Abstract
Spindle positioning and spindle elongation are critical for proper cell division. In human cells, an evolutionary conserved ternary complex (NuMA/LGN/Gαi) anchors dynein at the cortex during metaphase, thus ensuring correct spindle positioning. Whether this complex contributes to anaphase spindle elongation is not known. More generally, the mechanisms coupling mitotic progression with spindle behaviour remain elusive. Here, we uncover that levels of cortical dynein markedly increase during anaphase in a NuMA-dependent manner. We demonstrate that during metaphase, CDK1-mediated phosphorylation at T2055 negatively regulates NuMA cortical localization and that this phosphorylation is counteracted by PPP2CA phosphatase activity. We establish that this tug of war is essential for proper levels of cortical dynein and thus spindle positioning during metaphase. Moreover, we find that upon CDK1 inactivation in anaphase, the rise in dephosphorylated NuMA at the cell cortex leads to cortical dynein enrichment, and thus to robust spindle elongation. Our findings uncover a mechanism whereby the status of NuMA phosphorylation coordinates mitotic progression with proper spindle function.
Keywords: CDK1, dynein, NuMA, mitotic progression, protein phosphatases
Introduction
Correct placement of the cleavage furrow and faithful segregation of the genetic material during mitosis relies notably on accurate spindle positioning and robust spindle elongation. In metazoan organisms, spindle positioning requires the function of an evolutionary conserved ternary complex that comprises a large coiled-coil protein, a GoLoCo domain-containing protein and a heterotrimeric G protein α-subunit (reviewed in Gönczy, 2008; Knoblich, 2008; Siller and Doe, 2009; Morin and Bellaïche, 2011). Members of this ternary complex are evolutionary conserved and the available evidence indicates that they serve to anchor the minus end-directed motor protein complex dynein (hereafter referred to as dynein for simplicity) at the cell cortex below the plasma membrane.
Dynein is a multisubunit complex that comprises an AAA ATPase heavy chain motor protein, intermediate and light chains, as well as several additional factors that are needed for dynein activity, including the dynactin complex (reviewed in Kardon and Vale, 2009). Dynein is fundamental for a number of cellular processes across eukaryotic evolution, including organelle distribution, spindle assembly and kinetochore function. In the case of spindle positioning, dynein located at the cell cortex is thought to generate pulling forces on astral microtubules that emanate from the spindle poles, thus positioning the mitotic spindle (Nguyen-Ngoc et al, 2007; Couwenbergs et al, 2007; Park and Rose, 2008; Woodard et al, 2010; Kiyomitsu and Cheeseman, 2012). In human cells, support for this model comes notably from experiments in which dynein anchored artificially to the membrane in a manner that no longer relies on the ternary complex directs spindle positioning during metaphase (Kotak et al, 2012). Moreover, the Polo-like kinase Plk1 negatively regulates cortical dynein levels during metaphase by negatively regulating its interaction with NuMA/LGN (Kiyomitsu and Cheeseman, 2012). Besides directing metaphase spindle positioning, dynein has also been postulated to be necessary for the increase in distance between the two spindle poles as the spindle elongates during anaphase B (Cande and Wolniak, 1978; Aist and Berns, 1981; Cande, 1982; Fink et al, 2006; Collins et al, 2012). However, direct evidence for such an involvement is lacking in human cells. More generally, the mechanisms by which the distribution of cortical dynein is regulated as cells progress through mitosis remain elusive.
In human cells, the members of the ternary complex that anchor dynein to the cell cortex during metaphase are NuMA, the GoLoCo protein LGN and the three Gα proteins Gαi1–3. NuMA is a large protein that is essential for the assembly and maintenance of the mitotic spindle (Yang and Snyder, 1992; Merdes et al, 1996). During interphase, NuMA is present in the nucleus, whereas following nuclear envelope breakdown (NEBD), the protein becomes enriched in metaphase in the vicinity of the two spindle poles, as well as slightly at the cell cortex above them (Yang et al, 1992; Du and Macara, 2004; Woodard et al, 2010). NuMA comprises globular head and tail domains separated by a large coiled-coil (Yang et al, 1992). An N-terminal fragment of NuMA can interact with dynein, whereas the tail domain contains microtubules and LGN-binding sites, as well as a nuclear localization signal (NLS) (Gueth-Hallonet et al, 1996; Haren and Merdes, 2002; Du et al, 2002; Kotak et al, 2012). Moreover, NuMA harbours four consensus sites in its tail domain for the CDK1/CyclinB kinase complex (hereafter referred to as CDK1 for simplicity) and can be phosphorylated by CDK1 in Xenopus egg extracts (Yang et al, 1992; Compton and Luo, 1995; Gehmlich et al, 2004). CDK1 activity augments drastically when cells enter mitosis and decreases sharply due to sudden degradation of the CyclinB moiety when cells progress into anaphase (reviewed in King et al, 1996; Lindqvist et al, 2009). CDK1-dependent phosphorylation of the four sites in NuMA has been proposed to regulate its localization to spindle poles at the onset of mitosis (Gehmlich et al, 2004). Accordingly, a NuMA mutant bearing a nonphosphorylatable residue at one of these predicted CDK1 target sites (T2055A) exhibits diminished localization at spindle poles and concomitantly increased localization at the plasma membrane in hamster BHK21 cells (Compton and Luo, 1995). However, it remains to be investigated whether CDK1 directly phosphorylates NuMA and whether such potential phosphorylation is of biological significance.
In this study, we uncover that dynein becomes enriched at the cortex as cells progress from metaphase to anaphase in a NuMA-dependent manner. Moreover, we demonstrate that a tug of war between CDK1 and PPP2CA phosphatase on T2055 of NuMA dictates the extent of cortical NuMA and dynein levels. We establish that this ensures that low levels of cortical dynein are present during metaphase, which is essential for proper spindle positioning. Furthermore, our findings indicate that upon anaphase onset, CDK1 inactivation results in increased levels of nonphosphorylated T2055 NuMA at the cell cortex, and thus cortical dynein, which drives robust spindle elongation.
Results
NuMA-dependent cortical dynein enrichment during anaphase
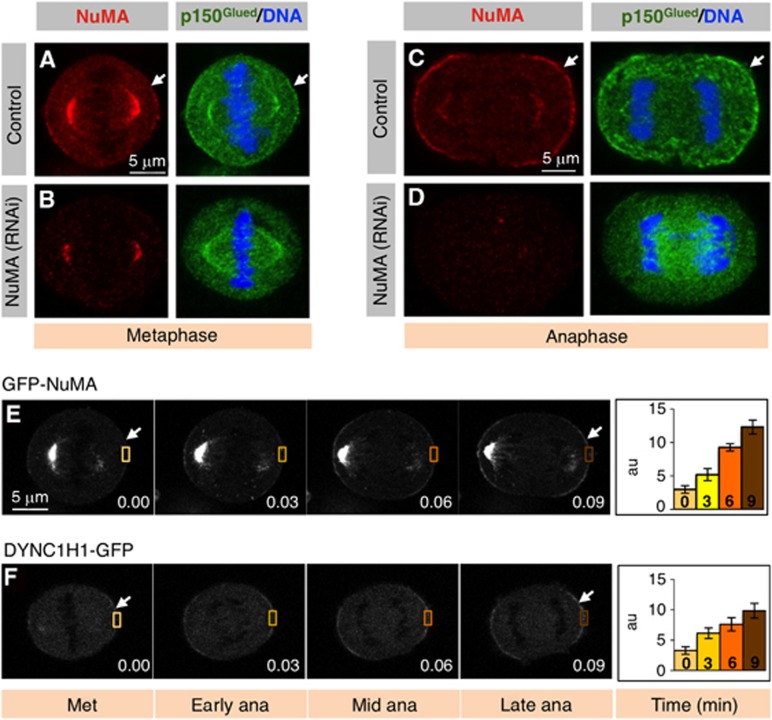
During metaphase in HeLa cells, the dynein complex, as well as the dynein-interacting dynactin component p150Glued, are known to localize weakly to the cell cortex above the spindle poles in a NuMA-dependent manner (Figure 1A and B) (Woodard et al, 2010; Kiyomitsu and Cheeseman, 2012; Kotak et al, 2012). We discovered that cortical levels of NuMA and p150Glued are markedly higher in anaphase than in metaphase (Figure 1C, compare to Figure 1A; see also Supplementary Figure S1A–F for NuMA/p150Glued localization throughout the cell cycle). Analogous conclusions were reached by conducting time-lapse recordings of HeLa cells expressing GFP fused to NuMA or to the dynein heavy chain DYNC1H1 (Figure 1E and F and Supplementary Movies S1 and S2). In addition, we found that cortical NuMA levels are likewise markedly enriched during anaphase in osteosarcoma U2OS cells, in breast cancer MCF7 cells, in nontransformed RPE1 cells as well as in primary mouse embryonic fibroblasts (MEFs) (Supplementary Figure S2A–D). These results are consistent with and extend findings showing that cortical dynein levels transiently increase during mitosis in LLC-Pk1 cells (Collins et al, 2012). Our results prompted us to test whether NuMA is required for cortical dynein localization during anaphase. As shown in Figure 1D, we found that treating HeLa cells with siRNAs directed against NuMA abolishes the presence of cortical p150Glued in anaphase, as it does in metaphase (Figure 1B; see also line scans in Supplementary Figure S2E–H).
