Abstract
The development of clinically useful histone deacetylase inhibitors has expanded greatly. In a preclinical study, we showed that panobinostat (LBH589) inhibits cell cycle progression of human head and neck squamous cell carcinoma (HNSCC) cell lines at G2/M and an associated decrease in expression of particular genes required for passage through G2 and mitosis. In this study we sought to analyse the mechanistic underpinnings of panobinostat-induced growth arrest. HNSCC cell lines were synchronised and progression through mitosis monitored. We demonstrate that panobinostat causes a marked G2 delay and mitotic defects. A loss of G2-specific Plk1 and Cyclin B1 expression and co-incident increase in p21Waf1/Cip1 expression is also shown. Furthermore, we show a significant loss of E2F1 recruitment to the promoters of these genes in response to panobinostat treatment. These data provide mechanistic evidence of panobinostat-induced cell cycle arrest and highlight its potential as a chemotherapeutic agent for HNSCC.
Squamous cell carcinoma of the head and neck (HNSCC) is a disease of considerable morbidity and mortality with an estimated 560,000 cases per year and 300,000 deaths worldwide, and approximately 50,000 new cases and 11,000 deaths in the United States annually1,2. HNSCC occur in the upper aerodigestive tract from the nasopharynx and oral cavity to the larynx with varying metastatic rates for each anatomic site. Treatment depends upon the disease location, the extent of the primary tumour and the presence of metastatic disease. Conventional treatment usually includes surgery and adjuvant radiation therapy, with or without chemotherapy. Any of these therapies can, and do, produce morbidities affecting speech, swallowing and overall quality of life. Despite these interventions, recurrence of the disease is observed in about 50% of patients with high rates of associated mortality3. Even though there has been only a modest improvement in overall survival over the past decades, it has been questioned whether a large percentage of patients are over-treated, incurring additional morbidity2. Therefore, strategies must be developed for the most appropriate treatment for any given patient and new treatment paradigms need to be defined.
Histone deacetylases (HDACs) are a group of enzymes that remove acetyl groups from histone proteins, thereby compacting chromatin and silencing the expression of genes. There are now 18 known HDACs grouped into four classes, which can also deacetylate numerous non-histone target proteins and alter their function. Indeed, some HDACs have been shown to alter the transcriptional activity of proteins such as p53, E2F and c-Myc4,5,6. HDAC inhibition is generally thought to increase the expression of repressed genes, including tumor suppressor genes, causing dramatic effects on cell growth and survival and because of this normal cells may be ten-fold or more resistant to HDAC inhibitor (HDACi) induced cell death than transformed cells7. The isolation of potent HDAC inhibitors (HDACi) has been the focus of numerous anti-cancer drug discovery efforts and the testing and development of clinically useful HDACi via clinical trials has expanded greatly. Some of these drugs inhibit selective classes of HDACs and some, like panobinostat (LBH589), a member of the hydroxamate class, inhibit multiple HDAC classes7.
Panobinostat has been shown to be effective in the treatment of hematologic malignancies such as Hodgkin lymphoma, multiple myeloma and cutaneous T-cell lymphoma8,9,10,11,12 and its use has also been tested for the treatment of epithelial neoplasms either alone or in combination with other chemotherapeutic agents13,14,15,16,17,18,19. While these are very early clinical trials, some of the initial results are promising. In a preclinical study, we showed that panobinostat inhibits cell cycle progression of human HNSCC cell lines ultimately resulting in cell death in vitro20. The cell cycle arrest occurred at G2/M and has been observed in other studies using a variety of cell types including the prostate cancer cell lines, PC3 and PC9 and an epithelial lung cancer cell line A54921,22. In our study, we showed that the G2/M arrest was associated with a substantial decrease in the expression of a particular sub-set of genes that are required for the passage through the G2 checkpoint and into mitosis20. The objectives of the present study were to define more specifically the cell cycle arrest caused by panobinostat and to elucidate the mechanistic underpinnings.
Results
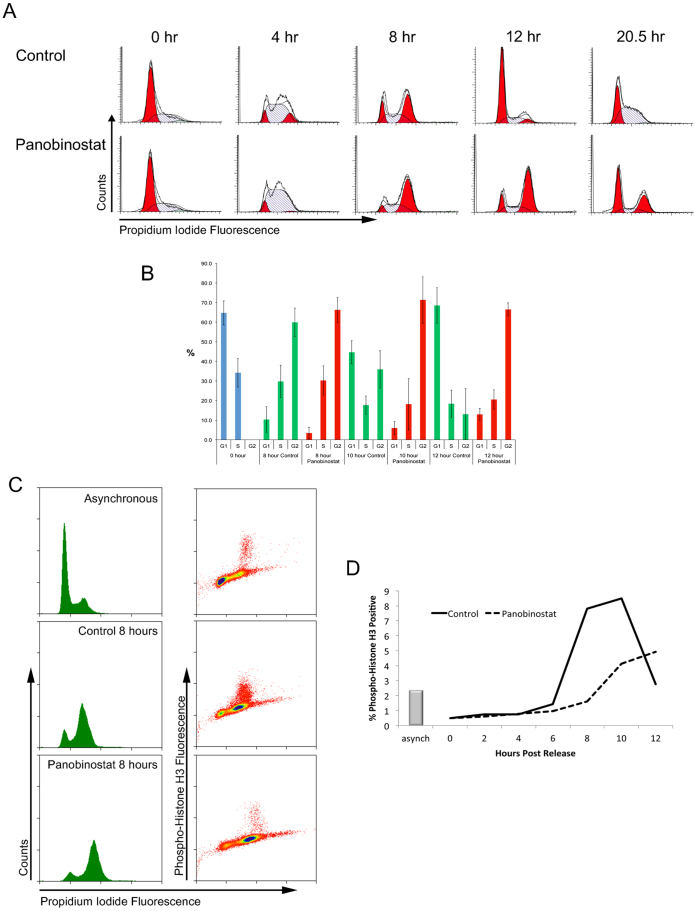
Previously we showed that panobinostat results in a significant arrest of the cells at G2/M within 12 h post treatment20. To characterise this arrest further and study potential defects in DNA replication, cells were synchronised by double thymidine block and released into medium containing 100 nM panobinostat or excipient control. At 0 h, cells are synchronised at the G1/S boundary (figure 1A). Control treated cells progressed into S phase 4 h post release and passaged through G2/M and into G1 phase by 8 h. By 12 h, the majority of the cells had passed through mitosis and entered G1 phase. Cells subsequently re-entered S phase (20.5 h), showing completion of one cell cycle. Cells released into 100 nM panobinostat entered and passed through S phase with similar kinetics as the control cell population, but in contrast to control cells, displayed a significant and prolonged G2/M arrest at 8 and 12 h post release (figure 1A and B). By 20.5 h, some cells remained arrested at G2/M but entry into G1 phase was apparent, suggesting that treatment with panobinostat induces a prolonged cell cycle delay, but not a complete arrest. Similar effects of panobinostat on cell cycle progression were observed in UMSCC-1 and UMSCC-47 cells (oral cavity, obtained from Dr. Thomas Carey, University of Michigan23), with a significant increase in G2/M cells 11.5 hours post release into medium containing 100 nM panobinostat compared to cells released into control medium (11.5% (p = 0.02) and 21.7% (p = 0.0006) increase in the G2/M fraction in UM-SCC-1 and UMSCC-47 cells, respectively). To support the notion that this increase is due to a G2 arrest, a significant decrease in G1 entry was also observed with this experiment (12.4% (p = 0.009) and 19.1% (p = 0.01) decrease in G1 fraction on UMSCC-1 and UMSCC-47 cells, respectively). These data provide evidence that the cell cycle arrest caused by panobinostat treatment is not cell type specific (supplementary figure 1).
Figure 1. Analysis of panobinostat-induced growth arrest.
