Abstract
In rheumatoid arthritis (RA), macrophage is one of the major sources of inflammatory mediators. Macrophages produce inflammatory cytokines through toll-like receptor (TLR)-mediated signalling during RA. Herein, we studied macrophages from the synovial fluid of RA patients and observed a significant increase in activation of inositol-requiring enzyme 1α (IRE1α), a primary unfolded protein response (UPR) transducer. Myeloid-specific deletion of the IRE1α gene protected mice from inflammatory arthritis, and treatment with the IRE1α-specific inhibitor 4U8C attenuated joint inflammation in mice. IRE1α was required for optimal production of pro-inflammatory cytokines as evidenced by impaired TLR-induced cytokine production in IRE1α-null macrophages and neutrophils. Further analyses demonstrated that tumour necrosis factor (TNF) receptor-associated factor 6 (TRAF6) plays a key role in TLR-mediated IRE1α activation by catalysing IRE1α ubiquitination and blocking the recruitment of protein phosphatase 2A (PP2A), a phosphatase that inhibits IRE1α phosphorylation. In summary, we discovered a novel regulatory axis through TRAF6-mediated IRE1α ubiquitination in regulating TLR-induced IRE1α activation in pro-inflammatory cytokine production, and demonstrated that IRE1α is a potential therapeutic target for inflammatory arthritis.
Keywords: inflammation, IRE1, TRAF6, ubiquitination
Introduction
Rheumatoid arthritis (RA) is a chronic debilitating disease characterized by leukocyte infiltration, hyper-proliferation of synovial cells, and bone destruction. The elevated levels of toll-like receptors (TLRs) and their endogenous ligands significantly contribute to the initiation and progression of RA. TLRs were found to be highly expressed in macrophages and synovial fibroblasts from patients with RA (Seibl et al, 2003; Proost et al, 2004; Varoga et al, 2006; Ospelt et al, 2008; Yavuz et al, 2008; Kawai and Akira, 2011). Endogenous TLR ligands, such as fibrinogen, heat shock protein 22 (HSP22) (Roelofs et al, 2006), HSP60 (Ohashi et al, 2000), HSP70 (Asea et al, 2002), HSP96 (Huang et al, 2009) and the high mobility group box chromosomal protein 1 (HMGB-1) (Pisetsky et al, 2008; van Beijnum et al, 2008; Wahamaa et al, 2011), are induced or released in the synovial tissue of patients with RA. Upon recognizing specific ligands, TLR signalling transduction is initiated by individual TLRs that recruit adaptor proteins, namely, myeloid differentiation primary response gene 88 (MyD88) or TIR (Toll/IL-1 receptor) domain-containing adaptor inducing interferon (IFN)-β (TRIF). MyD88 is recruited by most of the TLRs to transmit signals culminating in NF-κB and mitogen-activated protein kinase (MAPK) activation and the induction of inflammatory cytokines. Additionally, TLR3 and TLR4 utilize TRIF to activate an alternative pathway that leads to the activation of NF-κB and IRF3 as well as the induction of type I IFN and inflammatory cytokines, such as TNF-α, IL-1β, and IL-6. TLR4 is the only TLR that activates two distinct signalling pathways: the MyD88-dependent and TRIF-dependent pathways (Cao et al, 2008; Gilliet et al, 2008; Egan et al, 2009). Targeting of TLRs to treat RA has shown promise (O’Neill, 2003). Indeed, Chaperonin 10, a mitochondrial protein that inhibits TLRs through blocking their binding to DAMPs, was clinically effective in a small-scale study with RA patients (Vanags et al, 2006).
The endoplasmic reticulum (ER) is an intracellular organelle responsible for protein folding and assembly, lipid and sterol biosynthesis, and calcium storage. Pathophysiological states that increase the demand for protein folding, or stimuli that disrupt protein folding, create an imbalance between the protein-folding load and capacity of the ER, causing the accumulation of unfolded or misfolded proteins in the ER lumen, a condition referred to as ‘ER stress’ (Kaser et al, 2011). To cope with ER stress, the ER has evolved a group of signal transduction pathways, that is, the unfolded protein response (UPR) pathways, mediated by three major ER transmembrane protein factors, including PKR-like ER kinase (PREK), inositol-requiring enzyme 1α (IRE1α), and activating transcription factor 6 (ATF6) (Zhang and Kaufman, 2006; Lee and Glimcher, 2009; Hotamisligil, 2010). IRE1α is the most conserved UPR transducer that is known to function as a protein kinase and endoribonuclease (RNase). During ER stress, IRE1α is homo-dimerized and auto-phosphorylated to render its RNase activity. Activated IRE1α executes an unconventional splicing of the mRNA encoding X-box binding protein 1 (XBP-1) by removing a 26-base intron (Shen et al, 2001; Calfon et al, 2002). Spliced XBP-1 mRNA encodes a potent transcription factor that activates the expression of several genes involved in protein folding, secretion, degradation, as well as inflammation and metabolism (Sha et al, 2011). During ER stress, the cytosolic domain of activated IRE1α recruits tumour necrosis factor receptor-associated factor 2 (TRAF2), which leads to the activation of c-JUN N-terminal kinase (JNK) through apoptosis signal-regulating kinase 1 (ASK1) (Urano et al, 2000).
Although recognition of unfolded proteins is generally regarded as the primary activation model for the IRE1α-mediated UPR, recent studies suggested that IRE1α signalling might not rely solely on ER-luminal sensing of unfolded proteins. In fact, TLR4 and TLR2 can specifically activate IRE1α and its downstream target XBP-1 to augment pro-inflammatory cytokine production in macrophages in the absence of an ER-stress response (Martinon et al, 2010; Hetz et al, 2011; Martinon and Glimcher, 2011). Moreover, high levels of fructose can activate the IRE1α-XBP-1 UPR branch to promote hepatic de novo lipogenesis without inducing classic ER stress-response pathways (Lee et al, 2008). These findings suggest that an unconventional, physiological UPR that is mediated through IRE1α might regulate pathophysiologic processes in specialized cells or organs.
Notably, a recent study showed a link between TLR signalling and activation of XBP-1, the UPR transducer IRE1α, in facilitating the production of pro-inflammatory cytokines by macrophages after acute bacterial infection (Martinon et al, 2010). XBP-1 deficiency resulted in a greater bacterial burden in mice infected with the TLR2-activating human intracellular pathogen Francisella tularensis. However, it is not known whether IRE1α is required for TLR-initiated macrophage inflammation, and if so, how TLR signalling activates IRE1α-mediated UPR. To study those processes, we generated a mouse strain with a genetic deletion of IRE1α in myeloid cells and demonstrated that TLR-mediated IRE1α activation is required for optimal pro-inflammatory cytokine production by macrophages during inflammation and progression of inflammatory RA. Importantly, mice with myeloid IRE1α deletion have reduced inflammatory arthritis. An IRE1α-specific inhibitor suppressed TLR-induced pro-inflammatory cytokine production and protected mice from inflammatory arthritis. Furthermore, our findings provide evidence that TLR signalling promotes IRE1α interaction with TRAF6, a key adaptor protein of TLR2/4 signalling pathways, and that TRAF6 catalyses IRE1α ubiquitination. Interestingly, TRAF6-mediated ubiquitination inhibits the recruitment of protein phosphatase 2A (PP2A), a ubiquitously expressed serine threonine phosphatase that dephosphorylates many key molecule players in cell proliferation, signal transduction, and apoptosis (Millward et al, 1999). In this study, we revealed a molecular mechanism by which IRE1α is involved in TLR signalling-mediated pro-inflammatory cytokine production in macrophages and identified IRE1α as a therapeutic target for the treatment of inflammatory arthritis.
Results
IRE1α is an important element in the development of inflammatory arthritis
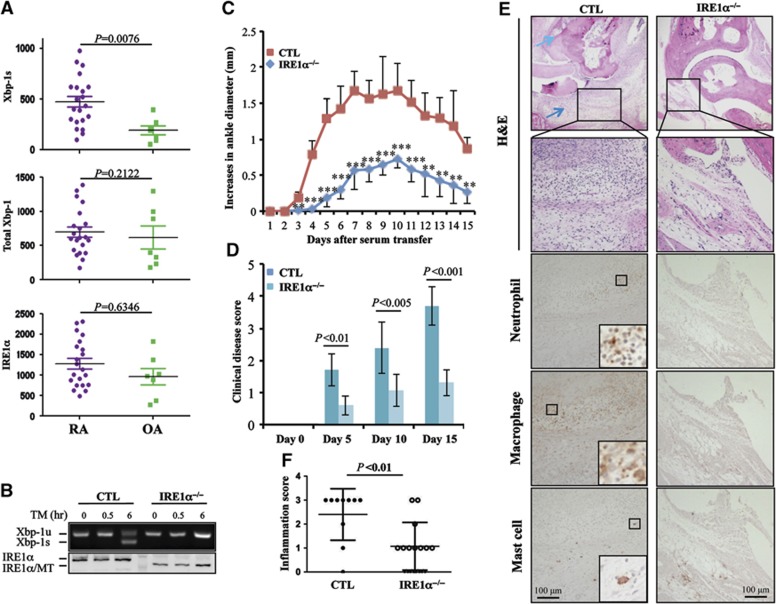
To investigate the involvement of the UPR signalling pathway in human with RA, we obtained synovial fluids from patients clinically diagnosed with either acute or chronic RA. The synovial fluids from patients with osteoarthritis (OA) were used as controls. The synovial fluids were cultured for 2 h to select adherent cells. Adherent cells were gently dislodged and analysed by flow cytometry for expression of macrophage markers, including CD11b and the macrophage mannose receptor CD206 (Chang et al, 2004). An average of >93% of adherent cells were CD11b and CD206 double positive cells, indicating that the majority of these adherent cells from both RA and OA synovial fluids are human macrophages (Supplementary Figure S1A and B). We next isolated total RNA for quantitative real-time RT–PCR analysis. The level of mRNA encoding the UPR trans-activator, spliced XBP-1, in cells from the synovial fluid of RA patients was significantly higher than those from control patients with OA (Figure 1A, top panel). We found that the average XBP-1s mRNA levels were comparable between patients with acute versus chronic RA (Supplementary Figure S1C). As the UPR transducer IRE1α is the sole enzyme that catalyses XBP-1 mRNA splicing (Zhang et al, 2011), the increased production of spliced XBP-1 mRNA reflects the elevated IRE1α activation in macrophages from RA patients. Moreover, levels of total XBP-1 and IRE1α mRNA in synovial fluid cells were comparable between RA and OA patients (Figure 1A, middle and bottom panels), suggesting that the increased production of spliced XBP-1 mRNA was due to enhanced activation, but not expression levels, of IRE1α in the synovial fluid cells from human RA patients. Therefore, the increased IRE1α activation in the synovial fluid cells from human RA patients suggests that IRE1α-mediated signalling might be involved in RA development.