Figure 1.
Cortical NuMA distribution increases during anaphase. (A–D) HeLa cells in metaphase (A, B) or anaphase (C, D) transfected with control siRNAs (A, C) or NuMA siRNAs (B, D), fixed 72 h thereafter and stained for NuMA (red) as well as p150Glued (green). In this and other figures, DNA is visualized in blue and arrows point to cortical localization. Only 3% and 5% of NuMA siRNA-treated cells exhibited some cortical p150Glued signal during metaphase (Met) and anaphase (ana), respectively, compared to 98% and 100% in control conditions, respectively (n>100 in all cases). (E, F) Images from time-lapse microscopy of HeLa Kyoto cells either transfected with GFP-NuMA (E) (see also corresponding Supplementary Movie S1) or stably expressing mouse dynein heavy chain (DYNC1H1-GFP; F) (see also corresponding Supplementary Movie S2). Quantification of GFP cortical enrichment (right) determined by calculating the mean intensity of cortical signal (area drawn) and subtracting from this value the mean intensity value in the cytoplasm (similar area) and correcting for background signal. Time is indicated in h.min. Stages of mitosis are indicated below the panels (n=5 cells, error bars: s.e.m.).
Dynein-mediated anaphase spindle elongation
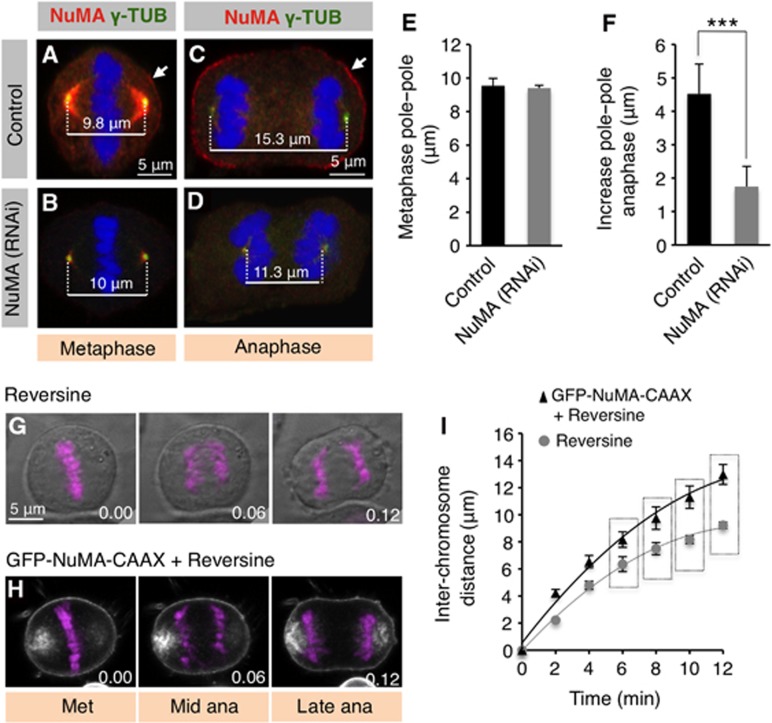
What is the biological significance of the presence of cortical dynein in anaphase? As mentioned in the introduction, one possibility is that cortically anchored dynein exerts pulling forces on astral microtubules, thus contributing to spindle elongation during anaphase. Analysing spindle elongation by live imaging upon NuMA depletion would be well suited to address this question. However, NuMA is difficult to deplete thoroughly (see Materials and methods), so that live imaging experiments would be complicated to interpret unambiguously because the extent of depletion in each analysed cell could not be ascertained. To circumvent this issue, we analysed fixed specimens instead, focussing on those cells in which anaphase cortical NuMA levels are severely impaired. We first measured the distance between the two spindle poles in control cells and compared them to that in cells depleted of NuMA. Whereas there is no significant difference in pole–pole distance between the two conditions in metaphase (Figure 2A, B and E), we found that NuMA depletion severely impairs spindle elongation in anaphase (Figure 2C, D and F). We also noted that the increase in chromosome–chromosome distance that normally occurs during anaphase is likewise affected upon NuMA depletion (compare Figure 2C and D), consistent with the fact that spindle elongation is coupled with chromosome separation in human cells (Roostalu et al, 2010).
Figure 2.
NuMA-dependent cortical dynein promotes spindle elongation during anaphase B. (A–D) Metaphase (A, B) and anaphase (C, D) cells transfected with control siRNAs (A, C) or NuMA siRNAs (B, D), fixed 96 h thereafter and stained for NuMA (red) as well as γ-tubulin (green). Pole-to-pole distances are indicated. (E, F) Quantification of pole-to-pole distances in metaphase (E) and increase over metaphase distances during anaphase (F). Two-tailed Student’s t-tests show that the anaphase pole-to-pole distance upon NuMA depletion is significantly different from that of control (***P<0.0005; n=50 for metaphase (Met) and n=43 for anaphase (ana), respectively). (G, H) Images from time-lapse microscopy of HeLa cells stably expressing mCherry-H2B and treated with Reversine (1 μM) (G) or transfected with GFP-NuMA-CAAX and treated with Reversine (H) (see also corresponding Supplementary Movies S5 and S6). The mCherry signal is shown in pink, overlaid on the phase image (G) or overlaid on the GFP image (H). Time is indicated in h.min. (I) Chromosome–chromosome distance in the conditions shown in (G) and (H). The significance determined using Student’s t-test is P<0.01 between the values highlighted by a vertical rectangle. Time is indicated in min (n=5 cells for each condition at each time point; error bars: s.d.).
We reasoned that if loss of cortical dynein during anaphase impairs spindle elongation, then experimental elevation of anaphase cortical dynein levels might enhance this process. To address this possibility, we sought to utilize GFP-NuMA(1–705)-CAAX (referred as GFP-NuMA-CAAX) that recruits excess dynein to the cell cortex in metaphase (Kotak et al, 2012). We found that GFP-NuMA-CAAX likewise recruits excess cortical dynein during anaphase (Supplementary Figure S3A–D). Therefore, we set out to address the consequence on spindle elongation of such excess cortical dynein using live imaging of HeLa cells expressing mCherry-H2B. Expression of GFP-NuMA-CAAX is accompanied by chromosomal abnormalities during metaphase (Kotak et al, 2012); most likely because of this, a majority of cells did not undergo the metaphase to anaphase transition (30/32 cells). Importantly, however, the two cells that did undergo this transition exhibited a marked increase in chromosome–chromosome distance (∼15 μm compared to ∼11 μm in control conditions; Supplementary Movies S3 and S4). To obtain more quantitative data, we repeated these experiments in the presence of the MPS1 inhibitor Reversine, which inactivates the spindle assembly checkpoint (Santaguida et al, 2010), reasoning that this should enable us to more readily analyse anaphase cells expressing GFP-NuMA-CAAX. Importantly, we found by live imaging that cells expressing GFP-NuMA-CAAX and treated with Reversine exhibit a significant increase in chromosome–chromosome distance during anaphase compared to cells treated with Reversine alone (Figure 2G, H and I; Supplementary Movies S5 and S6). As a second means to test the contribution of dynein to spindle elongation, we utilized the specific dynein inhibitor Ciliobrevin A, which is expected to impair dynein function throughout the cell (Firestone et al, 2012). Time-lapse microscopy of HeLa cells expressing mCherry-H2B and subjected to acute inhibition of dynein by Ciliobrevin A revealed an impairment of chromosome–chromosome distance during anaphase (Supplementary Figure S3E–G and Supplementary Movies S7 and S8).