(A) FaDu cells were synchronised by double thymidine block and released in medium containing 100 nM panobinostat or excipient control. Cells were harvested at the indicated times post release, fixed and stained with propidium iodide before analysis by flow cytometry. The histograms represent propidium iodide fluorescence versus counts. (B) The proportion of cells in G1, S, or G2/M phases from cells synchronised by double thymidine block (blue) and 8, 10 and 12 hours post release with (red) or without (green) 100 nM panobinostat was calculated using ModFit LT 3.0 software (Verity Software House). The data represent the mean and standard deviation of at least 5 independent experiments. (C) Cells were fixed and stained with propidium iodide (left panel) and a combination of propidium iodide and Alexa 488 conjugated anti-phospho-histone H3 antibody (right panel). The data shown are from asynchronously growing cells and are representative of data collected at all time points post release. (D) The percentage of mitotic cells post release from a double thymidine block was calculated using Summit software (Dako) from the phos-histone-H3 positive population of cells.
To determine whether the cells were delayed at G2 phase or in mitosis, phosphorylation of histone H3 (Serine 10) was assessed (figure 1C). Histone H3 phosphorylation in control treated cells peaked at around 9% at 9 h and quickly fell to below 3% by 12 h, indicating mitotic entry and exit between 8 and 12 h post release (figure 1D). In contrast, panobinostat-treated cells displayed a delay in mitotic entry, reaching a peak of only 4.5% phosphorylated histone H3 at 12 h, 4 h later than the control population. This strongly suggests that panobinostat results in a significant delay at G2 phase and reduced mitotic entry.
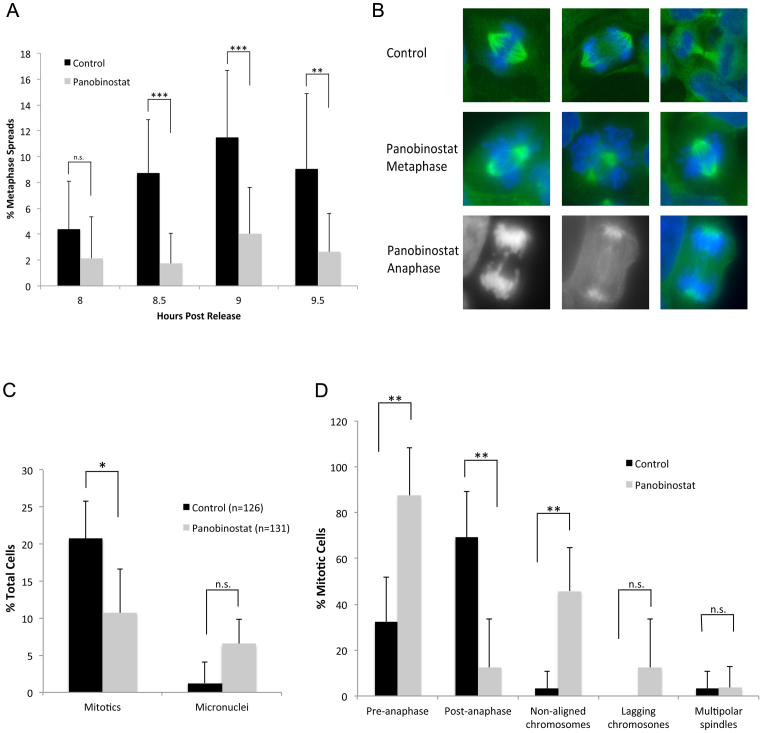
To confirm these data, we prepared metaphase spreads of synchronised cells at various times post release (figure 2A). The number of metaphases present in the control population peaked at 9 h (11.5%) while cells cultured in 100 nM panobinostat showed a highly significant reduction in metaphase cells (p < 0.0001), confirming the reduced mitotic entry observed by histone H3 phosphorylation. Although there is reduced entry into mitosis in cells treated with panobinostat, some cells clearly enter mitosis. We therefore sought to determine whether mitosis progressed normally in panobinostat-treated cells. Synchronised cells were fixed 9 h post release, and the mitotic spindles and DNA stained (figure 2B). The results confirmed reduced mitotic entry (Figure 2C; 10% compared to 20% in control-treated cells, p < 0.01). In addition, the percentage of the total mitotic cells in pre-anaphase (prophase and metaphase) was significantly increased by nearly three fold following panobinostat treatment (figure 2D; 87.5% compared to 32% in control cells, p < 0.002), with a concomitant decrease in the percentage of mitotic cells in late stage mitosis (anaphase and telophase, p < 0.001). Many mitotic cells at metaphase had non-aligned chromosomes, where cells had appeared to form a normal bipolar spindle but the chromosomes had not congressed to the metaphase plate (3.5% in control cells compared to 46% in panobinostat treated cells p < 0.001). In the few cells that were found to have progressed into anaphase, many had lagging chromosomes (12.5%, compared to 0% in control cells) although this was not statistically significant (p = 0.2), likely because very few cells are able to progress into anaphase.
Figure 2. Reduced mitotic entry and mitotic abnormalities caused by panobinostat.
FaDu cells were synchronised by double thymidine block and released in medium containing 100 nM panobinostat or excipient control. (A) Cells were harvested at the indicated times post release and metaphase spreads prepared. The percentage of metaphases was scored for each total cell population (n > 200). The data shown represents the mean and standard deviation of each sample. (B) 9 hours post release, cells were fixed and stained with anti-α-tubulin antibody (green) and the DNA stained with DAPI (blue and grey). Cells were visualised using an Axiovert 200 (Zeiss) microscope fitted with a 63× objective and images were captured using a Camera Hall 100 and the Applied Spectral Imaging software. The top panel shows representative images of mitotic cells in excipient control. Normal chromosome congression at metaphase and chromosome segregation at anaphase is observed. The middle panel shows representative images of metaphase cells treated with panobinostat. Failure of chromosome congression (non-aligned chromosomes) is common in these cells. The lower panel shows a representative image of a cell that has progressed to anaphase, showing lagging chromosomes (left DNA, middle α-tubulin and right merge). (C) The percentage of mitotic cells and micronuclei in control (n = 126) or panobinostat treated (n = 131) cell populations was determined by microscopic analysis. The data shown represent the mean and standard deviation for each sample. (D) The mitotic cells in control or panobinostat treated samples were scored for various phenotypes as indicated in the bar chart and expressed as a percentage of the number of mitotic cells counted. The data shown represent the mean and standard deviation for each sample.
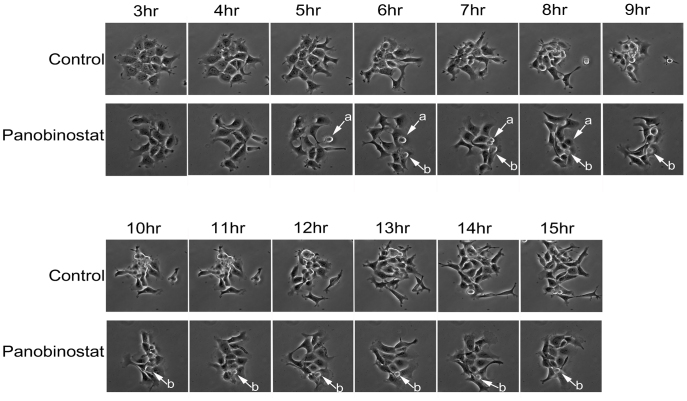
To visualize the G2 delay and mitotic failure, we imaged live cells by timelapse microscopy to observe the progression of panobinostat treated cells into and through mitosis (figure 3 and supplementary movies 1 and 2). While synchronised control cells entered mitosis 8–9 h post release and cells successfully completed mitosis within 1–2 h, panobinostat-treated cells had far fewer successful mitoses (one indicated by arrow (a) in the field), and some cells appeared to enter mitosis (arrow (b)) but remain rounded, for over 10 h in this case, and failed to complete mitosis.
Figure 3. Analysis of mitotic entry by live cell imaging.