Figure 1.
IRE1α activation promotes inflammatory arthritis in humans and mice. (A) Analysis of IRE1α activation in macrophages from patients with RA. Synovial fluids from patients with RA (N=21) or OA (N=7) were cultured in 6-well plates for 2 h. Total RNA from the adherent cells was extracted. The levels of spliced XBP-1 (XBP-1s), total XBP-1, and IRE1α mRNA were determined by quantitative real-time RT–PCR using β-actin as an internal control. Student’s t-test was used for the statistical analysis, and P-values are indicated. (B) Bone marrow-derived macrophages from control and IRE1α conditional knockout mice were stimulated with TM (5 g/ml) for different time intervals. The expression levels of IRE1α protein were determined by western blotting (bottom panel). The mRNA levels of unspliced XBP-1 and spliced XBP-1 were determined by semi-quantitative RT–PCR. (C–F) Control (CTL, N=5) and IRE1αflox/floxLysM-Cre+ (IRE1α−/−, N=8) mice were treated with serum from K/BxN mice by intraperitoneal injection (200 μl/mouse). The diameter of the ankle joints was measured daily. (C) The increases in the ankle joint diameters in millimeters and (D) the disease scores (described in Materials and methods) are shown. (E, F) Mice were euthanized at day 15 after anti-GPI treatment, tissue sections of the ankle joints were analysed by H&E staining, and the representative images are shown (top two panels). The light blue arrow indicates cartilage erosion and dark blue arrow shows the immune cell infiltration. The infiltration of neutrophils (third panel), macrophages (fourth panel), and mast cells (bottom panel) was characterized by IHC staining as described in Materials and methods (E). The inflammation of joint sections was scored (F). Student’s t-test was used for the statistical analysis. **P<0.01, ***P<0.005; error bars represent standard deviation (s.d.).
Source data for this figure is available on the online supplementary information page.
To investigate the roles of IRE1α in arthritis, we generated conditional IRE1α knockout mice by breeding IRE1αflox/flox mice with lysozyme M-Cre (LysM-Cre) transgenic mice (IRE1αflox/floxLysM-Cre mice), in which the IRE1α gene was specifically deleted in myeloid cells, including macrophages, monocytes, and neutrophils (Clausen et al, 1999). Western blot analysis confirmed that IRE1α activity in macrophages was efficiently disrupted in the IRE1α conditional knockout mice; the full-length IRE1α protein was not detected in bone marrow-derived macrophages from IRE1αflox/floxLysM-Cre mice (Figure 1B). Because the exons 16–17 of the IRE1α gene are flanked by two loxP sites, the Cre-mediated deletion resulted in a truncated, non-functional IRE1α protein production (Zhang et al, 2011). Moreover, disruption of IRE1α activity was evidenced by the defect in splicing XBP-1 mRNA in macrophages derived from IRE1αflox/floxLysM-Cre mice after tunicamycin (TM) treatment (Figure 1B). The non-functional IRE1α mutant protein was detected in CD11b+F4/80+ macrophages, but neither in CD3+ T cells and B220+ B cells, from the conditional knockout mice, nor in the cells from the wild-type mice (Supplementary Figure S2A), thus confirming the specificity of the IRE1α gene deletion in the myeloid lineage. Similarly, IRE1α mutant protein was detected in peritoneal macrophages from the conditional knockout mice (Supplementary Figure S2B). As a consequence, >95% reduction in the TM-induced XBP-1s levels was detected in the peritoneal macrophages from IRE1α conditional knockout mice (Supplementary Figure S2C and D). We then used the IRE1α conditional knockout and control mice to establish a K/BxN serum transfer-induced arthritis model as previously reported (Scatizzi et al, 2010; Mavers et al, 2012). In this study, mice with the following genotypes, IRE1αflox/floxLysM-Cre−, IRE1αflox/+LysM-Cre−, or IRE1α+/+LysM-Cre+, were used as controls because both the expression level and the activity of IRE1α in the macrophages from these mice were indistinguishable from those of wild-type mice. The development of arthritis in those mice, with increased swelling of the ankle joints, was observed in control mice from day 2 after the K/BxN serum transfer. The inflammation reached its peak around day 7 and slightly reversed by day 10 as previously reported (Korganow et al, 1999). In contrast, myeloid-specific deletion of the IRE1α gene in mice delayed arthritis onset and significantly reduced the swelling in the ankle joints (Figure 1C). The clinical RA scores in IRE1α conditional knockout mice were significantly lower than those of the control mice (Figure 1D). Mice were euthanized at day 15 and their joint tissues were characterized by haematoxylin and eosin (H&E) staining (Figure 1E). In the control mice, we observed large amounts of leukocyte infiltration and bone destruction (indicated by arrows) in the ankle joints, whereas in the IRE1α conditional knockout mice only modest inflammation was observed in <40%, and no inflammation in >60%, of ankle joint tissues, thus confirming that deletion of the IRE1α gene in myeloid cells inhibited joint inflammation in mice (Figure 1F). These results indicate that myeloid-specific deletion of the IRE1α gene inhibits the development of inflammatory arthritis in mice.
It is known that lysozyme M promoter drives Cre recombinase expression in myeloid cells, including monocytes, macrophages, mast cells, and neutrophils (Clausen et al, 1999). In addition to macrophages (Bruhns et al, 2003), both mast cells and neutrophils play important roles in the inflammatory arthritis (Wipke and Allen, 2001; Corr and Crain, 2002; Lee et al, 2002). Therefore, we analysed their infiltration in the inflamed joints in mice. A significant amount of both macrophages and neutrophils were detected in the joint sections from K/BxN serum-treated wild-type control, but not IRE1α−/− mice. This result indicates that myeloid IRE1α gene deletion represses macrophage and neutrophil accumulation and infiltration in the joint during inflammatory arthritis. In contrast, only few mast cells were detected in the joints of both IRE1α knockout and control mice at the time point analysed (Figure 1E). While the majority of cells are macrophages in the synovial fluid from RA patient, we also detected 2–3% CD11b+CD206− cells, which are likely neutrophils. In addition to some dead cells, the CD11b−CD206− cells are possibly fibroblasts and a small number of lymphocytes. It is possible that IRE1α activation in these minor populations may contribute to human RA.
We then analysed the expression levels of chemokine receptors including CXCR2 (Jacobs et al, 2010) and CCR9 (Schmutz et al, 2010) (Supplementary Figure S3), which have been shown to be involved in the myeloid cell trafficking into the inflamed joints in mice after anti-GPI sera administration. Our data indicate that IRE1 gene deletion did not affect their expression either on the surface of macrophages or on neutrophils, excluding the possibility that IRE1 gene deletion inhibits myeloid cell trafficking during anti-GPI induced inflammation. In addition, it has been shown that the cell surface expression levels of the Fc receptor CD16 and the complement C5a receptor (C5aR) on myeloid cells are critical for K/BxN serum-induced arthritis (Ji et al, 2002). Therefore, we compared their expression levels between IRE1−/− and control wild-type macrophage and neutrophils, and the results show that the expression levels of both CD16 and C5aR are indistinguishable between wild-type and IRE1 knockout cells (Supplementary Figure S3).
IRE1α promotes TLR-induced inflammatory cytokine production by macrophages and neutrophils
To gain insight into how inflammatory arthritis is suppressed in conditional IRE1α-null mice, we first analysed the percentages of macrophages, mast cells, and neutrophils in IRE1αflox/floxLysM-Cre+ mice. IRE1α is not required for development of myeloid cells, as the percentages of CD11b+F4/80+ macrophages, the FcεR+CD117+ mast cells, and Gr-1+CD11b+ neutrophils were comparable in the peripheral lymphoid organs of IRE1αflox/floxLysM-Cre+ and control mice (Supplementary Figures S4A and S7A). Moreover, neither CD4+/CD8+ T cells nor FoxP3+ Tregs in thymus and spleens were affected by the myeloid-specific deletion of the IRE1α gene (Supplementary Figure S5A–D). In addition, myeloid-specific IRE1α deletion did not alter the percentages of NK1.1+ NK cells, NK1.1+CD3+ NKT cells, B220+ B cells, and CD11c+ dendritic cells in the spleen, and the B-cell subsets in bone marrow from mutant mice are also normal (Supplementary Figure S5E–G).
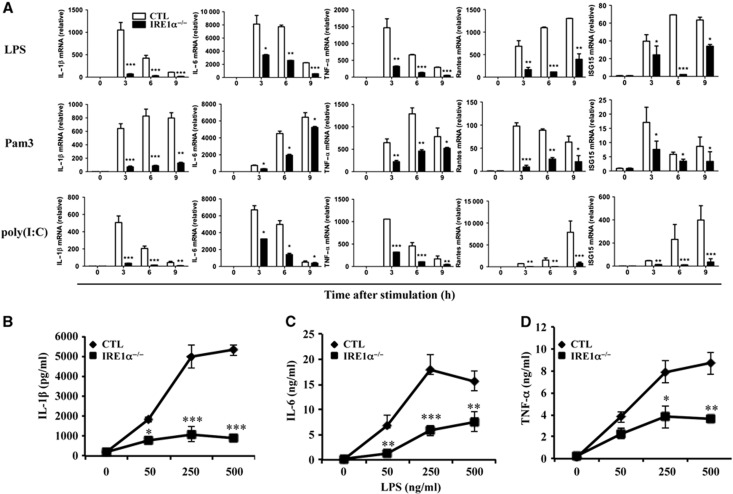
When bone marrow-derived macrophages from conditional IRE1α knockout mice were stimulated with TLR agonists, including lipopolysaccharide (LPS, an agonist of TLR4), Pam3CysSK4 (Pam3, an agonist of TLR2/1), and polyinosinic-polycytidylic acid (poly(I:C), an agonist of TLR3), the production of inflammatory cytokines, including IL-1β, IL-6, TNF-α RANTES, and ISG15, was significantly reduced, compared with control macrophages, as determined by quantitative real-time RT–PCR (Figure 2A) and ELISA (Figure 2B–D). These results indicate that IRE1α is required for TLR-mediated pro-inflammatory cytokine production in macrophages. The suppression of inflammatory cytokine production by IRE1α deficiency in macrophages was not caused by increased cell death, as the percentages of Annexin V-positive cells in IRE1α-null and control macrophages were comparable after stimulation with different TLR agonists (Supplementary Figure S6A). Moreover, the expression of TLR2, TLR3, and TLR4 was not affected by IRE1α deficiency (Supplementary Figure S6B), thus excluding the possibility that impaired cytokine production was caused by reduced TLR expression in IRE1α-null macrophages. Similarly, while the percentages and total numbers of neutrophils in the peritoneal cavity of wild-type and IRE1α−/− mice are comparable (Supplementary Figures S7A and B), a significant reduction in TNF-α by IRE1α-null neutrophils was detected (Supplementary Figure S7C). Therefore, our study demonstrated that IRE1α is required for pro-inflammatory cytokine production by both macrophages and neutrophils after acute inflammatory challenges. The reduced production of pro-inflammatory cytokines from the IRE1α-null macrophages and neutrophils might account for the resistance to inflammatory arthritis of the conditional IRE1α-null mice.