Overall, we conclude that NuMA is important for recruiting dynein to the cell cortex in anaphase, and that the motor protein plays a critical role in driving robust spindle elongation.
CDK1 phosphorylates NuMA at T2055 in vitro and in vivo
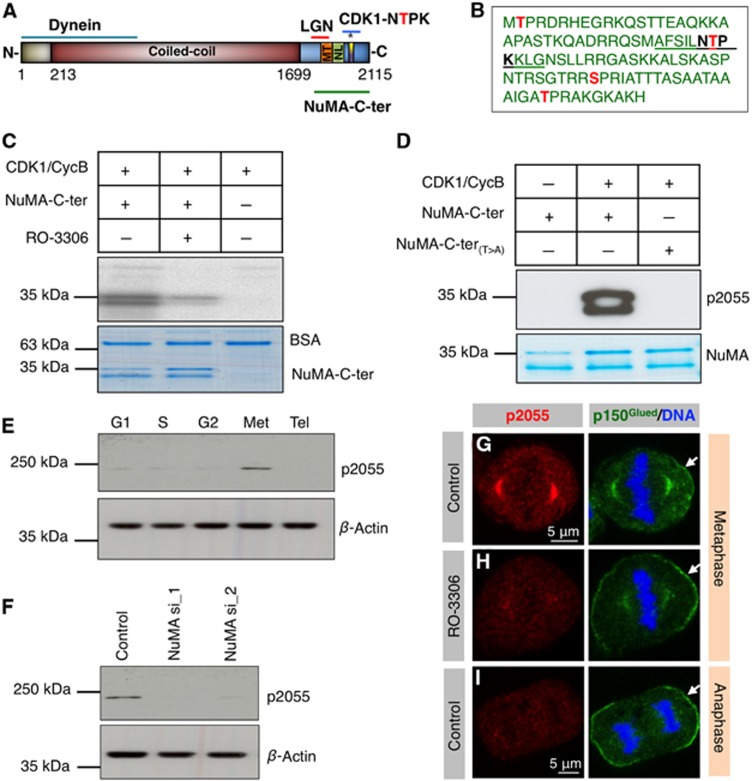
We next addressed whether CDK1-mediated regulation of NuMA might be at the root of the change in the distribution of cortical NuMA, and thus of dynein, between metaphase and anaphase (Figure 1). To test this possibility, we performed in vitro kinase assays and found that CDK1 can phosphorylate a C-terminal fragment of NuMA, whereas this phosphorylation is severely impaired in the presence of the CDK1 inhibitor RO-3306 (Figure 3C). To identify the phosphorylated residue(s), we performed mass spectrometry analysis which established that phosphorylation occurs at T2015, T2055, S2087 and T2106, corresponding to the four CDK1 consensus sites (Figure 3B). We conclude that CDK1 can directly phosphorylate NuMA in vitro.
Figure 3.
CDK1 phosphorylates NuMA at T2055 during metaphase. (A) Schematic representation of NuMA, with coiled-coil domain and regions mediating interaction with dynein, LGN and microtubules (MT); the nuclear localization signal (NL) and the CDK1 consensus sequence encompassing T2055 are also depicted. (B) Stretch in the C-terminal part of NuMA (amino acids 2014–2115) that contains the four residues (red) identified by mass spectrometry (MS) to be in vitro phosphorylated by CDK1. The peptide used to raise phospho-T2055 antibodies is underlined. (C) In vitro kinase assay with C-terminal fragment of NuMA (NuMA-C-ter, see (A), amino acids 1876–2115) incubated with CDK1/CyclinB plus [γ-32P]-ATP either in the presence of 0.1% DMSO or the CDK1 inhibitor RO-3306 (9 μM) and analysed by autoradiography (top). Equal loading is shown by Coomassie staining (bottom); BSA serves as a negative control. In (C–F), molecular mass is indicated in kDa. Note that bacterially expressed NuMA-C-ter is unstable, thus explaining the presence of two species. (D) In vitro kinase assay of wild-type or T2055A NuMA-C-ter (NuMA-C-ter(T>A)), as indicated, incubated with CDK1/CyclinB plus cold ATP and analysed by western blot using phospho-T2055 antibodies. (E) Western blot with phospho-T2055 antibodies of lysates from cells synchronized in the indicated stages of the cell cycle (G1, S, G2, metaphase (Met) and Telophase (Tel)). (F) Western blot with phospho-T2055 antibodies of lysates from cells treated with either control siRNAs or two independent siRNAs against NuMA (NuMA si_1 and NuMA si_2) and synchronized in metaphase. (G–I) HeLa cells in metaphase (G, H) or anaphase (I) stained with phospho-T2055 antibodies (red) and p150Glued (green). The cell in (H) has been treated in addition with RO-3306 (9 μM) for 5 min.
Because Threonine 2055 is the only amino acid among these four whose mutation to Alanine impairs NuMA localization at spindle poles and promotes its presence at the plasma membrane (Compton and Luo, 1995), we focussed further analysis on this residue. We generated phospho-specific antibodies against T2055 (phospho-T2055; Figure 3B). As shown in Figure 3D, these antibodies recognize the wild-type C-terminal fragment of NuMA following in vitro phosphorylation by CDK1, but not a T2055A mutant version (NuMA-C-ter(T>A)). Moreover, western blot analysis of whole-cell lysates from synchronized populations revealed that phospho-T2055 antibodies recognize a single band at the expected size, primarily during metaphase (Figure 3E). This band disappears in metaphase cells treated with siRNAs against NuMA or incubated with the CDK1 inhibitor RO-3306 (Figure 3F and Supplementary Figure S4A), indicating specificity for the phosphorylated form of T2055. Immunofluorescence analysis uncovered phospho-T2055 accumulation in the nucleus just before NEBD in prophase (Supplementary Figure S1G), mirroring the distribution of active CDK1 (Gavet and Pines, 2010). Importantly in addition, we found that phospho-T2055 is enriched at spindle poles in prometaphase and metaphase, but not at the cell cortex, in contrast to total NuMA (compare Figure 3G and Supplementary Figure S1H–I with Supplementary Figure S1B and C). Moreover, we found that phospho-T2055 is essentially absent during anaphase and telophase, when CDK1 is inactive (Figure 3I and Supplementary Figure S1J–K). Furthermore, brief incubation with the CDK1 inhibitor RO-3306 drastically reduces phospho-T2055 staining in metaphase (compare Figure 3H with Figure 3G). Overall, we conclude that CDK1 phosphorylates NuMA at T2055 in vivo during metaphase and that a nonphosphorylated T2055 NuMA species is present at the cell cortex, weakly during metaphase and more prominently during anaphase.