Cells growing in glass bottom dishes were synchronised by double thymidine block and released in panobinostat or excipient control medium. 3 hours following release, cells were placed in a heated chamber and imaged using a 20× objective every 2 minutes. The representative images shown were taken at the stated time points following release.
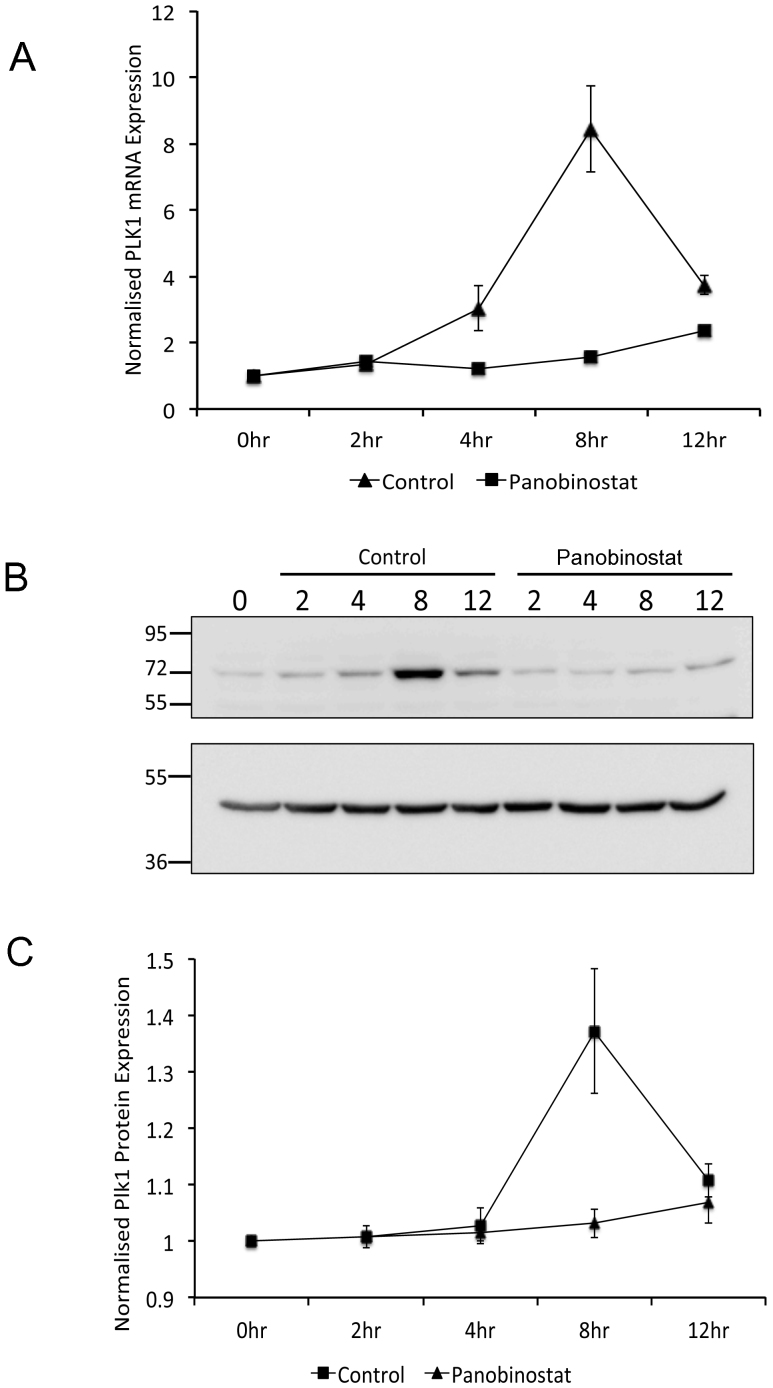
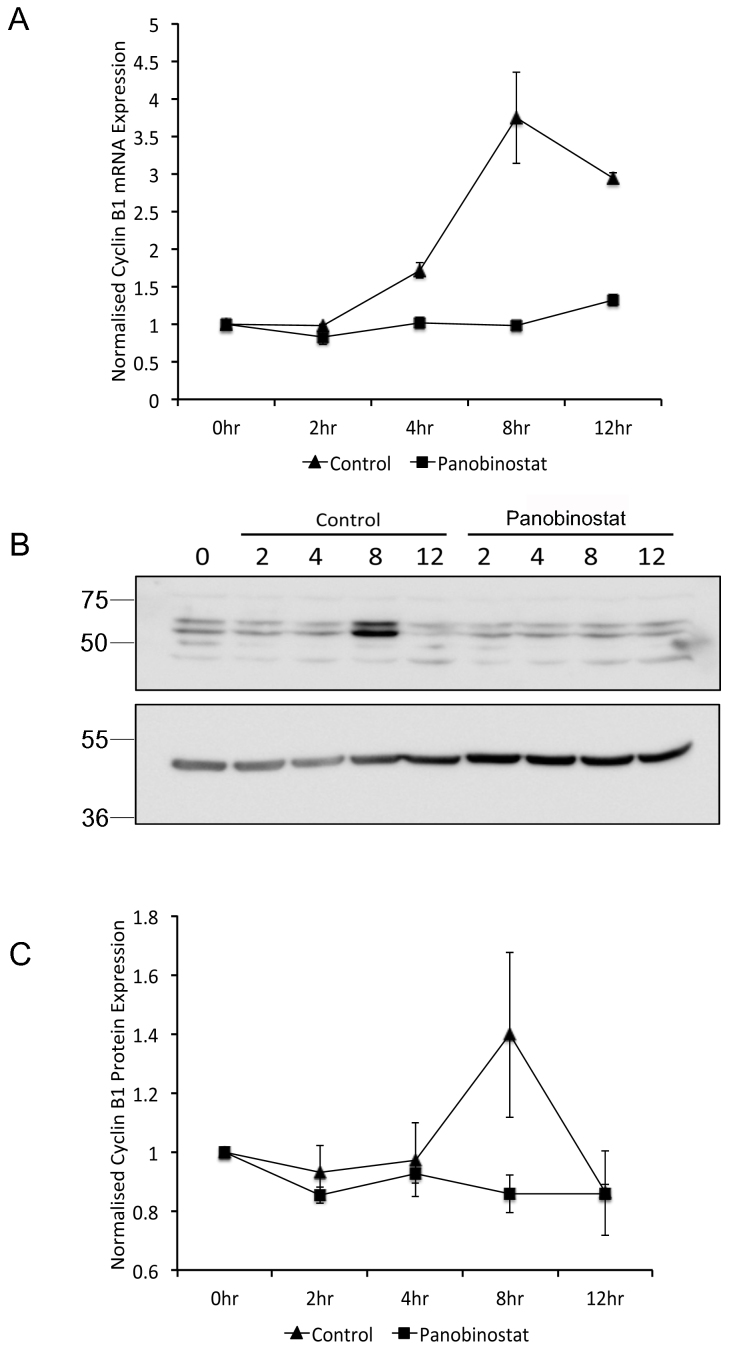
Our previous report showed that panobinostat causes a dramatic reduction in genes required for cell cycle progression including Polo-like kinase 1 (PLK1)20. We therefore hypothesised that the mitotic abnormalities observed are due to a loss of cell cycle dependent Plk1 expression. To test this, RNA and protein were prepared from synchronised cells cultured in the presence or absence of panobinostat for 2, 4, 8 and 12 h post release. Plk1 mRNA and protein expression dramatically increased by 8 h post release in control cells, however no significant increase in PLK1 mRNA or protein was observed in panobinostat-treated cells (figure 4). In addition, microarray analysis of gene expression in synchronised FaDu cells released for 8 h in panobinostat or excipient control revealed a significant reduction in the expression of Cyclin B1 mRNA (data not shown). More careful analysis of Cyclin B1 mRNA and protein expression showed an increase in expression between 4 and 8 h post release in control cells, concomitant with mitotic entry, but a loss of cell cycle dependent up-regulation in panobinostat-treated cells (figure 5). The loss of cell cycle dependent upregulation of Plk1 and Cyclin B1 protein expression was also assessed in two further oropharyngeal squamous cell carcinoma cell lines (UMSCC-1 and UMSCC-47; supplementary figures 2 and 3), confirming that these effects of panobinostat treatment are not specific to FaDu cells.