Figure 2.
Loss of IRE1α function impairs inflammatory cytokine production by macrophages. Bone marrow cells from wild-type control and IRE1αf/fLysM-Cre+ mice were isolated and cultured for 6 days following a standard protocol for bone marrow-derived macrophage polarization. (A) Cells were stimulated with or without LPS (100 ng/ml, top panels), Pam3 (100 ng/ml, middle panels), or poly(I:C) (10 g/ml, bottom panels) for 0, 3, 6, or 9 h. Total RNA was extracted from the stimulated cells. The mRNA levels (relative to β-actin) of IL-6, IL-1β, TNF-α, RANTES, and ISG15 were determined by quantitative real-time RT–PCR. Relative fold changes of the mRNA levels were calculated after normalization to the mRNA levels in the wild-type and mutant bone marrow-derived macrophages without stimulation. (B–D) Macrophages were stimulated with different doses of LPS for 24 h. The levels of IL-1β (B), IL-6 (C), and TNF-α (D) were analysed by ELISA. Error bars represent data from three independent experiments. Student’s t-test was used for the statistical analysis. *P<0.05, **P<0.01, ***P<0.005; error bars represent standard deviation (s.d.).
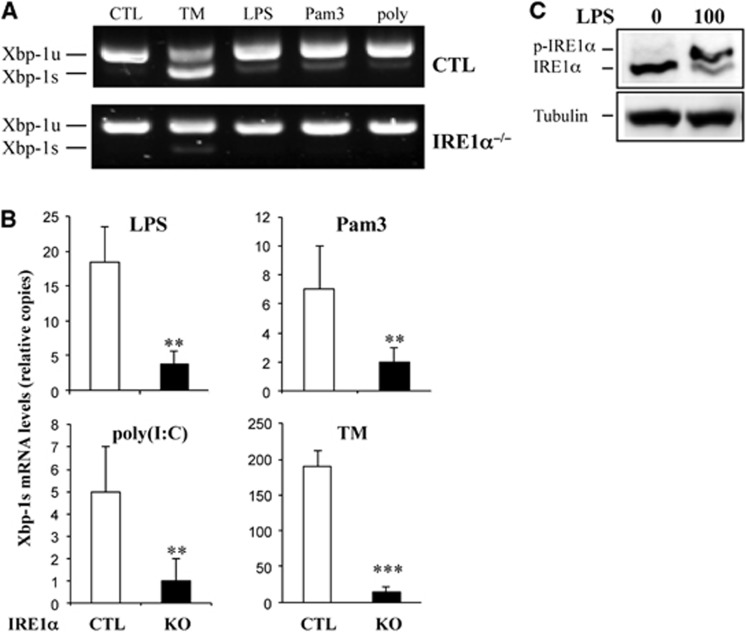
As TLR signalling induces XBP-1 mRNA splicing for full-scale pro-inflammatory cytokine production (Martinon et al, 2010), we compared the levels of TLR-induced XBP-1 mRNA splicing between wild-type and IRE1α-null macrophages. As expected, loss of IRE1α significantly impaired TLR-mediated XBP-1 splicing, as determined by semi-quantitative RT–PCR and quantitative real-time RT–PCR (Figure 3A and B). As a positive control, TM induced a significant amount of spliced XBP-1 in wild-type macrophages, whereas only a background level of spliced XBP-1 mRNA was detectable in IRE1α-null macrophages. Note that the background level of spliced XBP-1 mRNA was likely due to contamination by other cells in bone marrow-derived macrophages, as only 93–95% of cells were CD11b+F4/80+ (Supplementary Figure S4B). Additionally, we demonstrated that LPS stimulation promoted IRE1α phosphorylation in macrophages (Figure 3C). Taken together, these results indicate that TLR signalling induces XBP-1 mRNA splicing through IRE1α activation.
Figure 3.
TLR signalling-induced XBP-1 mRNA splicing requires IRE1α. (A, B) Control (CTL) and IRE1α-knockout bone marrow-derived macrophages were stimulated with tunicamycin (5 g/ml) or LPS (100 ng/ml), Pam3 (100 ng/ml), or poly(I:C) (10 g/ml) for 6 h. Total RNA was isolated from these stimulated cells, and the levels of XBP-1 mRNA (unspliced: XBP-1u, spliced: XBP-1s) were analysed by semi-quantitative RT–PCR (A) and quantitative real-time RT–PCR (B). (C) RAW264.7 cells were stimulated with LPS (100 ng/ml) for 16 h before IRE1α activation was analysed using the phos-tag gel approach. Student’s t-test was used for the statistical analysis. **P<0.01, ***P<0.005; error bars represent standard deviation (SD).
Source data for this figure is available on the online supplementary information page.
To further confirm TLR-mediated IRE1α activation, we isolated macrophages from ER stress-activated indicator (ERAI) transgenic mice (Iwawaki et al, 2004; Mao et al, 2004), and measured XBP-1 spliced product upon TLR stimuli. The ERAI transgenic mice carry an engineered XBP-1 gene fused with the venus gene, a variant of green fluorescent protein (GFP) (Iwawaki et al, 2004; Mao et al, 2004). Upon induction of the UPR, XBP-1 is transcribed and then spliced and result in the production of an XBP-1–venus fusion protein that can be detected by flow cytometry. After stimulation with TLR agonist, the green fluorescence in the ERAI macrophages increased with the stimulation time (Supplementary Figure S8A and B), confirming our conclusion that TLR stimuli promote XBP-1 mRNA splicing. In addition, TNF-α stimulation also induced ERAI signals in macrophages, suggesting that TNF-α can also activates IRE1α. Indeed, the phosphorylation of IRE1α was confirmed in macrophages stimulated with TNF-α (Supplementary Figure S8C). Therefore, our studies show that both TLR and TNF-α can active IRE1α-XBP-1 pathways, suggesting that the elevated IRE1α activation in RA macrophages is possibly induced by multiple inflammatory factors.
On the basis of the profiles of pro-inflammatory cytokines (Figure 2A), it appears that IRE1 deletion led to more profound impairment in the production of inflammatory cytokines by macrophages, compared with XBP-1 deletion (Martinon et al, 2010). This suggests that additional IRE1α targets, rather than XBP-1, may be involved in TLR-induced inflammatory cytokine productions. Indeed, reconstitution of XBP-1s expression in IRE1-null mouse embryonic fibroblasts (MEFs) partially rescued IL-6 expression (Supplementary Figure S9), confirming that additional factors exist in IRE1-mediated cytokine production. Recent studies suggest that the regulated Ire1-dependent decay (RIDD) of mRNA is involved in regulating ER stress responses and other physiological functions (Hollien and Weissman, 2006; Han et al, 2009; Hollien et al, 2009). However, the mRNA levels of Hgnat, Pmp22, Scara3, Col6, and PdgfR, the well-defined RIDD target genes, are indistinguishable between wild-type and IRE1-null macrophages in the presence or absence of the inflammatory stimuli (Supplementary Figure S10). In addition, it has been reported that IRE1α activates the JNK to mediate ER stress-induced cell death (Urano et al, 2000). As JNK regulates activation of the transcription factor AP-1 in cytokine productions, the robust reduction in the pro-inflammatory cytokine productions in the absence of IRE1 may be due to impaired JNK activation, in addition to lack of functional XBP-1. However, both the phosphorylated and total JNK protein levels, as well as the activation of the MAPKs p38 and Erk1/2, were comparable between wild-type and IRE1α-null macrophages (Supplementary Figure S11A). Moreover, activation of the other branches of ER stress pathways, including PERK/eIF2α and ATF6 pathways, in macrophages was not affected by IRE1α deficiency (Supplementary Figure S11B).
TRAF6 is required for TLR-mediated IRE1α activation
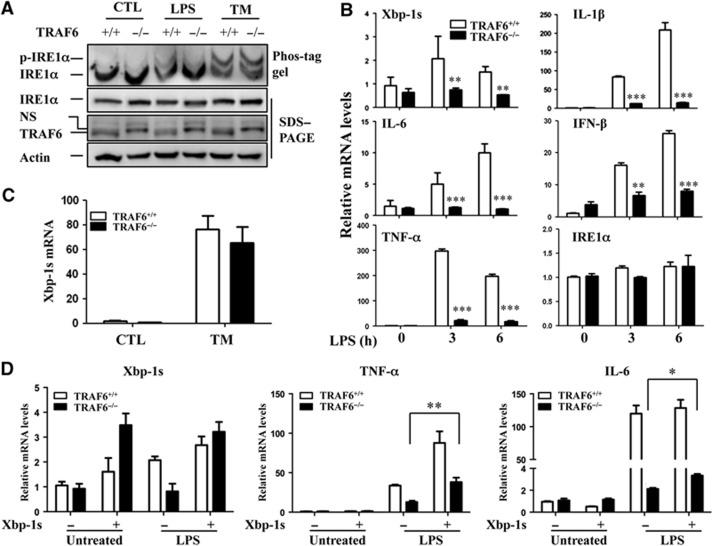
Knockdown of intracellular adaptor proteins of the TLR pathway, including MyD88, TRIF, and TIRAP, has been shown to partially eliminate TLR-induced IRE1α activation (Martinon et al, 2010). In fact, we found that TRAF6 is essential for TLR-mediated IRE1α activation, as IRE1α phosphorylation and XBP-1 mRNA splicing were attenuated in TRAF6-null MEFs after LPS stimulation. Importantly, TLR-independent TM-induced IRE1α activation was not affected by the loss of TRAF6 functions (Figure 4A–C). This result suggests that TLR and ER stress activate IRE1α through different mechanisms. Consistent with previous observations (Lomaga et al, 1999), the loss of TRAF6 remarkably inhibited TLR-induced production of inflammatory cytokines, including IL-1β, IL-6, TNF-α, and IFN-β (Figure 4B). Interestingly, compared with the wild-type control MEFs, the TRAF6−/− MEFs expressed significantly reduced levels of the splice XBP-1 mRNA in response to 16-h LPS stimulation (Supplementary Figure S12), indicating the requirement of TRAF6 in LPS-triggered IRE1 activation. Notably, TRAF6 deficiency did not affect IRE1α mRNA expression between control and TRAF6-null MEFs (Figure 4B). Together, our findings suggest that a crosstalk between TRAF6 and IRE1α is involved in TLR-induced IRE1α activation that regulates pro-inflammatory cytokine production.