The phosphorylation status at T2055 governs NuMA distribution during mitosis
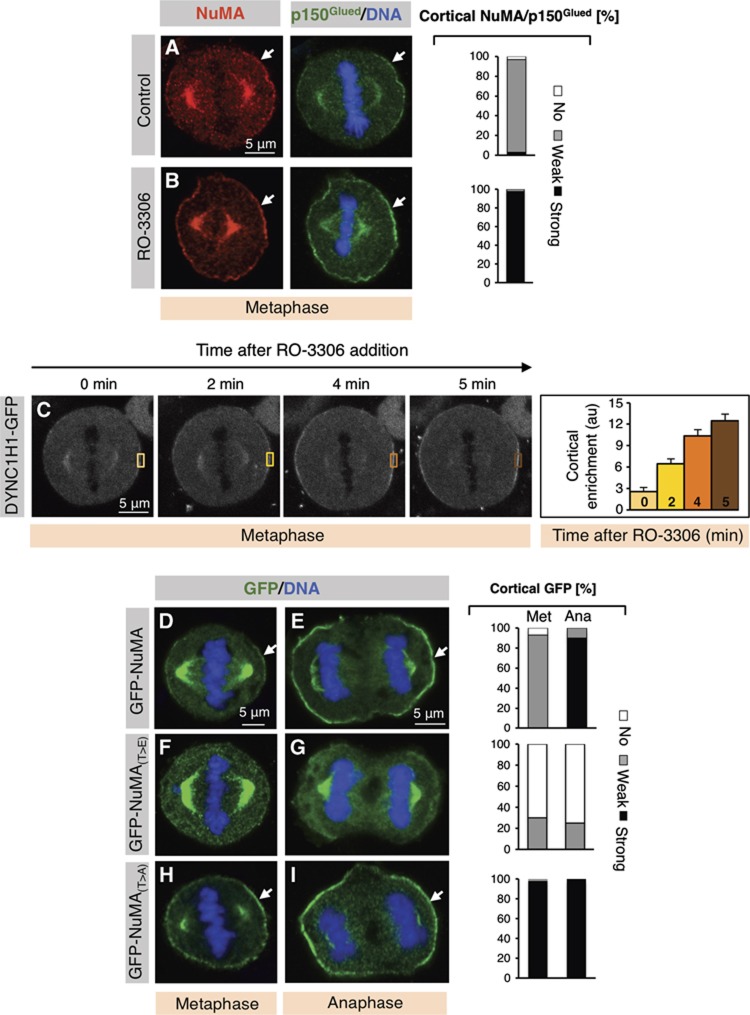
We set out to address the importance of NuMA phosphorylation by CDK1. Importantly, we found that inhibiting CDK1 using RO-3306 results in excess cortical localization of NuMA and p150Glued during metaphase (Figure 4B, compare with Figure 4A). Similar results were obtained with RO-3306 in MEFs (data not shown), as well as by using Roscovitine, a distinct CDK1 inhibitor, in HeLa cells (Supplementary Figure S4C, compare with Supplementary Figure S4B). In addition, we found that cortical DYNC1H1-GFP enrichment also increases following RO-3306 treatment (Figure 4C).
Figure 4.
CDK1 negatively regulates NuMA/dynein cortical distribution by phosphorylating NuMA at T2055. (A, B) Metaphase HeLa cells treated with 0.1% DMSO (Control) (A) or RO-3306 (9 μM) (B) and stained for NuMA (red) as well as p150Glued (green). Stacked columns on the right show the extent of cortical NuMA/p150Glued, which was either absent (‘No’), ‘Weak’ (as for instance in (A)), or ‘Strong’ (as for instance in (B)) (n>100 in each condition; NuMA and p150Glued systematically colocalize and are thus reported together). (C) Time-lapse recording of HeLa cells expressing DYNC1H1-GFP and treated with RO-3306 (9 μM) for 5 min. Quantification of DYNC1H1-GFP cortical enrichment (right) determined by calculating the mean intensity of cortical signal (area drawn) and subtracting from this value the mean intensity value in the cytoplasm (similar area) and correcting for background signal (n=5, error bars: s.e.m.). (D–I) Metaphase and anaphase HeLa cells, as indicated, transfected with GFP-NuMA (D, E), GFP-NuMA(T>E) (F, G) or GFP-NuMA(T>A) (H, I), fixed 36 h thereafter and stained for GFP (green). Stacked columns on the right show the extent of cortical GFP localization in metaphase (Met) and anaphase (Ana), as mentioned above (n>60 in each condition except anaphase GFP-NuMA(T>E), where n=46).
To further investigate the importance of NuMA phosphorylation at T2055 by CDK1, we generated fusion proteins between GFP and nonphosphorylatable (T>A) or phosphomimetic (T>E) mutants of NuMA for the 2055 residue, and compared their distribution to that of GFP fused to the wild-type protein. Interestingly, we found that in contrast to GFP-NuMA (Figure 4D and E), in the majority of cells GFP-NuMA(T>E) does not localize to the cortex in either metaphase or anaphase (Figure 4F and G). In addition, cells expressing GFP-NuMA(T>E) exhibit strong GFP signal at spindle poles as well as mitotic abnormalities, including chromosome congression defects (see Figure 4F). The lack of cortical localization of GFP-NuMA(T>E) is reminiscent of phospho-T2055 (compare Figure 3G with Figure 4F), further confirming that NuMA phosphorylated at T2055 does not localize to the cortex. Conversely, GFP-NuMA(T>A) is strongly enriched at the cortex already in metaphase, and remains strongly enriched at that location in anaphase (Figure 4H and I).
Overall, the premature strong cortical accumulation of GFP-NuMA(T>A) in metaphase and the lack of cortical localization of GFP-NuMA(T>E) in anaphase indicate that CDK1-mediated phosphorylation at T2055 acts as a switch that modulates the levels of cortical NuMA as cells progress through mitosis.
Balanced levels of cortical NuMA/dynein is achieved by opposing CDK1 and PPP2CA phosphatase activities
How can the pool of nonphosphorylated NuMA that localizes weakly to the cell cortex in metaphase be shielded from the action of CDK1? As shown in Figure 4C, we found that brief incubation with RO-3306 leads to a rapid increase in cortical dynein in metaphase cells, indicating that there is an activity counteracting CDK1 during metaphase.
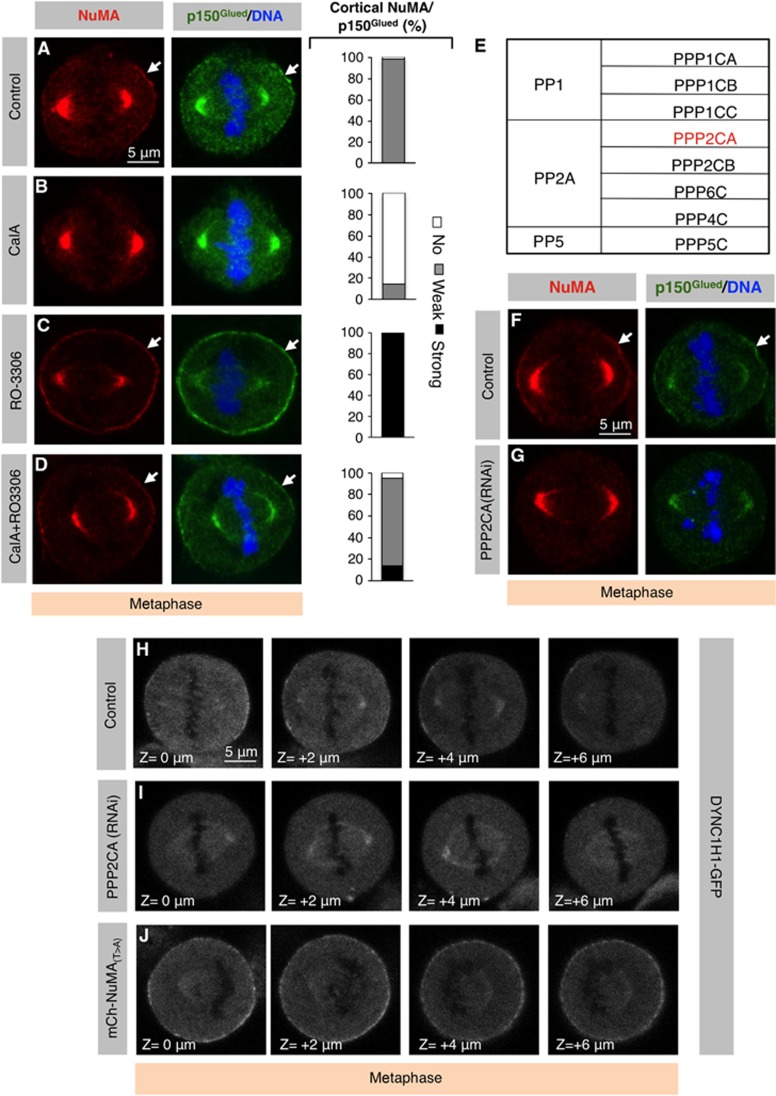
We explored whether this activity corresponds to a counteracting phosphatase. To this end, we first assayed the consequence of inhibiting phosphatase activity by briefly exposing metaphase cells to Calyculin A (CalA), an inhibitor of PP1 and PP2A protein phosphatases. As can be appreciated by comparing Figure 5B with Figure 5A, we found that cortical localization of NuMA/p150Glued is severely impaired in cells treated with CalA. In principle, CalA could influence NuMA/p150Glued cortical localization directly through 2055 or instead indirectly through another residue, perhaps even on another protein. To distinguish between these possibilities, we treated cells expressing GFP-NuMA(T>A) with CalA and analysed GFP cortical localization. As shown in Supplementary Figure S5A, we found that GFP-NuMA(T>A) expressing cells treated with CalA exhibit an enriched cortical signal analogous to that of untreated cells (see Figure 4H), demonstrating that CalA directs NuMA localization through T2055. In line with these results, we found that the strong cortical enrichment observed in metaphase cells briefly treated with the CDK1 inhibitor RO-3306 is alleviated by the simultaneous treatment with CalA (Figure 5D). Similarly, western blot analysis indicates that simultaneous inactivation of CDK1 and of phosphatases results in levels of phospho-T2055 analogous to that of control conditions, without altering total NuMA levels (Supplementary Figure S4A).