Figure 4. Reduction of G2-specific Plk1 expression following panobinostat treatment.
FaDu cells were synchronised by double thymidine block and released in medium containing 100 nM panobinostat or excipient control. Cells were harvested at the stated time points post release and RNA (A) and protein (B and C) harvested as described in the materials and methods. (A) PLK1 mRNA expression was assessed by qRT-PCR. The data shown represent PLK1 mRNA levels normalised to 0 hour time point and are shown as the mean and standard deviation of three independent experiments. (B) 60 μg protein from each extract was separated by SDS-PAGE on two separate gels run using identical conditions. The upper panel was probed with Plk1 antibody and the lower panel probed with β-actin antibody. Full-length digital images of these western blots can be viewed in the supplementary information (supplementary figure 5). (C) Plk1 protein levels from three independent experiments were quantified using ImageGauge v4.21 software and normalised to β-actin levels from the same experiment. The data shown represent the mean and standard deviation.
Figure 5. Reduction of G2-specific Cyclin B1 expression following panobinostat treatment.
FaDu cells were synchronised by double thymidine block and released in medium containing 100 nM panobinostat or excipient control. Cells were harvested at the stated time points post release and RNA (A) and protein (B and C) harvested as described in the materials and methods. (A) Cyclin B1 mRNA expression was assessed by qRT-PCR. The data shown represent Cyclin B1 mRNA levels normalised to 0 hour time point and are shown as the mean and standard deviation of three independent experiments. (B) 60 μg protein from each extract was separated by SDS-PAGE on two separate gels that were run using identical conditions. Cyclin B1 (upper panel) and β-actin (lower panel) protein levels were assessed by Western blot. Full-length digital images of these western blots can be viewed in the supplementary information (supplementary figure 5). (C) Cyclin B1 protein levels from three independent experiments were quantified using ImageGauge v4.21 software and normalised to β-actin levels from the same experiment. The data shown represent the mean and standard deviation.
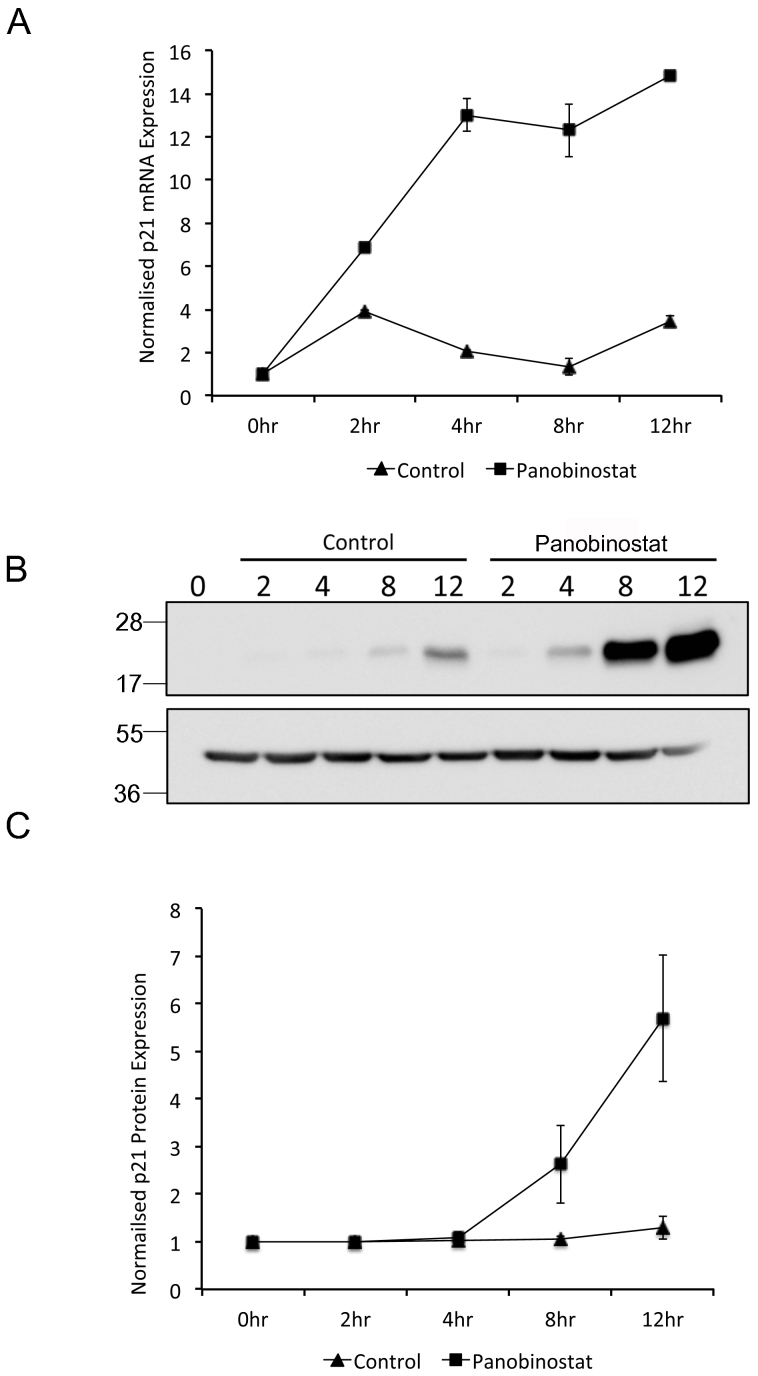
Panobinostat, like many HDACi, results in a rapid and significant increase in p21Waf1/Cip1 protein expression24. Furthermore, p21Waf1/Cip1 overexpression inhibits the transcription of PLK1, mediated by the cell cycle-dependent element (CDE) and cell cycle genes homology region (CHR) within the Plk1 promoter25,26. Cyclin B1 expression has also been shown to be regulated by p21Waf1/Cip127. We therefore hypothesized that the loss of cell cycle dependent PLK1 and Cyclin B1 expression in panobinostat treated cells was due to panobinostat-induced increase in p21Waf1/Cip1 expression prior to mitotic entry. p21Waf1/Cip1 RNA and protein levels following panobinostat treatment of synchronised cells were analysed as before (figure 6). A dramatic increase in p21Waf1/Cip1 mRNA and protein expression was observed in panobinostat treated cells that was not observed in the control population. The timing of the upregulation of p21Waf1/Cip1 was of particular significance. p21Waf1/Cip1 mRNA was double that observed in control cells by 2 hours and increased 10-fold by 4 h. More importantly, p21Waf1/Cip1 protein increased minimally by 4 h post release (1.2-fold) and significantly (3-fold) at 8 h post release in panobinostat treated cells whereas no increase in p21Waf1/Cip1 protein was observed in control treated cells (figure 6C).
Figure 6. Panobinostat induced p21Waf1/Cip1 expression.
FaDu cells were synchronised by double thymidine block and released in medium containing 100 nM panobinostat or excipient control. Cells were harvested at the stated time points post release and RNA (A) and protein (B and C) harvested as described in the materials and methods. (A) p21Waf1/Cip1 mRNA expression was assessed by qRT-PCR. The data shown represent p21Waf1/Cip1 mRNA levels normalised to 0 hour time point and are shown as the mean and standard deviation of three independent experiments. (B) 60 μg protein from each extract was separated by SDS-PAGE on two separate gels runs using identical conditions and p21Waf1/Cip1 (upper panel) and β-actin (lower panel) protein levels were assessed by Western blot. Full-length digital images of these western blots can be viewed in the supplementary information (supplementary figure 5). (C) p21Waf1/Cip1 protein levels from three independent experiments were quantified using ImageGauge v4.21 software and normalised to β-actin levels from the same experiment. The data shown represent the mean and standard deviation.