Figure 4.
TRAF6 is required for TLR-induced IRE1α activation. (A) TRAF6+/+ and TRAF6−/− MEFs were stimulated with LPS (100 ng/ml) or tunicamycin (TM, 5 g/ml) for 16 h. IRE1α phosphorylation was determined by pho-tag gel (top panel). The parallel-prepared samples were subjected to SDS–PAGE and western blot analysis for IRE1α (second panel), TRAF6 (third panel), and β-actin (bottom panel). (B) Total RNA from LPS-treated MEFs was purified, and the levels of XBP-1s, IL-1β, IL-6, and IFN-β were analysed by quantitative real-time RT–PCR. Error bars represent data from a triplicate analysis (mean+s.d.), and representative results from three independent experiments are shown. (C) TRAF6+/+ and TRAF6−/− MEFs were stimulated with tunicamycin (5 g/ml) for 6 h. Levels of spliced XBP-1 mRNA were determined by quantitative real-time RT–PCR. (D) Wild-type and TRAF6-null MEFs were infected with adenovirus expressing spliced XBP-1 protein. Two days after infection, cells were stimulated with or without LPS for an additional 6 h. The levels of XBP-1s, TNF-α, and IL-6 mRNAs were characterized by quantitative real-time RT–PCR. Student’s t-test was used for the statistical analysis. *P<0.05, **P<0.01, ***P<0.005; error bars represent standard deviation (s.d.).
Source data for this figure is available on the online supplementary information page.
To test whether the XBP-1 can rescue the defect in inflammatory cytokine production due to impaired IRE1 activation in TRAF6-null cells, we next expressed a spliced form of XBP-1 in TRAF6-null MEFs. Ectopic expression of spliced XBP-1 only partially rescued TNF-α and IL-6 production by TRAF6-null MEFs, thus confirming that additional factors are required for IRE1α-mediated inflammatory cytokine productions. Importantly, the partial rescue effect of the functional XBP-1 is dependent on TLR stimulation, as expression of spliced XBP-1 failed to promote inflammatory cytokine production in the absence of LPS treatment, even in the wild-type MEFs (Figure 4D). This result indicates that XBP-1s requires other inflammatory factors, which are presumably driven by TLR stimuli, to promote gene transcription.
TRAF6 interacts with IRE1α in macrophages
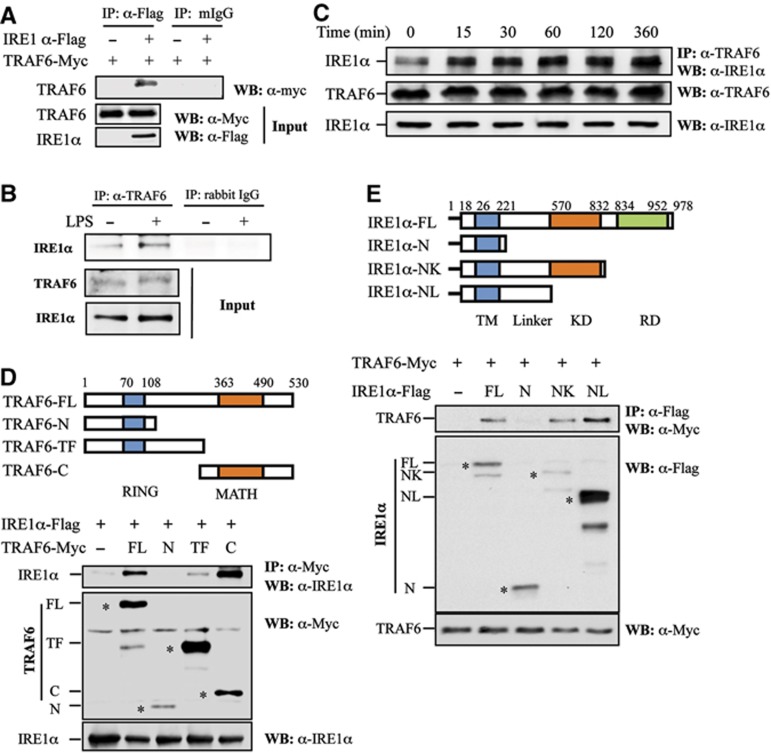
To further delineate the molecular mechanism by which TRAF6 regulates TLR-mediated IRE1α activation, we tested the interaction between IRE1α and TRAF6 in the presence or absence of inflammatory stimuli. Immunoprecipitation (IP)–western blot analysis with the HEK293 cells transiently expressing exogenous IRE1 and TRAF6 showed that IRE1 protein can interact with TRAF6 (Figure 5A). Further, we demonstrated the interaction between endogenous TRAF6 and IRE1α in primary mouse bone marrow-derived macrophages also by IP (Figure 5B). Importantly, LPS stimulation significantly enhanced TRAF6–IRE1α interaction in macrophages (Figure 5B). A dynamic analysis in the mouse monocyte/macrophage cell line RAW264.7 further confirmed that LPS stimulation significantly enhanced the interaction between IRE1α and TRAF6 (Figure 5C), indicating that the interaction between IRE1α and TRAF6 in macrophages is regulated by TLR signalling. To gain insights into the mechanism underlying IRE1 and TRAF6 interaction, we generated truncated mutations for both TRAF6 and IRE1α proteins to map their interaction domains. Co-IP–western blot analysis revealed that the C-terminal meprin-associated TRAF homology (MATH) domain of TRAF6 was required for its interaction with IRE1α (Figure 5D). Moreover, we defined that the linker region between the transmembrane domain and the kinase domain of IRE1α is required for IRE1α interaction with TRAF6 (Figure 5E). We reasoned that because PP2A interacts with IRE1α through the same region of IRE1α (Qiu et al, 2010) TRAF6 might compete with PP2A to activate IRE1α.
Figure 5.
TRAF6 interacts with IRE1α. (A) Flag-tagged IRE1α and Myc-tagged TRAF6 plasmids were co-transfected into HEK293 cells. IRE1α protein in the lysates of transfected cells was immunoprecipitated with an anti-Flag antibody or with normal mouse IgG (mIgG) as a control. The bound TRAF6 was determined by western blotting using an anti-Myc antibody (top panel). The expression levels of TRAF6 and IRE1α in whole cell lysates were confirmed by western blot analysis using anti-Myc (middle panel) and anti-Flag (bottom panel) antibodies, respectively. (B) Mouse primary macrophages derived from bone marrow cells were stimulated with or without LPS (1 g/ml) for 16 h. The interaction between TRAF6 and IRE1α was analysed by western blotting. (C) The interaction between TRAF6 and IRE1α in RAW cells stimulated with LPS (1 g/ml) under a time course was determined by co-immunoprecipitation and western blot analysis. (D) Schematic representation of TRAF6 and its truncated mutants. TRAF6 carries an N-terminal RING finger domain and a C-terminal MATH domain (top panel). FL: full-length structure, N: N-terminal RING finger domain, TF: trans-membrane domain, C: C-terminal MATH domain. IRE1α was co-transfected with TRAF6 or its mutants into HEK293 cells. The interactions between IRE1 and TRAF6 or its mutants were determined as described in (A). The expression of the full-length and truncated TRAF6 protein was indicated by the symbol ‘*’. (E) Schematic representation of IRE1α and its truncated mutants. IRE1α contains an N-terminal trans-membrane (TM) domain, kinase domain (KD), and a C-terminal RNase domain (RD) (top panel). FL: full length, N: N-terminal trans-membrane domain, NK: N-terminus and the kinase domain, NL: N-terminal linker domain. TRAF6 was co-transfected with IRE1α or its mutants into HEK293 cells. The interactions between TRAF6 and IRE1α or its mutants were determined as described in (A). The expression of the full-length and truncated IRE1α protein was indicated by the symbol ‘*’.
Source data for this figure is available on the online supplementary information page.