Figure 5.
A balance of CDK1 kinase and PPP2CA phosphatase activities determines the extent of cortical NuMA/dynein localization. (A–D) Metaphase HeLa cells treated with 0.1% DMSO (Control) (A), Calyculin A (CalA) (50 nM) (B), RO-3306 (9 μM) (C) or CalA plus RO-3306 (D), stained for NuMA (red) as well as p150Glued (green). Stacked column on the right shows the extent of cortical NuMA/p150Glued localization as explained in the legend of Figure 4 (n>100 in each condition). (E) The eight human PPP family catalytic subunits fall into three subgroups (PP1, PP2A and PP5). (F, G) Metaphase cells transfected with control siRNAs (F) or siRNAs against PPP2CA (G), fixed 72 h thereafter and stained for NuMA (red) as well as p150Glued (green). For quantification see Supplementary Figure S5B. (H–J) Z-projection of images that are 2 μm apart of metaphase cell stably expressing DYNC1H1-GFP and transfected with control siRNAs (H), PPP2CA siRNAs (I) or transfected with mCherry-NuMA(T>A) (J). Note loss of cortical DYNC1H1-GFP in cells transfected with siRNAs against PPP2CA and enrichment of DYNC1H1-GFP in cells expressing mcherry-NuMA(T>A). For each condition, 30 cells were analyzed and representative results are shown.
In order to identify the relevant phosphatase(s) needed for NuMA/dynein cortical localization during mitosis, we depleted the catalytic subunit of each phospho-protein phosphatase (PPP) family member. This includes PP1 and PP2A phosphatases, which are both affected by low concentrations of CalA as used in our analysis, as well as PP5 phosphatase, which can be affected by higher drug concentrations (Figure 5E) (Picard et al, 1989; Axton et al, 1990; Felix et al, 1990). We depleted each of these components using two independent siRNAs which revealed that only the depletion of PPP2CA causes loss of NuMA/p150Glued cortical staining in metaphase cells (Supplementary Figure S5B; compare Figure 5G with Figure 5F; Supplementary Table S1). Similarly, we found that PPP2CA depletion causes loss of cortical DYNC1H1-GFP (compare Figure 5I with Figure 5H). We found that PPP2CA depletion also results in other mitotic defects, including chromosome congression and segregation errors, as reported previously (Neumann et al, 2010; data not shown). This complicated conducting a thorough analysis of the effect of PPP2CA depletion on NuMA/dynein localization during anaphase. Although Reversine would have potentially allowed us to circumvent this limitation, instead we examined the dependency of cortical NuMA accumulation on PPP2CA in conditions where CDK1 is prematurely inactivated during metaphase using RO-3306, thus mimicking the anaphase situation. Interestingly, we found that brief inactivation of CDK1 by RO-3306 in cells depleted of PPP2CA results in a reduction of cortical NuMA (compare Supplementary Figure S5D with Supplementary Figure S5C). Furthermore, we found that CalA-treated cells exhibit diminished levels of cortical NuMA during anaphase (Supplementary Figure S5E and F).
Taken together, we conclude that balanced activities of CDK1 kinase and PPP2CA phosphatase are essential for proper distribution of cortical NuMA/dynein during metaphase. In addition, these results suggest that the mere inhibition of CDK1 as cells progress into anaphase is not sufficient for achieving cortical NuMA localization, but must be accompanied by sustained phosphatase activity.
Balanced level of cortical NuMA is essential for dynein-dependent metaphase spindle positioning
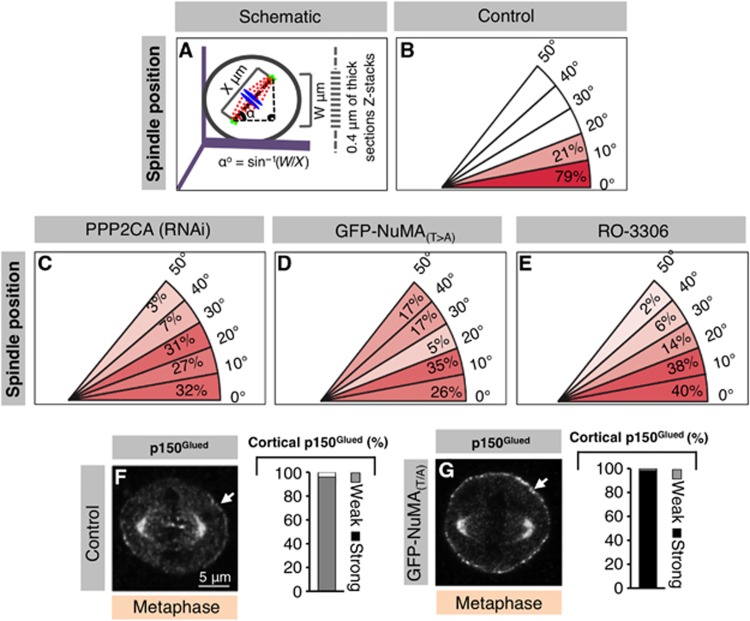
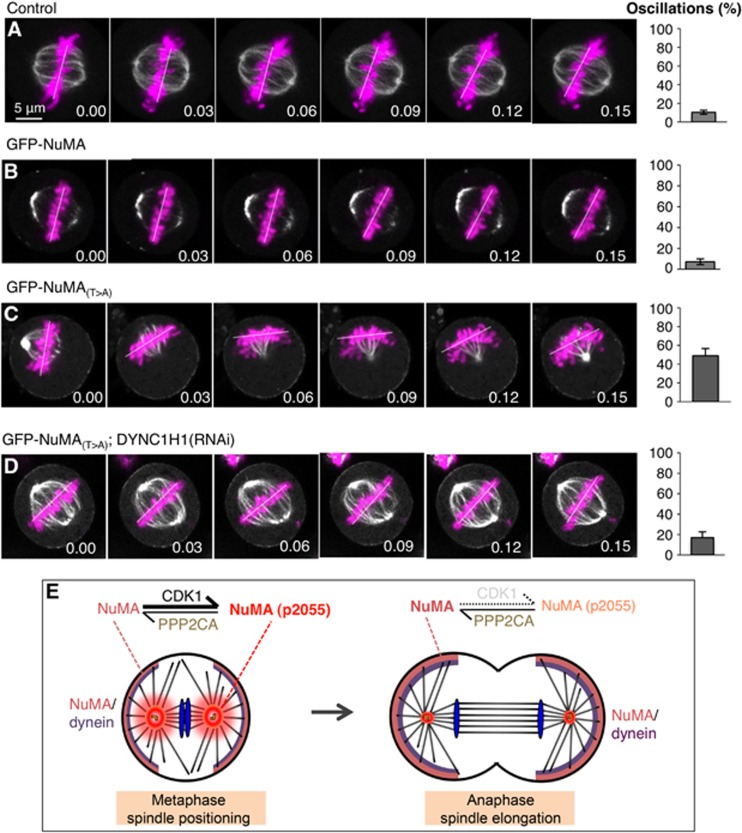
We set out to address the significance of the balanced activities of CDK1 and PPP2CA during metaphase. To this end, we monitored spindle positioning using coverslips uniformly coated with fibronectin that usually directs the metaphase spindle to align parallel to the substratum in control conditions (Toyoshima and Nishida, 2007) (Figure 6A and B). In contrast, we found that siRNA-mediated depletion of PPP2CA results in the spindle adopting a significantly more random position (Figure 6C). Analogous results were obtained by briefly treating cells with CalA (compare Supplementary Figure S5H with Supplementary Figure S5G). In addition, spindle positioning is also affected in cells depleted of endogenous NuMA and expressing the phosphomimetic form GFP-NuMA(T>E) that does not localize to the cell cortex (Supplementary Figure S5I). Next, we tested whether GFP-NuMA(T>A), which results in excess cortical NuMA/dynein (compare Figure 4H with Figure 4D; Figure 5J with Figure 5H; and Figure 6G with Figure 6F), interferes with this process. In stark contrast to control cells, we found that expression of GFP-NuMA(T>A) also causes a near randomization of spindle position (Figure 6D). Similarly, brief exposure of metaphase cells to the CDK1 inhibitor RO-3306 also randomizes spindle position (Figure 6E). In addition, live imaging established that whereas the metaphase spindle does not exhibit much oscillations in control cells or in cells expressing GFP-NuMA (Figure 7A and B; Supplementary Movies S9 and S10), dramatic oscillations result from GFP-NuMA(T>A) expression (Figure 7C; Supplementary Movie S11). To address whether excess dynein is causative of these excess oscillations, we partially depleted the dynein heavy chain DYNC1H1 in cells expressing GFP-NuMA(T>A). Importantly, we found that such depletion abrogates the exaggerated spindle oscillations observed upon GFP-NuMA(T>A) expression (Figure 7D, compare with Figure 7C; Supplementary Movie S12).