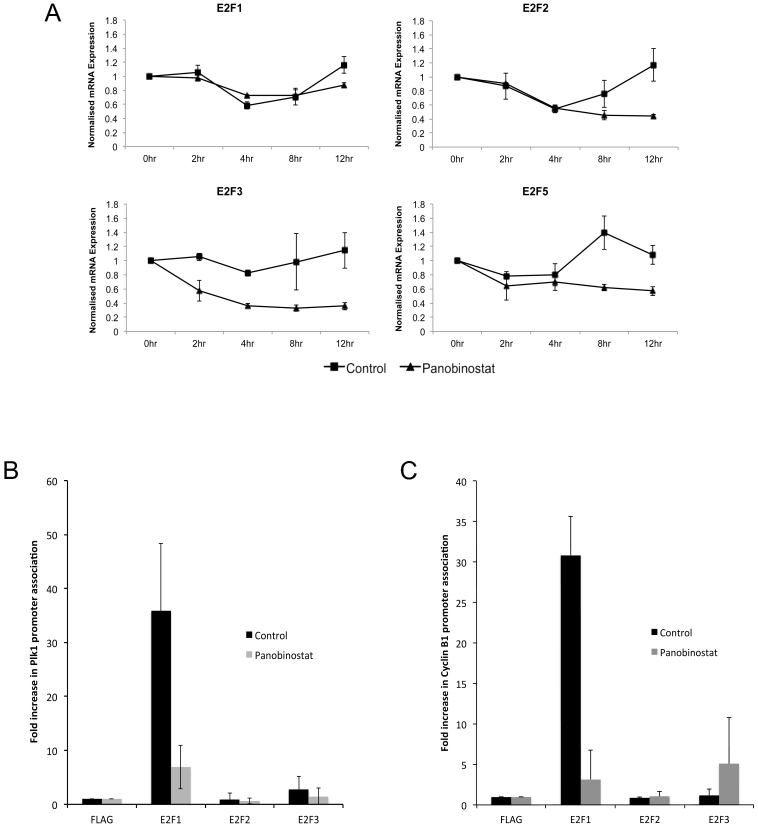
p21Waf1/Cip1 protein binds to the transcriptional activator E2F1 and prevents E2F-dependent activation of the PLK1 promoter26. In addition, the PLK1 and Cyclin B1 promoters are positively regulated by E2F family members28,29. Therefore, we determined the effect of panobinostat treatment on the expression of E2F family members in synchronised cells (figure 7A). mRNA expression of all E2Fs tested is reduced following treatment with panobinostat, with the greatest effect on E2F3, which was repressed at least 2-fold compared to control cells at 4 h post release. However, no significant reduction in the protein levels of E2F1, E2F2 and E2F3 were observed by western blot (data not shown). To explore the possibility that treatment with panobinostat results in reduced association of the activating E2F family members, E2Fs 1, 2 and 3, with the PLK1 and Cyclin B1 promoters and therefore loss of G2-specific activation of PLK1 and Cyclin B1 expression, we measured association of these proteins with the endogenous PLK1 and Cyclin B1 promoters by ChIP assay. Treatment of cells with panobinostat for 6 h caused a dramatic reduction in the association of E2F1 with both the PLK1 and Cyclin B1 promoters (figures 7B and C). In contrast, E2F2 and E2F3 were detected at both promoters at similar levels in control and panobinostat treated cells.
Figure 7. Loss of E2F1 recruitment to the Plk1 and Cyclin B1 promoters in response to panobinostat.
(A) FaDu cells were synchronised by double thymidine block and released in medium containing 100 nM panobinostat or excipient control. Cells were harvested at the stated time points post release and RNA. E2F1, E2F2, E2F3 and E2F5 mRNA expression was assessed by qRT-PCR. The data shown represent mRNA levels normalised to 0 hour time point and are shown as the mean and standard deviation of three independent experiments. (B and C) Asynchronous FaDu cells were incubated with 100 nM panobinostat or excipient control for 12 hours and crosslinked. Chromatin was then extracted and sheared and clarified chromatin preparations were immunoprecipitated with anti-FLAG (negative), E2F1, E2F2 or E2F3 antibody. The percentage of bound PLK1 promoter (B) or Cyclin B1 promoter (C) was then assessed by qRT-PCR and shown as the mean and standard deviation of three independent experiments.
Discussion
We and others have shown that panobinostat results in a cell cycle arrest at G2/M20,21,22,30. The use of synchronised cell populations has now enabled us to show definitively that the cells progress through S phase with normal kinetics, followed by a significant delay at the G2 checkpoint and decreased mitotic entry. Cells that eventually enter mitosis display significant abnormalities such as misaligned and lagging chromosomes. Live cell imaging confirms that successful completion of mitosis in the presence of panobinostat is a rare event. The experiments presented here have been carried out primarily in FaDu cells, but the G2 arrest and changes in Plk1 and Cyclin B1 expression following panobinostat treatment of synchronised cells have been confirmed in UMSCC-1 and UMSCC-47 (20 and supplementary figures 1, 2 and 3), demonstrating that the G2 arrest caused by panobinostat treatment observed in our studies is not specific to a particular cell line. It is interesting to note that FaDu and UMSCC-1 cells are HPV negative while UMSCC-47 cells are HPV positive, indicating that the HPV status of the cells used in these experiment does not affect the response to panobinostat.
It should be noted that our data are in contrast to a study by Pettazzoni et al.21, which demonstrates an increase in histone H3 acetylation and phosphorylation at serine 10 following treatment with panobinostat. While we did not specifically look at acetylation of histone H3, the study by Pettazzoni et al. indicates that phosphorylation at serine 10 is not affected by increased acetylation, indicating that increased acetylation is not responsible for the loss of histone H3 phosphorylation in our study (figure 1). Pettazzoni et al. also show an increase in mitotic cells, normal Cyclin B1 protein expression and a subtle decrease in Cdc2 phosphorylation at threonine 14 and tyrosine 15 in an asynchronous population following treatment with panobinostat. From these experiments, it was concluded that panobinostat treatment pushes cells into mitosis, increasing the proportion of cells at G2/M, followed by subsequent mitotic failure resulting in cell death. These experiments were performed in prostate cancer cells (LNCaP, DU145 and PC3) and while this difference alone could account for the differential response to panobinostat treatment, all of the experiments reported by Pettazzoni et al. were also performed at low concentrations of panobinostat (10 nM). Indeed, a study by Brazelle et al.22 in which various non-small cell lung cancer cells were treated with 40 nM panobinostat documented a G2/M arrest but also noted a subtle increase in the phosphorylation of histone H3 at serine 10 and an increase in multinucleated cells that indicates mitotic catastrophe. It is possible that different cell types respond differently to panobinostat exposure with either arrest of the cells prior to mitotic entry (our study), or mitotic catastrophe due to inappropriate mitotic entry21,22, or that different effects on the cell cycle may be caused by different concentrations of drug used. In order to address this question, we have carried out an experiment in which synchronized FaDu cells were treated with 25 nM panobinostat and progression through the cell cycle compared to cells in excipient control or 100 nM panobinostat (supplementary figure 4). These experiments show that in agreement with the experiments presented in Figures 1–3, cells in excipient control pass through the S, G2 and mitotic checkpoints normally and cells treated with 100 nM panobinostat are almost completely arrested at the G2/M peak of the flow analysis with minimal slippage through mitosis and into G1. However, cells released into medium containing 25 nM panobinostat pass through mitosis more frequently and by 12 hours post release, an almost 100% increase in G1 cells is observed when compared to cells treated with 100 nM panobinostat (12.39% G1 with 100 nM panobinostat compared to 23.12% G1 cells with 25 nM panobinostat). It will be interesting to determine whether other cells derived from HNSCC behave in the same way as prostate and lung cancer cells at low dose panobinostat and rather than arresting at G2, pass through this checkpoint and into mitosis with catastrophic consequences. The potential differential effects with varying panobinostat concentration are likely to be very important in the clinical use of panobinostat and should be taken into consideration when designing new clinical trials. During a phase I clinical trial in which intravenous panobinostat was administered, it was demonstrated that the half-life of panobinostat is in the order of 10–14 hours and serum concentrations of 400–700 nM are achievable at doses that are well tolerated31. These data indicate that the experiments carried out in this study have relevance to the clinical use of panobinostat and that the dose administered needs to be carefully assessed for optimal performance of this drug in treating different malignancies.