TRAF6 ubiquitinates IRE1α and inhibits its interaction with the PP2A
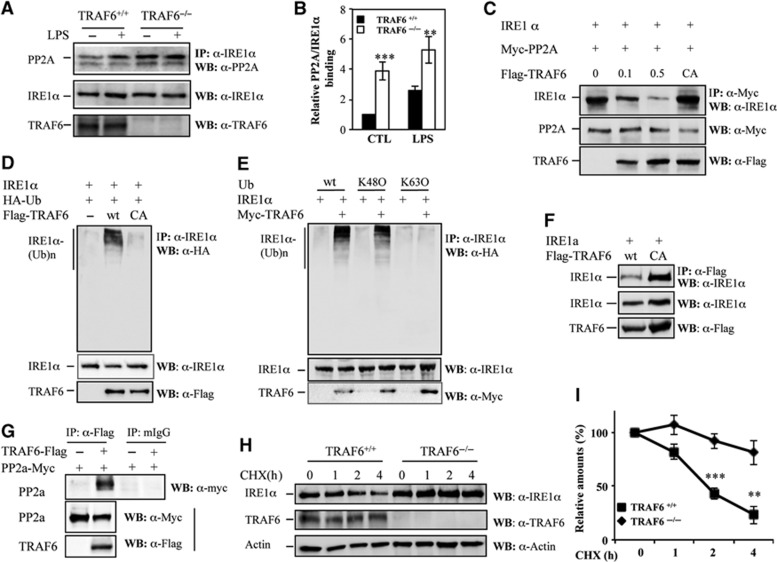
Next, we determined how TRAF6 regulates IRE1α activation in which PP2A is involved. A recent report showed that, in pancreatic β cells, the PP2A interacts with the IRE1α linker region through its adaptor protein RACK1 to suppress IRE1α phosphorylation, the key prerequisite for IRE1α RNase activity (Qiu et al, 2010). Because we found that the IRE1α linker region mediates its interaction with TRAF6 (Figure 5E), we speculated that TRAF6 might activate IRE1α by inhibiting the recruitment of PP2A. Indeed, we detected a significant increase in IRE1α–PP2A interaction in TRAF6-null MEFs (Figure 6A and B). In contrast to LPS-induced IRE1α–PP2A interaction in wild-type MEFs, constitutive interaction between IRE1α and PP2A was observed in TRAF6-null MEFs. It is possible that metabolic signals, such as glucose, growth factors, and amino acids, all of which exist in the culture media and have been shown to activate IRE1 (Lee et al, 2008; Zhang and Kaufman, 2008; Zhang, 2010), may trigger basal IRE1/PP2A interaction in the absence of TRAF6. Indeed, only background levels of PP2A/IRE1 interaction could be detected in both wild-type and TRAF6-null MEFs under glucose and serum starvation. In cells cultured with normal culture media, PP2A/IRE1 interaction was detected, and loss of TRAF6 further enhanced their interaction (Supplementary Figure S13). These results indicate that TRAF6 suppresses the recruitment of PP2A to IRE1 upon extracellular stimuli, such as LPS and the metabolic factors. Supporting this conclusion, over-expression of TRAF6 inhibited IRE1α interaction with PP2A in a dose-dependent manner (Figure 6C). However, the presence of the E3 ligase catalytic-inactive C70A mutant of TRAF6 (TRAF6/CA) failed to suppress the interaction between IRE1α and PP2A, even though the C70A mutation did not affect the interaction between TRAF6 and IRE1 (Figure 6F). These results indicate that the E3 ligase ubiquitin activity of TRAF6 is required for its suppressive effect on the IRE1α–PP2A interaction, and that TRAF6 inhibits IRE1α–PP2A interaction not through its competition for their binding regions in IRE1α with PP2A. These three proteins possibly form a complex because the interaction of TRAF6 with PP2A was also detected (Figure 6G). Furthermore, the expression levels of PP2A in TRAF6−/− MEFs were comparable to those in the control wild-type MEFs, thus excluding the possibility that increased IRE1α–PP2A interaction is due to the altered PP2A expression levels in TRAF6-null cells (Supplementary Figure S15).
Figure 6.
TRAF6 catalyses IRE1α ubiquitination to suppress IRE1α interaction with PP2A. (A) TRAF6+/+ and TRAF6−/− MEFs were treated with 500 ng/ml LPS for 30 min. The interaction between IRE1α and PP2A was determined by co-immunoprecipitation using an anti-IRE1α antibody and western blotting using an anti-PP2A antibody (top panel). The expression levels of IRE1α (middle panel) and TRAF6 (bottom panel) were determined by western blot analysis. (B) The intensities of IRE1α bands that interacted with PP2A were quantified. The relative band intensities are shown. (C) HEK293 cells were transfected with IRE1α (1 g), Myc-PP2A (1 g), and different amounts of Flag-TRAF6 (0, 0.1, and 0.5 g) or its CA mutant (1 g) plasmids. The interaction between IRE1α and PP2A was determined by co-immunoprecipitation and western blot analysis as described in (A). (D) IRE1α, TRAF6, and HA-Ubiquitin expression plasmids were co-transfected into HEK293 cells as indicated. IRE1α ubiquitination was determined by immunoprecipitation using the anti-IRE1α antibody and western blotting using the anti-HA antibody (top panel). The expression level of IRE1α and TRAF6 in whole cell lysates was confirmed (middle and bottom panels). (E) Expression plasmids of IRE1α, TRAF6, and each HA-ubiquitin were co-transfected into HEK293 cells. IRE1α ubiquitination was examined as described in (D). (F) The interactions between IRE1α and TRAF6 or its C70A mutant were determined by co-immunoprecipitation and western blot analysis. (G) Flag-tagged TRAF6 and Myc-tagged PP2a plasmids were co-transfected into HEK293 cells. TRAF6 protein in the lysates of transfected cells was immunoprecipitated with anti-Flag antibody or normal mouse IgG as a control; the bound PP2a was determined by western blotting with anti-Myc Abs (top panel). The expression levels of PP2a and TRAF6 in whole cell lysates were confirmed by western blotting with anti-Myc antibody (middle panel) and anti-Flag antibody (bottom panel), respectively. (H) TRAF6+/+ and TRAF6−/− MEFs were treated with 100 μg/ml cycloheximide (CHX) for the indicated time intervals. The protein levels of IRE1α (top panel) and β-actin (bottom panel) in the lysates of treated cells were examined by western blot analysis. (I) The band densities of western blot analysis were quantified, and the relative levels were calculated. Error bars represent data from three independent experiments (mean±s.d.), **P<0.01, ***P<0.005.
Source data for this figure is available on the online supplementary information page.
E3 ubiquitin ligases are known to promote ubiquitination of their binding proteins. We speculated that TRAF6 might inhibit the IRE1α–PP2A interaction by catalysing IRE1α ubiquitination. Indeed, transient expression of TRAF6, but not the E3 ligase catalytic activity-negative mutant TRAF6/C70A, enhanced IRE1α ubiquitination, indicating that TRAF6 is an E3 ubiquitin ligase of IRE1α (Figure 6D). This finding implies that TRAF6 inhibits IRE1α–PP2A interaction by specifically catalysing IRE1α ubiquitination. TRAF6 often catalyses K63-linked polyubiquitin conjugation onto its substrates (Deng et al, 2000; Wang et al, 2001; Yang et al, 2009). To characterize TRAF6-mediated ubiquitination of IRE1α, we co-expressed IRE1α with a mutant ubiquitin isoform that carries a single lysine residue at position 63 (K63O) or 48 (K48O) in HEK293 cells (Figure 6E). K63O and K48O ubiquitin mutants carry a single lysine residue, residues 48 and 63, respectively, which allows us to determine the topology of polyubiquitin chains. When Ub/K48O mutant is expressed, IRE1α ubiquitination was detected in a similar level to that in cells expressing wild-type ubiquitin. In contrast, when the Ub/K63O mutant was co-transfected, only a low level of IRE1α ubiquitination was detected. Therefore, the poly-ubiquitin chain conjugation onto IRE1α protein requires the lysine residue 48 (K48) but not the K63, indicating that TRAF6 catalyses IRE1α K48-linked, but not K63-linked polyubiquitination. The weak K63-linkage ubiquitination might have been catalysed by other endogenous E3 ubiquitin ligases, because TRAF6 co-expression did not affect K63-linked IRE1α ubiquitination.
K48-linked polyubiquitination usually mediates protein degradation, whereas K63-linked polyubiquitination regulates the functions of target proteins. As TRAF6 enhances K48-linked polyubiquitination of IRE1α, we tested whether TRAF6-mediated ubiquitination enhances IRE1α protein degradation. As shown in Figure 6H and I, the half-life of IRE1α in wild-type MEFs is about 1.8 h, but it was significantly prolonged by TRAF6 deficiency. Therefore, TRAF6 promotes IRE1α degradation by catalysing K48-linked polyubiquitination. Since TLR stimulation enhances IRE1/TRAF6 interaction, we asked whether LPS stimulation promotes IRE1 degradation. However, neither the half-life nor the levels of newly synthesized IRE1 protein were altered in macrophages upon LPS stimulation for up to 4 h (Supplementary Figure S14A and B). This is likely because TRAF6-mediated degradation compromises the new synthesis. Cultivation of macrophages with LPS for a longer period, such as 8–16 h, increased the expression levels of IRE1, clearly indicating that LPS induces IRE1 protein expression and this induction succeeds TRAF6-mediated degradation during later time points (Supplementary Figure S14C and D). Supporting this speculation, both the newly synthesized IRE1 protein levels and their stability (half-life) were significantly increased in TRAF6-null cells, compared to that in the wild-type cells (Supplementary Figure S14E and F). Therefore, our studies reveal a previously uncharacterized molecular pathway responsible for TLR-induced IRE1α activation: TLR signalling promotes TRAF6 interaction with and ubiquitinates IRE1α, which blocks the recruitment of IRE1α PP2A and subsequently benefits the activation of IRE1 through auto-phosphorylation. Additionally, the interaction between TRAF6 and IRE1 also contributes to the regulation of IRE1α protein stability under inflammatory stress conditions. The detailed molecular mechanism and dynamics for the fine-tune regulation of IRE1 activation or stability by TRAF6 is an intriguing question to be further elucidated in the future.
IRE1α-specific inhibitor protects mice from inflammatory arthritis
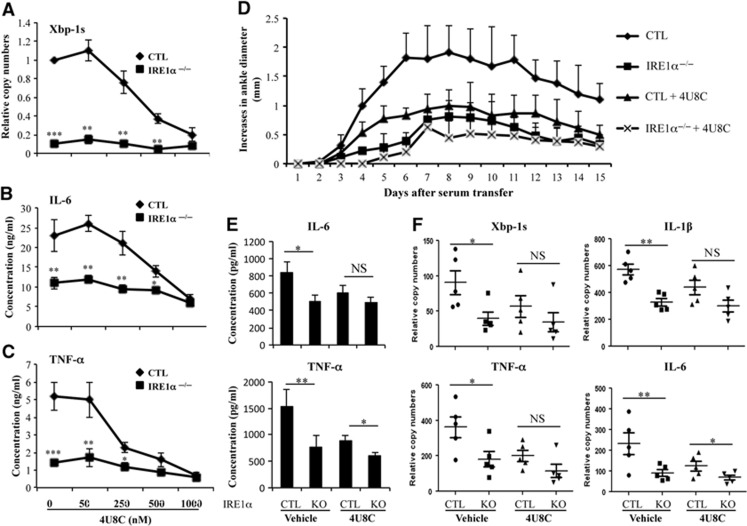
Because elevated IRE1α activation in synovial fluid cells is associated with RA in humans and because deletion of myeloid-specific IRE1α attenuates anti-GPI-induced inflammatory arthritis in mice, IRE1α might be a potential therapeutic target for treating inflammatory arthritis. Recently, 8-formyl-7-hydroxy-4-methylcoumarin (4U8C), which specifically binds to the lysine residue in the ribonuclease catalytic pocket and blocks IRE1α RNase activity, was identified as a novel IRE1α inhibitor (Cross et al, 2012). We tested the ability of 4U8C to suppress TLR-induced IRE1α activation and cytokine production in macrophages. 4U8C significantly inhibited LPS-induced splicing of XBP-1 mRNA with a 50% inhibition efficacy (IE50) of about 150 nM in macrophages from control mice (Figure 7A). Notably, 4U8C treatment significantly inhibited LPS-induced production of IL-6 (Figure 7B) and TNF-α by macrophages (Figure 7C). In IRE1α-null macrophages, 4U8C treatment did not further reduce pro-inflammatory cytokine genes, suggesting that the effect of 4U8C on TLR-induced pro-inflammatory cytokine production is through suppression of IRE1α activity.