Figure 6.
Appropriate levels of dynein mediated by opposing CDK1 and PPP2CA activities are necessary for proper metaphase spindle positioning. (A) Analysis of metaphase spindle positioning performed by computing the distance (X, μm) between the two spindle poles. The angle was calculated by imaging Z-stacks of 0.4 μm-thick sections and calculating the angle using an inverse trigonometric function, as illustrated. (B–E) Distribution of metaphase spindle angles with respect to the fibronectin substratum in cells transfected with Control siRNAs (B), PPP2CA siRNAs (C), cells transfected with GFP-NuMA(T>A) (D) or cells treated with RO-3306 (9 μM) (E). A minimum of 50 cells were analysed for each condition and the significance determined using a two-tailed Student’s t-test (P <0.0005 in C–E compared to B). (F, G) Untransfected control HeLa cells (F) or HeLa cells transfected with GFP-NuMA(T>A) (G) and stained for p150Glued (grey). Stacked columns on the right show the extent of cortical p150Glued localization in metaphase, similar to Figure 4 (n>100 in each condition).
Figure 7.
Excess cortical NuMA causes dynein-dependent metaphase spindle oscillations. (A–D) Images from time-lapse recordings of metaphase HeLa Kyoto cells stably expressing GFP-α-tubulin as well as mCherry-H2B (A) and transfected with GFP-NuMA (B), or transfected with either GFP-NuMA(T>A) (C) or GFP-NuMA(T>A) and treated with DYNC1H1 siRNAs in addition (D) (see also corresponding Supplementary Movies S9–S12). The white line indicates the position of chromosomes. For each condition, 10 cells were imaged. The bar graphs on the right represent the frequency at which chromosome position changes >10° between two frames, along with the s.e.m. Two-tailed Student’s t-tests show that the extent of spindle oscillations upon overexpression of GFP-NuMA(T>A) is statistically different from that observed either in control conditions (P<0.0001) or upon overexpression of GFP-NuMA (P<0.0005), and that in GFP-NuMA(T>A) plus DYNC1H1 (RNAi) statistically different from the GFP-NuMA(T>A) condition (P<0.0005). Time is indicated in h.min. (E) Model for cortical localization of NuMA/dynein during metaphase and anaphase. During metaphase (left), the counteracting influences of CDK1 and PPP2CA phosphatases on T2055 results in the presence of moderate levels of unphosphorylated NuMA, and thus of dynein, at the cell cortex; such moderate levels are critical for ensuring proper spindle positioning. During anaphase, upon CDK1 inactivation and continued PPP2CA phosphatase activity, the balance is shifted towards unphosphorylated NuMA, thus leading to higher levels of cortical dynein, which are critical for robust spindle elongation during anaphase B.
We conclude that a balance of CDK1 and PPP2CA activities in metaphase is crucial to ensure proper NuMA and dynein levels at the cell cortex and thus accurate spindle positioning.
Discussion
Elucidation of the mechanisms that govern spindle positioning during metaphase and spindle elongation during anaphase is important for a thorough understanding of the spatial and temporal control of cell division. The work reported here reveals a novel mechanism whereby levels of cortical dynein are regulated by the phosphorylation status of NuMA at T2055 that serves to coordinate cell cycle progression with proper spindle behaviour during mitosis (Figure 7E).
Correct spindle positioning dictates the relative sizes of daughter cells and ensures the appropriate segregation of cell-fate determinants during cell division (reviewed in Gönczy, 2008; Knoblich, 2008). Therefore, spindle positioning is critical notably for asymmetric cell division during development and in stem cell lineages and, when aberrant, can lead to tumour development (Caussinus and Gonzalez, 2005; reviewed in Knoblich, 2010, Inaba and Yamashita, 2012). Studies in a number of systems established that a ternary complex, which in human cells comprises NuMA/LGN/Gαi1–3, recruits dynein to the cortex, where the motor protein is thought to direct spindle positioning by promoting pulling on astral microtubules (reviewed in Gönczy, 2008; Knoblich, 2008; Siller and Doe, 2009; Morin and Bellaïche, 2011). These findings notwithstanding, the mechanisms governing the spatial and temporal regulation of dynein distribution as cells progress through mitosis remain poorly understood.
Counteracting activity of CDK1 and PPP2CA on NuMA regulate cortical dynein levels in mitosis
The polo-like kinase Plk1 negatively regulates cortical dynein distribution during metaphase by inhibiting its interaction with NuMA/LGN (Kiyomitsu and Cheeseman, 2012). Our work demonstrates that, in addition, NuMA must remain nonphosphorylated at T2055 to be present at the cell cortex during metaphase and anaphase. We establish that during metaphase, cortical levels of nonphosphorylated NuMA are kept low, resulting in likewise low levels of cortical dynein. We further demonstrate that NuMA levels are kept in check by a balance of CDK1 and PPP2CA activity. Since CDK1 activity is high during metaphase, the majority of NuMA is phosphorylated at T2055 and only a small fraction of molecules is dephosphorylated by PPP2CA. Premature CDK1 inactivation or loss of PPP2CA dramatically alter levels of nonphosphorylated cortical NuMA and thus dynein in an antagonistic manner; in both cases, spindle positioning is perturbed. Using a chemical-based approach, together with RNAi, we show that brief inactivation of CDK1 in metaphase cells depleted of PPP2CA causes dramatic diminutions (Supplementary Figure S5D), but not complete loss, of cortical NuMA, as upon PPP2CA depletion alone (Figure 5G). This suggests that CDK1 could also be involved in negatively regulating PP2A activity during metaphase, as previously postulated (Schmitz et al, 2010; reviewed by Wurzenberger and Gerlich, 2011; Mochida and Hunt, 2012). Interestingly, the CDK1 consensus sequence within the tail of NuMA is conserved in vertebrates (Supplementary Figure S4D), suggesting that the CDK1/PPP2CA-mediated mechanism may operate in other species. Because CDK1 is activated first in the vicinity of centrosomes (reviewed in Basto and Pines, 2007), we propose that a source of CDK1 kinase activity emanating from the centrosomes is opposed by uniform PPP2CA phosphatase activity, generating a gradient of nonphosphorylated NuMA away from the spindle poles. Our findings argue against Plk1 being involved in spindle positioning through the regulation of PPP2CA, because Plk1 acts on dynein but not on NuMA (Kiyomitsu and Cheeseman, 2012). In contrast, we show here that both NuMA and dynein are affected by the phosphatase inhibitor CalA or PPP2CA depletion. In addition, the Abl1 kinase phosphorylates NuMA on Tyrosine 1774 to maintain its localization during metaphase, but whether this also influences cortical dynein distribution has not been investigated (Matsumura et al, 2012). Overall, it is likely that multiple regulatory mechanisms contribute to the homeostasis of cortical dynein and thus proper spindle positioning.