In agreement with our study, cells treated with various HDACi (although not panobinostat) were delayed at the G2 checkpoint and progressed slowly through mitosis32. In this study, cells arrested for an extended period in prometaphase with infrequent progression to metaphase and anaphase. Mitotic defects similar to those observed in our study were also demonstrated along with a significant increase in transit time through mitosis. Interestingly, combined treatment of cells with the HDACi suberohydroxamic acid (SBHA) and nocodazole, to inactivate the spindle assembly checkpoint, resulted in delayed mitotic entry followed by mitotic exit with similar kinetics to untreated cells. This provides evidence that SBHA treated cells override the spindle assembly checkpoint and exit mitosis with aberrant chromosome segregation.
It is well documented that p21Waf1/Cip1 protein expression is induced by numerous HDACi (reviewed by33). The activation of p21Waf1/Cip1 by various HDACi has been demonstrated in p53 null or mutant cells such as the FaDu cells used in this study, which have a homozygous mutation in p53 at codon 248 that results in an arginine to leucine substitution rendering p53 incapable of activating transcription from a variety of promoters34,35. The activation of p21Waf1/Cip1 by HDACi in cells with mutated or null p53 protein has been studied and in this context activation of p21Waf1/Cip1 requires acetylation of H2A.X at the p21 promoter36. p21Waf1/Cip1 has pleiotropic effects on cell cycle progression, mediated primarily by association with and inhibition of Cyclin dependent kinases (CDKs) leading to G1, S or G2-specific growth arrest. The kinetics of the increase in p21Waf1/Cip1 protein expression observed in this study are in agreement with that seen by Maiso et al. who document an increase in p21Waf1/Cip1 protein 3 h following treatment with 100 nM panobinostat24. We speculated that this dramatic increase in p21Waf1/Cip1 protein levels, which coincides with mitotic entry in our experimental system results in the repression of other p21Waf1/Cip1 regulated genes that are essential for mitotic entry and exit, although why a co-incident G1 arrest is not observed in asynchronously growing cells from HNSCC treated with panobinostat has not been resolved. It is worth noting that p21Waf1/Cip1 can inactivate proliferating cell nuclear antigen (PCNA) thereby inhibiting PCNA-dependent DNA replication37. However, DNA replication appears to progress normally in the presence of panobinostat leading us to conclude that this does not contribute to the growth arrest caused by panobinostat in our experiments.
We attribute the panobinostat-mediated G2 delay and downstream mitotic defects to aberrant Plk1 and Cyclin B1 expression since reduction of Plk1 expression by RNA interference produces similar mitotic and cytokinesis defects, and Cyclin B1 knockdown causes a G2 delay and slower mitotic transition with similar abnormalities to those observed in this study38,39. It has been demonstrated that both the PLK1 and Cyclin B1 promoters are repressed by the over expression of p21Waf1/Cip1 25,26,27. A role for the E2F family of transcription factors in the positive regulation of PLK1 expression has also been reported. E2Fs 1, 2 and 3 act directly on the PLK1 promoter to stimulate transcription activation28. Furthermore, p21Waf1/Cip1 has been shown to control the activity of E2F1 and prevent E2F1-dependent activation of promoters that contain E2F binding sites. This is at least in part independent of Retinoblastoma (Rb) protein function and is reported to be through a direct interaction between p21Waf1/Cip1 and E2F140. We have assessed the occupancy of the endogenous Plk1 and Cyclin B1 promoters by E2Fs 1, 2 and 3 by ChIP assay and show a dramatic and significant decrease in E2F1 protein associated with both of these promoters. This was somewhat surprising since the mRNA expression of E2F1 is least affected by treatment with panobinostat, however, the PLK1 promoter at least is most responsive to E2F128 and small changes in protein levels or activity could have dramatic effects on the expression of downstream activated genes. Nonetheless, these findings support our hypothesis that panobinostat-induced p21Waf1/Cip1 protein expression results in the E2F1 dependent repression of G2-specific genes. Whether this is regulated by post-translational modification including acetylation of E2F1 or p21Waf1/Cip1, changes in sub-cellular localization of E2F1, or direct interaction of p21Waf1/Cip1 and E2F1 following panobinostat treatment are hypotheses we are currently exploring.
Our study provides evidence that panobinostat treatment causes a G2 delay and abnormal mitotic progression through p21Waf1/Cip1-mediated suppression of E2F1 activation of Cyclin B1 and PLK1 expression, which are required for passage through the G2 checkpoint and normal mitotic transition. Our findings will inform future clinical studies designed to explore how panobinostat might be used in combination with other therapies to improve outcomes in patients with epithelial tumours, for example, through its sequential delivery with other mitosis blocking agents such as taxanes. Given that there is now considerable interest in exploring how epigenetic priming may make cancer cells more sensitive to radiotherapy, there is a compelling case for adding panobinostat to the list of HDAC inhibitors currently under evaluation as potential modulators of radiation sensitivity41,42,43. We are currently addressing these possibilities to optimise the use of panobinostat in future pre-clinical trials designed to determine whether these combinations would be more effective and less toxic than current regimens.
Methods
Cell culture
FaDu cells (ATCC #HTB-43), are an HPV negative cell line derived from a squamous cell carcinoma of the hypopharynx, were cultured at 37°C, 5% CO2 in RPMI 1640, containing 10% fetal bovine serum and 1 mM sodium pyruvate. UMSCC-1 cells originate from a tumour from the floor of the mouth in a male patient and are HPV negative. UMSCC-47 cells are derived from a tumour of the lateral tongue in a male patient and are HPV16 positive. Both of these cell lines were approved and cultured as previously described23,44. Cells are routinely tested for mycoplasma infection using a MycoAlert kit (Lonza).
Panobinostat
Panobinostat (LBH589; obtained from Peter Atadja, Novartis) was dissolved in d5W to 4.6 mM, the stock was then diluted to 100 nM in culture medium. Excipient control was prepared at the same d5W dilution.
Cell cycle synchronisation
2 mM thymidine was added to 50% confluent cells, which were incubated for 15 h. Cells were then reseeded at 2 × 106 cells/10 cm plate and left to recover for 12 h. 2 mM thymidine was added again and the cells were incubated for 14 h. Cells were either harvested immediately (0 h) or released in medium containing panobinostat or D5W alone (control) and harvested at the indicated times.
Flow cytometry
Cells were trypsinised and fixed in 1 ml of ice cold 70% ethanol. Cells were then washed in 1% BSA/PBS, and incubated for 30 min in BSA/PBS containing 50 μg/ml propidium iodide and 20 μg/ml RNase A. Samples were analysed using a FACSCanto II (Becton Dickinson) flow cytometer equipped with a 488 nm laser. Emission was detected using a 585/42 nm band pass filter. Data were analysed using ModFit LT 3.0 software (Verity Software House).
For immunofluorescent staining of phosphorylated histone H3, cells were fixed with 1% formaldehyde in PBS before fixing in ethanol. Cells were resuspended in 1% BSA/PBS containing with Alexa 488-conjugated anti-phospho-histone H3 (Ser10) antibody (Cell Signaling 9708) and incubated for 1 h before staining with propidium iodide. Cells were analysed as described above with the additional measurement of fluorescence emission using 530/30 nm band pass filter.