Figure 7.
IRE1α inhibitor suppresses inflammatory cytokine production and protects mice from inflammatory arthritis. (A) Bone marrow-derived macrophages were stimulated with 200 ng/ml LPS in the presence of the IRE1α inhibitor 4U8C (concentrations 0–1000, nM) for 24 h. Total RNA from the stimulated cells was isolated, and the levels of spliced XBP-1 mRNA were analysed by quantitative real-time RT–PCR. (B, C) The levels of IL-6 (B) and TNF-α (C) in the supernatants of stimulated cells in (A) were determined by ELISA. Error bars represent data from three independent experiments (mean+s.d.). (D) Inflammatory arthritis was induced in IRE1α−/− and control (CTL) mice by intraperitoneal injection of 200 μl K/BxN serum at day 0. Mice were treated daily with 4U8C at 10 mg/kg/day from day 1. The diameters of their ankle joints were measured daily. Error bars represent data from five mice per group (mean+s.d.). (E) Sera from mice in (D) were collected at day 15. The levels of IL-6 (top panel) and TNF-α (bottom panel) were examined by ELISA. (F) The mice in (D) were euthanized, their ankle joints were collected and homogenized, and total RNA was isolated. The levels of XBP-1s, IL-1β, IL-6, and TNF-α mRNAs were determined by quantitative real-time RT–PCR. Student’s t-test was used for the statistical analysis. *P<0.05, **P<0.01, ***P<0.005. NS, non-significant; error bars represent the standard deviation (s.d.).
To test the therapeutic potential of 4U8C in a mouse model of inflammatory arthritis, we administered 4U8C at a dose of 10 mg per kg body weight per day via intraperitoneal injection. This dose exerted a potent suppressive effect on anti-GPI-induced joint inflammation in control mice. Although myeloid-specific deletion of the IRE1α gene resulted in significantly reduced disease severity in mice after the anti-GPI serum transfer, treatment of IRE1α conditional knockout mice with 4U8C did not further inhibit joint inflammation after anti-GPI serum transfer (Figure 7D), suggesting that the suppressive effect of 4U8C on inflammatory arthritis relies on its suppression of IRE1α. Moreover, the levels of inflammatory cytokines in the sera of mice receiving anti-GPI serum were inhibited by 4U8C treatment (Figure 7E). The activation of IRE1α in the joints of 4U8C-treated mice was inhibited, as measured by the levels of spliced XBP-1 mRNA (Figure 7F). A significant reduction in expression of TNF-α, IL-1β, and IL-6 mRNAs in the joint tissues from 4U8C-treated mice was also detected (Figure 7F). These results indicate that the IRE1α inhibitor 4U8C has promising therapeutic potential for treating inflammatory RA.
Discussion
Our studies demonstrated previously unknown molecular mechanisms underlying TLR-induced activation of IRE1α and identified IRE1α as a potential therapeutic target for inflammatory arthritis. These conclusions are supported by the following discoveries: first, IRE1α is required for optimal production of inflammatory cytokines by macrophages after TLR stimulation; second, elevated IRE1α activation is associated with RA in humans, and myeloid-specific IRE1α deletion largely protects mice from inflammatory arthritis; third, TRAF6 is essential for TLR-mediated IRE1α activation as it catalyses IRE1α ubiquitination and blocks the recruitment of PP2A; and fourth, treatment with the IRE1α-specific inhibitor 4U8C reduces the symptoms of experimental inflammation in mice.
It is well documented that TLR signalling promotes cytokine production by activating downstream transcription factors, including NF-κB and IFN regulatory factors (IRFs), during innate immune responses to pathogens (Mills, 2011). Here, we discovered that IRE1α activation mediates an additional pathway that is required for TLR-induced cytokine production in macrophages. Although a recent study showed that XBP-1, the target of the IRE1α-mediated UPR pathway, is essential for maximizing TLR-mediated innate immune responses (Martinon et al, 2010), our study showed that IRE1α deficiency did not result in a phenotype identical to that of XBP-1 deletion in macrophages. IRE1α deletion impaired the production of a broader spectrum of inflammatory cytokines, and led to more profound inflammatory defects than XBP-1 deletion (Martinon et al, 2010). Indeed, it has been shown that IRE1α executes its functions through mechanisms that are independent of XBP-1 in B-cell development and in insulin-producing β cells (Zhang et al, 2005; Lee et al, 2011). While IRE1α is known to be involved in activation of JNK (Urano et al, 2000), a key player that mediates pro-inflammatory cytokine production through activating transcription factor AP-1 (Brenner et al, 1989), the activation of JNK, as well as other MAPKs including p38 and Erk1/2, was not affected by the IRE1α gene deletion. Therefore, additional XBP-1s-independent IRE1α activity in facilitating TLR-induced cytokine production likely exists. A recent study indicates that IRE1 is involved in inflammasome functions (Oslowski et al, 2012). As IRE1 deficiency impairs the production of IL-1β, a cytokine that is regulated by inflammasome, it is possible that IRE1 regulates IL-1β production partially through the inflammasome pathway, an interesting question to be elucidated in the future. In addition to macrophages, neutrophils play critical roles in inflammatory arthritis in both human patients and experimental animal models. In fact, we observed that IRE1α gene deletion dramatically inhibited production of TNF-α by neutrophil and infiltration of neutrophil into the inflamed joints. Therefore, reduced activation of both macrophages and neutrophils contributes to the suppression of K/BxN serum-induced arthritis by myeloid IRE1α gene deletion in mice.
Our findings reveal that TRAF6 activates IRE1α through a linker region between the ER transmembrane domain and the kinase domain of IRE1α. It was recently reported that the PP2A, as well as its adaptor protein RACK1, interacts with IRE1α through this linker region to suppress IRE1α autophosphorylation, a prerequisite for IRE1α activation in pancreatic β cells (Qiu et al, 2010). An increase in PP2A–IRE1α interaction was observed in TRAF6-null cells, which suggests that TRAF6 may activate IRE1α by suppressing the recruitment of the IRE1α inhibitor PP2A. The interaction between IRE1α and PP2A can be detected in TRAF6−/− MEFs, even without TLR stimulation. This is possibly due to basal physiological stimuli, such as glucose, which has been shown to activate IRE1α (Qiu et al, 2010), in the culture media may sufficiently activate IRE1α and PP2A interaction in the absence of TRAF6. Moreover, the TRAF6-mediated suppression of PP2A–IRE1α interaction likely occurs through a K48-linked ubiquitination, as the ubiquitin ligase-inactive mutant, TRAF6/C70A, failed to suppress PP2A recruitment to IRE1α without affecting IRE1α–TRAF6 interaction. Additionally, TRAF6 appears to promote IRE1α protein degradation, because loss of TRAF6 functions resulted in a significant increase in IRE1α protein stability. Protein ubiquitination has been shown to play dual roles, in both activation and activator destruction (Salghetti et al, 2001). TRAF6-mediated polyubiquitination appears to positively regulate IRE1α activation by suppressing PP2A recruitment and IRE1α protein degradation.
Our study suggests that TRAF6 is also involved in IRE1α protein degradation, because loss of TRAF6 functions resulted in a significant increase in IRE1α protein stability. Recent studies indicated that ubiquitination-mediated degradation and activation of the same substrate occurs. For example, the E3 ubiquitin ligase Met30-mediated ubiquitination of the transcription activation domain of VP16 transcription factor activates VP16 followed by VP16 protein destruction, suggesting that ubiquitination serves as a dual signal for activation and activator destruction (Salghetti et al, 2001). Similarly, we show here that TRAF6-mediated ubiquitination not only suppresses IRE1/PP2A interaction to enhance IRE1 activation but also catalyses IRE1 protein degradation. Likely, TRAF6 ubiquitinates IRE1 to block the interaction with IRE1 inhibitor PP2A for IRE1 activation at the early phase of TLR stimulation. This is followed by IRE1 protein destruction at the late phase of stimulation to terminate the signalling transduction. TRAF6-mediated polyubiquitination appears to positively regulate IRE1α activation by suppressing PP2A recruitment and IRE1α protein degradation. In addition, we reproducibly detected that LPS stimulation promotes IRE1 interaction both with TRAF6 and with PP2A, which presumably promotes IRE1α ubiquitination and degradation, while the levels of IRE1α protein were not reduced. One explanation is that LPS facilitates IRE1α degradation but also induces its protein expression. Indeed, both the levels (time 0) and stability (half-life) of the newly synthesized IRE1 protein in TRAF6-null cells significantly increased compared to that in the wild-type cells. Nevertheless, the detailed dynamics of IRE1/PP2A interaction in the presence or absence of TRAF6 under inflammatory stress needs to be further elucidated.
It is important to note that reconstitution of the functional/spliced XBP-1 only partially rescued cytokine production by TRAF6-null MEFs upon LPS stimulation. Interestingly, expression of spliced XBP-1 in wild-type or TRAF6-null cells failed to stimulate cytokine production in the absence of LPS stimulation. Clearly, XBP-1s protein alone is not sufficient to promote production of the inflammatory cytokines. One speculation is that XBP-1s may need a crosstalk with other TRAF6 downstream transcription factors, such as NF-kB, AP-1, and IRFs, in driving expression of the pro-inflammatory cytokine genes. It is possible that, under ER stress, XBP-1s binds to the promoter of ER stress responsive genes; in contrast, upon TLR stimuli, the downstream transcription factors, such as NF-kB, AP-1, and IRFs, are activated and interact with the spliced XBP-1 to elicit their trans-activation effects on pro-inflammatory gene expression. Further studies are needed to test these scenarios.