Cortical dynein regulates spindle elongation
The importance of spindle elongation during anaphase B has been documented in several systems (reviewed in Roostalu et al, 2010). For instance, Saccaromyces cerevisiae, Schizosaccharomyces pombe and Ustilago maydis all utilize strictly anaphase B for chromosome segregation (Funabiki et al, 1993; Straight et al, 1997; Fink et al, 2006). In contrast, most vertebrate cells rely on both anaphase A and anaphase B for segregating chromosomes (Aist et al, 1993). Despite their importance for genome integrity, the molecular underpinning of spindle elongation has remained elusive in human cells. Here, we demonstrate that dynein enrichment at the cell cortex increases markedly during anaphase in a NuMA-dependent manner. Importantly, our analysis further uncovers that NuMA-dependent cortical dynein localization is required for robust separation of the two sets of chromosomes and of the spindle poles, probably by exerting pulling forces on astral microtubules. In addition, our work reveals that when CDK1 becomes inactive at the onset of anaphase, the bulk of NuMA is dephosphorylated by PPP2CA. As a result, NuMA becomes significantly enriched at the cell cortex and recruits more cortical dynein, thus enabling robust spindle elongation. In Drosophila embryos, expression of nondegradable CyclinB to maintain high CDK1 activity in anaphase impairs spindle elongation (Parry and O'Farrell, 2001), and our work raises the possibility that one relevant substrate in this context could be the NuMA homologue Mud.
In conclusion, our study uncovers a novel mechanism by which NuMA serves as a critical linker between cell cycle progression and spindle behaviour to ensure the faithful execution of cell division.
Materials and methods
Cell culture, cell synchronization and transfection
HeLa cells expressing GFP-Centrin 1 (Piel et al, 2000), HeLa Kyoto cells expressing EGFP-α-tubulin and mCherry-H2B (Schmitz et al, 2010), HeLa Kyoto cells expressing mouse DYNC1H1-GFP (Hutchins et al, 2010) and HeLa cells expressing mCherry-H2B, MCF7, RPE1, U2OS as well as MEFs were cultured in high-glucose DMEM with GlutaMAX (Invitrogen) media supplemented with 10% fetal calf serum (FCS) in a humidified 5% CO2 incubator at 37°C. For monitoring spindle positioning in fixed specimens, cells were grown on coverslips uniformly coated with fibronectin (BD Bioscience, 354088) and synchronized using a double thymidine block. In brief, cells were incubated with 2 mM thymidine for 17 h, released for 8 h and again incubated with 2 mM thymidine for 17 h. Cells were then released and fixed after 10.5 h, when the maximum of cells were in mitosis. For obtaining cell lysates at different stages of cell cycle, a double thymidine block was performed as mentioned above and cells were lysed after 3 h (for S phase), 8 h (for G2), 10.5 h (for Met), 11.5 h (for Tel) and 15 h (for G1), respectively. For siRNA experiments, ∼100 000 cells were seeded in 6-well plates. Then, 6 μl of 20 μM siRNAs in 100 μl OptiMEM medium (Invitrogen) and 4 μl of Lipofectamine RNAiMAX (Invitrogen) in 100 μl OptiMEM were incubated in parallel for 5 min, mixed for 20 min and then added to 2.5 ml medium per well. For siRNA experiments in which synchronized lysates were collected, cells were synchronized starting 6 h after transfection with a double thymidine block as indicated above. For plasmid transfections, cells were seeded at 80–90% confluency. Then, 4 μg of plasmid DNA in 100 μl OptiMEM and 4 μl of Lipofectamine 2000 (Invitrogen) in 100 μl OptiMEM were incubated in parallel for 5 min, mixed for 20 min and added to each well. Transfection efficiency of 80% was routinely achieved as monitored by GFP staining in all the cases except for cells expressing GFP-NuMA(T>E), where transfection efficiency was ∼50%.
Plasmids and RNAi
All NuMA clones were constructed using full-length NuMA as a template with appropriate PCR primer pairs (the sequences of all primers is available upon request). The amplified products were subcloned into pcDNA3-GFP (Merdes et al, 2000). Site-directed mutagenesis to create nonphosphorylatable (T2055A) and phosphomimetic (T2055E) clones of NuMA were performed using site-directed mutagenesis kit (Agilent Technologies, 210515) with appropriate primers.
Double-stranded siRNA oligonucleotides were synthesized with the sequences: 5′-AAGGGCGCAAACAGAGCACUA-3′ (NuMA-siRNA (NuMA si_1), Qiagen); 5′-CAGCGCCAACUCAUCGUUCUA-3′ (NuMA-siRNA (NuMA si_2), Qiagen); 5′-CCAGACAGCGCCAACUCAUCGUUCU-3′ (NuMA-siRNA Stealth (Life Technologies)); and 5′-CAGGUGGGUGUACAUUACGAA-3′ (DYNC1H1-siRNA, Qiagen). Information for siRNAs used for PPP catalytic subunit depletion are provided in Supplementary Table S1. To generate a siRNA-resistant allele of NuMA, the sequence 5′-CCAGACAGCGCCAACTCATCGTTC-3′ was changed to 5′-CCTGATTCCGCAAACTCCAGCTTT-3′ by introducing silent mutations.
For NuMA depletion, cells were treated with siRNAs a second time 48 h after the initial transfection and analysed 48 h thereafter. Despite such double RNAi treatment, 52% of anaphase cells still possessed weak NuMA signal at the cell cortex detected by immunostaining; such cells were excluded for the pole–pole measurements reported in Figure 2F. For PPP depletion, cells were treated with given siRNA species for 72 h (see Supplementary Table S1 for siRNA information).
Dynein inhibition was performed by treating mCherry-H2B cells with 500 μM Ciliobrevin A in 0.5% of DMSO for 15 min (Firestone et al, 2012) (Sigma-Aldrich, H4541); cells undergoing the metaphase to anaphase transition were then analysed. For MPS-1 inhibition, cells were treated with 1 μM Reversine in 0.1% of DMSO (Santaguida et al, 2010) (Santa Cruz, sc-203236) for 15 min, and mCherry-H2B cells undergoing the metaphase to anaphase transition were analysed by time-lapse microscopy.
CDK1 inhibition was performed by treating metaphase synchronized cells for 5 min with either 9 μM RO-3306 in 0.1% DMSO (Vassilev et al, 2006) (Santa Cruz, sc-358700) or 10 μM Roscovitine in 0.1% DMSO (LC Laboratories, R-1234). For PP1/PP2A inhibition, metaphase synchronized cells were exposed for 10 min to 50 nM CalA in 0.1% DMSO (LifeTechnologies, PHZ1044) and either metaphase or early anaphase cells were scored 10 min later for cortical NuMA/p150Glued localization. Similarly, simultaneous inhibition of CDK1 and PP1/PP2A was performed by exposing cells synchronized in metaphase for 10 min with RO-3306 and CalA at the above-mentioned concentrations.
Spindle positioning assay, pole–pole measurements, chromosome–chromosome distance and line scans
The angle of the metaphase spindle with respect to the fibronectin substratum was determined as described previously (Toyoshima and Nishida, 2007). In brief, cells were stained with γ-tubulin antibodies to mark spindle poles and counterstained with 1 μg/ml Hoechst 33342 (Sigma-Aldrich) to mark DNA. Stacks of confocal images 0.4 μm apart were acquired and the distances between the two spindle poles in Z and in XY determined using Imaris (Bitplane Inc.); the spindle angle to the substratum was calculated using inverse trigonometry.
Pole–pole distances were calculated using Imaris (Bitplane Inc.) on metaphase and anaphase cells cultured on uniform fibronectin, stained with γ-tubulin antibodies and imaged as described above. Chromosome–chromosome distance was calculated using Imaris (Bitplane Inc.) on cells expressing mCherry-H2B and undergoing metaphase to anaphase transition. The measurements were performed from the centre of mass of the separating chromosomes in the fluorescence channel.
Line scans of pixel intensity were computed in ImageJ.
