Abstract
We used a mouse model of the schizophrenia-predisposing 22q11.2 microdeletion to evaluate how this genetic lesion affects cortical neural circuits at the synaptic, cellular, and molecular levels. Guided by cognitive deficits, we demonstrated that mutant mice display robust deficits in high-frequency synaptic transmission and short-term plasticity (synaptic depression and potentiation), as well as alterations in long-term plasticity and dendritic spine stability. Apart from previously reported reduction in dendritic complexity of layer 5 pyramidal neurons, altered synaptic plasticity occurs in the context of relatively circumscribed and often subtle cytoarchitectural changes in neuronal density and inhibitory neuron numbers. We confirmed the pronounced DiGeorge critical region 8 (Dgcr8)-dependent deficits in primary micro-RNA processing and identified additional changes in gene expression and RNA splicing that may underlie the effects of this mutation. Reduction in Dgcr8 levels appears to be a major driver of altered short-term synaptic plasticity in prefrontal cortex and working memory but not of long-term plasticity and cytoarchitecture. Our findings inform the cortical synaptic and neuronal mechanisms of working memory impairment in the context of psychiatric disorders. They also provide insight into the link between micro-RNA dysregulation and genetic liability to schizophrenia and cognitive dysfunction.
Introduction
Carriers of deletions in chromosome 22q11.2, which predominantly occur de novo, exhibit a spectrum of cognitive deficits in children (Kates et al., 2007) and develop into schizophrenia (SCZ) in adulthood at a rate of 25–30% (Karayiorgou et al., 2010). Recurrent 22q11.2 deletions account for as many as 1–2% of cases of sporadic SCZ (Karayiorgou et al., 1995; Xu et al., 2008) and exemplify the important contribution of rare mutations to disease susceptibility (Xu et al., 2008, 2012a; Rodriguez-Murillo et al., 2012). Importantly, there are no major clinical differences in the core SCZ phenotype between individuals with SCZ who are 22q11.2 microdeletion carriers and those who are not (Bassett et al., 1998, 2003; Karayiorgou et al., 2010).
Individuals with SCZ show deficits in specific cognitive domains, and severity of cognitive dysfunction is the most critical determinant of functional outcome in SCZ, even more so than the severity of psychotic symptoms (Barch and Ceaser, 2012). One common denominator to such deficits may be impaired function and connectivity of the prefrontal cortex (PFC), a brain area that affects multiple cognitive functions (Arguello and Gogos, 2012; Barch and Ceaser, 2012). Diminished PFC function may also increase susceptibility to psychopathology by failing to buffer disease effects on perception and affect. Our previous behavioral analysis implicated PFC dysfunction and disconnectivity in a mouse model of the 22q11.2 deletion [Df(16)A+/− mice; Stark et al., 2008; Sigurdsson et al., 2010], and analysis of the PFC of mice deficient for the DiGeorge critical region 8 (Dgcr8) gene, a micro-RNA (miRNA) processor gene located within the 22q11.2 locus, revealed robust deficits in high-frequency short-term synaptic plasticity (Fénelon et al., 2011). However, a comprehensive analysis of the effect of the 22q11.2 deletion or any other genuine SCZ-predisposing mutation on the structure and function of PFC circuitry remains forthcoming (Arguello and Gogos, 2006, 2012; Karayiorgou et al., 2012).
Here we show that layer 5 (L5) pyramidal neurons from mutant mice display robust alterations in short-term synaptic depression (STD) and potentiation (STP) after physiologically relevant high-frequency stimulation, mimicking a pattern observed previously in the PFC of mutant Dgcr8 mice. Unlike Dgcr8-deficient mice, cortical neurons from Df(16)A+/− mice show modest alterations in long-term potentiation (LTP), and they also demonstrate a marked dendritic spine instability. Changes in synaptic and structural plasticity are accompanied by specific and modest alterations in local cytoarchitecture, which for the most part could not be accounted for by Dgcr8 deficiency. We confirmed that there are pronounced deficits in primary miRNA (pri-miRNA) processing and identified additional changes in gene expression and splicing that may mediate the effects of this deletion on neuronal structure and function. Our results strongly suggest that alterations in synaptic plasticity (including high-frequency short-term plasticity) and structural connectivity within the PFC may be primary contributors to the 22q11.2-related cognitive and psychiatric impairments. Notably, although alterations in short-term synaptic plasticity can be accounted for by reduction in the levels of the Dgcr8 gene, alterations in long-term plasticity as well as structural alterations do not depend primarily on Dgcr8.
Materials and Methods
Animals.
Experiments were performed on Df(16)A+/− male mice and their wild-type (WT) littermates. The generation of the Df(16)A+/− mice has been described previously (Stark et al., 2008). All mice were backcrossed to a C57BL/6J background for at least ten generations.
Novel object recognition test.
Naive adult male littermate mice [n = 14 WT, n = 14 Df(16)A+/−] were subjected to a one-trial novel object recognition (NOR) test (as described by Kvajo et al., 2008). In summary, mice were given a 10 min exposure to an empty novel testing box (standard rat cage) on the first day. On the second day, mice were exposed, for 5 min, to two identical objects located on the northeast and northwest corners of the cage. One hour later, each mouse was exposed to both the original object and a new object (N1), using the same two corners of the cage where the objects had been originally placed. An additional test was then performed. Four hours after their original exposure to the objects, mice were again exposed to two objects for 5 min. In this instance, it was again the original object, as well as another novel object (N2). Object locations were counterbalanced to minimize any preference for one side of the cage. The objects used in the experiment were all plastic: a Pokemon toy, a towel ring, and a brick-shaped toy. Objects were similar in size and complexity and had been tested previously pairwise to show that there was not an innate preference for one object over the other. Test sessions were videotaped and later manually scored by an experimenter blind to genotype. An animal was determined to be exploring the object if its nose was within 1 cm of the object. The percentage time exploring either the original object or the novel object was then calculated and compared between genotypes [(time exploring novel or original object/total exploration time) × 100)].
Latent inhibition assay.
For the latent inhibition (LI) assay, we implemented the protocol described by Yee et al. (2006) that is based on a fear-conditioning paradigm. Briefly, mice were tested in sound-attenuated fear conditioning chambers. The test consisted of four phases: (1) preexposure; (2) conditioning; (3) contextual test; and (4) tone test. Within each genotype group, adult male mice were randomly chosen for the preexposed (PE) group [n = 10 WT, n = 10 Df(16)A+/−] or the non-preexposed (NPE) group [n = 9 WT, n = 10 Df(16)A+/−]. In the PE group, mice received 40 presentations of a 30 s, 86 dB tone [conditioned stimulus (CS)] at a variable interstimulus interval (ISI) of 40 ± 30 s. NPE mice were confined to the testing chamber for an equivalent period of time. During this phase and in the following conditioning phase, the chambers were scented with rum scent, which was dabbed onto paper towels placed beneath the chamber floors. Conditioning began immediately after preexposure, without removing the animals from the chambers. Conditioning consisted of three trials of CS–unconditioned stimulus (US) pairings. Each trial began with the 30 s tone stimulus, followed immediately by a 1 s footshock set at 1.0 mA. Each trial was preceded by and followed by a 180 s interval. Twenty-four hours later, the contextual test took place. Mice were returned to the same chambers, and, in the absence of any discrete stimulus, they were observed for 480 s. The test of conditioned response to the CS was conducted after another 24 h. Mice then had a 180 s acclimation period, followed by 480 s of the CS tone. The tone test was conducted in a different context. White and pink plastic inserts were used to cover the walls and the floors, and a different scent than the one used previously (orange) was placed beneath the floors. Freezing was measured on all days of the experiment. Digital cameras mounted directly above the chambers were used to capture images of the animals. Successive frames were compared to evaluate the freezing behavior. The percentage of time spent freezing was then compared between genotypes.
Immunohistochemical analysis of laminar organization.
We used brains from 8-week-old males, perfused with PBS and 4% paraformaldehyde (PFA), that were postfixed in PFA overnight. Vibratome sections (60 μm thickness) were washed three times in PBS and blocked in PBS with 0.4% Triton X-100 and goat serum at room temperature for 3 h. Primary antibodies were incubated overnight at 4°C. After washing, samples were incubated with appropriate secondary antibodies for 3 h at room temperature. Primary antibodies used included the following: NeuN (mouse, 1:200; BD Bioscience), parvalbumin (PV) (rabbit, 1:500; Swant), and calbindin (CB) (rabbit, 1:300; Swant). All secondary antibodies (goat; Invitrogen) were used at a concentration of 1:500. Confocal images were obtained blind to genotype with the LSM 510 using a Carl Zeiss 20× objective lens.
Images acquired for the NeuN puncta analysis were taken with sequential acquisition setting at 2048 × 2048 pixel resolution as single image from the prelimbic area of medial PFC (mPFC). A total of 16 images from six male mice were analyzed for each genotype. The cortical layers were identified by NeuN immunostaining and Nissl staining (Invitrogen), and the cortical thickness was divided into seven bins (bin 1, marginal zone and L1; bin 2, L2; bin 3, L3; bins 4 and 5, L5; bins 6 and 7, L6). Some bins in same layer were evenly divided. The region of interest was defined by a 500 μm line parallel to the cortical layer. The particle measurement feature was then used, with a same setting of minimal puncta size and threshold, to count the number of discrete puncta in the image.
Images acquired for the PV and CB puncta analysis were taken with a z-series projection of 13 images, with 3.0 μm depth intervals, using the same settings for pinhole size, brightness, and contrast. Data were analyzed by counting the number of puncta in a z-series projection images. A total of 16 images from six male mice were analyzed for each genotype. The cortical layers were identified by NeuN immunostaining and Nissl staining (Invitrogen), and the cortical thickness was divided into seven bins (bin 1, marginal zone and L1; bin 2, L2; bin 3, L3; bins 4 and 5, L5; bins 6 and 7, L6). Some bins in same layer were evenly divided. The region of interest was defined by a 500 μm line parallel to the cortical layer. The particle measurement feature was then used, with a same setting of minimal puncta size and threshold, to count the number of discrete puncta in the image.
Analysis of dendritic architecture and spine morphology of pyramidal cells in the mPFC.
Df(16)A+/− mice were crossed to the Thy1–GFP/M mouse line. Adult male littermates, Df(16)A+/−;Thy1–GFP/M and WT;Thy1–GFP/M, at 2–3 months of age, were anesthetized and transcardially perfused with 1× PBS, followed by 4% PFA in PBS. Brains were postfixed in 4% PFA and then sectioned coronally at 100 μm on a vibratome (Leica). Sections were mounted and images were obtained on a Carl Zeiss LSM 510 laser-scanning confocal microscope. L5 pyramidal neurons were almost exclusively labeled in the mPFC of the mice, throughout the cell body and the dendritic tree. Ten times, 20×, 40×, and 63× oil-immersion objectives were used, and stack images of GFP-labeled neurons were obtained. GFP-labeled neurons were measured for a number of morphological variables as described previously (Lai et al., 2006). Traced dendritic images were imported into NIH ImageJ (http://rsb.info.nih.gov/ij/). Quantification of spine density, length, and width was performed first with auto tracing, followed by manual correction using Neuron Studio Software (Wearne et al., 2005). The length of the entire spine (including head and neck) was measured as the distance of a straight line with the shortest distance from the farthest tip of the spine head to the dendritic shaft. Spine width was measured as the distance of a straight line drawn across the widest part of the spine head. Total spine density was assessed by Student's t test. Distributions of the length and width of mushroom spines were compared using the Kolmogorov–Smirnov test.
