB, Rachubinski RA (2009) Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J Cell Biol 187: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA (2006) The peroxisomal membrane protein Inp2p of the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell 10: 587–600 [DOI] [PubMed] [Google Scholar]
- Fagarasanu A, Mast FD, Knoblach B, Jin Y, Brunner MJ, Logan MR, Glover JN, Eitzen GA, Aitchison JD, Weisman LS, Rachubinski RA (2009) Myosin-driven peroxisome partitioning in S. cerevisiae. J Cell Biol 186: 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Mast FD, Knoblach B, Rachubinski RA (2010) Molecular mechanisms of organelle inheritance: lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol 11: 644–654 [DOI] [PubMed] [Google Scholar]
- Fagarasanu M, Fagarasanu A, Tam YYC, Aitchison JD, Rachubinski RA (2005) Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J Cell Biol 169: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Morrell JC, Jones JM, Gould SJ (2004) PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol 164: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau WH, Erdmann R (1998) Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol 18: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Girzalsky W, van den Berg M, Erdmann R, Distel B (2000) Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J 19: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF (2005) Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122: 85–95 [DOI] [PubMed] [Google Scholar]
- Hoepfner D, van den Berg M, Phillipsen P, Tabak HF, Hettema EH (2001) A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol 155: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J (2006) The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol 173: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ (2006) Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci 119: 3994–4001 [DOI] [PubMed] [Google Scholar]
- Lam SK, Yoda N, Schekman R (2011) A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci USA 107: 21523–21528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Hettema EH (2007) Yeast peroxisomes multiply by growth and division. J Cell Biol 178: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck JM, Motley AM, Nuttall JM, Hettema EH (2009) A dual function for Pex3p in peroxisome formation and inheritance. J Cell Biol 187: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Shibata H, Nakano H, Matsuzono Y, Kashiwayama Y, Kobayashi Y, Fujiki Y, Imanaka T, Kato H (2008) Characterization of the interaction between recombinant human peroxin Pex3p and Pex19p: identification of TRP-104 in Pex3p as a critical residue for the interaction. J Biol Chem 283: 6136–6144 [DOI] [PubMed] [Google Scholar]
- Sato Y, Shibata H, Nakatsu T, Nakano H, Kashiwayama Y, Imanaka T, Kato H (2010) Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p. EMBO J 29: 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R (2005) Peroxisomes: another branch of the secretory pathway? Cell 122: 1–2 [DOI] [PubMed] [Google Scholar]
- Schmidt F, Treiber N, Zocher G, Bjelic S, Steinmetz MO, Kalbacher H, Stehle T, Dodt G (2010) Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J Biol Chem 285: 25410–25417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat Biotech 19: 773–776 [DOI] [PubMed] [Google Scholar]
- Tam YYC, Fagarasanu A, Fagarasanu M, Rachubinski RA (2005) Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem 280: 34933–34939 [DOI] [PubMed] [Google Scholar]
- Thoms S, Harms I, Kalies KU, Gartner J (2012) Peroxisome formation requires the endoplasmic reticulum channel protein Sec61. Traffic 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Chan H, Rachubinski RA (2000) Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J Cell Biol 148: 29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (1998) Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol 18: 2789–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A, Braakman I, Tabak HF (2010) Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell 21: 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A, Gent J, Braakman I, Tabak HF (2012) Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 [DOI] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger A, Voeltz GK (2011) A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol 193: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CG, Magliery TJ, Regan L (2004) Detecting protein-protein interactions with GFP-fragment reassembly. Nat Methods 1: 255–262 [DOI] [PubMed] [Google Scholar]
- Yan M, Rachubinski DA, Joshi S, Rachubinski RA, Subramani S (2008) Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol Biol Cell 19: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.