Recent studies suggest that ER stress is a driving force for, or a consequence of, inflammatory disease (Turner et al, 2005; Colbert et al, 2009; Harama et al, 2009; Nundlall et al, 2010; Yoo et al, 2012). However, a direct connection between IRE1α activation and inflammatory RA had not been established. In light of the facts that TLR signalling activates IRE1α and endogenous TLR ligands are considered to be pathogenic factors of RA, it is not surprising that IRE1α activation is elevated in synovial fluid cells from RA patients. In addition to TLR signalling, we show that TNF-α stimulation can directly activate IRE1α-mediated XBP-1 mRNA splicing in macrophages. In addition, metabolic and cardiovascular risk factors, including hypertension, obesity, high glucose, and dyslipidaemia, some of which like glucose can activate IRE1, are prevalent in patients with RA (Rostom et al, 2013). These factors, raised under the RA microenvironment, may also be triggers of the IRE1/XBP-1 pathway and contribute to RA development. This is supported by our finding that myeloid-specific deletion of IRE1α largely protected mice from inflammatory arthritis triggered by anti-GPI serum from K/BxN mice. The involvement of IRE1α in RA development was further confirmed by the effect of the IRE1α inhibitor 4U8C: it suppressed TLR-induced macrophage inflammation and attenuated the clinical symptoms of inflammatory arthritis in mice. However, it has been recently shown that TLR2 knockout mice develop enhanced arthritis upon the challenge of K/BxN serum (Huang et al, 2013). One of the mechanisms underlying the RA model of TLR2 knockout mice is that TLR2 deletion leads to the reduced IL-10 production. We show here that TLR2 signalling activates IRE1α activation in macrophages and subsequently promotes inflammatory arthritis in mice.
The IRE1α inhibitor 4U8C is a synthetic coumarin derivative that attains its selectivity by forming an unusually stable Schiff base with lysine 907 in the IRE1α endonuclease domain. This inhibitor blocks substrate access to the active site of IRE1 and selectively inactivates both XBP-1 splicing and IRE1α-mediated mRNA degradation (Cross et al, 2012). The coumarin compounds have been widely studied for their potent anti-inflammatory activities in both murine and human inflammation (Yesilada et al, 2001; Pochet et al, 2004; Kontogiorgis et al, 2006; Pan et al, 2010; Shin et al, 2010). Several clinical trials are currently investigating coumarin compounds for treatments of inflammatory diseases, in particular RA. Previous studies showed that coumarin compounds could target several signalling pathways, including NF-κB, MAPK, and protein kinase C in vivo (Shin et al, 2008; Li et al, 2011; Yao et al, 2011). Our studies suggest that 4U8C suppresses TLR-induced pro-inflammatory cytokine production through IRE1α, as it had no further inhibitory effect on the production of inflammatory cytokines in IRE1α-null macrophages. Further studies are needed to identify additional in vivo targets of 4U8C to gain a better understanding of the molecular mechanisms of 4U8C in anti-inflammation therapy.
Materials and methods
Cells, reagents, and mouse strains are described in the Supplementary Data.
Bone marrow-derived macrophage culture
Bone marrow cells were collected from the femurs and tibias of control and IRE1αflox/floxLysM-Cre+ mice and cultivated in RPMI medium supplemented with 10% (vol/vol) FCS, M-CSF (5 ng/ml), penicillin (100 U/ml), and streptomycin (100 U/ml). After 1 day of culture, non-adherent precursors were plated for 6 days in 6-well plates at a density of 1 × 106 cells per well in RPMI medium supplemented with 10% FCS, 30 ng/ml M-CSF (eBioscience), and antibiotics. On day 6 or 7, the cells were stimulated with TLR agonist or tunicamycin (Invitrogen), and gene transcription and cytokine production were analysed.
Phos-tag gel analysis of IRE1α phosphorylation
Preparation of the phosphor-tag gel was performed according to the manufacturer’s instructions (Phos-tag acrylamide AAL-107, Wako Pure Chemical Industries). The detailed protocol was followed as previously described (Yang et al, 2010; Qi et al, 2011). In brief, 15–30 g of whole cell lysates was boiled for 5 min prior to loading onto a 25-μM phos-tag 5% SDS–PAGE gel at 100 V for 3 h. The proteins were then transferred onto a PVDF membrane at 100 V for 90 min. Both phosphorylated and non-phosphorylated IRE1 were detected with anti-IRE1 antibody (14C10; Cell Signaling).
Anti-GPI-induced arthritis in mice
Experimental arthritis was induced in control or IRE1 conditional knockout (C57BL/6 genetic background) mice, using the K/BxN serum-transfer model (Scatizzi et al, 2010; Mavers et al, 2012). Mice (8–10 weeks old) were injected intraperitoneally with anti-GPI serum (200 μl/mouse). The diameter of each rear ankle joint was measured daily and the increases were determined based on the size measured the day before serum transfer. Clinical scores were determined as follows: grade 0=no swelling; grade 1=mild but definite redness and swelling of an ankle, wrist, or digits; grade 2=moderate redness and swelling of ankle and wrist; grade 3=severe redness and swelling of entire paw, including digits; and grade 4=maximally inflamed limb with involvement of multiple joints. Mice were euthanized 15 days after serum transfer, and their ankle joints were harvested for histological analysis. Joints were fixed in 4% paraformaldehyde for 24 h and decalcified. Tissue sections were characterized by H&E staining, and the inflammation of joint sections was scored as: 0=no inflammation; 1=modest inflammation; 2=severe inflammation, and 3=severe inflammation with bone or cartilage destruction.
The collection of all human synovial fluid samples used in this study has been approved by the Institutional Review Board of the Harbin Medical University and the Third Military Medical University.
For a full description of Materials and methods used in this work, see Supplementary Data.
Supplementary Material
Acknowledgments
We thank Sinyi Kong for critically reading the manuscript and laboratory members for critical discussion. This work was supported by National Institutes of Health (NIH) grants (AI079056, DK083050 and their supplemental grants) to DF, NIH grants (DK090313 and ES017829) and an American Heart Association grant (09GRNT2280479) to KZ, NIH grant CA154377 to DZ, and NIH grants HL057346 and DK042394 to RJK.
Author contributions: QQ, KZ, Z Zheng, LC, ZL, Y-SZ, AD, MG, XL, TZ, Z Zhang, QK, SC, AC, and W-LY performed the study. QQ and DF analysed the data. QQ, HL, H-KL, DZ, HP, RK, DF, KZ, and ET wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Asea A, Rehli M, Kabingu E, Boch JA, Bare O, Auron PE, Stevenson MA, Calderwood SK (2002) Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J Biol Chem 277: 15028–15034 [DOI] [PubMed] [Google Scholar]
- Brenner DA, O'Hara M, Angel P, Chojkier M, Karin M (1989) Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature 337: 661–663 [DOI] [PubMed] [Google Scholar]
- Bruhns P, Samuelsson A, Pollard JW, Ravetch JV (2003) Colony-stimulating factor-1-dependent macrophages are responsible for IVIG protection in antibody-induced autoimmune disease. Immunity 18: 573–581 [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B (2008) Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol 9: 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Hsu TL, Lin HH, Chio CC, Chiu AW, Chen NJ, Lin CH, Hsieh SL (2004) Modulation of macrophage differentiation and activation by decoy receptor 3. J Leukoc Biol 75: 486–494 [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8: 265–277 [DOI] [PubMed] [Google Scholar]
- Colbert RA, DeLay ML, Layh-Schmitt G, Sowders DP (2009) HLA-B27 misfolding and spondyloarthropathies. Adv Exp Med Biol 649: 217–234 [DOI] [PubMed] [Google Scholar]
- Corr M, Crain B (2002) The role of FcgammaR signaling in the K/BxN serum transfer model of arthritis. J Immunol 169: 6604–6609 [DOI] [PubMed] [Google Scholar]
- Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP (2012) The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA 109: E869–E878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Egan CE, Sukhumavasi W, Butcher BA, Denkers EY (2009) Functional aspects of Toll-like receptor/MyD88 signalling during protozoan infection: focus on Toxoplasma gondii. Clin Exp Immunol 156: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliet M, Cao W, Liu YJ (2008) Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 8: 594–606 [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR (2009) IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138: 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harama D, Koyama K, Mukai M, Shimokawa N, Miyata M, Nakamura Y, Ohnuma Y, Ogawa H, Matsuoka S, Paton AW, Paton JC, Kitamura M, Nakao A (2009) A subcytotoxic dose of subtilase cytotoxin prevents lipopolysaccharide-induced inflammatory responses, depending on its capacity to induce the unfolded protein response. J Immunol 183: 1368–1374 [DOI] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH (2011) The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev 91: 1219–1243 [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107 [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140: 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QQ, Koessler RE, Birkett R, Perlman H, Xing L, Pope RM (2013) TLR2 deletion promotes arthritis through reduction of IL-10. J Leukoc Biol 93: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QQ, Sobkoviak R, Jockheck-Clark AR, Shi B, Mandelin AM 2nd, Tak PP, Haines GK 3rd, Nicchitta CV, Pope RM (2009) Heat shock protein 96 is elevated in rheumatoid arthritis and activates macrophages primarily via TLR2 signaling. J Immunol 182: 4965–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M (2004) A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med 10: 98–102 [DOI] [PubMed] [Google Scholar]
- Jacobs JP, Ortiz-Lopez A, Campbell JJ, Gerard CJ, Mathis D, Benoist C (2010) Deficiency of CXCR2, but not other chemokine receptors, attenuates autoantibody-mediated arthritis in a murine model. Arthritis Rheum 62: 1921–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ohmura K, Mahmood U, Lee DM, Hofhuis FM, Boackle SA, Takahashi K, Holers VM, Walport M, Gerard C, Ezekowitz A, Carroll MC, Brenner M, Weissleder R, Verbeek JS, Duchatelle V, Degott C, Benoist C, Mathis D (2002) Arthritis critically dependent on innate immune system players. Immunity 16: 157–168 [DOI] [PubMed] [Google Scholar]
- Kaser A, Flak MB, Tomczak MF, Blumberg RS (2011) The unfolded protein response and its role in intestinal homeostasis and inflammation. Exp Cell Res 317: 2772–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S (2011) Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34: 637–650 [DOI] [PubMed] [Google Scholar]
- Kontogiorgis CA, Savvoglou K, Hadjipavlou-Litina DJ (2006) Antiinflammatory and antioxidant evaluation of novel coumarin derivatives. J Enzyme Inhib Med Chem 21: 21–29 [DOI] [PubMed] [Google Scholar]