Electrophysiology in mPFC slice preparations.
Experiments were performed on 4- to 6-week-old mice. Isoflurane was used to anesthetize mice that were then decapitated. After a skull incision, the brain was removed and placed in ice-cold dissecting solution (in mm): 195 sucrose, 10 NaCl, 2.5 KCl, 1 NaH2PO4, 25 NaHCO3, 10 glucose, 4 MgSO4, and 0.5 CaCl2. The cerebellum and part of the hippocampus (HPC) were removed, and coronal brain sections were cut on a vibratome (Leica VT1200S). The freshly cut mPFC slices were immediately transferred to an interface chamber and allowed to recover for at least 2 h at 34–36°C. During all recordings, the slices were continuously perfused with artificial CSF (aCSF) (bubbled with 5% CO2/95% O2) that had the following composition (in mm): 124 NaCl, 2.5 KCl, 1 NaH2PO4, 25 NaHCO3, 10 glucose, 1 MgSO4, and 2 CaCl2. The aCSF was maintained at 34–36°C and fed by gravity at a rate of 2–3 ml/min. Field EPSPs (fEPSPs) were recorded via a glass microelectrode (3–5 MΩ) filled with aCSF and placed in L5 of the mPFC (600–700 μm from midline).The stimulation site was always aligned ∼200 μm away from the recording site along the axis perpendicular to the pial surface. Basic synaptic transmission was characterized at 0.033 Hz, with stimulation intensities of 3–24 V (pulse duration, 0.1 ms). The subsequent experiments were performed at the stimulus intensity that generated a fEPSP one-third of the maximum fEPSP obtained at 24 V. Short-term synaptic facilitation was induced using a paired-pulse protocol with ISIs of 50, 100, 200, 400, and 800 ms. To assess STD, fEPSPs were evoked by using a 40-pulse train at 5, 10, 20, 40, and 50 Hz (pulse duration, 0.1 ms). LTP was induced by one 800-ms (40 pulses), 50-Hz train after a stable 10 min baseline and monitored during 15 min. Then, 15 min after the first tetanus, four additional 50 Hz trains (separated by 10 s) were applied. The fEPSPs were then monitored for 40 min. Signals were acquired using the pClamp10 software (Molecular Devices), the Digidata 1440A (Molecular Devices), and an extracellular amplifier (Cygnus Technologies). Fiber volley was quantified by measuring the amplitude of the first peak negativity of the field responses, and the fEPSPs were quantified by measuring the initial slope of the second peak negativity of the responses. Statistical analyses were done using the SigmaPlot and Statview software. A t test or a two-way repeated-measures ANOVA followed by post hoc testing was used to compare differences between genotypes. Data are presented as means ± SEM. N indicates number of animals, and n indicates number of slices. All recordings and the majority of data analyses were done blind to the genotype.
Two-photon microscopy.
One-month-old (P30 ± 1) male mice were used in the experiments. Spine formation and elimination were examined by imaging the mouse cortex through a thinned-skull window as described previously (Yang et al., 2010). Briefly, 1-month-old male mice expressing YFP were anesthetized with ketamine and xylazine (20 and 3 mg/ml, respectively, i.p., in saline; 6 μl/g body weight). Thinned-skull windows were made in head-fixed mice with high-speed microdrills. Skull thickness was reduced to ∼20 μm. A two-photon microscope tuned to 920 nm (60× water-immersion lens; numerical aperture, 1.1) was used to acquire images. For reimaging of the same region, thinned regions were identified on the basis of the maps of the brain vasculature. Microsurgical blades were used to re-thin the region of interest until a clear image could be obtained. The area of the imaging region was 200 × 200 μm in the frontal association cortex. The centers of the imaging regions were as follows: +2.8 mm bregma, +1.0 mm midline.
RNA isolation and expression profiling.
We dissected a total of 20 frontal cortices from 10 WT mice and 10 Df16(A)+/− mice, all 8-week-old male littermates. Total RNA was isolated from the brain tissues using miRNeasy kit (Qiagen) according to the instructions of the manufacturer. RNA was suspended in RNase-free water. The concentration and purity of each sample was determined by spectrophotometer (ND1000; Nanodrop) and confirmed by Microchip Gel Electrophoresis (Agilent), using Agilent 2100 Bioanalyzer Chip according to the instructions of the manufactures. RNA integrity numbers of all samples were more than seven. Transcriptional profiling was performed using Affymetrix Gene expression microarrays using the Gene 1.1 ST Array plates processed on the GeneTitan instrument at Hudson Alpha Institute.
Microarray data analyses.
We analyzed initial microarray images obtained by our GeneChip (Affymetrix) analysis by using Microarray Suite version 5 (Affymetrix) to extract intensity values for each probe set. We then imported the .CEL files into R software. The raw data were first normalized by the robust multichip average (RMA) method implemented in Bioconductor R package. Normalized data were then log transformed before any analysis was conducted using Limma package to identify genes that showed genotype-dependent differential expression. Benjamini and Hochberg false discovery rate (FDR) was used to control false positives attributable to multiple testing.
Atp2a2 (also called Serca2) mRNA and protein levels analyses.
Total RNA was isolated from brain or culture cells using the miRNeasy mini kit (Qiagen) according to instructions of the manufacturers. qRT-PCR was performed as described in detail previously (Stark et al., 2008). Atp2a2 TaqMan expression assay (Mm_01201431_m1; catalog #4331182; Applied Biosystems) was used. GAPDH (catalog #4352339E; Applied Biosystems) was used as internal control. Synaptosomal extracts were prepared as described previously (Kvajo et al., 2011) by homogenizing dissected tissue in 100 μl of 5 mm HEPES/10% sucrose, pH 7.5. Homogenates were spun down at 1000 × g, and the supernatant was further centrifuged at 12,000 × g. The pellet was resuspended in 50 μl (PFC) or 100 μl (HPC) of the same buffer, and 20 μg of protein was analyzed by Western blotting. After the transfer, nitrocellulose membranes were probed with 1:200 anti-SERCA2 antibody (sc-8095; Santa Cruz Biotechnology) and then with 1:1000 peroxidase-conjugated secondary antibody (PI-9500; Vector Laboratories). As loading control, the membranes were probed with 1:1000 anti-GAPDH antibody (ab9484; Abcam) and then with 1:1000 peroxidase-conjugated secondary antibody (A9044; Sigma). The washed membrane was incubated with HRP substrate, and chemiluminescence images were obtained using an Alpha imaging system.
Analysis of gene coexpression networks using weighted-gene coexpression network analysis.
Data analysis was performed using APT software (Affymetrix), R software, and Bioconductor packages. Raw expression data were normalized by RMA and log2 transformed using APT software. Each probe set was processed at the exon level and gene level using the APT software. Robustness of each probe set was determined by detected above background (DABG) p value and a set of filters described by Lockstone (2011). Only probes with DABG p value <0.05 in at least half of the samples in either genotype group were included. At the gene level, only genes with log2 intensity values >6 in at least one sample were analyzed further (16,438 genes). Unsigned coexpression networks were built using the weighted-gene coexpression network analysis (WGCNA) package in R according to the tutorial (http://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/Rpackages/WGCNA/#tutorials) based on the assumption that genes whose expression profiles are highly correlated are components of a functional module. For such modules, one can summarize the module expression profile by one representative gene, the module “eigengene.” The module eigengene is defined as the first principal component of a given module and can be considered a representative of the gene expression profiles in a module. A more detailed definition can be found in the study by Langfelder and Horvath (2007). Networks were constructed using “one-step, automatic network construction and module detection” procedure. The power of 6 was chosen using scale-free topology criterion for all samples. Minimum module size was set to 40 genes, and minimum height for merging modules was set at 0.1.
Functional annotation analyses.
The differentially expressed gene lists were imported into DAVID gene functional annotation database. For individual differentially expressed coding genes, 97% (31 of 32) of upregulated genes and all 14 downregulated genes were mapped in the DAVID database. For the genes in each WGCNA module and genes predicted to contain alternative splicing usage, each gene list was imported into the DAVID database and analyzed separately using the functional annotation analysis of the program with default settings.
Analysis of alternative exon usage.
We used AltAnalyze software to identify expressed genes that exhibit differential exon usage between Df(16)A+/− and WT mice (Emig et al., 2010). The AltAnalyze software implements the splicing index (SI) and Microarray Detection of Alternative Splicing (MiDAS) methods and calculates the likelihood and extent of alternative splicing for all Ensembl genes with one or more constitutive probe sets. Two probability estimates for alternative exon regulation are calculated with a one-way ANOVA model. AltAnalyze also produced a MiDAS (Gardina et al., 2006) p value, by interfacing with APT (version 1.6.0) and SI and performing a t test of the normalized exon expression values (exon probe set expression divided by constitutive expression) for the control and experimental sample groups. The primary filters for identifying alternative exons were a conservative absolute log2 SI fold change >1 (equivalent to a twofold difference in expression relative to constitutive expression levels), an SI t test p value <0.05, or a MiDAS p value <0.05.