Indirect immunofluorescence and time-lapse imaging of HeLa cells
For immunofluorescence, cells were fixed in −20°C methanol for 7–10 min and washed in PBS-0.05% Triton X-100 (PBST). After blocking in 1% bovine serum albumin (BSA) in PBST for 1 h, cells were incubated with primary antibodies overnight at 4°C. Following three washes in PBST for 5 min each, cells were incubated with secondary antibodies for 1 h at room temperature, counterstained with 1 μg/ml Hoechst 33342, washed three times for 5 min in PBST and mounted. Primary antibodies were 1:200 rabbit anti-NuMA (Santa Cruz, sc-48773), 1:200 mouse anti-p150Glued (Transduction Laboratories, 612709), 1:2000 mouse anti-γ-tubulin (GTU88, Sigma-Aldrich), 1:300 mouse anti-GFP (MAB3580, Millipore) and 1:500 rabbit anti-GFP (gift from V Simanis). Phosphospecific antibodies against T2055 residue (p2055) were raised against AFSILNT(PO3H2)PKKLG. Secondary antibodies were Alexa488 anti-mouse, Alexa488 anti-rabbit, Alexa568 anti-mouse and Alexa568 anti-rabbit, all used in a 1:500 dilution (Invitrogen). Confocal images were acquired on a Zeiss LSM 710 confocal microscope equipped with a Axiocam MRm (B/W) CCD camera using a 63 × NA 1.0 oil objective and processed in ImageJ and Adobe Photoshop, maintaining relative image intensities. For qualitative assessment of cortical levels, the entire cell was taken into consideration by focussing through it; for illustration purpose, a single plane is shown where the signal intensity is maximal.
Time-lapse microscopy was conducted on a Zeiss LSM 700 confocal microscope equipped with a Axiocam MRm (B/W) CCD camera using a 40 × NA 1.3 oil objective, using a Hi Q4 dish (Ibidi) at 5% CO2, 37°C, 90% humidity. Images were acquired every 3 min, capturing 4–5 sections 7 μm apart at each time point. For acute CDK1 inactivation with RO-3306 in cells expressing DYNC1H1-GFP, images were acquired every 1 min, capturing 4 sections 2 μm apart. Time-lapse figures and movies were generated using a single confocal section of the Z-stack at each time point.
The in vitro kinase assays and immunoblotting
To assay CDK1/CyclinB kinase activity, His-CDK1/GST-CyclinB (Millipore, 14–450) were incubated with bacterially expressed NiNTA beads (Qiagen) purified C-ter NuMA (wild type) as well as C-ter NuMA (T2055A) at room temperature for 30 min in kinase assay buffer (20 mM Hepes, pH 7.8; 10 mM MgCl2, 15 mM KCl, 1 mM EGTA and 0.1 mg/ml BSA) supplemented with 200 μM ATP and 2 μCi of 32P-ATP (Hartmann Analytic), with or without 3 μM RO-3306; the samples were analysed by autoradiography after SDS–PAGE.
The specificity of phospho-specific antibodies (p2055) was determined by conducting an analogous in vitro phosphorylation reaction using cold ATP and then immunoblotting with p2055 antibodies, using 1:1000 rabbit anti-p2055 and 1:10 000 mouse anti-actin (ABNOVA, MAB8172).
Supplementary Material
Acknowledgments
We thank Andreas Merdes, Daniel Gerlich, Oliver Hantschel, Anthony Hyman and Arnaud Echard for providing plasmid and cell lines. We are thankful to Viesturs Simanis, Fernando R Balestra, Virginie Hachet, Benita Wolf, Zoltan Spiro and Sveta Chakrabarti for critical comments on the manuscript. We thank Daniel Gerlich for his invaluable advice on phosphatases. We are grateful to the EPFL School of Life Sciences microscopy core facility (PT-BiOP) for imaging advice. SK held a postdoctoral fellowship from the EMBO (ALTF-366-2009). This study was also supported by a grant to PG from the Swiss National Science Foundation (3100A0-122500/1).
Author contribution: SK and PG conceived and designed the experiments; SK and CB executed experiments; SK and PG interpreted the results and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aist JR, Berns MW (1981) Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J Cell Biol 91(2 Pt 1): 446–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aist JR, Liang H, Berns MW (1993) Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J Cell Sci 104(Pt 4): 1207–1216 [DOI] [PubMed] [Google Scholar]
- Axton JM, Dombradi V, Cohen PT, Glover DM (1990) One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell 63: 33–46 [DOI] [PubMed] [Google Scholar]
- Basto R, Pines J (2007) The centrosome opens the way to mitosis. Dev Cell 12: 475–477 [DOI] [PubMed] [Google Scholar]
- Cande WZ (1982) Inhibition of spindle elongation in permeabilized mitotic cells by erythro-9-[3-(2-hydroxynonyl)] adenine. Nature 295: 700–701 [DOI] [PubMed] [Google Scholar]
- Cande WZ (1982) Permeabilized cell models for studying chromosome movement in dividing PtK1 cells. Methods Cell Biol 25(Pt B): 57–68 [DOI] [PubMed] [Google Scholar]
- Cande WZ, Wolniak SM (1978) Chromosome movement in lysed mitotic cells is inhibited by vanadate. J Cell Biol 79(2 Pt 1): 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E, Gonzalez C (2005) Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet 37: 1125–1129 [DOI] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL (2012) Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell 23: 3380–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Luo C (1995) Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J Cell Sci 108(Pt 2): 621–633 [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M (2007) Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol 179: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Macara IG (2004) Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119: 503–516 [DOI] [PubMed] [Google Scholar]
- Du Q, Taylor L, Compton DA, Macara IG (2002) LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr Biol 12: 1928–1933 [DOI] [PubMed] [Google Scholar]
- Felix MA, Cohen P, Karsenti E (1990) Cdc2 H1 kinase is negatively regulated by a type 2A phosphatase in the Xenopus early embryonic cell cycle: evidence from the effects of okadaic acid. EMBO J 9: 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G (2006) Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J 25: 4897–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O'Donnell M, Gelfand VI, Kapoor TM, Chen JK (2012) Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 484: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121: 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J (2010) Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol 189: 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehmlich K, Haren L, Merdes A (2004) Cyclin B degradation leads to NuMA release from dynein/dynactin and from spindle poles. EMBO Rep 5: 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P (2008) Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol 9: 355–366 [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet C, Weber K, Osborn M (1996) NuMA: a bipartite nuclear location signal and other functional properties of the tail domain. Exp Cell Res 225: 207–218 [DOI] [PubMed] [Google Scholar]
- Haren L, Merdes A (2002) Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J Cell Sci 115(Pt 9): 1815–1824 [DOI] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A et al. (2010)) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yamashita YM (2012) Asymmetric stem cell division: precision for robustness. Cell Stem Cell 11: 461–469 [DOI] [PubMed] [Google Scholar]
- Kardon JR, Vale RD (2009) Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW (1996) How proteolysis drives the cell cycle. Science 274: 1652–1659 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM (2012) Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol 14: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA (2008) Mechanisms of asymmetric stem cell division. Cell 132: 583–597 [DOI] [PubMed] [Google Scholar]
- Knoblich JA (2010) Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gönczy P (2012) Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol 199: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Rodriguez-Bravo V, Medema RH (2009) The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol 185: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S, Hamasaki M, Yamamoto T, Ebisuya M, Sato M, Nishida E, Toyoshima F (2012) ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat Commun 3: 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW (2000) Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW (1996) A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87: 447–458 [DOI] [PubMed] [Google Scholar]
- Mochida S, Hunt T (2012) Protein phosphatases and their regulation in the control of mitosis. EMBO Rep 13: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Bellaïche Y (2011) Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell 21: 102–119 [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R et al. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gönczy P (2007) Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol 9: 1294–1302 [DOI] [PubMed] [Google Scholar]
- Park DH, Rose LS (2008) Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev Biol 315: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, O'Farrell PH (2001) The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr Biol 11: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard A, Capony JP, Brautigan DL, Doree M (1989) Involvement of protein phosphatases 1 and 2A in the control of M phase-promoting factor activity in starfish. J Cell Biol 109(6 Pt 2): 3347–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M (2000) The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol 149: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Schiebel E, Khmelinskii A (2010) Cell cycle control of spindle elongation. Cell Cycle 9: 1084–1090 [DOI] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A (2010) Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol 190: 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, Hyman AA, Mechtler K, Peters JM, Gerlich DW (2010) Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol 12: 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Doe CQ (2009) Spindle orientation during asymmetric cell division. Nat Cell Biol 11: 365–374 [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277: 574–578 [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E (2007) Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J 26: 1487–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L (2006) Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA 103: 10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH (2010) Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol 30: 3519–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW (2011) Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 12: 469–482 [DOI] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Snyder M (1992) NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol 116: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Snyder M (1992) The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol Biol Cell 3: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.