- Korganow AS, Ji H, Mangialaio S, Duchatelle V, Pelanda R, Martin T, Degott C, Kikutani H, Rajewsky K, Pasquali JL, Benoist C, Mathis D (1999) From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity 10: 451–461 [DOI] [PubMed] [Google Scholar]
- Lee AH, Glimcher LH (2009) Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci 66: 2835–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH (2011) Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci USA 108: 8885–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH (2008) Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DM, Friend DS, Gurish MF, Benoist C, Mathis D, Brenner MB (2002) Mast cells: a cellular link between autoantibodies and inflammatory arthritis. Science 297: 1689–1692 [DOI] [PubMed] [Google Scholar]
- Li ZP, Hu JF, Sun MN, Ji HJ, Zhao M, Wu DH, Li GY, Liu G, Chen NH (2011) Effect of compound IMMLG5521, a novel coumarin derivative, on carrageenan-induced pleurisy in rats. Eur J Pharmacol 661: 118–123 [DOI] [PubMed] [Google Scholar]
- Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR et al. (1999) TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev 13: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Dong D, Little E, Luo S, Lee AS (2004) Transgenic mouse model for monitoring endoplasmic reticulum stress in vivo. Nat Med 10: 1013–1014 author reply 1014 [DOI] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, Glimcher LH (2010) TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 11: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Glimcher LH (2011) Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr Opin Immunol 23: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavers M, Cuda CM, Misharin AV, Gierut AK, Agrawal H, Weber E, Novack DV, Haines GK 3rd, Balomenos D, Perlman H (2012) Cyclin-dependent kinase inhibitor p21, via its C-terminal domain, is essential for resolution of murine inflammatory arthritis. Arthritis Rheum 64: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KH (2011) TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol 11: 807–822 [DOI] [PubMed] [Google Scholar]
- Millward TA, Zolnierowicz S, Hemmings BA (1999) Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci 24: 186–191 [DOI] [PubMed] [Google Scholar]
- Nundlall S, Rajpar MH, Bell PA, Clowes C, Zeeff LA, Gardner B, Thornton DJ, Boot-Handford RP, Briggs MD (2010) An unfolded protein response is the initial cellular response to the expression of mutant matrilin-3 in a mouse model of multiple epiphyseal dysplasia. Cell Stress Chaperones 15: 835–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA (2003) Therapeutic targeting of Toll-like receptors for inflammatory and infectious diseases. Curr Opin Pharmacol 3: 396–403 [DOI] [PubMed] [Google Scholar]
- Ohashi K, Burkart V, Flohe S, Kolb H (2000) Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J Immunol 164: 558–561 [DOI] [PubMed] [Google Scholar]
- Oslowski CM, Hara T, O'Sullivan-Murphy B, Kanekura K, Lu S, Hara M, Ishigaki S, Zhu LJ, Hayashi E, Hui ST, Greiner D, Kaufman RJ, Bortell R, Urano F (2012) Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metab 16: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospelt C, Brentano F, Rengel Y, Stanczyk J, Kolling C, Tak PP, Gay RE, Gay S, Kyburz D (2008) Overexpression of toll-like receptors 3 and 4 in synovial tissue from patients with early rheumatoid arthritis: toll-like receptor expression in early and longstanding arthritis. Arthritis Rheum 58: 3684–3692 [DOI] [PubMed] [Google Scholar]
- Pan R, Gao XH, Li Y, Xia YF, Dai Y (2010) Anti-arthritic effect of scopoletin, a coumarin compound occurring in Erycibe obtusifolia Benth stems, is associated with decreased angiogenesis in synovium. Fundam Clin Pharmacol 24: 477–490 [DOI] [PubMed] [Google Scholar]
- Pisetsky DS, Erlandsson-Harris H, Andersson U (2008) High-mobility group box protein 1 (HMGB1): an alarmin mediating the pathogenesis of rheumatic disease. Arthritis Res Ther 10: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochet L, Frederick R, Masereel B (2004) Coumarin and isocoumarin as serine protease inhibitors. Curr Pharm Des 10: 3781–3796 [DOI] [PubMed] [Google Scholar]
- Proost P, Verpoest S, Van de Borne K, Schutyser E, Struyf S, Put W, Ronsse I, Grillet B, Opdenakker G, Van Damme J (2004) Synergistic induction of CXCL9 and CXCL11 by Toll-like receptor ligands and interferon-gamma in fibroblasts correlates with elevated levels of CXCR3 ligands in septic arthritis synovial fluids. J Leukoc Biol 75: 777–784 [DOI] [PubMed] [Google Scholar]
- Qi L, Yang L, Chen H (2011) Detecting and quantitating physiological endoplasmic reticulum stress. Methods Enzymol 490: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Mao T, Zhang Y, Shao M, You J, Ding Q, Chen Y, Wu D, Xie D, Lin X, Gao X, Kaufman RJ, Li W, Liu Y (2010) A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal 3: ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs MF, Boelens WC, Joosten LA, Abdollahi-Roodsaz S, Geurts J, Wunderink LU, Schreurs BW, van den Berg WB, Radstake TR (2006) Identification of small heat shock protein B8 (HSP22) as a novel TLR4 ligand and potential involvement in the pathogenesis of rheumatoid arthritis. J Immunol 176: 7021–7027 [DOI] [PubMed] [Google Scholar]
- Rostom S, Mengat M, Lahlou R, Hari A, Bahiri R, Hajjaj-Hassouni N (2013) Metabolic syndrome in rheumatoid arthritis: case control study. BMC Musculoskelet Disorders 14: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP (2001) Regulation of transcriptional activation domain function by ubiquitin. Science 293: 1651–1653 [DOI] [PubMed] [Google Scholar]
- Scatizzi JC, Hutcheson J, Pope RM, Firestein GS, Koch AE, Mavers M, Smason A, Agrawal H, Haines GK 3rd, Chandel NS, Hotchkiss RS, Perlman H (2010) Bim-Bcl-2 homology 3 mimetic therapy is effective at suppressing inflammatory arthritis through the activation of myeloid cell apoptosis. Arthritis Rheum 62: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C, Cartwright A, Williams H, Haworth O, Williams JH, Filer A, Salmon M, Buckley CD, Middleton J (2010) Monocytes/macrophages express chemokine receptor CCR9 in rheumatoid arthritis and CCL25 stimulates their differentiation. Arthritis Res Ther 12: R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibl R, Birchler T, Loeliger S, Hossle JP, Gay RE, Saurenmann T, Michel BA, Seger RA, Gay S, Lauener RP (2003) Expression and regulation of Toll-like receptor 2 in rheumatoid arthritis synovium. Am J Pathol 162: 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H, He Y, Yang L, Qi L (2011) Stressed out about obesity: IRE1alpha-XBP1 in metabolic disorders. Trends Endocrinol Metab 22: 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107: 893–903 [DOI] [PubMed] [Google Scholar]
- Shin EM, Zhou HY, Guo LY, Kim JA, Lee SH, Merfort I, Kang SS, Kim HS, Kim S, Kim YS (2008) Anti-inflammatory effects of glycyrol isolated from Glycyrrhiza uralensis in LPS-stimulated RAW264.7 macrophages. Int Immunopharmacol 8: 1524–1532 [DOI] [PubMed] [Google Scholar]
- Shin YJ, Han SH, Kim DS, Lee GH, Yoo WH, Kang YM, Choi JY, Lee YC, Park SJ, Jeong SK, Kim HT, Chae SW, Jeong HJ, Kim HR, Chae HJ (2010) Autophagy induction and CHOP under-expression promotes survival of fibroblasts from rheumatoid arthritis patients under endoplasmic reticulum stress. Arthritis Res Ther 12: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MJ, Sowders DP, DeLay ML, Mohapatra R, Bai S, Smith JA, Brandewie JR, Taurog JD, Colbert RA (2005) HLA-B27 misfolding in transgenic rats is associated with activation of the unfolded protein response. J Immunol 175: 2438–2448 [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- van Beijnum JR, Buurman WA, Griffioen AW (2008) Convergence and amplification of toll-like receptor (TLR) and receptor for advanced glycation end products (RAGE) signaling pathways via high mobility group B1 (HMGB1). Angiogenesis 11: 91–99 [DOI] [PubMed] [Google Scholar]
- Vanags D, Williams B, Johnson B, Hall S, Nash P, Taylor A, Weiss J, Feeney D (2006) Therapeutic efficacy and safety of chaperonin 10 in patients with rheumatoid arthritis: a double-blind randomised trial. Lancet 368: 855–863 [DOI] [PubMed] [Google Scholar]
- Varoga D, Paulsen F, Mentlein R, Fay J, Kurz B, Schutz R, Wruck C, Goldring MB, Pufe T (2006) TLR-2-mediated induction of vascular endothelial growth factor (VEGF) in cartilage in septic joint disease. J Pathol 210: 315–324 [DOI] [PubMed] [Google Scholar]
- Wahamaa H, Schierbeck H, Hreggvidsdottir HS, Palmblad K, Aveberger AC, Andersson U, Harris HE (2011) High mobility group box protein 1 in complex with lipopolysaccharide or IL-1 promotes an increased inflammatory phenotype in synovial fibroblasts. Arthritis Res Ther 13: R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Wipke BT, Allen PM (2001) Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol 167: 1601–1608 [DOI] [PubMed] [Google Scholar]
- Yang L, Xue Z, He Y, Sun S, Chen H, Qi L (2010) A Phos-tag-based approach reveals the extent of physiological endoplasmic reticulum stress. PLoS One 5: e11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK (2009) The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325: 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Fu Y, Li S, Tu L, Zeng X, Kuang N (2011) Regulatory effect of daphnetin, a coumarin extracted from Daphne odora, on the balance of Treg and Th17 in collagen-induced arthritis. Eur J Pharmacol 670: 286–294 [DOI] [PubMed] [Google Scholar]
- Yavuz S, Elbir Y, Tulunay A, Eksioglu-Demiralp E, Direskeneli H (2008) Differential expression of toll-like receptor 6 on granulocytes and monocytes implicates the role of microorganisms in Behcet's disease etiopathogenesis. Rheumatol Int 28: 401–406 [DOI] [PubMed] [Google Scholar]
- Yesilada E, Taninaka H, Takaishi Y, Honda G, Sezik E, Momota H, Ohmoto Y, Taki T (2001) In vitro inhibitory effects of Daphne oleoides ssp. oleoides on inflammatory cytokines and activity-guided isolation of active constituents. Cytokine 13: 359–364 [DOI] [PubMed] [Google Scholar]
- Yoo SA, You S, Yoon HJ, Kim DH, Kim HS, Lee K, Ahn JH, Hwang D, Lee AS, Kim KJ, Park YJ, Cho CS, Kim WU (2012) A novel pathogenic role of the ER chaperone GRP78/BiP in rheumatoid arthritis. J Exp Med 209: 871–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2010) Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int J Clin Exp Med 3: 33–40 [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ (2006) The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology 66: S102–S109 [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ (2008) From endoplasmic-reticulum stress to the inflammatory response. Nature 454: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ (2011) The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J 30: 1357–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ (2005) The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest 115: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.