Results
Cognitive profile of Df(16)A+/− mice
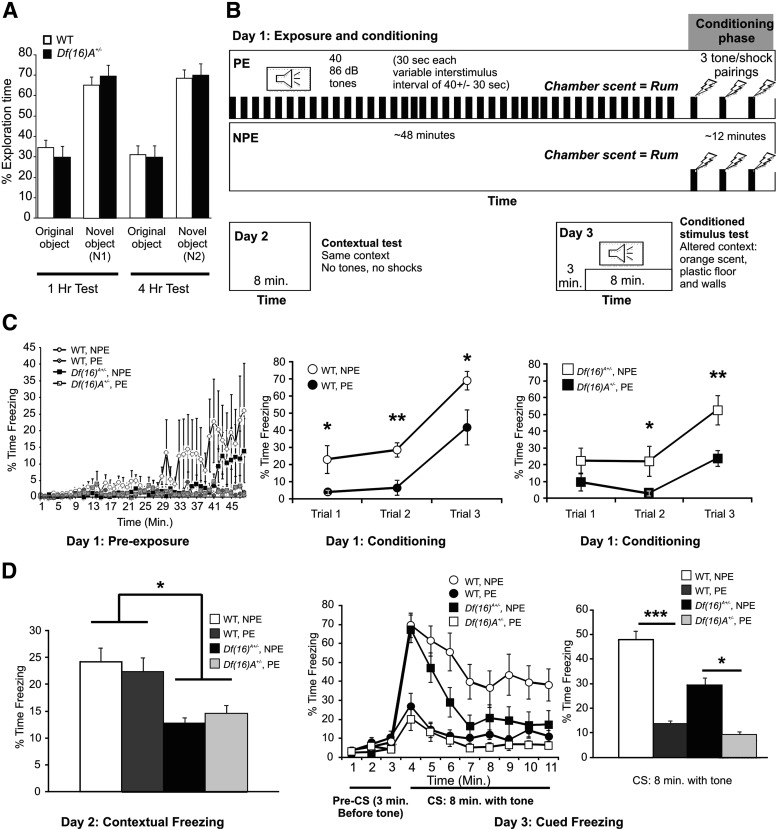
The profile of deficits in mechanistically well understood cognitive tasks that isolate specific cognitive processes can inform disease-related neurobiological mechanisms (Arguello and Gogos, 2006, 2010, 2012). Our previous work on the Df(16)A+/− mice has highlighted specific cognitive alterations, which collectively indicate a dissociation between primarily PFC-dependent tasks requiring intact working memory (WM) and HPC-dependent tasks requiring integral long-term reference memory as well as a robust impairment in associative fear memory (Stark et al., 2008; Drew et al., 2011). We confirmed the highly specific pattern by which the modeled disease risk allele affects cognition using two additional tests: the NOR task and the LI assay. The NOR task (Kvajo et al., 2008) is a non-PFC-dependent, nonspatial and nonassociative form of learning that requires intact HPC. In this task, mutant mice did not show deficits in long-term recognition memory as indicated by their similarity to their WT littermates in the time spent exploring novel objects relative to familiar objects to which they had been exposed to 1 or 24 h before testing (Fig. 1A). LI is a non-PFC-dependent attention phenomenon (Joel et al., 1997) reflecting the proactive interference of repeated but non-reinforced preexposure to a stimulus with subsequent performance on a learning task involving that stimulus. LI assays confirmed the robust deficits in associative fear learning in mutant mice but failed to reveal any genotype effect of non-reinforced preexposure to the CS (Fig. 1B). During the preexposure phase on day 1 [Fig. 1B, the time period before the tone/shock pairings were administered (∼48 min)], mice in the NPE group tended to move less (freeze more) over time than those in the PE group (Fig. 1C, left; repeated-measures ANOVA, main effect of condition, p = 0.05210). There was no main effect of genotype on freezing (p = 0.2266) nor any significant interaction of genotype × condition (NPE or PE) (p = 0.2112) during this phase of the assay (Fig. 1C, left). During the conditioning phase (pairing of sound and shocks) on day 1, there was a significant effect of preexposure on freezing. Mice in the NPE group froze significantly more (p < 0.0001) than mice that had been exposed previously to the sound (PE mice), demonstrating an LI effect. LI was demonstrated by the fact that the PE group exhibited less learning (reduced freezing) compared with the NPE group. There was no main effect of genotype (p = 0.1684) nor interaction of genotype × condition (p = 0.7931) during this conditioning phase (Fig. 1C, left). However, there was a significant interaction of genotype × conditioning over time (p = 0.0459). This suggests that the genotypes were adapting to the tone/shock pairings differently. Therefore, we examined the genotypes separately. LI was seen across the three trials of CS–US pairing for both genotypes [repeated-measures ANOVA, p = 0.0150 for Df(16)A+/− mice, p = 0.0006 for WT mice]. However, when examined on a point-by-point basis (Fig. 1C, middle and right), we found that Df(16)A+/− mice were slower to learn the association across the three trials (p = 0.1835, 0.0503, and 0.0089, respectively) than were the WT mice (p = 0.0246, 0.0026, and 0.0359 respectively). On day 2, when mice were reexposed to the same chamber for 8 min (contextual test), there was a significant main effect of genotype (p = 0.0344) in which Df(16)A+/− froze less than WT mice (Fig. 1D, left). There was no effect of the previous experience (PE vs NPE) on freezing (p = 0.9976) nor was there a significant interaction of genotype × previous experience (p = 0.6812). On the final day of the test (day 3), there were no significant effects of genotype, previous exposure, or exposure × genotype interaction during the 3 min pre-CS period when mice were placed in the chamber (all p > 0.05) (Fig. 1D, middle). However, during the 8 min CS test, there was a very significant effect of condition (p < 0.0001) and a significant effect of genotype (p = 0.0295), with Df(16)A+/− mice freezing less than WT mice (Fig. 1D, middle and right). Both groups displayed a significant increase in freezing with the NPE condition compared with the PE condition [p = 0.0100 for Df(16)A+/− mice, p = 0.0002 for WT mice], with no significant interaction of genotype × condition (p = 0.1683). Overall, although we clearly demonstrated an LI effect (effect of previous exposure during the CS test; Fig. 1D, middle and right (PE vs NPE), as well as during the CS–US pairings on day1], our results show that Df(16)A+/− and WT mice displayed equal levels of LI. Notably, we observed a delay in learning in the Df(16)A+/− mice during the pairing of CS and US at day 1, as well as a deficit in contextual conditioning at day 2. Because the LI testing paradigm includes both contextual (Fig. 1D, left) and cued (Fig. 1D, middle and right) fear conditioning tests, these observations replicate previous findings that Df(16)A+/− mice have deficits in both the contextual and cued fear conditioning (Stark et al., 2008). Overall, normal performance in the new behavioral tests corroborates previous findings and suggests that the profile of learning deficits observed in Df(16)A+/− mice is highly specific and is driven in part by a prominent effect of the mutation on PFC function and connectivity.
Figure 1.
NOR and LI assays of Df(16)A+/− mice. A, In the NOR test, Df(16)A+/− mice and their WT littermates spent a similar amount of time exploring novel objects relative to familiar objects to which they had been exposed to 1 or 4 h before testing [n = 14 WT, 14 Df(16)A+/−; all p > 0.05]. B, LI test design scheme. C, Left, During the preexposure phase, NPE mice tended to move less over time. Middle and right, During the conditioning phase of the LI test, LI was demonstrated across three trials of CS–US pairing for both Df(16)A+/− and WT mice. The Df(16)A+/− mice exhibited a delay in learning on day 1. D, Left, On day 2, Df(16)A+/− mice demonstrated a deficit in contextual fear conditioning. Middle, Time course of the freezing response during the pre-CS and CS test. Both Df(16)A+/− and WT mice demonstrated LI (decreased freezing in PE mice vs NPE mice). Right, Bar graph of the 8 min CS (cued) part of the test demonstrating LI in both genotypes on day 3, as well as decreased overall freezing in the Df(16)A+/−mice. *p ≤ 0.05, **p < 0.01, ***p < 0.001. Data are shown as means ± SEM.
We showed previously that animals lacking a single-copy Dgcr8 display similar WM deficits as the Df(16)A+/− mice without, however, affecting associative fear memory (Stark et al., 2008), suggesting that the WM deficits observed in Df(16)A+/− mice arise, at least in part, as a result of deficiency of Dcgr8 and miRNA biogenesis. Notably, morphological and electrophysiological analysis of PFC pyramidal neurons from Dgcr8+/− mutant mice showed modest cytoarchitectural changes and identified changes in high-frequency STD and STP at the synapses between the superficial afferents and L5 pyramidal neurons (Fénelon et al., 2011) as two major consequences of Dgcr8 haploinsufficiency. Therefore, we examined local cytoarchitecture and synaptic dynamics in the PFC of Df(16)A+/− mice and compared these observations with the pattern of cortical dysfunction observed in Dgcr8+/− mice to identify the neural substrates of cognitive phenotypes.
Laminar organization and cytoarchitecture in the cortex of Df(16)A+/− mice
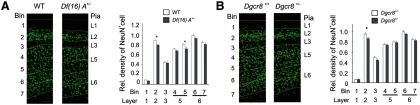
We looked for changes in the positional arrangement and neuronal density of the cortical layers in the prelimbic area of mPFC of 8-week-old Df(16)A+/− mice using the pan-neuronal marker NeuN. Although there were no overt alterations in laminar organization (Fig. 2A, left), we observed a modestly diminished NeuN cell density in L2 (Fig. 2A, right) [bin 2 (L2): 10.3% reduction, p < 0.01; n = 16 per genotype] as well as in L5 [bin 5 (deeper L5): 10.6% reduction, p < 0.05; n = 16 per genotype]. To determine whether the observed alterations in neuronal density can be accounted for by Dgcr8 deficiency, we looked for changes in the positional arrangement and neuronal density of cortical layers in the same area of 8-week-old Dgcr8+/− mice. Using the pan-neuronal marker NeuN, we did not observe any overt alterations in laminar organization (Fig. 2B, left). However, we detected a modest reduction in NeuN cell frequency only in L2 (Fig. 2B, right) [bin 2 (L2): 9.0% reduction, p < 0.05; n = 16 per genotype] but not in L5.
Figure 2.
Cytoarchitectural changes in the cortex of Df(16)A+/− mice. A, Left, Schematic representation of probe locations for quantifying density of neurons labeled with a pan-neuronal marker (NeuN, green) in the prelimbic area of mPFC of 8-week-old WT and Df(16)A+/− mice. Right, NeuN-labeled cells in mPFC of WT and Df(16)A+/− mice. The relative density of NeuN-labeled cells across seven bins from pia to white matter at the mPFC is shown. B, Left, Schematic representation of probe locations for quantifying density of neurons labeled with a pan-neuronal marker (NeuN, green) in the prelimbic area of mPFC of 8-week-old WT and Dgcr8+/− mice. Right, NeuN-labeled cells in mPFC of WT and Dgcr8+/− mice. The relative density of NeuN-labeled cells across seven bins from pia to white matter at the mPFC is shown. Data are shown as mean ± SEM. *p < 0.05. Scale bars, 100 μm.
Using Df(16)A+/− mice intercrossed with a Thy1–GFP/M+/− reporter strain (Feng et al., 2000; Mukai et al., 2008), we showed previously normal soma size but an overall decrease of both the total number and total length of basal dendritic branches of L5 neurons in the Df(16)A+/− mice, which could be accounted for primarily by a significant reduction in the number and length of primary basal dendrites (Xu et al., 2013). Similar analysis of the apical dendritic tree did not reveal significant differences in the total number of apical dendrites [WT, 13.20 ± 1.47 and Df(16)A+/−, 13.25 ± 2.19, t test, p = 0.9846] or in the total length of the apical dendrites [WT, 702.63 ± 79.85 μm and Df(16)A+/−, 657.38 ± 88.36 μm, t test, p = 0.7093] between genotypes. However, there was a significant decrease in the length of the primary apical dendrite (i.e., the distance from the soma to the apical bifurcation) in the Df(16)A+/−;Thy1–GFP/M+/− mice [WT, 314.06 ± 9.95 μm and Df(16)A+/−, 256.81 ± 11.95 μm, t test, p = 0.0004]. Soma size did not differ between genotypes [WT, 192.73 ± 8.06 μm and Df(16)A+/−, 181.87 ± 9.78 μm, t test, p = 0.3989]. Notably, the alterations in dendritic complexity contrast results from a similar analysis of the Dgcr8+/− mice, which did not reveal any changes in the basal or apical dendritic complexity in the mPFC (Fénelon et al., 2011).