Chromosome analysis and immunofluorescence
Cells were synchronised and released into culture medium, followed by standard hypotonic treatment and fixation in methanol/acetic acid. For immunofluorescence, panobinostat treated and control cells were grown on glass coverslips and fixed in ice cold methanol for 10 min. Cells were then incubated with 5% goat serum followed by incubation with mouse anti α-tubulin (DM1A, Sigma; 1:1000) and detection with anti mouse Alexa-488 (Invitrogen; 1:1000). The cells were counterstained with DAPI and imaged with an Axiovert 200 microscope (Zeiss) with an oil immersion lens (60×, NA 1.35).
Live cell imaging
Time-lapse microscopy was performed with a 20×/0.5 NA phase contrast objective on an inverted microscope (NIKON TE2000-E) at 37°C and 5% CO2. Red LED illumination (CoolLED) and a CCD camera (Andor iXon), controlled by Micro-Manager software were used to acquire one image every 2 min45.
RNA extraction
Total RNA was isolated using TRIzol (Invitrogen). RNA was quantified using a Nanodrop 2000 and quality was confirmed using an Agilent Bioanalyzer and Agilent RNA 6000 Nanochips. All had RNA Integrity Numbers ≥9.2.
Quantitative real time PCR
qRT-PCR was performed using the Taqman RNA-to-CT 1-Step Kit (Applied Biosystems) and Gene Expression Assays; CDKN1A (p21) (Hs00355782_m1), PLK1 (Hs00153444), CCNB1 (Cyclin B1) (Hs01030097_m1) and GAPDH (Hs99999905_m1). Individual reactions were set up in triplicate from a master mix (32 μl Taqman 2× RT-PCR mix, 3.2 μl RNA (100 ng/μl), 3.2 μl primers (18 μM), 1.6 μl enzyme, 24 μl water). qRT-PCR measurements were performed on a StepOnePlus RT-PCR instrument (Applied Biosystems), using the following program: 48°C for 15 min, 95°C for 10 min and 40 cycles of 95°C for 15 s, 60°C for 1 min. The comparative threshold (CT) was determined for each gene as the average of three separate experiments.
Normalized values (ΔCT) were calculated by subtracting the CT for GAPDH from the CT of each gene. ΔΔCT values for each time point/condition was generated by subtracting the ΔCT for each time sample from the ΔCT of the 0 hour sample for each experiment. The power (2, ΔΔCT) was used to determine the fold change of the gene of interest in each sample relative to the 0 hour sample for each experiment.
Protein sample preparation and quantification
Cells were lysed in ice cold RIPA buffer (1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 2 mM EDTA, 25 mM Tris-HCl, pH 7.5 containing 1× Complete protease inhibitors (Roche)). The genomic DNA was sheared by passing the lysate 3× through a 21-gauge needle and lysates cleared by high-speed centrifugation at 4°C. Protein concentration was determined by BCA assay (Pierce).
Western blotting
Membranes were probed with anti-Plk1 antibody (Abcam Ab70697, 1:1000), p21Waf1/Cip1 (Dako SX118, 1:1000), Cyclin B1 (Santa Cruz H-433, 1:1000) or H2AX (Bethyl A300-081, 1:1000) and either anti-mouse or anti-rabbit secondary antibody conjugated to HRP (Jackson Laboratories. 1:10,000). Detection was performed using Supersignal Dura (ThermoFisher Scientific) and images captured on a Fujifilm LAS3000 digital detection system. Protein levels were quantified using ImageGauge.
Chromatin immunoprecipitation
Cells were treated with 100 nM panobinostat for 6 h before crosslinking with 1% formaldehyde for 30 min. 120 mM Glycine (final concentration) was added for 5 min before washing with PBS. 300 μl lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS and 1× protease inhibitor cocktail) was added and the cells harvested by scraping. Samples were sonicated and centrifuged at 14,000 rpm, 4°C. Supernatants were diluted 10-fold in ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.0, 0.01% SDS, 1.1% Trition-X-100, 1.2 mM EDTA, 167 mM NaCl, 1× protease inhibitor cocktail) and to 1 ml of each sample, 10 μl 50% slurry Protein G magnetic sepharose beads (Invitrogen) and 2 μg of antibody (FLAG M2 (Sigma, F1804), E2F1 (Abcam, ab94888), E2F2 (Abcam, ab65222) or E2F3 (Abcam, ab74180)) was added. Following incubation at 4°C for 2 h with nutation, beads were washed in low salt buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 2 mM EDTA, 0.5% Triton-X-100), high salt buffer (20 mM Tris-HCl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP40, 1% Na-deoxycholate), and TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). 100 μl elution buffer (1% SDS, 100 mM NaHCO3) was then added to each sample and incubated for 30 min. NaCl concentration was made up to 200 mM and the samples incubated overnight at 65°C. 10 mM EDTA, 40 mM Tris-HCl, pH 8.0 (final concentration) and RNase A (0.25 mg/ml) were added to each sample followed by incubation at 37°C for 1 h. Proteinase K (50 μg/ml final) was added and the samples incubated at 50°C for 1 h. The DNA was extracted with phenol-chloroform and ethanol precipitated. Pellets were resuspended in 20 μl water.
For qRT-PCR analysis a 10-fold serial dilution of input samples was created and 1 μl of each sample added to 24 μl Brilliant II SyBr Green mastermix (Agilent Technologies) containing 10 pmol of each primer for amplification of the PLK1 promoter (forward: 5′-GGTTTGGTTTCCCAGGCTAG-3′, reverse: 5′-GCTGGGAACGTTACAAAAGC-3′)28 or the Cyclin B1 promoter (forward: 5′-CGATCGCCCTGGAAACGCATTC-3′, reverse 5′-CCAGCAGCCACCAACAGCCGTTC-3′)46. DNA was amplified by incubation at 95°C for 10 min followed by 40 cycles of 95°C for 30 s, 57°C for 30 s and 72°C for 1 min. Ct values for each sample were calculated at a constant threshold and expressed as fold change over negative control.
Author Contributions
MBP, study design, interpretation of the data, drafting and revision of the manuscript, and financial support; KMF, NK, CM, MKSW, CWW, MA and OL, acquisition, analysis and interpretation of the data; JLP, study design, study supervision, acquisition, analysis and interpretation of the data, preparation of the manuscript and financial support.
Supplementary Material
Supplementary figures 1-5
Acknowledgments
Work by JP is funded by a Royal Society University Research Fellowship award. Experiments carried out by MBP were supported by the Department of Pathology, Albert Einstein College of Medicine / Montefiore Medical Center. We thank Professor Ciarán Woodman, University of Birmingham, UK, for informative discussion and critical reading of the manuscript.