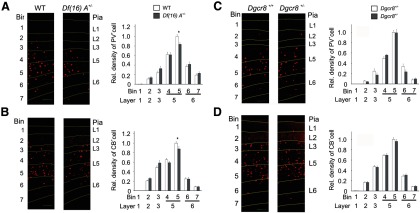
We also evaluated inhibitory neuronal density in the prelimbic area of mPFC using the inhibitory neuronal markers PV (Fig. 3A,C) and CB (Fig. 3B,D). PV-positive (PV+) cell density in Df(16)A+/− mice was modestly reduced in L5 [bin 5 (deeper L5): PV, 16.4% reduction, p < 0.01; n = 16 per genotype] (Fig. 3A). The density of CB+ neurons was also modestly reduced in L5 [bin 5 (deeper L5): CB, 12.1% reduction, p < 0.05; n = 16 per genotype] (Fig. 3B). Evaluation of the inhibitory neuronal density in the same area of 8-week-old Dgcr8+/− mice did not reveal significant changes in the frequency of PV+ (Fig. 3C) and CB+ (Fig. 3D) cells (n = 16 per genotype). Thus, Dgcr8 deficiency contributes only partly in a layer- and cell-type-specific manner to the observed neuronal density alterations in the PFC of Df(16)A+/− mice.
Figure 3.
The effect of Df(16)A on the density of cortical inhibitory interneurons. A, Left, Schematic representation of probe locations for quantifying density of inhibitory neurons labeled with a PV antibody (red) in the prelimbic area of mPFC of 8-week-old WT and Df(16)A+/− mice. Right, PV-labeled cells in mPFC of WT and Df(16)A+/− mice. The relative density of PV-labeled cells across seven bins from pia to white matter at the mPFC is shown. B, Left, Schematic representation of probe locations for quantifying density of inhibitory neurons labeled with a PV antibody (red) in the prelimbic area of mPFC of 8-week-old WT and Dgcr8+/− mice. Right, PV-labeled cells in mPFC of WT and Dgcr8+/− mice. The relative density of PV-labeled cells across seven bins from pia to white matter at the mPFC is shown. C, Left, Schematic representation of probe locations for quantifying density of inhibitory neurons labeled with a CB antibody (red) in the mPFC of 8-week-old WT and Df(16)A+/− mice. Right, CB-labeled cells in mPFC of WT and Df(16)A+/− mice. The relative density of CB-labeled cells across seven bins from pia to white matter at the mPFC is shown. D, Left, Schematic representation of probe locations for quantifying density of inhibitory neurons labeled with a CB antibody (red) in the mPFC of 8-week-old WT and Dgcr8+/− mice. Right, CB-labeled cells in mPFC of WT and Dgcr8+/− mice. The relative density of CB-labeled cells across seven bins from pia to white matter at the mPFC is shown. Data are shown as mean ± SEM. *p < 0.05. Scale bars, 100 μm.
Basic synaptic transmission in the mPFC is normal in Df(16)A+/− mice
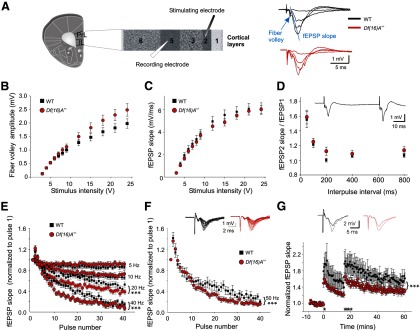
In both WT and Df(16)A+/− mice, field recordings were used to characterize the basic synaptic properties of cortical neurons in L5 while stimulating L2. Neurons and input fibers coming from L2 represent an important source of monosynaptic and glutamatergic inputs to the mPFC (Fig. 4A), and they form synapses with the apical and basal dendrites of pyramidal neurons in L5, the major output layer of the mPFC network (Szentágothai, 1978; Bannister, 2005). These synapses play a key role in WM and goal-directed behaviors (Fuster, 1991; Goldman-Rakic, 1995).
Figure 4.
The effect of Df(16)A on synaptic transmission and synaptic plasticity in cortical L5. A, Left, Schematic representation of a coronal mPFC slice including the prelimbic (PrL) and infralimbic (IL) areas. The stimulating electrode was placed on L2, and the recording electrode was placed in L5. Right, Sample traces obtained in response to increasing stimulation intensities and showing the fiber volley (arrow) as well as the fEPSP initial slope (blue line). B, Plot showing normal afferent volley amplitude in Df(16)A+/− mice (2-way repeated-measures ANOVA, p > 0.05). C, Plot showing normal stimulus–response curve across experiments in Df(16)A+/− mice (2-way repeated-measures ANOVA, p > 0.05). In both C and D, WT mice (black squares; N = 17, n = 40) and Df(16)A+/− mice (red circles; N = 16, n = 38). D, Plot showing that paired-pulse ratio is normal in Df(16)A+/− mice at ISIs of 50, 100, 200, 400, and 800 ms (2-way repeated-measures ANOVA, p > 0.05). Individual points represent mean ratio obtained at each ISI for both genotypes. WT mice (black squares; N = 7, n = 16) and Df(16)A+/− mice (red circles; N = 7, n = 16). A paired-pulse sample trace is shown at the top with an ISI of 50 ms. E, Plot showing the frequency dependence of STD of the fEPSPs between WT (black squares; N = 7, n = 16) and Df(16)A+/− (red circles; N = 6, n = 13) mice at 5 Hz (1st pair of black and red symbols), 10 Hz (2nd pair of black and red symbols), 20 Hz (3rd pair of black and red symbols), and 40 Hz (4th pair of black and red symbols). At 5 and 10 Hz, STD is similar between genotypes across experiments (2-way repeated-measures ANOVA, p > 0.05). At 20 and 40 Hz, both genotypes show a similar facilitation between the first two pulses, but the subsequent STD is significantly greater in Df(16)A+/− mice (N = 9, n = 18) compared with their WT controls (N = 9, n = 18; 2-way repeated-measures ANOVA, p < 0.0001 for 20 Hz × genotype interaction; p < 0.0004 for 40 Hz × genotype interaction). F, Plot showing that, at 50 Hz, the initial short-term facilitation is similar between genotypes but the following STD is significantly greater in Df(16)A+/− mice (red circles; N = 9, n = 20) compared with their WT littermates (black squares; N = 10, n = 20; 2-way repeated-measures ANOVA; p = 0.0003 for the 50 Hz × genotype interaction). Superimposed sample traces of individual fEPSPs evoked by 40 stimuli at 50 Hz in WT (black traces) and Df(16)A+/− (red traces) mice are shown. G, The plot shows synaptic potentiation in WT mice (black squares; N = 7, n = 12) and Df(16)A+/− mice (red circles; N = 7, n = 16). There is a significant difference in the degree of STP and LTP of fEPSPs over time (2-way repeated-measures ANOVA, p = 0.0215 for genotype and p < 0.0001 for time × genotype interaction). At the end of the first 50 Hz train (1st asterisk), the level of STP is significantly lower in the Df(16)A+/− mice [at 14.5 min: WT, 1.57 ± 0.14 vs Df(16)A+/−, 1.23 ± 0.06; post hoc test, p < 0.05]. Similarly, after the four consecutive 50 Hz trains (4 asterisks), post hoc testing revealed that the difference in potentiation lasts for the entire duration of the remaining testing period, affecting both STP and LTP [at 60 min: WT, 1.61 ± 0.12 vs Df(16)A+/−: 1.30 ± 0.05; post hoc test, p < 0.05]. fEPSPs traces obtained before [WT and Df(16)A+/− mice are black and red traces, respectively] and immediately after the first 50 Hz train [WT and Df(16)A+/− are gray and pink traces, respectively] are shown at the top. Values are normalized to slope of the first fEPSP in the train (D–F) or to the baseline (G).
Stimulation of the superficial cortical layers of the mPFC elicited a small initial nonsynaptic fiber volley, which had an onset latency typically of <2 ms from the stimulation artifact and was followed by a synaptic fEPSP (Fig. 4A). A stable baseline was obtained when the fEPSPs, evoked at low-stimulation frequency (0.033 Hz), were reproducible with a high signal-to-noise ratio. Basal synaptic transmission was then assessed by gradually increasing the stimulation intensity (Fig. 4A). At the stimulation intensities tested, the nonsynaptic fiber volley was not different between genotypes (Fig. 4B). The fEPSPs were quantified by measuring the initial slope of the linear rising phase (Fig. 4A, blue line). Overall, no genotypic differences were observed (Fig. 4C) [WT, N = 17, n = 40; Df(16)A+/−, N = 16; n = 38; two-way repeated-measures ANOVA, p > 0.05], indicating that basal synaptic transmission is not affected in a wholesale manner by Df(16)A when the same number of afferent fibers is activated.
High-frequency STD is altered in the mPFC of Df(16)A+/− mice
We next determined whether short-term synaptic plasticity was impaired in the mPFC of the Df(16)A+/− mice. To assess the fractional neurotransmitter release, we used a paired-pulse facilitation (PPF) protocol at various ISIs (Fig. 4D; WT: N = 7; n = 16; Df(16)A+/−: N = 7; n = 16). At ISIs shorter than 200 ms, a significant facilitation of the fEPSPs was observed in both genotypes as described previously (Fénelon et al., 2011), suggesting that, in both mutant and WT mice, the synapses between the superficial L2 afferents and the L5 mPFC neurons have low initial probability of neurotransmitter release. However, when compared at all tested ISIs (50–800 ms), PPF was unchanged in the PFC of Df(16)A+/− mice (two-way repeated-measures ANOVA, p > 0.05), indicating that Df(16)A did not affect the initial probability of neurotransmitter release.
During PFC-dependent WM tasks, PFC neurons can receive trains of inputs from neighboring cells in the 20–60 Hz frequency range (Miller et al., 1996). We determined whether Df(16)A affects synaptic transmission within this physiological range using trains of 40 pulses applied at 5, 10, 20, and 40 Hz (Fig. 4E) and 50 Hz (Fig. 4F). As can be seen in Figure 4E, in both genotypes, the 5 Hz stimulation train produced an ∼10% STD of the fEPSPs [last 10 pulses: WT, 0.89 ± 0.04 and Df(16)A+/−, 0.93 ± 0.04] that did not differ between genotypes [WT, N = 7, n = 16; Df(16)A+/−, N = 6, n = 13; two-way repeated-measures ANOVA, p > 0.05]. Similarly, at 10 Hz, the fEPSPs showed an ∼20–25% STD [last 10 pulses: WT, 0.73 ± 0.04 and Df(16)A+/−, 0.81 ± 0.04] that did not differ between WT and Df(16)A+/− mice [WT, N = 7, n = 16; Df(16)A+/−, N = 6, n = 13; two-way repeated-measures ANOVA, p > 0.05]. At 20 Hz, both the second and third fEPSPs were facilitated, followed by depressed fEPSPs, and, in this case, the Df(16)A+/− mice showed significantly more STD [Fig. 4E; WT, N = 9, n = 18; Df(16)A+/−, N = 9, n = 18; two-way repeated-measures ANOVA, p < 0.0001 for pulse number × genotype interaction]. Similarly, at 40 Hz, although both WT and Df(16)A+/− mice showed similar facilitation at the beginning of the train, the Df(16)A+/− mice showed significantly greater STD [Fig. 4E; WT, N = 9, n = 18; Df(16)A+/−, N = 9, n = 18; two-way repeated-measures ANOVA, p = 0.05 for genotype and p < 0.0004 for 40 Hz × genotype interaction]. Finally, at 50 Hz [Fig. 4F; WT, N = 10, n = 20; Df(16)A+/−, N = 9, n = 20], WT and Df(16)A+/− mice showed similar facilitation between the first two pulses of the 50 Hz train [Fig. 4F; WT, 1.41 ± 0.48; Df(16)A+/−, 1.35 ± 0.45; post hoc test, p > 0.05], which was followed by an STD of the fEPSPs that was significantly greater in the Df(16)A+/− mice (Fig. 4F; two-way repeated-measures ANOVA, p = 0.0003 for the pulse number × genotype interaction).
Synaptic potentiation is decreased in the cortex of Df(16)A+/− mice
Both brief and sustained forms of synaptic potentiation can be induced in PFC networks in acute slices (Hirsch and Crepel, 1990; Hempel et al., 2000; Gemperle et al., 2003). Although the mechanistic and temporal relationships between STP and LTP are still under debate (Gustafsson and Wigström, 1990; Hanse and Gustafsson, 1994; Schulz and Fitzgibbons, 1997; Volianskis and Jensen, 2003), STP has been defined as a decremental potentiation lasting 10–30 min (Schulz and Fitzgibbons, 1997; Volianskis and Jensen, 2003). When STP is pharmacologically occluded, LTP can still be elicited without any decremental component (Schulz and Fitzgibbons, 1997). Thus, here STP refers to the early decremental potentiation and LTP refers to the non-decremental potentiation. We used a 50 Hz stimulation train to induce potentiation and compare its level in L5 of the mPFC of WT (N = 7, n = 12) and Df(16)A+/− (N = 7, n = 16) mice. After a stable 10–15 min baseline was obtained, a single 50 Hz stimulation was applied to L2, which induced STP (Fig. 4G, single asterisk). This STP, monitored every 30 s for 15 min, was consistently lower in the Df(16)A+/− mice and became significantly different at 6 min [WT, 1.73 ± 0.17; Df(16)A+/−, 1.37 ± 0.07; t test, p = 0.04]. Notably, Dgcr8-deficient mice also show robust deficits in STP (Fénelon et al., 2011). At the end of the 15 min, four trains of 50 Hz separated by 10 s were applied (Fig. 4G, four asterisks) to induce LTP, and the fEPSP slopes were monitored for 40 min after the tetani. Potentiation induced by this second round of stimulation was modestly but significantly different between genotypes, starting at 16.5 min [WT, 1.95 ± 0.20; Df(16)A+/−, 1.58 ± 0.06; t test, p = 0.05] and remained different for the entire testing period (two-way repeated-measures ANOVA, p = 0.0215 for genotype and p < 0.0001 for time × genotype interaction).
In conclusion, our analysis of PFC pyramidal neurons from Df(16)A+/− mice and their WT littermates found that, similarly to the Dgcr8+/− mice, L5 pyramidal neurons from mutant mice showed normal basal synaptic transmission during activation of superficial layer afferents but displayed a greater level of STD (at frequencies >20 Hz) and less STP after physiologically relevant high-frequency stimulation. Unlike Dgcr8+/− mice, L5 pyramidal neurons from mutant mice showed modest but significant impairment in LTP.
Cortical spine stability in Df(16)A+/− mice
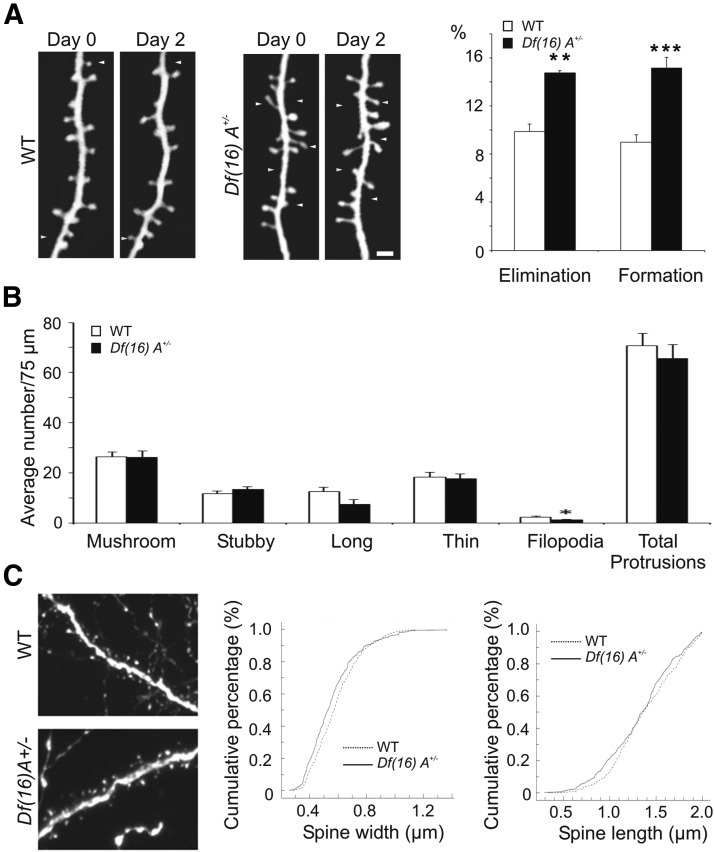
A very tight structure–function relationship exists between dendritic spines and synaptic forms of plasticity over long timescales (such as LTP). A shorter than normal spine lifespan at any age might affect the proper functioning of synapses during learning and memory formation (De Roo et al., 2008; Kasai et al., 2010). Indeed, it is well established that spine remodeling in recruited frontal cortical regions plays an important role in associative fear memory (Lai et al., 2012), a process that is affected in Df(16)A+/− but not Dgcr8+/− mice. Therefore, we used Df(16)A+/−;Thy1–YFP/H+/− compound heterozygous mice to examine the formation and elimination rate of dendritic mushroom spines on dendrites of L5 pyramidal neurons in the frontal cortex by using in vivo transcranial two-photon imaging microscopy. We imaged Df(16)A+/−;Thy1–YFP/H/+/− and WT;Thy1–YFP/H+/− mice over a 2 day period to examine the turnover rate of dendritic spines. We found that the rate of spine formation and spine elimination over the course of 2 d were both significantly higher in Df(16)A+/−;Thy1–YFP/H+/− mice compared with the age-matched WT Thy1-YFP/H+/− littermates. The formation and elimination rate were 15.1 ± 0.9 and 14.8 ± 0.2%, respectively, in Df(16)A+/−;Thy1–YFP/H+/− mice (n = 5) compared with 9.0 ± 0.6 and 9.9 ± 0.6% in WT littermates (n = 4; p < 0.02; Fig. 5A). Together, these results demonstrate that the Df(16)A leads to heightened dendritic spine turnover.
Figure 5.
Altered spine turnover in the cortex of Df(16)A+/− mice. A, In juvenile mice (P30 ± 1), using in vivo two-photon microscopy, Df(16)A+/−;Thy1–YFP/H+/− mice were shown to have significantly greater spine elimination, as well as significantly greater spine formation, relative to WT;Thy1–YFP/H+/− mice (t test, p = 0.003 and p = 0.0009, respectively). B, Except for a small decrease in filopodia in the Df(16)A+/−;Thy1–GFP/M+/− mice, no significant differences were found in the density of apical spine types between the genotypes. C, The width of apical mushroom spines was decreased (by 4.9%) in the Df(16)A+/−;Thy1–GFP/M+/− mice. No significant differences were found in apical mushroom spine length between genotypes. Data are shown as means ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001.
To test whether increase spine turnover is associated with alterations in spine density, we used high-magnification representative images of apical dendritic spines of EGFP+ cortical pyramidal neurons in the Df(16)A+/−;Thy1–GFP/M+/− and WT;Thy1–GFP/M+/− mice (Fig. 5C, left) to examine mushroom spines in the L5 apical dendrites. We found no differences in the total number of protrusions or in the density of the different spine types (Fig. 5B). However, we found a significant decrease in the width (4.9%; Kolmogorov–Smirnov test, p = 0.009; Fig. 5C, middle) but not the length (Kolmogorov–Smirnov test, p = 0.131; Fig. 5C, right) in Df(16)A+/−;Thy1–GFP/M+/− mice. Interestingly, previous analysis of spines in basal dendrites of PFC pyramidal neurons of Df(16)A+/−;Thy1–GFP/M+/− mice revealed a significant decrease in mushroom spine density (Xu et al., 2013). These findings may reflect differences in the relative rates of spine formation and elimination at the apical or basal dendritic fields, possibly related to local differences in the excitatory inputs because numerous excitatory cell types target the basal dendritic arborization almost exclusively (Bannister, 2005).
Molecular pathology in the frontal cortex of Df(16)A+/− mice
Characterizing the transcriptional networks and signaling cascades that mediate the effects of copy number variants (CNVs) on neuronal structure and function is an important step toward elucidating the affected biological processes (Kvajo et al., 2010; Karayiorgou et al., 2010). We showed previously that 22q11.2 deletions lead to miRNA dysregulation, which is shaped by the combined effect of two disrupted genes: Dgcr8, whose hemizygosity leads to modest dysregulation in the production of up to 20% of miRNAs in the brain of mutant mice (Stark et al., 2008), and miR-185, whose mature form is reduced to levels <50% attributable to a combined effect of hemizygosity and impaired maturation of the pri-miR-185 transcript as a result of the reduction in Dgcr8 levels (Xu et al., 2013). Reduction in miR-185 levels results in de-repression of a novel neuronal regulatory gene, 2310044H10Rik/Mirta22, that represents the most robust change in gene expression during postnatal development in both PFC and HPC of Df(16)A+/− mice. We showed that the 2310044H10Rik/Mirta22 gene encodes a protein that is located in the Golgi apparatus and in vesicles and tubular-like extensions in dendrites and mediates, at least in part, the effects of the 22q11.2 deletions on dendrite and spine formation (Rosso et al., 2005; Evans et al., 2011).
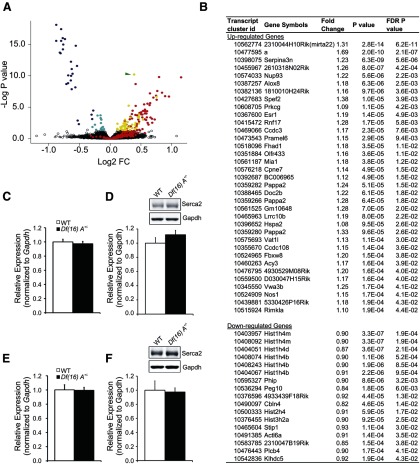
To examine the effect of Df(16)A on transcriptional networks in more detail, we used high-resolution expression profiling arrays (Affymetrix Gene 1.1 ST Genechips), which offer a wide representation of pri-miRNA transcripts and also afford a detailed analysis of mRNA splicing alterations (data has been deposited at Gene Expression Omnibus Repository, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vbsfhuiisowigjs&acc=GSE45935). We confirmed the impaired miRNA processing in the PFC of Df(16)A+/− mice (Stark et al., 2008) and provided a more accurate picture of its relative impact on the PFC transcriptional profile of mutant mice. As shown in Figure 6A together with the expected downregulation of the hemizygously-deleted 22q11.2 genes (blue dots), upregulation of unprocessed pri-miRNA transcripts (red dots) represents a major component of the altered PFC transcriptional profile (FDR, p < 0.05). There are 318 expressed miRNA genes annotated in Gene 1.1 ST microarray data. Among these miRNAs, 51 are significantly altered using strict statistical criteria (FDR, p < 0.05), and all of them are upregulated. There is very good correlation with previously reported data: elevated pri-miRNA forms are observed for 23 of the 25 mature miRNAs reported as significantly reduced in the PFC of Df16(A)+/− mice (Stark et al., 2008). Significantly upregulated (yellow dots) and downregulated (cyan dots) coding genes comprise the remaining two components. Upregulated genes are enriched in genes related to neuronal function (Fig. 6B) and likely include miRNA targets. Consistent with this expectation, at the top of this list is 2310044H10Rik/Mirta22. In contrast, we failed to observe any upregulation in the levels of Atp2a2, another recently proposed target of 22q11.2-associated miRNA dysregulation (Earls et al., 2012). Atp2a2 is not included in the list of top upregulated genes, and we could not detect elevation of either the transcript levels or protein levels in synaptosomal extracts (Fig. 6C–F; see Materials and Methods).