References
- Ferlay J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 127, 2893–2917 (2010). [DOI] [PubMed] [Google Scholar]
- Jemal A. et al. Global cancer statistics. CA: A Cancer Journal for Clinicians. 61, 69–90 (2011). [DOI] [PubMed]
- Takes R. et al. Markers for assessment of nodal metastasis in laryngeal carcinoma. Archives of Otolaryngology-Head & Neck Surgery. 412–419 (1997). [DOI] [PubMed] [Google Scholar]
- Boutillier A., Trinh E. & Loeffler J. Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J. Neurochem. 84, 814–828 (2003). [DOI] [PubMed] [Google Scholar]
- Patel J. H. et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24, 10826–10834 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Packman K., Jeffrey R. & Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 12, 482–491 (2005). [DOI] [PubMed] [Google Scholar]
- Shankar S. & Srivastava R. K. Histone deacetylase inhibitors: Mechanisms and clinical significance in cancer - HDAC inhibitor-induced apoptosis. Adv. Exp. Med. Biol. 615, 261–298 (2008). [DOI] [PubMed] [Google Scholar]
- Ellis L. et al. Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin. Can. Res. 14, 4500–4510 (2008). [DOI] [PubMed] [Google Scholar]
- Maiso P. et al. The synergy of panobinostat plus doxorubicin in acute myeloid leukemia suggests a role for HDAC inhibitors in the control of DNA repair. Leukemia. 23, 2265–2274 (2009). [DOI] [PubMed] [Google Scholar]
- Dickinson M. et al. Preliminary evidence of disease response to the pan deacetylase inhibitor panobinostat (LBH589) in refractory Hodgkin Lymphoma. Brit. J. of Haematol. 147, 97–101 (2009). [DOI] [PubMed] [Google Scholar]
- Ocio E. M. et al. In vitro and in vivo rationale for the triple combination of panobinostat (LBH589) and dexamethasone with either bortezomib or lenalidomide in multiple myeloma. Haematologica. 95, 794–803 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez E. et al. The histone deacetylase inhibitor LBH589 enhances the anti-myeloma effects of chemotherapy in vitro and in vivo. Leukemia Research. 35, 373–379 (2011). [DOI] [PubMed]
- Hainsworth J. D. et al. A phase II trial of panobinostat, a histone deacetylase inhibitor, in the treatment of patients with refractory metastatic renal cell carcinoma. Cancer Invest. 29, 451–455 (2011). [DOI] [PubMed] [Google Scholar]
- Catalano M. G. et al. Cytotoxic activity of the histone deacetylase inhibitor panobinostat (LBH589) in anaplastic thyroid cancer in vitro and in vivo. Int. J. Cancer. 130, 694–704 (2011). [DOI] [PubMed] [Google Scholar]
- Crisanti M. C. et al. The HDAC inhibitor panobinostat (LBH589) inhibits mesothelioma and lung cancer cells in vitro and in vivo with particular efficacy for small cell lung cancer. Mol. Cancer Ther. 8, 2221–2231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathkopf D. et al. A phase I study of oral panobinostat alone and in combination with docetaxel in patients with castration-resistant prostate cancer. Cancer Chemother Pharmacol. 66, 181–189 (2010). [DOI] [PubMed] [Google Scholar]
- LaBonte M. J. et al. The dual EGFR/HER2 inhibitor lapatinib synergistically enhances the antitumor activity of the histone deacetylase inhibitor panobinostat in colorectal cancer models. Cancer Res. 71, 3635–3648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. P. et al. Antitumor activities and on-target toxicities mediated by a TRAIL receptor agonist following cotreatment with panobinostat. Int. J. Cancer. 128, 2735–2747 (2010). [DOI] [PubMed] [Google Scholar]
- Jones S. F. et al. A phase I study of panobinostat in combination with gemcitabine in the treatment of solid tumors. Clin. Adv. Hematol. Oncol. 9, 225–230 (2011). [PubMed] [Google Scholar]
- Prystowsky M. B. et al. The histone deacetylase inhibitor LBH589 inhibits expression of mitotic genes causing G2/M arrest and cell death in head and neck squamous cell carcinoma cell lines. J. Pathol. 218, 467–477 (2009). [DOI] [PubMed] [Google Scholar]
- Pettazzoni P. et al. Induction of cell cycle arrest and DNA damage by the HDAC inhibitor panobinostat (LBH589) and the lipid peroxidation end product 4-hydroxynonenal in prostate cancer cells. Free Radic. Biol. Med. 50, 313–322 (2011). [DOI] [PubMed] [Google Scholar]
- Brazelle W. et al. Histone Deacetylase Inhibitors Downregulate Checkpoint Kinase 1 Expression to Induce Cell Death in Non-Small Cell Lung Cancer Cells. PLoS ONE. 5, e14335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. J. et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 29, 163–188 (2007). [DOI] [PubMed] [Google Scholar]
- Maiso P. The histone deacetylase inhibitor LBH589 is a potent antimyeloma agent that overcomes drug resistance. Cancer Res. 66, 5781–5789 (2006). [DOI] [PubMed] [Google Scholar]
- Lin Y.-C., Sun S. H. & Wang F.-F. Suppression of Polo like kinase 1 (PLK1) by p21Waf1 mediates the p53-dependent prevention of caspase-independent mitotic death. Cell. Signal. 23, 1816–1823 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu H., Chang B.-D., Uchiumi T. & Roninson I. Identification of promoter elements responsible for transcriptional inhibition of Polo-like Kinase 1 and Topoisomerase II alpha genes by p21WAF1/CIP1/SDI1. Cell cycle. 1, 59–66 (2002). [PubMed] [Google Scholar]
- Archer S. Y. et al. The histone deacetylase inhibitor butyrate downregulates cyclin B1 gene expression via a p21/WAF-1-dependent mechanism in human colon cancer cells. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G696–703 (2005). [DOI] [PubMed] [Google Scholar]
- Tategu M. et al. Transcriptional regulation of human polo-like kinases and early mitotic inhibitor. J. gen genom. 35, 215–224 (2008). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Regulation of a novel androgen receptor target gene, cyclin B1, through androgen-dependent E2F family member switching. Mol Cell Biol. 32, 2454–2466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuto A. et al. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph- acute lymphoblastic leukemia cells. Blood. 111, 5093–5100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles F. et al. A phase I study of intravenous LBH589, a novel cinnamic hydroxamic acid analogue histone deactylase inhibitor, in patients with refractory hematologic malignancies. Cancer Therapy: Clinical. 12, 4628–4635. [DOI] [PubMed] [Google Scholar]
- Stevens F. E., Beamish H., Warrener R. & Gabrielli B. Histone deacetylase inhibitors induce mitotic slippage. Oncogene. 27, 1345–1354 (2008). [DOI] [PubMed] [Google Scholar]
- Ocker M. & Schneider-Stock R. Histone deacetylase inhibitors: Signalling towards p21cip1/waf1. Int. J. Biochem. Cell Biol. 39, 1367–1374 (2007). [DOI] [PubMed] [Google Scholar]
- Somers K. D. et al. Frequent p53 mutation in head and neck cancer. Cancer Res. 52, 5997–6000 (1992). [PubMed] [Google Scholar]
- Campomenosi P. et al. p53 mutants can often transactivate promoters containing a p21 but not Bax or PIG3 responsive elements. Oncogene. 20, 3573–3579 (2001). [DOI] [PubMed] [Google Scholar]
- Bellucci L., Dalvai M., Kocanova S., Moutahir F. & Bystricky K. Activation of p21 by HDAC inhibitors requires acetylation of H2A.Z. PLoS One 8, e54102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T. & Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 9, 400–414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Lénárt P. & Peters J.-M. Polo on the Rise—from Mitotic Entry to Cytokinesis with Plk1. Devel. Cell. 14, 646–659 (2008). [DOI] [PubMed] [Google Scholar]
- Soni D. V., Sramkoski R. M., Lam M., Stefan T. & Jacobberger J. W. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle. 7, 1285–1300 (2008). [DOI] [PubMed] [Google Scholar]
- Delavaine L. & La Thangue N. Control of E2F activity by p21(Waf1/Cip1). Oncogene. 18, 5381–5392 (1999). [DOI] [PubMed] [Google Scholar]
- Chung Y. L., Lee M. Y. & Pui N. N. M. Epigenetic therapy using the histone deacetylase inhibitor for increasing therapeutic gain in oral cancer: prevention of radiation-induced oral mucositis and inhibition of chemical-induced oral carcinogenesis. Carcinogenesis. 30, 1387–1397 (2009). [DOI] [PubMed] [Google Scholar]
- Miller A. C., Cohen S., Stewart M., Rivas R. & Lison P. Radioprotection by the histone deacetylase inhibitor phenylbutyrate. Radiat Environ Biophys. 50, 585–596 (2011). [DOI] [PubMed] [Google Scholar]
- Brown S. L., Kolozsvary A., Liu J., Ryu S. & Kim J. H. Histone deacetylase inhibitors protect against and mitigate the lethality of total-body irradiation in mice. Radiat. Res. 169, 474–478 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. C. et al. Genotyping of UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 32, 417–426 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R. & Stuurman N. Current Protocols in Molecular Biology. Ausubel F. M., et al. editors. Hoboken, NJ, USA: John Wiley & Sons, Inc.; (2001).
- Zhu W., Giangrande P. & Nevins J. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23, 4615–4626 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures 1-5