Figure 6.
Molecular abnormalities in the cortex of Df(16)A+/− mice. A, Volcano plot of the FDR-corrected log p values (y-axis) and the corresponding log2-fold change of each gene (x-axis) of the PFC gene expression profile of Df16(A)+/− mice. Blue spots represent genes within the Df16(A) deficiency. Red spots represent primary transcripts of miRNA genes. Cyan spots represent downregulated protein encoding genes. Yellow spots represent upregulated protein encoding genes. Green arrow indicates 2310044H10Rik/Mirta22 (18) (B). List of genes differentially expressed in the PFC region of Df(16)A+/− mice. C–F, Atp2a2 mRNA and protein levels in PFC and HPC of adult Df(16)A+/− mice. Atp2a2 mRNA expression levels in PFC (C) and HPC (E) of adult Df(16)A+/− mice (n = 10 for PFC, n = 7 for HPC) and their WT littermates (n = 10 for PFC, n = 5 for HPC), as assayed by qRT-PCR. p = 0.59 (PFC), p = 0.73 (HPC), Student's t test. D, F, Atp2a2 protein expression levels in synaptosomal preparations from PFC (C) and HPC (E) of adult Df(16)A+/− mice and their WT littermate mice. Top, Representative Western blot assays of Atp2a2 in PFC (C) and HPC (E) synaptosomal samples prepared from Df(16)A+/− animals and WT littermates. GAPDH is used as loading control. Bottom, Quantification of Atp2a2 protein levels in PFC (D) and HPC (F) of Df(16)A+/− and WT animals (n = 5 each genotype). p = 0.27 (PFC), p = 0.87 (HPC), Student's t test. Expression levels in mutant animals were normalized to their respective WT littermates. Results are expressed as mean ± SEM.
Other notable known upregulated genes that may modulate the observed alterations in structural and synaptic plasticity include Fbxw8 (F-box and WD-40 domain protein 8, a E3 ubiquitin ligase that, like Mirta22, localizes to the Golgi complex in mammalian brain neurons and modulates Golgi morphology and dendritic patterning; Litterman et al., 2011), Ttyh1 (tweety homolog 1, a chloride channel that is localized in the presynaptic active zone and maybe involved in the short- and long-term modulation of presynaptic plasticity; Morciano et al., 2009), Nos1 (neuronal nitric oxide synthase 1; Barañano et al., 2001), Vat1l (vesicle amine transport protein 1 homolog-like), Prkcc (protein kinase Cγ, a protein kinase with well established role in synaptic pruning and long-term synaptic plasticity; Shuvaev et al., 2011), and Rimkla [ribosomal modification protein rimK-like family member A, which along with Rimklb acts as N-acetylaspartylglutamate (NAAG) synthetase and modulates levels of NAAG, an agonist of group II metabotropic glutamate receptors; Lodder-Gadaczek et al., 2011]. Dgcr8 may contribute in part to the observed transcript upregulation by contributing to miRNA dysregulation and to a lesser extent by directly targeting and destabilizing some of these transcripts (Knuckles et al., 2012). In addition to Mirta22, at least three other top upregulated genes (Vat1l, Ttyh1, and Rimkla) contain experimentally validated miRNA binding sites according to the CLIP-seq database (Yang et al., 2011).
Downregulated genes are enriched in genes involved in chromosome organization and remodeling, including Actl6a, Hist1h4m, Hist1h4b, Hist2h4, and Hist3h2a (Fig. 6B). Their contribution remains to be determined, but it is noteworthy that we have recently described an overrepresentation of developmentally regulated genes involved in chromatin remodeling among targets of de novo mutations in SCZ (Gilman et al., 2012; Xu et al., 2012a).
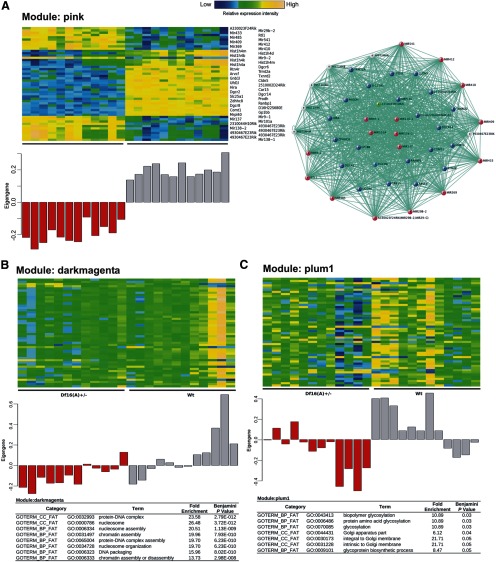
To integrate individual expression differences and investigate potentially disrupted biological processes as a whole instead of individual genes, we applied WGCNA (Zhang and Horvath, 2005) on the entire gene expression dataset. We constructed coexpression networks, identified discrete groups of coexpressed genes (modules), and asked whether there are modules related to specific biological functions that show transcriptional differences between the PFC of Df16(A)+/− and WT mice. Among a total of 54 modules identified, we detected 13 modules whose eigengenes were highly correlated with genotypes (p < 0.05; Table 1). The top module (“pink module”) consists of 468 known genes and shows a highly significant enrichment for the genes residing within the deleted region as well as miRNA genes (Fig. 7A). The high eigengene value of this module reflects highly correlated changes in gene expression attributable to downregulation of 22q11.2 orthologs and upregulation of pri-miRNA transcripts (Fig. 7A) and serves as a positive control validating our analytical procedure. A subset of downregulated histone-related genes are also part of this module (Fig. 7A). Interestingly, Gene Ontology (GO) enrichment analysis identified one more module (“darkmagenta”) enriched for functions related to protein–DNA interactions, nucleosome and chromatin assembly (p < 2.45E-09) (Fig. 7B). Genes with the highest degree of connectivity within this module include Hist1h2af, Hist1h2ag, and Hist2h3c1, all downregulated in mutant brains (Fig. 7B, darkmagenta module). Additional GO enrichment analysis identified five additional modules that contain genes enriched in specific GO categories, such as “ribonucleoprotein complex,” “RNA/ncRNA processing,” “nucleotide binding,” “establishment of protein localization,” and “glycosylation/Golgi apparatus part” (Table 1). This latter observation (Fig. 7C) extends our finding that Golgi-related genes are causally linked to the 22q11.2-related pathophysiology and many of them represent miR-185 targets (Xu et al., 2013).
Table 1.
List of modules identified by WGCNA analysis highly correlated with genotype
| Module | Module trait correlation | Module trait correlation p value | Significantly enriched GO terms |
|---|---|---|---|
| Pink | −0.968 | 1.15E-014 | |
| Darkolivegreen | −0.549 | 5.49E-003 | |
| Darkgrey | −0.530 | 7.77E-003 | Ribosomal protein; ribonucleoprotein complex |
| White | −0.529 | 7.91E-003 | |
| Plum1 | −0.527 | 8.12E-003 | Biopolymer glycosylation; Golgi apparatus part |
| Purple | −0.522 | 8.94E-003 | Cellular protein localization; mitochondrion |
| Violet | −0.489 | 1.54E-002 | RNA processing; noncoding RNA processing |
| Darkmagenta | −0.464 | 2.24E-002 | Protein–DNA complex; chromatin assembly |
| Salmon4 | −0.433 | 3.44E-002 | |
| Cyan | −0.427 | 3.73E-002 | Nucleotide binding; ribonucleoprotein |
| Darkorange2 | −0.419 | 4.14E-002 | |
| Bisque4 | −0.414 | 4.42E-002 | Establishment of protein localization; protein transport |
| Lightgreen | 0.405 | 4.93E-002 |
Figure 7.
Disrupted biological processes in the cortex of Df(16)A+/− mice. A, Left, Gene expression heat map of the top 50 genes included in the top module (pink) identified by WGCNA analysis (top) and the module eigengene values (y-axis) for each sample (x-axis). Right, Correlation expression network of the top module. Blue spots indicate genes within the Df16(A) deficiency. Red spots indicate primary transcripts of miRNA genes. Cyan spots represent downregulated protein encoding genes. Yellow spots depict upregulated protein encoding genes. The green spot indicates 2310044H10Rik/Mirta22 (Xu et al., 2013). B, C, Indicated are gene expression heat maps of the top 50 genes included in two modules that are highly correlated with genotype and show statistically significant GO term enrichment (top) and the module eigengene values (y-axis) for each sample (x-axis).
Finally, to identify additional molecular targets that may underlie the observed cellular and synaptic phenotypes, we used individual exon expression data, as provided by the Gene 1.1 ST Genechip, to search for disturbed alternative exon usage in Df16(A)+/− mice using the AltAnalyze software. One hundred seventy-eight unique genes were predicted to show differential exon usage between genotypes (SI t test, p < 0.05 or MiDAS, p < 0.05 and absolute log2 SI >1; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vbsfhuiisowigjs&acc=GSE45935). Top GO categories included “ATP binding,” “motor activity,” “extracellular matrix part,” “ionic channel,” and “GTPase regulator activity” (Table 2). The observed enrichment for altered splicing events among genes involved in cytoskeletal organization, extracellular matrix, as well as genes encoding ionic channels (including voltage-gated calcium, potassium and chloride channels) may be related to abnormal cortical functional and structural plasticity in Df16(A)+/− mice.
Table 2.
Top GO Terms enriched among genes with alternative exon usage
| Annotation cluster | Enrichment score | Category | Term | Gene count | Fold enrichment | Benjamini p value | |
|---|---|---|---|---|---|---|---|
| 1 | 10.00 | GOTERM_MF_FAT | GO:0005524 | ATP binding | 44 | 3.14 | 6.0E-10 |
| 2 | 6.90 | GOTERM_MF_FAT | GO:0003774 | Motor activity | 14 | 10.45 | 3.4E-08 |
| GOTERM_CC_FAT | GO:0043232 | Intracellular non-membrane-bounded organelle | 43 | 2.21 | 1.9E-05 | ||
| 3 | 3.47 | GOTERM_CC_FAT | GO:0044420 | Extracellular matrix part | 10 | 10.70 | 2.5E-05 |
| GOTERM_MF_FAT | GO:0005201 | Extracellular matrix structural constituent | 7 | 24.04 | 1.0E-05 | ||
| 4 | 3.24 | GOTERM_CC_FAT | GO:0016459 | Myosin complex | 11 | 17.75 | 9.4E-08 |
| GOTERM_BP_FAT | GO:0006936 | Muscle contraction | 10 | 16.90 | 4.9E-06 | ||
| 5 | 3.18 | GOTERM_MF_FAT | GO:0003774 | Motor activity | 14 | 10.45 | 3.4E-08 |
| 6 | 2.29 | GOTERM_MF_FAT | GO:0005262 | Calcium channel activity | 7 | 10.60 | 1.1E-03 |
| 7 | 1.96 | GOTERM_MF_FAT | GO:0005083 | Small GTPase regulator activity | 11 | 5.08 | 1.3E-03 |
| 8 | 1.78 | GOTERM_MF_FAT | GO:0004672 | Protein kinase activity | 14 | 2.47 | 4.2E-02 |
| 9 | 0.79 | GOTERM_MF_FAT | GO:0005509 | Calcium ion binding | 18 | 2.21 | 3.1E-02 |
Discussion
This study was designed to evaluate how the 22q11.2 microdeletion, a bona fide pathogenic mutation that predisposes to SCZ, affects the structure and function of cortical neural circuits at the synaptic, cellular, and molecular levels. We elucidated the effect of the mutation by applying an array of behavioral, morphological, neurophysiological, and gene expression assays to an etiologically valid mouse model of this mutation.
Behavioral assays
Using new behavioral tests, we corroborated and expanded on previous findings (Stark et al., 2008; Drew et al., 2011) that Df(16)A+/− mice show normal performance in learning tasks that do not require intact PFC. Df(16)A also results in robust deficits in fear learning that are not, however, differentially sensitive to the non-reinforced preexposure to the CS.
Assays of synaptic plasticity
Guided by the overall cognitive profile, we demonstrated that mutant mice display robust changes in synaptic plasticity that could affect PFC function. Comparison with the results from a similar analysis on the PFC of Dgcr8+/− mice, which model one aspect of miRNA dysregulation attributable to 22q11.2 deletions, suggests that overall the effects of Df(16)A on synaptic plasticity are more widespread but inclusive of the effects observed in Dgcr8+/− mice, consistent with a contribution of several deleted genes to the overall phenotype (Karayiorgou et al., 2010). In that context, and given the only partial overlap in the cognitive profile between the Df(16)A+/− mouse strain and heterozygous Dgcr8-deficient mice, it is instructive to point out that both the Dgcr8 and the Df(16)A mutations robustly affect the expression of short-term forms of synaptic plasticity in response to high-frequency stimulation, including large deficits in STD and STP. Computational models and in vivo recordings of PFC neural activity during a WM task have highlighted the importance of synaptic facilitation and depression in determining temporally precise functional connectivity between cell ensembles (Fujisawa et al., 2008). Based on our finding that both Dgcr8+/− and Df(16)A+/− mouse mutants, which share WM deficits, also share altered STD and STP during sustained activation of afferent fibers in the PFC, we propose that such changes contribute to altered circuit dynamics in vivo during WM performance (Sigurdsson et al., 2010; Kvajo et al., 2011; Arguello and Gogos, 2012). The observed alteration in short-term dynamics is likely to be presynaptic (Fénelon et al., 2011), possibly because of altered synaptic vesicle cycling and neurotransmitter release.
Assays of spine stability
Examination of formation and elimination of dendritic spines, the postsynaptic sites of the majority of excitatory synapses, of L5 pyramidal neurons in the frontal cortex of adult Df(16)A+/− mice (Pan and Gan, 2008; Bhatt et al., 2009) revealed increased spine turnover (i.e., both increased formation and elimination of dendritic spines) and highlighted spine instability as a major abnormality of dendritic spine development and plasticity in the cortex of Df(16)A+/− mice. In turn, altered spine dynamics, density, and volume may be related to the long-term changes in synaptic plasticity observed in the cortex of Df(16)A+/− mice (De Roo et al., 2008; Yang et al., 2009; Kasai et al., 2010). Although it is possible that such changes may contribute to some extent to the WM deficits observed in the Df(16)A+/− mice, it is worth noting that Dgcr8+/− mice, which also show WM deficits, consistently demonstrate normal spine density in both HPC (basal dendrites of CA1 pyramidal neurons) and PFC (both apical and basal dendrites of L5 pyramidal neurons) (Stark et al., 2008; Fénelon et al., 2011). Conversely, it is well established that spine remodeling in recruited frontal cortical regions plays an important role during both the formation and the extinction of contextual fear memories (Lai et al., 2012). In that respect, the increased spine instability in the Df(16)A+/− mice may contribute to the robust changes observed in associative fear learning observed in Df(16)A+/− mice but not in Dgcr8+/− mice. To our knowledge, this is the first time that dendritic spine turnover has been evaluated in a genetic mouse model of SCZ.
Assays of cytoarchitecture
Changes in structural and functional synaptic plasticity occur in the context of relatively circumscribed and often modest cytoarchitectural changes in laminar organization, neuronal density, and dendritic architecture in the PFC. Most notable among them is the robust reduction of dendritic complexity in the basal dendritic tree of L5 pyramidal neurons (Xu et al., 2013). Evaluation of inhibitory neuronal density revealed small changes in PV+ and CB+ cell frequency in L5. Only changes in neuronal density in L2 but not changes in either dendritic complexity or inhibitory neuron density can be attributed to Dgcr8 deficiency. Alterations in pyramidal neuron density and dendritic complexity almost certainly contribute to cortical thinning reported in 22q11.2 deletion carriers, and cumulative disruptions in cortical circuitry may account to some extent for the reported correlations between regional gray matter volumes and cognitive functions (Bearden et al., 2009). Similarly the effect, if any, of the subtle decrease in inhibitory neuron density and whether it is accompanied by alterations in the functional properties of inhibitory neurons remains to be determined.
Assays of gene expression and splicing
Using higher-resolution expression profiling, we corroborated previous findings and provided a more accurate quantification of the pervasive role that impaired processing of pri-miRNA attributable to the 22q11.2 deletion has on the PFC transcriptional profile of mutant mice (Stark et al., 2008). In addition, our transcriptional profiling revealed for the first time potential alterations in chromosome remodeling and differential splicing emerging as a result of 22q11.2 deletions. Overall, our analysis highlighted a number of individual genes and convergent pathways that may underlie the observed cortical alterations. In particular, our network analysis identified a number of convergent molecular abnormalities in the form of distinct modules of coexpressed genes enriched in specific biological functions. Perturbations in at least some of these modules (“ribonucleoprotein complex,” “RNA/ncRNA processing”) can be partly accounted for by both canonical and noncanonical functions of Dgcr8 (Chong et al., 2010; Knuckles et al., 2012), whereas convergent alterations in Golgi-related genes have been attributed previously to depletion of miR-185. Thus, miRNA dysregulation appears to be a major driver of the observed molecular perturbations.
We provide evidence for a previously unknown effect of 22q11.2 deletion on differential splicing. The mechanistic basis of this effect remains unknown and requires additional analysis. Altered exon usage could be the direct effect on splicing of haploinsuffiency of one or more genes within the deleted genomic region, such as Dgcr8 or Dgcr14 (Jurica et al., 2002; Macias et al., 2012). Alternatively, altered splicing may be attributable to indirect effects of perturbation of chromatin structure (Schor et al., 2012) or to indirect effects of impaired processing of primary miRNA transcripts on the splicing of host genes (Janas et al., 2011). Independent of the mechanism, we show that many of the affected exons belong to genes involved in cytoskeleton and extracellular matrix-related functions as well as to genes encoding an array of ion channels.
Our results strongly suggest that robust alterations in synaptic properties within the PFC, which to a large extent but not entirely can be accounted for by Dgcr8 deficiency, is a primary contributor to the cognitive impairments observed in mouse models and possibly in patients with the 22q11.2 deletion. In particular, our results provide evidence for a novel mechanism of 22q11.2-associated WM dysfunction in which an inability of synapses to rapidly and efficiently adapt to high-frequency stimulation leads to disruptions of synaptic and network activities in PFC that are necessary to support WM and possibly other cognitive operations. We hypothesize that such an alteration in rapid synaptic dynamics is one common substrate that can be disrupted by various genetic or environmental insults contributing to impaired cognitive function in SCZ. Cognitive endpoints may shed light on other disease symptoms (such as delusions and hallucinations or negative symptoms), with less direct correlates in animal models (Arguello and Gogos, 2012; Lerner et al., 2012). Therefore, our results may offer more general insights into the nature of the neural substrates underlying 22q11.2-associated psychiatric phenotypes.
Finally, our findings are in line with accumulating evidence that miRNAs play an important role in the pathogenesis and pathophysiology of psychiatric disorders and cognitive dysfunction (Stark et al., 2008; Fénelon et al., 2011; Miller et al., 2012; Xu et al., 2010, 2012b, 2013), as well as with additional supporting evidence accumulating from parallel expression profiling studies in brains and peripheral blood of patients (Perkins et al., 2007; Beveridge et al., 2008; Lai et al., 2011; Moreau et al., 2011; Gardiner et al., 2012; Miller et al., 2012). Importantly, we recently provided evidence that rare de novo deleterious mutations in genes showing a prenatal expression bias and miRNA regulation are enriched in individuals with SCZ, especially those with prominent early prepsychotic, deviant behaviors (Gilman et al., 2012; Xu et al., 2012a). In that respect, the miRNA dysregulation observed in the Df(16)A+/− mice models a key molecular pathway underlying genetic liability of SCZ and is likely to provide more general insights into how mutations on a highly diverse set of miRNA regulated genes predispose to SCZ.
Footnotes
This work was supported by National Institute of Mental Health Grants MH67068 (M.K. and J.A.G.) and MH077235 (J.A.G.), the March of Dimes Foundation (M.K.), and the McKnight Endowment Fund for Neuroscience (M.K.). B.X. has been supported in part by a National Alliance for Research on Schizophrenia and Depression Young Investigator Award. K.L.S. was supported by an National Alliance for Research on Schizophrenia and Depression Suzanne and John Golden Young Investigator Award. We thank Yan Sun and Darshini Mahadevia for technical assistance.
References
- Arguello PA, Gogos JA. Modeling madness in mice: one piece at a time. Neuron. 2006;52:179–196. doi: 10.1016/j.neuron.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Arguello PA, Gogos JA. Cognition in mouse models of schizophrenia susceptibility genes. Schizophr Bull. 2010;36:289–300. doi: 10.1093/schbul/sbp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello PA, Gogos JA. Genetic and cognitive windows into circuit mechanisms of psychiatric disease. Trends Neurosci. 2012;35:3–13. doi: 10.1016/j.tins.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Bannister AP. Inter- and intra-laminar connections of pyramidal cells in the neocortex. Neurosci Res. 2005;53:95–103. doi: 10.1016/j.neures.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Barañano DE, Ferris CD, Snyder SH. Atypical neural messengers. Trends Neurosci. 2001;24:99–106. doi: 10.1016/S0166-2236(00)01716-1. [DOI] [PubMed] [Google Scholar]
- Barch DM, Ceaser A. Cognition in schizophrenia: core psychological and neural mechanisms. Trends Cogn Sci. 2012;16:27–34. doi: 10.1016/j.tics.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Hodgkinson K, Chow EW, Correia S, Scutt LE, Weksberg R. 22q11 deletion syndrome in adults with schizophrenia. Am J Med Genet. 1998;81:328–337. doi: 10.1002/(SICI)1096-8628(19980710)81:4<328::AID-AJMG10>3.0.CO;2-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R. The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry. 2003;160:1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, van Erp TG, Dutton RA, Lee AD, Simon TJ, Cannon TD, Emanuel BS, McDonald-McGinn D, Zackai EH, Thompson PM. Alterations in midline cortical thickness and gyrification patterns mapped in children with 22q11.2deletions. Cereb Cortex. 2009;19:115–126. doi: 10.1093/cercor/bhn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Bhatt DH, Zhang S, Gan WB. Dendritic spine dynamics. Annu Rev Physiol. 2009;71:261–282. doi: 10.1146/annurev.physiol.010908.163140. [DOI] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [Erratum (2010) 24: 2228] [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roo M, Klauser P, Garcia PM, Poglia L, Muller D. Spine dynamics and synapse remodeling during LTP and memory processes. Prog Brain Res. 2008;169:199–207. doi: 10.1016/S0079-6123(07)00011-8. [DOI] [PubMed] [Google Scholar]
- Drew LJ, Stark KL, Fénelon K, Karayiorgou M, Macdermott AB, Gogos JA. Evidence for altered hippocampal function in a mouse model of the human 22q11.2 microdeletion. Mol Cell Neurosci. 2011;47:293–305. doi: 10.1016/j.mcn.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earls LR, Fricke RG, Yu J, Berry RB, Baldwin LT, Zakharenko SS. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci. 2012;32:14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010:W755–W762. doi: 10.1093/nar/gkq405. 2010 38(Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SF, Irmady K, Ostrow K, Kim T, Nykjaer A, Saftig P, Blobel C, Hempstead BL. Neuronal brain-derived neurotrophic factor is synthesized in excess, with levels regulated by sortilin-mediated trafficking and lysosomal degradation. J Biol Chem. 2011;286:29556–29567. doi: 10.1074/jbc.M111.219675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénelon K, Mukai J, Xu B, Hsu PK, Drew LJ, Karayiorgou M, Fischbach GD, Macdermott AB, Gogos JA. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/S0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fujisawa S, Amarasingham A, Harrison MT, Buzsáki G. Behavior-dependent short-term assembly dynamics in the medial prefrontal cortex. Nat Neurosci. 2008;11:823–833. doi: 10.1038/nn.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JM. The prefrontal cortex and its relation to behavior. Prog Brain Res. 1991;87:201–211. doi: 10.1016/S0079-6123(08)63053-8. [DOI] [PubMed] [Google Scholar]
- Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, Davies C, Williams A, Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;27:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. 2012;17:827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperle AY, Enz A, Pozza MF, Luthi A, Olpe HR. Effects of clozapine, haloperidol and iloperidone on neurotransmission and synaptic plasticity in prefrontal cortex and their accumulation in brain tissue: an in vitro study. Neuroscience. 2003;117:681–695. doi: 10.1016/S0306-4522(02)00769-8. [DOI] [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Architecture of the prefrontal cortex and the central executive. Ann N Y Acad Sci. 1995;769:71–83. doi: 10.1111/j.1749-6632.1995.tb38132.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson B, Wigström H. Long-term potentiation in the hippocampal CA1 region: its induction and early temporal development. Prog Brain Res. 1990;83:223–232. doi: 10.1016/S0079-6123(08)61252-2. [DOI] [PubMed] [Google Scholar]
- Hanse E, Gustafsson B. Onset and stabilization of NMDA receptor-dependent hippocampal long-term potentiation. Neurosci Res. 1994;20:15–25. doi: 10.1016/0168-0102(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Hempel CM, Hartman KH, Wang XJ, Turrigiano GG, Nelson SB. Multiple forms of short-term plasticity at excitatory synapses in rat medial prefrontal cortex. J Neurophysiol. 2000;83:3031–3041. doi: 10.1152/jn.2000.83.5.3031. [DOI] [PubMed] [Google Scholar]
- Hirsch JC, Crepel F. Use-dependent changes in synaptic efficacy in rat prefrontal neurons in vitro. J Physiol. 1990;427:31–49. doi: 10.1113/jphysiol.1990.sp018159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA, Fisher DE, Shomron N, Levy C, Novina CD. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011;7:e1002330. doi: 10.1371/journal.pgen.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel D, Weiner I, Feldon J. Electrolytic lesions of the medial prefrontal cortex in rats disrupt performance on an analog of the Wisconsin Card Sorting Test, but do not disrupt latent inhibition: implications for animal models of schizophrenia. Behav Brain Res. 1997;85:187–201. doi: 10.1016/S0166-4328(97)87583-3. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/S1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, Eisen H, Childs B, Kazazian HH, Kucherlapati R, Antonarakis SE, Pulver AE, Housman DE. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Flint J, Gogos JA, Malenka RC Genetic and Neural Complexity in Psychiatry 2011 Working Group. The best of times, the worst of times for psychiatric disease. Nat Neurosci. 2012;15:811–812. doi: 10.1038/nn.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Kates WR, Krauss BR, Abdulsabur N, Colgan D, Antshel KM, Higgins AM, Shprintzen RJ. The neural correlates of non-spatial working memory in velocardiofacial syndrome (22q11.2deletion syndrome) Neuropsychologia. 2007;45:2863–2873. doi: 10.1016/j.neuropsychologia.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, Vogt MA, Lugert S, Milo M, Chong MM, Hautbergue GM, Wilson SA, Littman DR, Taylor V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat Neurosci. 2012;15:962–969. doi: 10.1038/nn.3139. [DOI] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB, Karayiorgou M, Gogos JA. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci U S A. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Gogos JA. Molecules, signaling, and schizophrenia. Curr Top Behav Neurosci. 2010;4:629–656. doi: 10.1007/7854_2010_41. [DOI] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Drew LJ, Lepagnol-Bestel AM, Xiao L, Levy RJ, Blazeski R, Arguello PA, Lacefield CO, Mason CA, Simonneau M, O'Donnell JM, MacDermott AB, Karayiorgou M, Gogos JA. Altered axonal targeting and short-term plasticity in the hippocampus of Disc1 mutant mice. Proc Natl Acad Sci U S A. 2011;108:E1349–E1358. doi: 10.1073/pnas.1012355108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Franke TF, Gan WB. Opposite effects of fear conditioning and extinction on dendritic spine remodelling. Nature. 2012;483:87–91. doi: 10.1038/nature10792. [DOI] [PubMed] [Google Scholar]
- Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, Huang YH, Hsiao PC, Hsiao CK, Liu CM, Yang PC, Hwu HG, Chen WJ. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Xu B, Westphal KG, Paterlini M, Olivier B, Pavlidis P, Karayiorgou M, Gogos JA. Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc Natl Acad Sci U S A. 2006;103:16906–16911. doi: 10.1073/pnas.0604994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner I, Bentin S, Shriki O. Excessive attractor instability accounts for semantic priming in schizophrenia. PLoS One. 2012;7:e40663. doi: 10.1371/journal.pone.0040663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litterman N, Ikeuchi Y, Gallardo G, O'Connell BC, Sowa ME, Gygi SP, Harper JW, Bonni A. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockstone HE. Exon array data analysis using Affymetrix power tools and R statistical software. Brief Bioinform. 2011;12:634–644. doi: 10.1093/bib/bbq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder-Gadaczek J, Becker I, Gieselmann V, Wang-Eckhardt L, Eckhardt M. N-acetylaspartylglutamate synthetase II synthesizes N-acetylaspartylglutamylglutamate. J Biol Chem. 2011;286:16693–16706. doi: 10.1074/jbc.M111.230136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Cáceres JF. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Erickson CA, Desimone R. Neural mechanisms of visual working memory in prefrontal cortex of the macaque. J Neurosci. 1996;16:5154–5167. doi: 10.1523/JNEUROSCI.16-16-05154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morciano M, Beckhaus T, Karas M, Zimmermann H, Volknandt W. The proteome of the presynaptic active zone: from docked synaptic vesicles to adhesion molecules and maxi-channels. J Neurochem. 2009;108:662–675. doi: 10.1111/j.1471-4159.2008.05824.x. [DOI] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, Karayiorgou M, Gogos JA. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Gan WB. Two-photon imaging of dendritic spine development in the mouse cortex. Dev Neurobiol. 2008;68:771–778. doi: 10.1002/dneu.20630. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Gogos JA, Karayiorgou M. The genetic architecture of schizophrenia: new mutations and emerging paradigms. Annu Rev Med. 2012;63:63–80. doi: 10.1146/annurev-med-072010-091100. [DOI] [PubMed] [Google Scholar]
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- Schor IE, Llères D, Risso GJ, Pawellek A, Ule J, Lamond AI, Kornblihtt AR. Perturbation of chromatin structure globally affects localization and recruitment of splicing factors. PLoS One. 2012;7:e48084. doi: 10.1371/journal.pone.0048084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz PE, Fitzgibbons JC. Differing mechanisms of expression for short- and long-term potentiation. J Neurophysiol. 1997;78:321–334. doi: 10.1152/jn.1997.78.1.321. [DOI] [PubMed] [Google Scholar]
- Shuvaev AN, Horiuchi H, Seki T, Goenawan H, Irie T, Iizuka A, Sakai N, Hirai H. Mutant PKCγ in spinocerebellar ataxia type 14 disrupts synapse elimination and long-term depression in Purkinje cells in vivo. J Neurosci. 2011;31:14324–14334. doi: 10.1523/JNEUROSCI.5530-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Szentágothai J. The Ferrier Lecture, 1977. The neuron network of the cerebral cortex: a functional interpretation. Proc R Soc Lond B Biol Sci. 1978;201:219–248. doi: 10.1098/rspb.1978.0043. [DOI] [PubMed] [Google Scholar]
- Volianskis A, Jensen MS. Transient and sustained types of long-term potentiation in the CA1 area of the rat hippocampus. J Physiol. 2003;550:459–492. doi: 10.1113/jphysiol.2003.044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012a;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Hsu PK, Karayiorgou M, Gogos JA. MicroRNA dysregulation in neuropsychiatric disorders and cognitive dysfunction. Neurobiol Dis. 2012b;46:291–301. doi: 10.1016/j.nbd.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan WB. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq andDegradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee BK, Balic E, Singer P, Schwerdel C, Grampp T, Gabernet L, Knuesel I, Benke D, Feldon J, Mohler H, Boison D. Disruption of a glycine transporter 1 restricted to forebrain neurons is associated with a procognitive and antipsychotic phenotypic profile. J Neurosci. 2006;26:3169–3181. doi: 10.1523/JNEUROSCI.5120-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]